Abstract
Purpose
Although normal function of the lacrimal gland is essential for vision (and thus for human well-being), the lacrimal gland remains rather poorly understood at a molecular level. The purpose of this study was to identify new genes and signaling cascades involved in lacrimal gland development.
Methods
To identify these genes, we used microarray analysis to compare the gene expression profiles of developing (embryonic) and adult lacrimal glands. Differential data were validated by quantitative RT-PCR, and several corresponding proteins were confirmed by immunohistochemistry and Western blot analysis. To evaluate the role of NOTCH signaling in lacrimal gland (LG) development, we used the NOTCH inhibitor DAPT and conditional Notch1 knockouts.
Results
Our microarray data and an in silico reconstruction of cellular networks revealed significant changes in the expression patterns of genes from the NOTCH, WNT, TGFβ, and Hedgehog pathways, all of which are involved in the regulation of epithelial-to-mesenchymal transition (EMT). Our study also revealed new putative lacrimal gland stem cell/progenitor markers. We found that inhibiting Notch signaling both increases the average number of lacrimal gland lobules and reduces the size of each lobule.
Conclusions
Our findings suggest that NOTCH-, WNT-, TGFβ-, and Hedgehog-regulated EMT transition are critical mechanisms in lacrimal gland development and morphogenesis. Our data also supports the hypothesis that NOTCH signaling regulates branching morphogenesis in the developing lacrimal gland by suppressing cleft formation.
Keywords: lacrimal gland, development, microarray analysis, NOTCH signaling, branching morphogenesis
The lacrimal gland, a tubuloacinar exocrine gland located in the superolateral aspect of each orbit, secretes the aqueous layer of tear film to help maintain the physiological function of the ocular surface.1,2 Dysfunction of the lacrimal gland leads to dry eye disease, which results in visual impairment if left untreated.3,4 Dry eye disease affects almost 5 million people per year in the United States alone, disproportionately afflicting people over the age of 50.2,3,5 While mild manifestations of the disease are treatable to a certain degree, severe forms of dry eye disease (e.g., due to complete loss of lacrimal gland function secondary to trauma or lacrimal gland carcinoma) are notoriously difficult to manage. A possible long-term solution might be to characterize, isolate, and transplant lacrimal gland stem/progenitor cells, and/or to bioengineer new lacrimal gland tissue from this and other stem cell sources.6 In a critical first step toward making this idea a reality, Hirayama and colleagues7,8 successfully bioengineered a functional lacrimal gland that efficiently maintained a healthy ocular surface in vivo. Unfortunately, this approach worked only with the fetal lacrimal gland, which contains a significant number of stem cells and progenitors; it was not shown to work with the adult lacrimal gland, which contains a limited number of still poorly defined stem cells/progenitors.6–8 With better techniques to identify stem cells/progenitors in the adult lacrimal gland and a deeper understanding of the signaling cascades that regulate lacrimal gland development, this problem could very likely be solved.
The use of stem cells/progenitors in lacrimal gland replacement therapy is an area of intense research. Three putative lacrimal gland stem cell/progenitor markers, c-kit, ABCG2, and ALDH1, have so far been described, but definitive evidence to support this claim is lacking.9–11 Our knowledge of the mechanisms that regulate lacrimal gland development remains similarly limited. It was shown that lacrimal gland development begins when Fgf10 in the periocular mesenchyme interacts with FGFR2b receptor in the ectoderm, inducing SOX9 expression.12 Sox9 regulates the initial budding and elongation of the lacrimal bud by activating heparan sulfate–synthesizing enzymes such as NDST1, NDST2, HS2ST, and HS6ST.12–15 Heparan sulfate regulates further Fgf10-dependent Fgfr2b activation, and also stabilizes Fgf10 levels in the mesenchyme by limiting Fgf10 diffusion.13 Sox9- and Fgfr2b-dependent activation of Sox10 is required for the further elongation and branching of the lacrimal bud, as well as for the formation of secretory acini.12 Other factors involved in the outgrowth of the lacrimal bud, branching, and acinar organization of the lacrimal gland include PAX6, BMP7, FOXC1, IGSF3, EGF, and Egfr.16–20 Nevertheless, lacrimal gland development cannot be sufficiently explained by activity of these signaling cascades alone. The purpose of this study was to identify key signaling cascades likely to be involved in lacrimal gland development and morphogenesis, as well as to identify new putative markers for a lacrimal gland stem/progenitor cell phenotype.
Methods
Animals
All experiments were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the ARVO Statement for the Use ofAnimals in Ophthalmic and Vision Research. The protocol was approved by the institutional animal care and use committee of the University of Miami (Miami, FL, USA). Notch1flox/flox mice (stock number 007181) and C57BL/6 J (stock number 000664) mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Mice were housed under standard conditions of humidity and temperature, with a 12-hour light/dark cycle and free access to food and water. We used 3-month-old mice and embryonic day (E) 15.5 and 16.5 fetuses. Animals were euthanized by CO2 inhalation under anesthesia.
RNA Extraction, Probe Preparation, and Array Hybridization
RNA samples were extracted from adult (3-month old) and embryonic (E16.5) lacrimal glands using the Absolutely RNA Microprep and Nanoprep kits, respectively (Agilent Technologies, Santa Clara, CA, USA). RNA samples were sent to Ocean Ridge Biosciences (Palm Beach Gardens, FL, USA) for analysis using mouse exonic evidence-based oligonucleotide (MEEBO) microarrays. Biotin-labeled complementary RNA was synthesized from the total RNA according to Van Gelder's protocol.21 Biotinylated complementary RNA samples were fragmented, diluted in a formamide-containing hybridization buffer, and loaded onto the MEEBO microarray chips enclosed in custom hybridization chambers. The slides were hybridized for 16 to 18 hours in a Model 400 hybridization oven (Scigene, Sunnyvale, CA, USA). After hybridization, the microarray slides were washed under stringent conditions, stained with Streptavidin-Alexa-647 (Invitrogen, Carlsbad, CA, USA), and scanned using an Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA).
Microarray Data Analysis
Spot intensities for each probe were calculated by subtracting median local background intensity from median local foreground intensity for each spot. The spot intensities were then normalized. After removing data for low-quality spots, the mouse probes' intensities were filtered to identify all probes with intensity above a normalized threshold. For statistical analysis, microarray data were examined for differences by 1-way ANOVA or Student's t-test. Values of P less than 0.05 were designated as statistically significant. The data files have been uploaded in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; in the public domain, http://www.ncbi.nlm.nih.gov/geo/) and are available through GEO series accession number GSE78062.
Quantitative RT-PCR Analysis
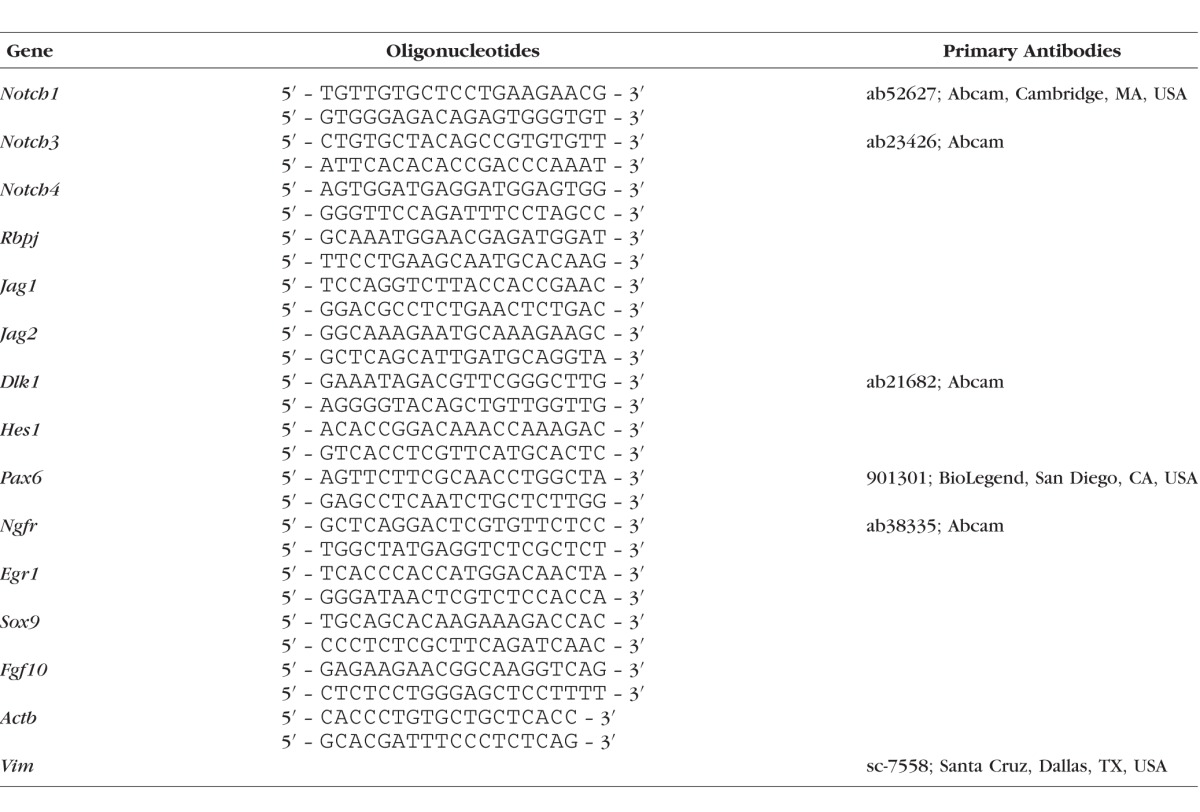
Quantitative RT-PCR analysis was performed as described previously22,23 using gene-specific primers (Table 1). Relative expression was calculated by comparison with a standard curve following normalization to expression of the housekeeping gene β-actin (Actb), chosen as a control. Data are presented as average ± SEM.
Table 1.
List of RT-PCR Primers and Primary Antibodies
Western Blot Analysis
The lacrimal glands were lysed with T-PER buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with complete protease inhibitor (Roche Applied Science, Indianapolis, IN, USA). An equal amount of total protein from each sample was resolved on SDS-PAGE gradient gels and transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific). Blots were blocked in 5% milk in Tris-buffered saline (TBS; pH 7.6); probed overnight with primary antibodies (listed in Table 1); washed in 0.15% Tween20 in TBS; and incubated for 1 hour with a secondary antibody (1:10,000; GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) diluted in TBS. Anti-β-actin antibodies were used as loading controls. Proteins were visualized using SuperSignal chemiluminescent substrates (Thermo Fisher Scientific).
Immunohistochemistry
Adult mice (the source of adult lacrimal glands) or pregnant mothers (the source of embryonic lacrimal glands) were transcardially perfused with 4% paraformaldehyde in PBS. The lacrimal glands were then removed and fixed in 4% paraformaldehyde for 20 minutes. While whole embryonic lacrimal glands were immunostained, adult LGs were cryoprotected with 30% sucrose, embedded in optimum cutting temperature (OCT) medium, and cryosectioned at 25-μm thickness. Prior to immunostaining, whole embryonic lacrimal glands or sections of adult lacrimal glands were permeabilized in 0.3% Triton X-100 in PBS for 30 minutes. Tissues were blocked with PBS containing 0.15% Tween 20, 2% BSA, and 5% serum at room temperature for 30 minutes. The samples were then incubated with primary antibodies (Table 1) for 16 hours in blocking solution, followed by species-specific secondary antibodies (AlexaFluor; Thermo Fisher Scientific). Control samples were incubated without primary antibodies. Imaging was performed with a Leica TSL AOBS SP5 confocal microscope (Leica Microsystems, Exton, PA, USA).
Lacrimal Gland Explant Ex Vivo Cultures, DAPT, and Ad-CMV-iCre Treatment
The lacrimal gland explant ex vivo cultures were prepared from E15.5 embryos prior to the onset of branching morphogenesis. To this end, the lacrimal glands were aseptically removed, washed in PBS, and plated on culture inserts (Transwell filters, Costar; Sigma-Aldrich Corp., St. Louis, MO, USA) pretreated with Matrigel (160 μL of a 1:30 dilution of Matrigel in Dulbecco's modified Eagle's medium [DMEM]/F12; BD Biosciences, Bedford, MA, USA). The inserts (each containing a lacrimal gland in DMEM/F12 supplemented with 10% FBS and 3.3% Matrigel) were placed into the culture wells of a 12-well plate (containing the same medium, without Matrigel). Subsequently, the desired concentration of DAPT (D5942; Sigma-Aldrich Corp.) was added to the media. To transduce lacrimal gland explants with Ad-CMV-iCre (Vector Biolabs, Malvern, PA, USA), the virus was added (5 × 107 genome copies per well) to the media. The 12-well plates were then transferred into a CO2 incubator, where they were kept for 72 hours at 37°C with 5% CO2.
Statistical Analysis
Statistical analysis was conducted with a 1-way ANOVA for multiple comparisons. For single comparisons, the Student t-test was applied. P values less than or equal to 0.05 were considered statistically significant.
Results
An In Silico Reconstruction of Cellular Networks Reveals Putative Activity of the NOTCH, WNT, TGFβ, and Hedgehog Signaling Cascades During Lacrimal Gland Development
To characterize and compare lacrimal glands isolated from embryonic and adult mice, RNA extracted from lacrimal glands of 3-month-old (adult) mice and E16.5 fetuses was used for microarray analysis. A total of three independent biological replicates of E16.5 lacrimal glands and three independent biological replicates of adult lacrimal glands were obtained for comparative profiling. The results of our microarray analysis showed that a total of 8188 genes passed the quality control criteria and were differentially expressed (P < 0.05) between the two studied groups (Table 2; Supplementary Table S1). The changes in gene expression detected by microarrays were further confirmed by quantitative RT-PCR, Western blot analysis, and immunohistochemistry for a group of selected genes (Figs. 1–4). Importantly, we detected expression of all genes that had been previously reported to be involved in lacrimal gland development (Table 3). Surprisingly, while expression of FGF10, FGFR2, SOX10, IGSF3, and EGFR was significantly upregulated in embryonic lacrimal glands, expression of PAX6, SOX9, and Egf was significantly upregulated in adult lacrimal glands (Table 3).
Table 2.
The Randomly Selected Genes Expressed Differently Between the Two Studied Groups
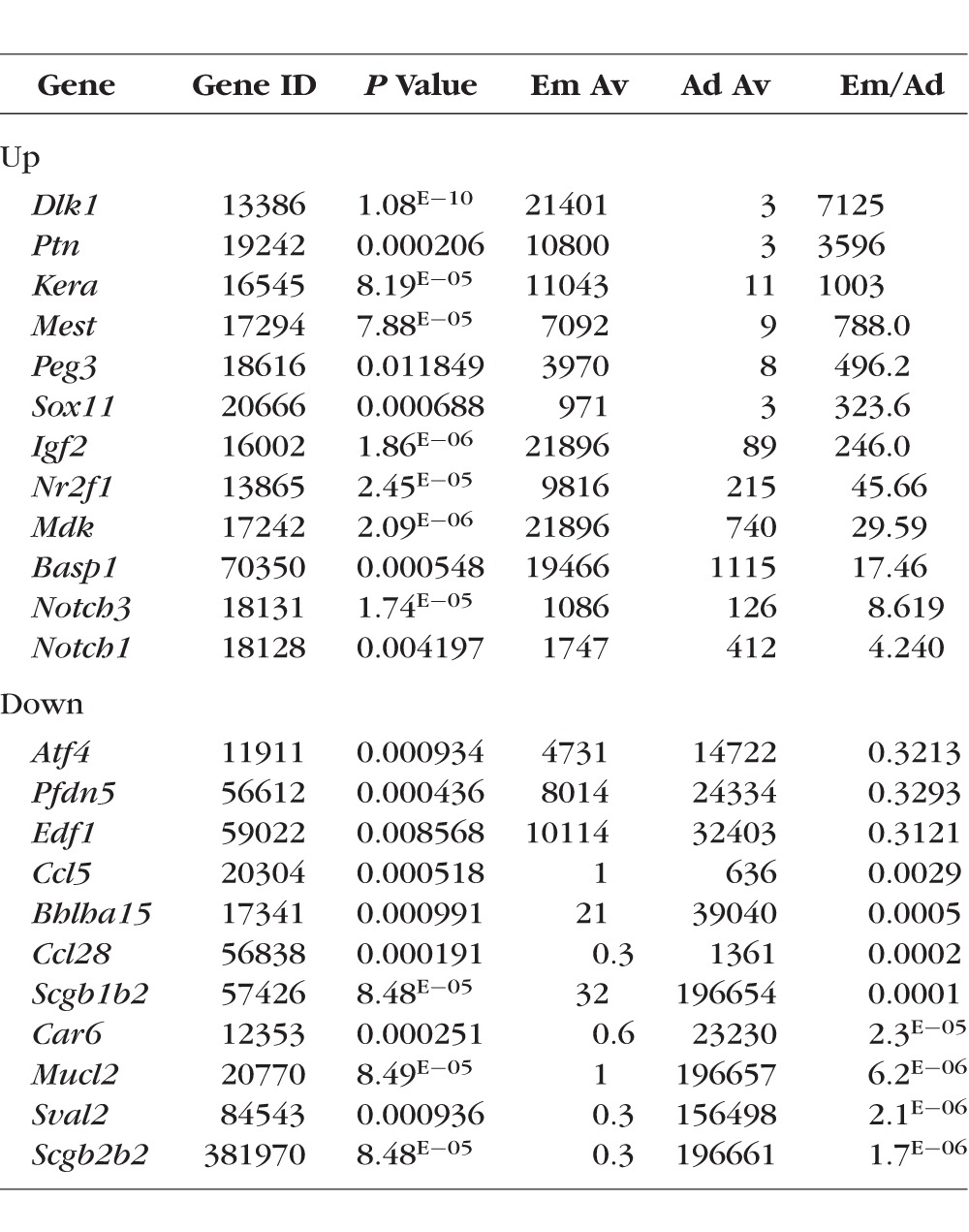
Figure 1.
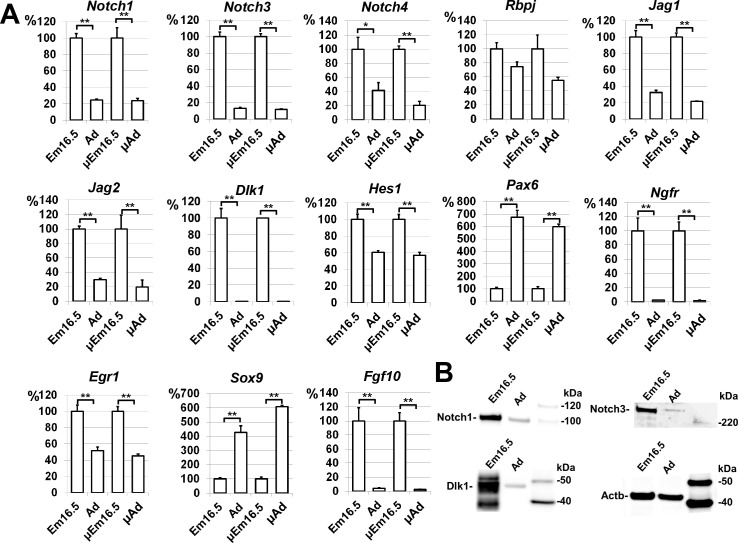
Differential gene expression between the embryonic and adult lacrimal glands revealed by microarray analysis was confirmed for a select group of genes by quantitative RT-PCR (A) and Western blot analysis (B) (*P < 0.05, **P < 0.01, μEm16.5 and μAd – microarray data).
Figure 2.
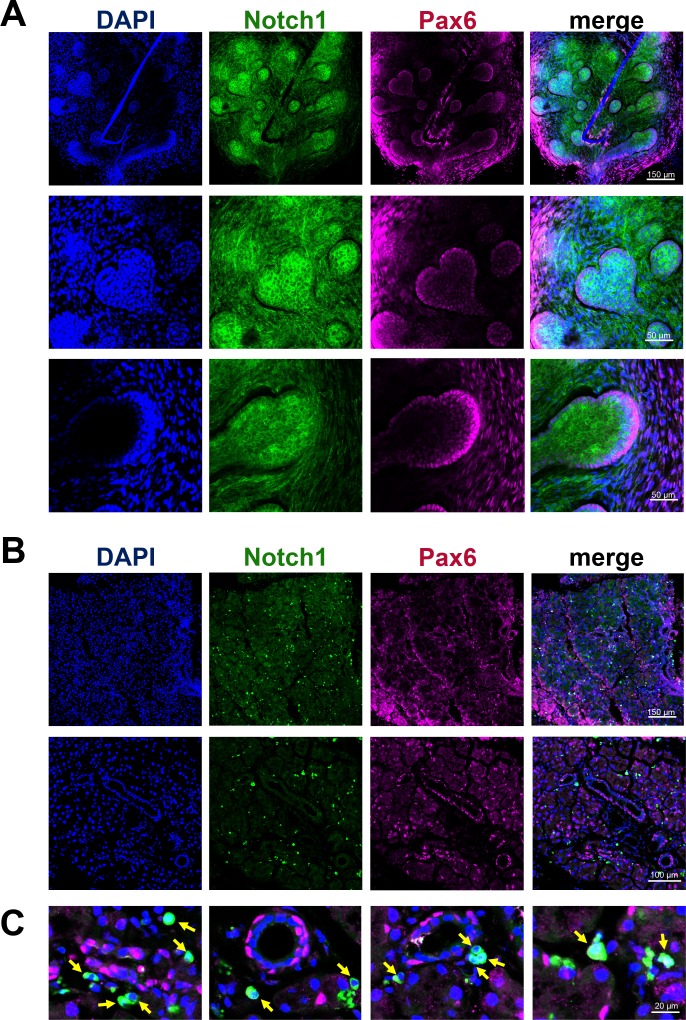
The Notch1 receptor is present in the embryonic and adult lacrimal gland at the protein level. While Notch1 was highly expressed in the epithelium and mesenchyme of E16.5 lacrimal glands (A), Notch1 was expressed only in single, isolated cells in adult lacrimal glands (B, C). Pax6 was used as a lacrimal gland marker. DAPI was used to visualize the nucleus of each cell.
Figure 3.
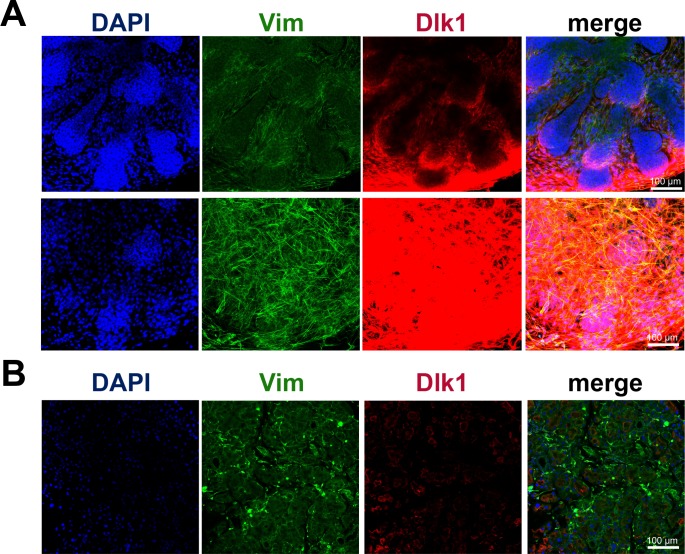
Immunohistochemistry showed significant accumulation of the noncanonical Notch ligand Dlk1 in embryonic lacrimal glands. (A) Significantly high Dlk1 expression was detected predominantly in mesenchymal cells of E16.5 LGs. (B) Levels of Dlk1 were insignificant in adult lacrimal glands. Anti-Vim antibodies were used to detect mesenchymal cells.
Figure 4.
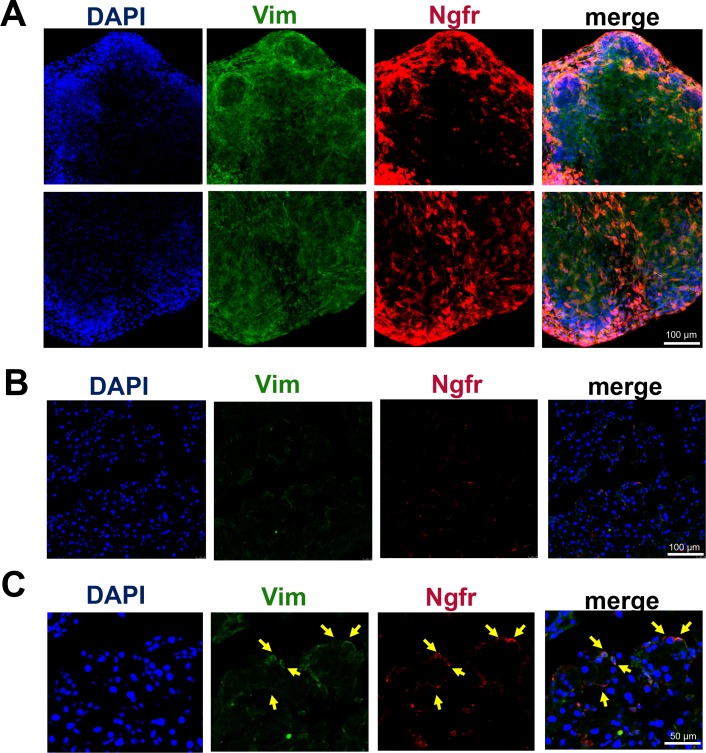
Ngfr expression in E16.5 and adult lacrimal glands. Ngfr was highly expressed in mesenchymal cells of embryonic lacrimal glands (A), but only in single, isolated cells in adult lacrimal glands (B, C). Anti-Vim antibodies were used to detect mesenchymal cells.
Table 3.
Genes Known to be Expressed (Before the Study) by Lacrimal Gland Tissue
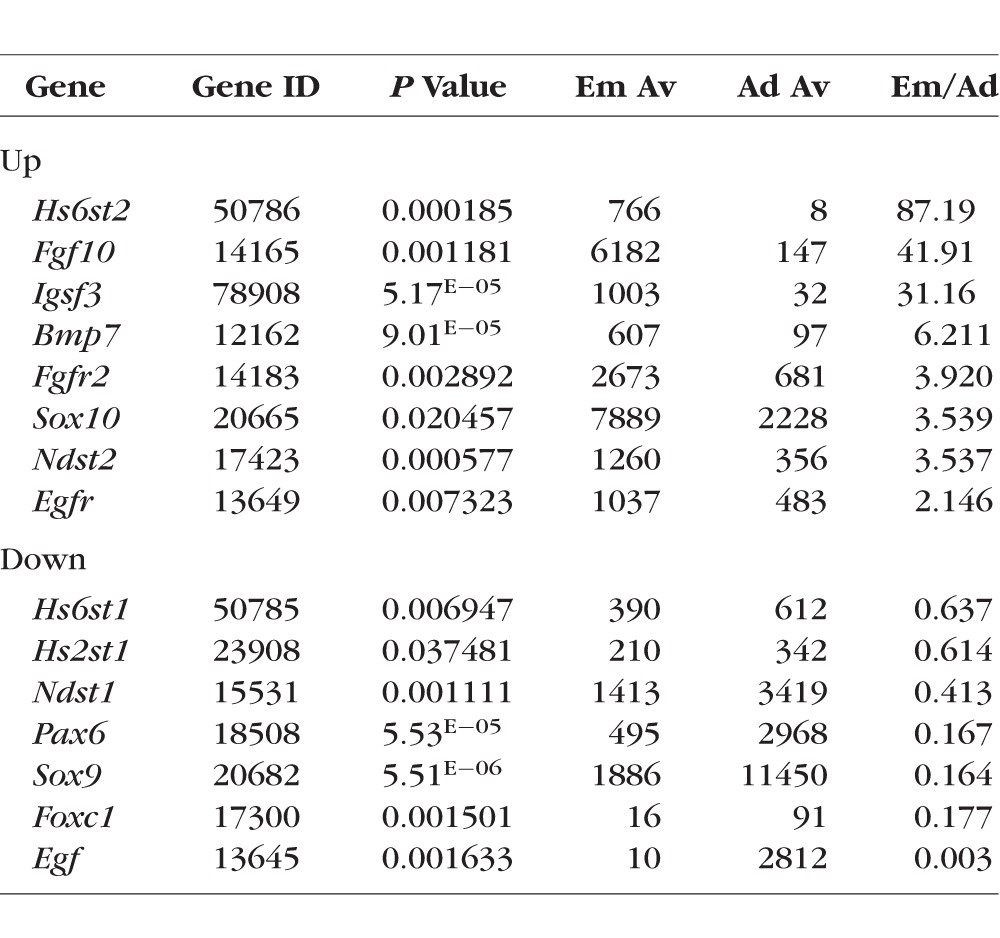
The significant differences in gene expression between embryonic and adult lacrimal glands created challenges in pathway analysis of our microarray data. Because our study focused predominantly on pathways involved in glandular development, we first analyzed the Gene Ontology (GO) database associated with “gland development,” and extracted a list of genes that passed the threshold level in our microarray data (Supplementary Table S2). We used DAVID functional annotation bioinformatic analysis tool to pinpoint enriched KEGG and PANTHER pathways (Supplementary Table S3). KEGG and PANTHER pathway mapping demonstrated that genes of the NOTCH, WNT, TGFβ, and Hedgehog signaling cascades were highly represented in our microarray data (Supplementary Table S3). This finding prompted us to investigate whether other genes in these pathways were also highly expressed. Using the KEGG PATHWAY database, we found that a significant number of genes from the NOTCH, WNT, TGFβ, and Hedgehog signaling pathways are expressed in both embryonic and adult lacrimal glands (Supplementary Table S4). Some of these genes were significantly upregulated in E16.5 lacrimal glands (NOTCH1, NOTCH3, JAG1, etc.), while others were significantly upregulated in adult lacrimal glands (RFNG, PSENEN, LEFTY2, etc.), suggesting that these signaling pathways may play a role in both lacrimal gland development and in the maintenance of adult lacrimal gland homeostasis (Supplementary Table S4). Because the NOTCH, WNT, TGFβ, and Hedgehog pathways all participate in the regulation of epithelial-to-mesenchymal transition (EMT), it is plausible that this biological process may play active roles in both embryonic and adult lacrimal glands.24,25 Our findings support this hypothesis, as genes from the GO database's EMT list were significantly represented in our microarray data (Supplementary Table S5). In addition, we tested for the presence of genes from the GO database's “mesenchymal to epithelial transition (MET)” list and found that these genes were also represented in our microarray data (Supplementary Table S5). Finally, we used these observations to generate the most comprehensive list of lacrimal gland–specific gene expression profile assembled to date (Supplementary Table S6).
Putative Lacrimal Gland Stem Cell/Progenitor Markers Were Revealed by Microarray Analysis
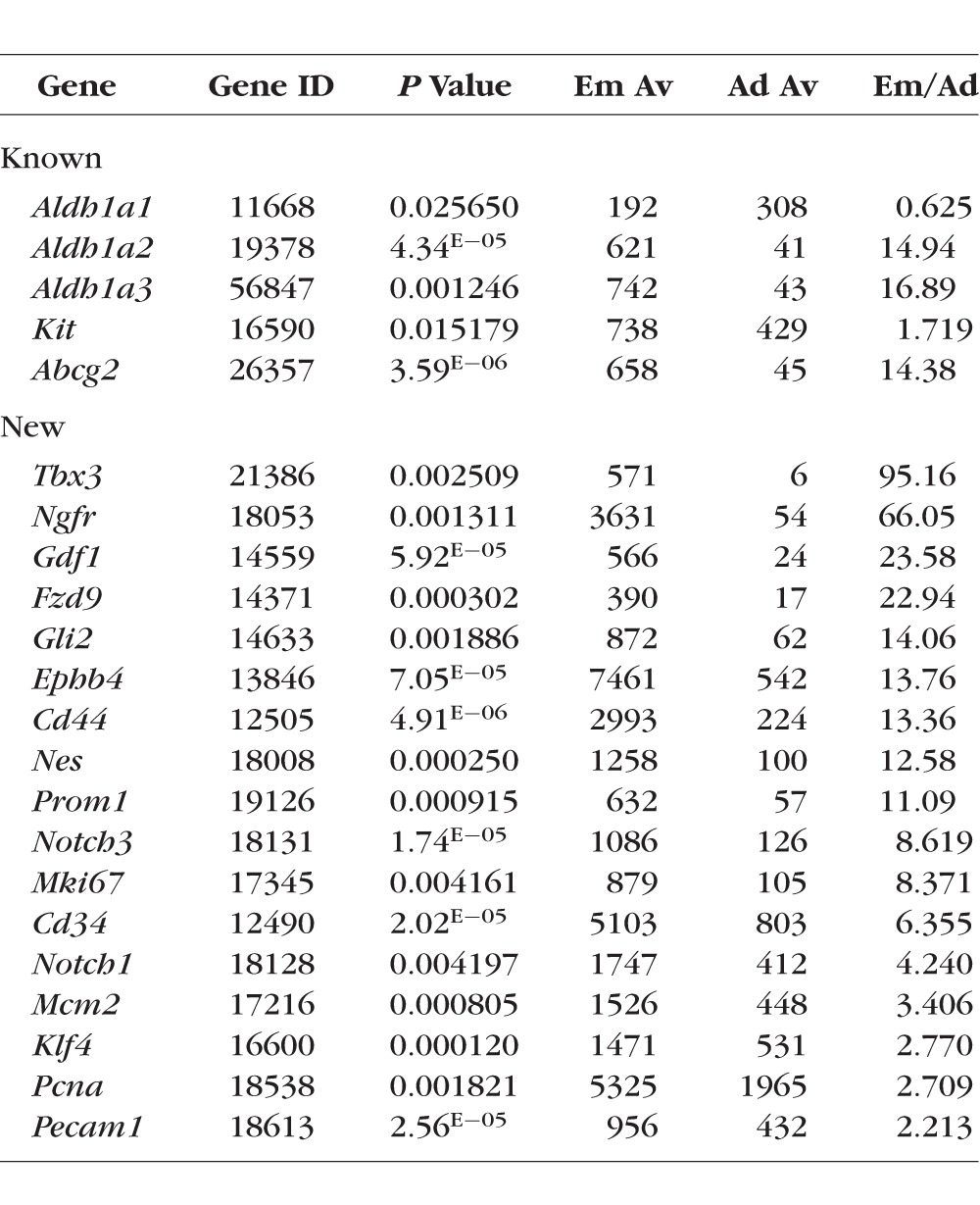
Two techniques—the use of adult stem cells in lacrimal gland regenerative therapy, and the transplantation of lacrimal gland–specific stem cells/progenitors—harbor the potential to restore lacrimal gland function and confer tremendous clinical benefits. Unfortunately, at the present time only c-kit (Kit), ALDH1 (Aldh1a1, Aldh1a2, and Aldh1a3), and ABCG2 (Abcg2) have been identified as putative lacrimal gland–specific stem cell/progenitor markers.9–11 Our data indicate that these markers are expressed in both embryonic and adult lacrimal gland tissue. All of them except Aldh1a1 were upregulated in E16.5 lacrimal glands compared with adult lacrimal glands (Table 4). To identify new potential lacrimal gland–specific stem cell/progenitor markers, we compared the results of our microarray data with a list of genes and proteins commonly associated with stem/progenitor cell states (Table 4). We found that expression of some of these genes was significantly upregulated in the E16.5 lacrimal gland compared with the adult lacrimal gland (Table 4). In addition, we employed immunohistochemistry assays to evaluate the distribution of two of the gene products, Ngfr/p75 and Notch1, in fixed embryonic and adult lacrimal glands. Expression of Ngfr and Notch1 was significantly higher in embryonic lacrimal glands compared with adult lacrimal glands (Table 4). Our immunohistochemistry data were consistent with these expression data (Figs. 2 and 4). In E16.5 lacrimal glands, Notch1-specific immunostaining was evident in both the epithelium and mesenchyme (Fig. 2), while substantial Ngfr-specific immunostaining was predominantly localized to the mesenchymal cells (Fig. 4). In the adult lacrimal glands, we found that Ngfr and Notch1 were expressed only in single, isolated cells (Figs. 2, 4). This observation led us to reason that these single cells may represent an adult lacrimal gland stem cell/ progenitors.
Table 4.
Known and Previously Undescribed Putative Lacrimal Gland Stem Cell/Progenitor Markers
NOTCH Signaling Regulates Average Lobule Size and Average Number of Lobules in the Lacrimal Gland
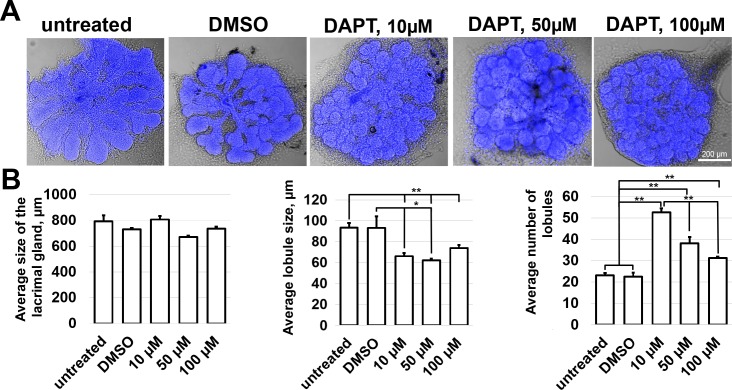
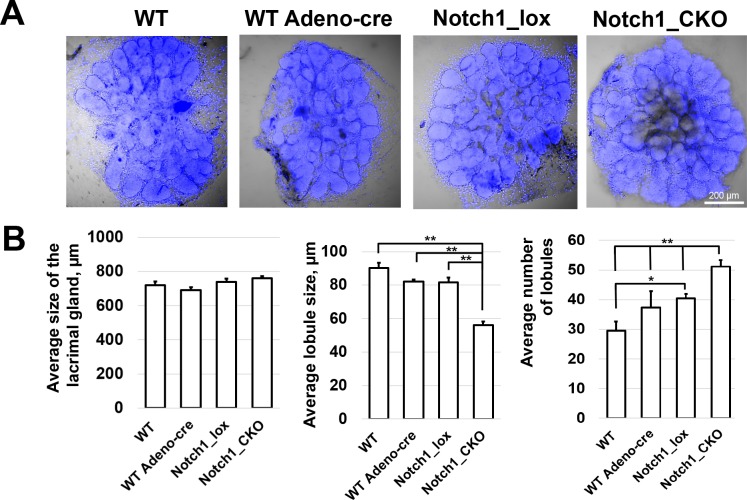
Our data uncovered several signaling pathways that may play important roles in lacrimal gland development and morphogenesis. From the large list of potential candidates, we chose to focus on one—the NOTCH signaling pathway—in order to directly evaluate its role in lacrimal gland development. We selected NOTCH because, as noted above, many genes of the NOTCH signaling pathway, including Notch1, Notch3, Hes1, Dlk1, Jag1, Jag2, and Rbpj, were expressed at significantly higher levels in the embryonic lacrimal gland than in the adult lacrimal gland (Figs. 1–3; Supplementary Table S4). We began our study of the NOTCH pathway by treating embryonic lacrimal glands with DAPT, a nonselective inhibitor of NOTCH signaling. We cultured lacrimal glands from E15.5 mice in the presence of 10, 50, and 100 μM of DAPT. Untreated lacrimal glands or lacrimal glands cultured in the presence of Dimethyl Sulfoxide (DMSO) (stock solutions of DAPT were prepared in DMSO) were saved as controls. Three days after treatment, the glands were collected and morphometrically analyzed. We found that while DAPT treatment did not affect significantly the overall size of the cultured embryonic lacrimal glands, it did exert a strong effect on average lobule size and average number of lobules (Fig. 5). DAPT treatment led to a reduction in the average size of lobules (93 ± 11 μm [DMSO], 66 ± 3 μm [10-μM DAPT], 60 ± 1 μm [50-μM DAPT], 73 ± 3 μm [100-μM DAPT]), but also to an increase in the average number of lobules (22 ± 2 [DMSO], 53 ± 2 [10-μM DAPT], 38 ± 4 [50-μM DAPT], 31 ± 1 [100-μM DAPT]; Fig. 5). We did not detect a difference between untreated and DMSO-treated lacrimal glands (Fig. 5). To demonstrate direct effect of Notch signaling inactivation, we used Notch1flox/flox mice. To this end, lacrimal glands isolated from Notch1flox/flox mice and wild-type animals were treated with Ad-CMV-iCre, the adenovirus constitutively expressing Cre recombinase that catalyzes site-specific recombination of DNA between loxP sites. Untreated Notch1flox/flox and wild-type lacrimal glands were saved as controls. Three days after Ad-CMV-iCre treatment, the glands were collected and morphometrically analyzed. In agreement with the DAPT data above, we found that Notch1 knockout in the embryonic lacrimal gland led to a reduction in the average size of lobules (90 ± 3 μm [wild-type untreated], 82 ± 1 μm [wild-type Ad-CMV-iCre treated], 81 ± 3 μm [Notch1flox/flox untreated], 56 ± 2 μm [Notch1flox/flox Ad-CMV-iCre treated]), but also to an increase in the average number of lobules (29 ± 3 [wild-type untreated], 37 ± 6 [wild-type Ad-CMV-iCre treated], 40 ± 2 [Notch1flox/flox untreated], 51 ± 2 [Notch1flox/flox Ad-CMV-iCre treated]), respectively (Fig. 6). To verify that the observed effect was due to reduced Notch1 expression, two groups of Notch1flox/flox untreated and Notch1flox/flox Ad-CMV-iCre–treated lacrimal glands were used to test Notch1 expression by quantitative RT-PCR. We found that Notch1 expression was reduced in Ad-CMV-iCre–treated lacrimal glands (100 ± 9% [Notch1flox/flox untreated] vs. 69 ± 3% [Notch1flox/flox Ad-CMV-iCre treated], P value = 0.02, n = 5).
Figure 5.
NOTCH signaling regulates the size and number of lobules in the developing LG: gamma secretase inhibition study. (A) Representative confocal images of DAPI-labeled developing lacrimal glands 3 days after treatment. The lacrimal glands were treated with a NOTCH signaling inhibitor (DAPT; 10, 50, and 100 μM). Controls were DMSO-treated or untreated. (B) Whole gland size, individual lobule size, and total number of lobules were measured and analyzed for each lacrimal gland (**P < 0.01, *P < 0.05, n = 5–7).
Figure 6.
NOTCH signaling regulates the size and number of lobules in the developing lacrimal gland: conditional Notch1 knockout study. (A) Representative confocal images of DAPI-labeled developing lacrimal glands 3 days after respective treatment. The lacrimal glands were treated with Cre-recombinase expressing adenovirus (Ad-CMV-iCre/Adeno-Cre) to induce Notch1 knockout. Controls included wild type (WT), virus treated wild types (WT Adeno-cre), and nonvirus treated Notchflox/flox glands (Notch1_lox). (B) Whole gland size, individual lobule size, and total number of lobules were measured and analyzed for each group of lacrimal glands (**P < 0.01, *P < 0.05, n = 5–7).
Discussion
Although the structure and function of the lacrimal gland have been studied extensively, the mechanisms behind lacrimal gland development, morphogenesis, and differentiation remain rather poorly understood. Enhancing our knowledge of these mechanisms will be critically important for the design of new treatment paradigms, such as postnatal lacrimal gland regeneration or lacrimal gland replacement therapies.6–8 In this study, comparing embryonic and adult lacrimal glands, we collected extensive data on lacrimal gland developmental biology, and further confirmed these findings in vitro. We leveraged this expression data to help identify new putative markers of a lacrimal gland stem/progenitor cell phenotype. Our findings also suggest that NOTCH-, WNT-, TGFβ-, and Hedgehog pathway-regulated EMT should be a critical mechanism in lacrimal gland development. In addition, our data indicate that the NOTCH signaling exert a strong effect on average lobule size and average number of lobules in the developing lacrimal gland.
This study has shed new light on the mechanisms of the lacrimal gland's development and morphology. Our data indicate that the majority of genes involved in the NOTCH-, WNT-, TGFβ-, and Hedgehog-signaling pathways are expressed in the lacrimal gland during both embryonic and adult stages. Such sustained expression lends credence to the hypothesis that the NOTCH-, WNT-, TGFβ-, and Hedgehog-signaling cascades play roles both in lacrimal gland development and in the maintenance of adult lacrimal gland homeostasis. Importantly, these cascades do not act independently, but instead maintain steady crosstalk between each other.25 We noted that the NOTCH, WNT, TGFβ, and Hedgehog pathways can activate regulators of EMT, and may therefore be involved in morphologic modulation of the developing gland.24–26 This observation led us to evaluate the expression of genes involved in EMT. In addition, we tested for expression of genes involved in MET. As expected, we found that many of these genes are indeed highly expressed in the lacrimal gland. Some of them (e.g., Zfhx1a, Twist1, Snai2, etc.) were highly upregulated in embryonic glands, while others (e.g., Snai1, Foxa1, Tcf4, etc.) were more highly expressed in the adult glands, suggesting that EMT and MET are involved in lacrimal gland development and in the maintenance of adult lacrimal gland homeostasis.26,27 Furthermore, our study revealed putative signaling cascades that may regulate lacrimal gland differentiation. As noted above, we found high embryonic expression of NOTCH pathway genes. Given the NOTCH pathway's essential role in preventing progenitor differentiation in the mammary gland (and many other tissues types), we suggest that the canonical NOTCH ligands Jag1/Jag2 and the noncanonical Dlk1 may activate Notch1/Notch3/Rbpj, preventing differentiation of all lacrimal gland cells expressing NOTCH receptors.23,28–32 Meanwhile, because Elf5 mediates mammary gland progenitor cell differentiation by inhibiting NOTCH signaling, we can suggest that a similar process may occur during lacrimal gland development, as Elf5 expression—while present in the embryonic gland—was significantly upregulated in the adult lacrimal gland.33
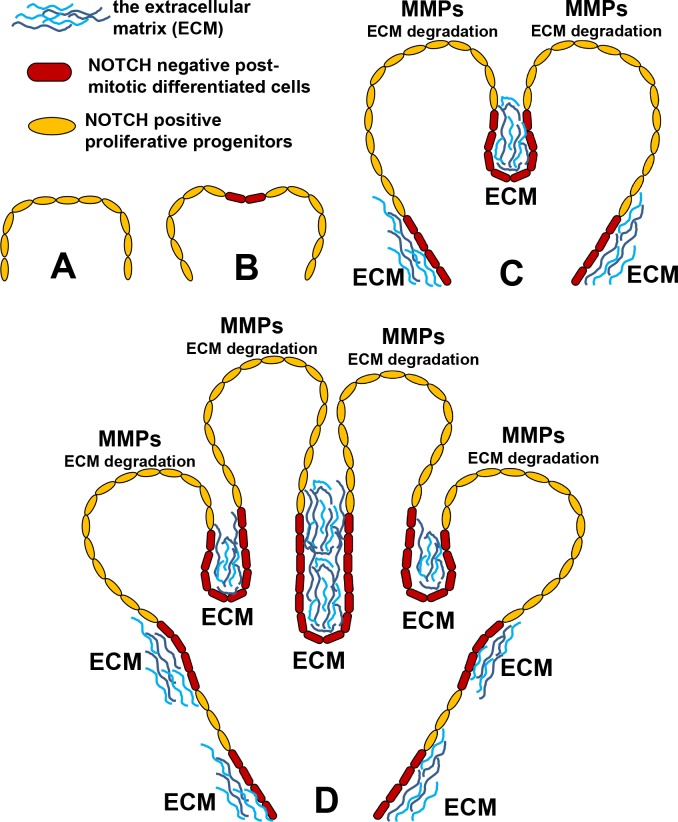
In order to directly evaluate the role of the identified pathways in lacrimal gland development, we chose to focus on the NOTCH signaling pathway. Our study allows us to link NOTCH signaling with lacrimal gland branching because inhibition of NOTH signaling significantly increased the average number of lobules and significantly reduced average lobule size observed. Branching processes are critical for the normal development of a number of organs, including the lung, kidney, pancreas, prostate, salivary gland, and lacrimal gland.34,35 The branching process in glands (including the lacrimal gland) begins with the formation of clefts within a single epithelial bud. As these clefts invaginate, they cleave the single bud into smaller buds, each of which grows to become a lobule. Recursive iteration of this process leads to the formation of an increasingly intricate glandular structure.34,35 Thus, our findings suggest that NOTCH signaling may regulate lacrimal gland branching by inhibiting cleft formation. We suspect that the observed results are associated with NOTCH signaling activity's control of progenitor cell cycle exit (into a postmitotic, differentiated phenotype). It was shown previously that extracellular matrix (ECM) and metalloproteases (MMPs; enzymes that degrade ECM) regulate the branching process in other glands.34–37 The ECM forms a barrier to gland growth, and if sufficiently high levels of ECM accumulate, a cleft is formed (as an analogy, imagine a balloon being inflated against a hard, narrow barrier; the balloon will distend around the barrier, creating the appearance of two separate “lobes” of one continuous structure). Metalloproteases activity degrades these ECM barriers, facilitating lobule formation and shaping the directionality of gland growth.34–37 If lacrimal gland progenitors highly express MMPs, while differentiated lacrimal cells predominantly produce ECM then NOTCH signaling inhibition should lead to an increased number of ECM-producing cells and, as a result, to an increased number of clefts (Fig. 7). Such a mechanism could explain how NOTCH-mediated distribution of progenitor and differentiated cells in the developing lacrimal gland may influence the branching process (Fig. 7). Future studies will be needed to confirm this proposed mechanism.
Figure 7.
NOTCH signaling may regulate lacrimal gland branching by controlling the transition from proliferative progenitor state into postmitotic differentiation state. (A) At the earliest stages of development, Notch positive lacrimal gland progenitors (yellow) proliferate, growing radially outward like an expanding balloon. (B) Cessation of Notch signaling causes some progenitors to exit the cell cycle and become Notch negative, differentiated cells (red). (C) Unlike Notch positive proliferating progenitors, Notch negative postmitotic cells secrete ECM (blue), which accumulates between them to form physical barriers (“clefts”). Progenitors express MMPs, enzymes that degrade ECM and allow the proliferating cells to continue their radial expansion. Consequently, the glandular tissue expands outward like a balloon being inflated against a solid structure. (D) Recursive iteration of this process gives rise to the complex branching and multilobular structure that is the mature lacrimal gland.
In conclusion, we have assembled here the most comprehensive list of putative lacrimal gland stem/progenitor cell markers to date (Table 4). In addition, available evidence appears to indicate important roles for the NOTCH-, WNT-, TGFβ-, and Hedgehog-signaling cascades in lacrimal gland development, morphogenic features, maintenance of phenotypes, and homeostasis. Our findings suggest a role of EMT and MET in both embryonic and adult lacrimal gland, possibly shedding new light on the biology of epithelial neoplasms in this tissue (i.e., adenoid cystic carcinoma). Finally, our data further suggest that NOTCH signaling may help regulate the lacrimal gland's branching morphogenesis. Thus, this study has allowed us to molecularly characterize, for the first time, the lacrimal gland as a whole, and to propose novel mechanisms that regulate its development.
Supplementary Material
Acknowledgments
The authors thank the Analytic Imaging Facility at the Bascom Palmer Eye Institute (BPEI). This study was possible due to the generous and visionary support of Nasser Ibrahim Al-Rashid, PhD.
Supported by grants in part from Nasser Al-Rashid Orbital Vision Research Center Grant (DP; Miami, FL, USA), GemCon Family Foundation Research Grant (DP; Palm Beach, FL, USA), National Eye Institute/National Institutes of Health (NIH) Grant R01 EY022348 (DI), and NIH Center Core Grant P30EY014801 (Bethesda, MD, USA).
Disclosure: G. Dvoriantchikova, None; W. Tao, None; S. Pappas, None; G. Gaidosh, None; D.T. Tse, None; D. Ivanov, None; D. Pelaez, None
References
- 1. Obata H. Anatomy and histopathology of the human lacrimal gland. Cornea. 2006; 25: S82–S89. [DOI] [PubMed] [Google Scholar]
- 2. Rocha EM,, Alves M,, Rios JD,, Dartt DA. The aging lacrimal gland: changes in structure and function. Ocul Surf. 2008; 6: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The epidemiology of dry eye disease: report of the Epidemiology Subcommittee of the International Dry Eye WorkShop (2007). Ocul Surf. 2007; 5: 93–107. [DOI] [PubMed] [Google Scholar]
- 4. Stern ME,, Beuerman RW,, Fox RI,, Gao J,, Mircheff AK,, Pflugfelder SC. The pathology of dry eye: the interaction between the ocular surface and lacrimal glands. Cornea. 1998; 17: 584–589. [DOI] [PubMed] [Google Scholar]
- 5. Schaumberg DA,, Sullivan DA,, Buring JE,, Dana MR. Prevalence of dry eye syndrome among US women. Am J Ophthalmol. 2003; 136: 318–326. [DOI] [PubMed] [Google Scholar]
- 6. Hirayama M,, Kawakita T,, Tsubota K,, Shimmura S. Challenges and strategies for regenerating the lacrimal gland. Ocul Surf. 2016; 14: 135–143. [DOI] [PubMed] [Google Scholar]
- 7. Hirayama M,, Tsubota K,, Tsuji T. Bioengineered lacrimal gland organ regeneration in vivo. J Funct Biomater. 2015; 6: 634–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirayama M,, Ogawa M,, Oshima M,, et al. Functional lacrimal gland regeneration by transplantation of a bioengineered organ germ. Nat Commun. 2013; 4: 2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tiwari S,, Ali MJ,, Balla MM,, et al. Establishing human lacrimal gland cultures with secretory function. PLoS One. 2012; 7: e29458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shatos MA,, Haugaard-Kedstrom L,, Hodges RR,, Dartt DA. Isolation and characterization of progenitor cells in uninjured, adult rat lacrimal gland. Invest Ophthalmol Vis Sci. 2012; 53: 2749–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. You S,, Kublin CL,, Avidan O,, Miyasaki D,, Zoukhri D. Isolation and propagation of mesenchymal stem cells from the lacrimal gland. Invest Ophthalmol Vis Sci. 2011; 52: 2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z,, Huang J,, Liu Y,, et al. FGF signaling activates a Sox9-Sox10 pathway for the formation and branching morphogenesis of mouse ocular glands. Development. 2014; 141: 2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qu X,, Pan Y,, Carbe C,, Powers A,, Grobe K,, Zhang X. Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development. 2012; 139: 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qu X,, Carbe C,, Tao C,, et al. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 2011; 286: 14435–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Y,, Carbe C,, Powers A,, et al. Bud specific N-sulfation of heparan sulfate regulates Shp2-dependent FGF signaling during lacrimal gland induction. Development. 2008; 135: 301–310. [DOI] [PubMed] [Google Scholar]
- 16. Makarenkova HP,, Ito M,, Govindarajan V,, et al. FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development. 2000; 127: 2563–2572. [DOI] [PubMed] [Google Scholar]
- 17. Dean C,, Ito M,, Makarenkova HP,, Faber SC,, Lang RA. Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development. 2004; 131: 4155–4165. [DOI] [PubMed] [Google Scholar]
- 18. Mattiske D,, Sommer P,, Kidson SH,, Hogan BL. The role of the forkhead transcription factor, Foxc1, in the development of the mouse lacrimal gland. Dev Dyn. 2006; 235: 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foster J, II,, Kapoor S,, Diaz-Horta O,, et al. Identification of an IGSF3 mutation in a family with congenital nasolacrimal duct obstruction. Clin Genet. 2014; 86: 589–591. [DOI] [PubMed] [Google Scholar]
- 20. Ueda Y,, Karasawa Y,, Satoh Y,, Nishikawa S,, Imaki J,, Ito M. Purification and characterization of mouse lacrimal gland epithelial cells and reconstruction of an acinarlike structure in three-dimensional culture. Invest Ophthalmol Vis Sci. 2009; 50: 1978–1987. [DOI] [PubMed] [Google Scholar]
- 21. Van Gelder RN,, von Zastrow ME,, Yool A,, Dement WC,, Barchas JD,, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990; 87: 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dvoriantchikova G,, Agudelo C,, Hernandez E,, Shestopalov VI,, Ivanov D. Phosphatidylserine-containing liposomes promote maximal survival of retinal neurons after ischemic injury. J Cereb Blood Flow Metab. 2009; 29: 1755–1759. [DOI] [PubMed] [Google Scholar]
- 23. Dvoriantchikova G,, Perea-Martinez I,, Pappas S,, et al. Molecular characterization of Notch1 positive progenitor cells in the developing retina. PLoS One. 2015; 10: e0131054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garg M. Epithelial-mesenchymal transition - activating transcription factors - multifunctional regulators in cancer. World J Stem Cells. 2013; 5: 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez DM,, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014; 7: re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lamouille S,, Xu J,, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tiwari N,, Tiwari VK,, Waldmeier L,, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013; 23: 768–783. [DOI] [PubMed] [Google Scholar]
- 28. Carvalho FL,, Simons BW,, Eberhart CG,, Berman DM. Notch signaling in prostate cancer: a moving target. Prostate. 2014; 74: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimojo H,, Ohtsuka T,, Kageyama R. Dynamic expression of notch signaling genes in neural stem/progenitor cells. Front Neurosci. 2011; 5: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bouras T,, Pal B,, Vaillant F,, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008; 3: 429–441. [DOI] [PubMed] [Google Scholar]
- 31. Leong KG,, Gao WQ. The Notch pathway in prostate development and cancer. Differentiation. 2008; 76: 699–716. [DOI] [PubMed] [Google Scholar]
- 32. Reedijk M. Notch signaling and breast cancer. Adv Exp Med Biol. 2012; 727: 241–257. [DOI] [PubMed] [Google Scholar]
- 33. Chakrabarti R,, Wei Y,, Romano RA,, DeCoste C,, Kang Y,, Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012; 30: 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gjorevski N,, Nelson CM. Branch formation during organ development. Wiley Interdiscip Rev Syst Biol Med. 2010; 2: 734–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakai T. Epithelial branching morphogenesis of salivary gland: exploration of new functional regulators. J Med Invest. 2009; 56 Suppl: 234–238. [DOI] [PubMed] [Google Scholar]
- 36. Moore KA,, Polte T,, Huang S,, et al. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Dev Dyn. 2005; 232: 268–281. [DOI] [PubMed] [Google Scholar]
- 37. Wiseman BS,, Sternlicht MD,, Lund LR,, et al. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003; 162: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.