SUMMARY
RNA-guided nucleases (RGNs) provide sequence-specific gene regulation through base-pairing interactions between a small RNA guide and target RNA or DNA. RGN systems, which include CRISPR-Cas9 and RNA interference (RNAi), hold tremendous promise as programmable tools for engineering and therapeutic purposes. However, pervasive targeting of sequences that closely resemble the intended target has remained a major challenge, limiting the reliability and interpretation of RGN activity and the range of possible applications. Efforts to reduce off-target activity and enhance RGN specificity have led to a collection of empirically derived rules, which often paradoxically include decreased binding affinity of the RNA-guided nuclease to its target. We consider the kinetics of these reactions and show that basic kinetic properties can explain the specificities observed in the literature and the changes in these specificities in engineered systems. The kinetic models described provide a foundation for understanding RGN targeting and a necessary conceptual framework for their rational engineering.
INTRODUCTION
Over the past decades, RNA-guided nucleases (RGNs) have emerged as powerful and versatile tools for genome editing and for manipulation of gene expression. RNAi, CRISPR-based genome editing, and ribozyme targeting utilize short oligonucleotides to guide enzymatic activity to particular DNA and RNA sequences through base-pairing interactions. The established rules for Watson-Crick base-pairing and their predictable energetics have led to the prevailing perspective that RGNs and related systems are readily programmable and inherently specific. Nevertheless, experimental studies have exposed limitations to RGN specificity, revealing persistent targeting of unintended sequences that resemble the target RNA or DNA sequence (‘off-targets’) (Jackson et al., 2003; Tsai and Joung, 2016). This limited specificity is one of several key challenges that must be overcome to allow RGN-based technologies to achieve their full impact in the laboratory, in engineering, and ultimately as therapies.
A series of successes in reducing off-target cleavage have been reported, most recently in the CRISPR-Cas9 field, by making changes in the nuclease or the RNA guide (Doench et al., 2016; Fu et al., 2014; Kleinstiver et al., 2016; 2015; Mali et al., 2013; Ran et al., 2013; Slaymaker et al., 2016; Tsai et al., 2014). A common theme that emerged from studies of CRISPR-Cas9 targeting is that specificity of targeting increases upon weakening interactions of the Cas9-RNA complex with the target DNA—e.g., by using truncated RNA guides (Fu et al., 2014; Tsai et al., 2015) or by introducing mutations to the protein-DNA interface (Kleinstiver et al., 2016; Slaymaker et al., 2016). This counterintuitive observation has been rationalized in terms of an “excess energy” model, postulating that binding energy beyond a certain threshold stabilizes binding to both correct and incorrect targets (Fu et al., 2014; Hu et al., 2016; Kleinstiver et al., 2016; Tsai et al., 2015; Tsai and Joung, 2016; Wyvekens et al., 2015). Similar observations have previously been made for RNAi, RNase H, and ribozyme targeting, and in the related fields of TALEN and zinc-finger-nuclease targeting (Guilinger et al., 2014a; Herschlag et al., 1991; Pattanayak et al., 2011; Pedersen et al., 2014; Østergaard et al., 2013). However, a mechanistic explanation for the “excess energy” phenomenon in CRISPR-Cas9 and RNAi has not been presented. Such a mechanistic explanation that takes into account the kinetics of individual steps of the targeting process will help transform isolated successes into generalizable approaches that will in turn direct the development of new rational strategies for improving the specificity of RGNs.
Here, we demonstrate the importance—and power—of kinetic considerations for determining and improving RGN specificity. RGN targeting can be described by a simple kinetic model that includes binding followed by a rapid downstream step such as cleavage, a model that is supported by published biochemical studies of RGNs and is consistent with the strategies utilized in the CRISPR-Cas9 and RNAi field to increase specificity. The concepts described here provide a conceptual framework for future engineering to enhance the specificity of RGN systems and for accurate predictions of RGN targets in vivo.
Kinetic regimes of enzyme specificity
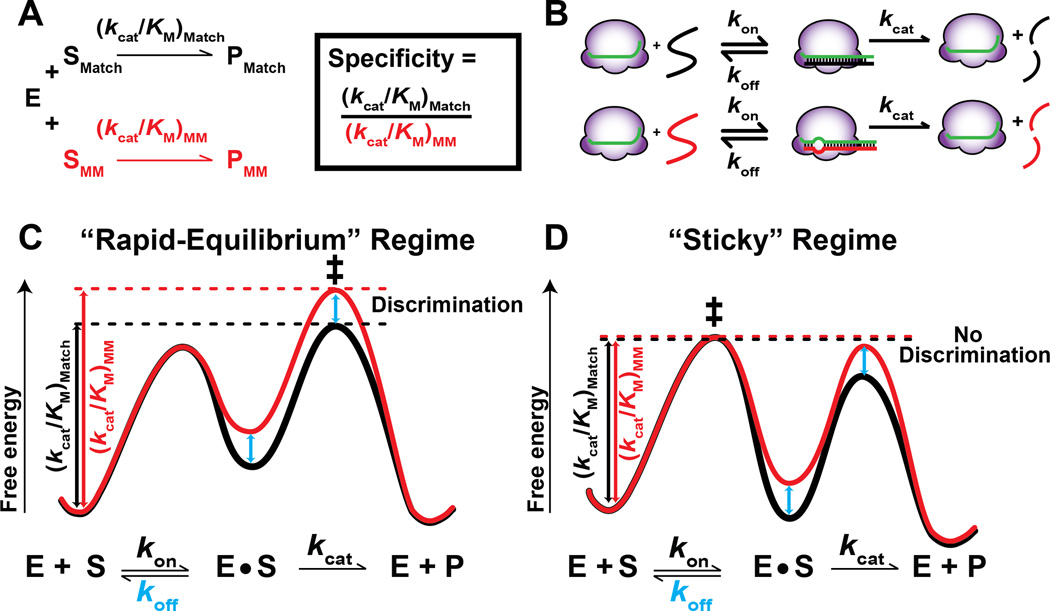
An intuitive approach to understanding and predicting RGN specificity is to use formalisms from enzyme kinetics. RGN specificity can be defined as the ratio of cleavage efficiencies for competing substrates (see Box 1). The prevailing perspective of the reaction catalyzed by RGNs, explicitly or implicitly, assumes rapid equilibrium of binding before the cleavage step (or other downstream steps (Wang et al., 2016)) (Box 1), an assumption also made in the original Michaelis-Menten kinetic formulation (Michaelis and Menten, 1913; Michaelis et al., 2011). In this “rapid equilibrium” kinetic regime, cleavage is slower than RGN dissociation (see Box 1), which allows RGN binding to reach equilibrium. Consequently, the difference in cleavage efficiencies (i.e. specificity) between a target and potential off-target is equal to the difference in binding affinity between the two sites. In an alternative kinetic regime, which we refer to as “sticky”, dissociation of the RGN from the target is slow relative to the downstream rate of cleavage (Briggs and Haldane, 1925) (Box 1). In this regime, binding equilibrium is not established, and the difference in cleavage efficiencies is less than the difference in binding affinity between two targets, In some cases, target and off-target sites can be cleaved with the same efficiency, regardless of differences in binding affinity, depending on the relative rate constants of dissociation and cleavage.
Box 1. Kinetic regimes of RGN specificity.
Specificity is determined by the ratio of kcat/KM values for a reaction with two different substrates, in this case—a matched (SMatch) versus a mismatched target (SMM) (Figure B1A; (Fersht, 1999)). kcat/KM represents the overall rate of the reaction and takes into account the rate constants with which the enzyme binds (kon), dissociates from (koff) and, for RGNs, cleaves a target (kcat) (Figure B1B). While here we discuss two directly competing substrates, the same concepts apply whether comparing two or many targets, with kcat/KM values, determined separately, dictating specificity for substrates in any mixture (Herschlag 1991). The relative magnitudes of the rate constants that comprise kcat/KM (i.e., kon, koff and kcat) determine the extent of discrimination, and two kinetic regimes can be defined based on the relative magnitudes of koff and kcat:
“Rapid-equilibrium“ regime: koff > kcat
Figure B1C illustrates the reaction using a free energy-reaction diagram, where the wells represent states and the peaks represent the barriers for transitions between those states. The heights of individual barriers are inversely proportional to the logarithm of the rate constant for each step, and kcat/KM corresponds to the free energy difference between the unbound state (E + S) and the highest reaction barrier, or transition state (‡). The RGN (‘E’) can bind the matched (black curve) or the mismatched (red curve) substrate. Once bound, there are peaks on either side the E•S complex: one for dissociation to E and S (koff) and one for cleavage to E + P (kcat). The central feature of “rapid-equilibrium” kinetics is that the peak for cleavage is higher relative to dissociation—both for the matched and the mismatched target (i.e., cleavage is the rate-limiting step). This means that the E•SMatch and E•SMM complexes dissociate faster than they are cleaved, such that E can equilibrate its binding before cleavage occurs. As a result, there is a preference for cleaving SMatch over SMM that matches the thermodynamic preference for binding SMatch over SMM.
“Sticky” regime: koff < kcat
In the “sticky” kinetic regime (Figure B1D), the barrier for dissociation of S from the E•S complex is higher than the barrier for cleavage. Thus, once either SMatch or SMM bind, they are ‘stuck’ to the enzyme and are cleaved essentially every time they bind. Because neither substrate has an opportunity to dissociate and achieve equilibration before cleavage, there is no discrimination between SMatch or SMM.
These models can be expanded to take into account more complex scenarios, including binding-site accessibility (e.g., determined by chromatin state), conformational changes, and non-cleavage activities of engineered RGNs (Hinz et al., 2015; Horlbeck et al., 2016; Isaac et al., 2016; Knight et al., 2015; Schirle et al., 2014; Sternberg et al., 2015; Wang et al., 2016).
Figure B1. RGN-catalyzed reaction and two regimes of specificity.
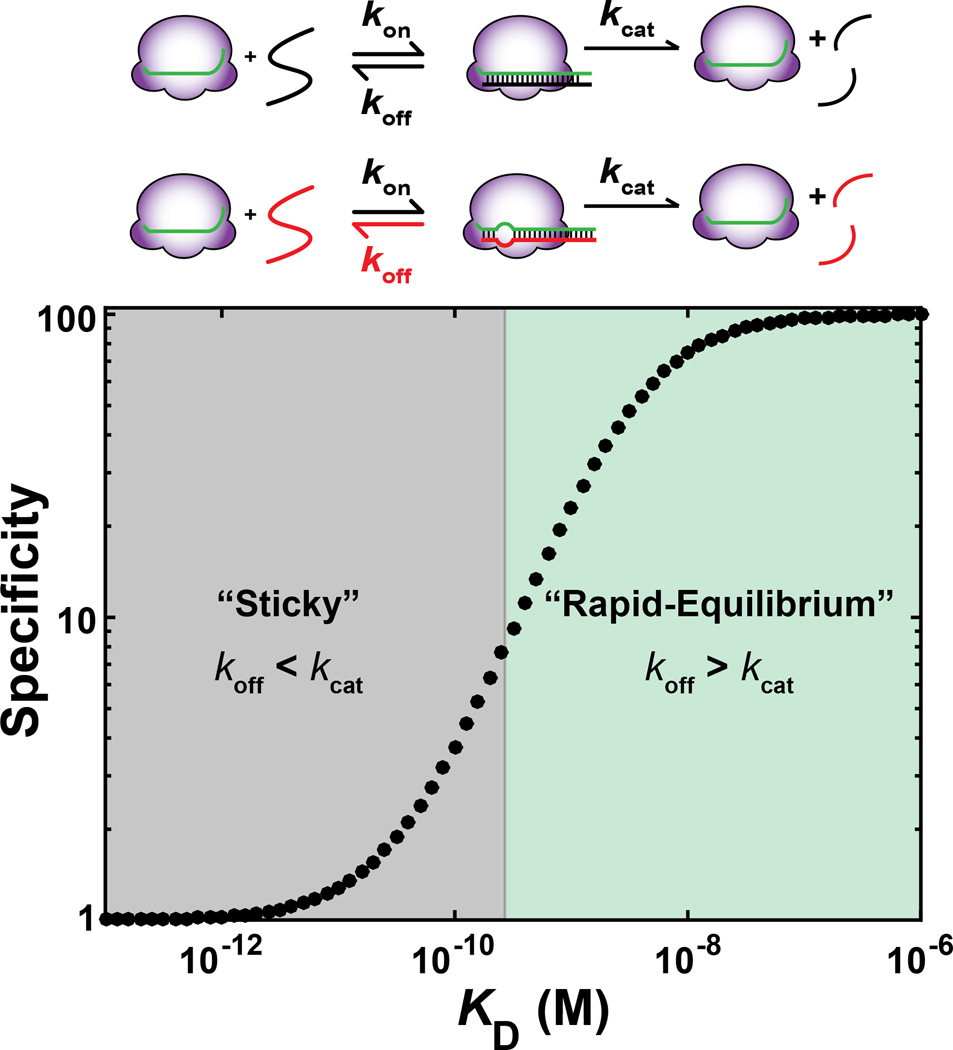
Figure 1 illustrates the predictions of a simple kinetic model for specificity between a target and a potential off-target. In this model, there are two steps, binding and cleavage, for both a target and off-target sequence. In the simplest scenario, we consider that the target and off-targets have the same on-rate (kon) and forward reaction rate (kcat) constant (Salomon et al., 2015; Wee et al., 2012), and that there is a 100-fold difference in dissociation rate constant (koff) between the target and off-target. As the absolute affinity for the target sequence is varied (e.g. by changing the GC content or length of the guide RNA), this model predicts two regimes of specificity. When targets have high binding affinities (i.e., slow dissociation rate constants), specificity between a target and potential off-target is absent or low (Figure 1, left). However, as the dissociation rates for both the target and the off-target increase (i.e., affinities decrease), specificity increases (Figure 1, right), eventually reaching the maximal level of 100-fold. Thus, the ‘excess energy’ model can be rationalized by a shift from the “sticky” enzyme regime to the “rapid-equilibrium” regime as the dissociation rate constant increases with concomitant improvement in specificity. While the example in Figure 1 used hypothetical kinetic values for illustrative purposes, we present evidence below that RGNs exist in a “sticky” enzyme regime and we discuss strategies for shifting RGN targeting to the more specific “rapid-equilibrium” regime.
Figure 1. Kinetic model for RGN targeting reaction.
Top. Two-step kinetic model illustrating a generic RGN targeting reaction with a matched (black) and mismatched (red) target. kon, koff, kcat: rate constants for target binding, dissociation and cleavage, respectively. Bottom. Prediction for specificity for the matched target assuming that the mismatched target is bound 100-fold less stably than the mismatched target, and that this difference is based on the dissociation rate (koff), while the association and cleavage rate constants are the same for both targets (kon = 1×107 M−1s−1, [E] = 1 nM, kcat = 3.5×10−2 s−1, based on values for mammalian AGO2 and the let7a miRNA and target (Salomon et al., 2015)). Specificity is defined as the ratio of kcat/KM values for matched versus mismatched target (Box 1). The KD values on the horizontal axis correspond to the equilibrium dissociation constants for the correct target (KD = koff/kon), where koff varies from 10−6 to 10 s−1.
RGNs in the “sticky” kinetic regime
Extensive biochemical studies have focused on the target binding and cleavage by Argonaute enzymes, and kinetic measurements of fly Ago2 and mammalian AGO2 enzymes support the model that these RGNs likely exist in the “sticky” kinetic regime (Salomon et al., 2015; Wee et al., 2012). In vitro, fly Ago2 cleaves a fully complementary target with a rate constant that is almost three orders of magnitude faster than dissociation (Table 1; kcat versus koff)(Wee et al., 2012). Thus, for every thousand fully complementary targets that bind, ~999 are cleaved and only one is expected to dissociate. The difference between cleavage (kcat) and dissociation (koff) is so large that even for targets with multiple mismatches, cleavage may occur before the target has an opportunity to dissociate.
Table 1.
Rate constants for fly Ago2 and mouse AGO2 in complex with let-7a guide RNA and fully complementary target RNA
| Enzyme | Temperature, °C |
Reference | koff (s−1) | kcat (s−1) | kcleave(s−1)a |
|---|---|---|---|---|---|
| Fly Ago2 | 25 | (Wee et al., 2012) | 8.8×10−5b | 6.1×10−2 | 4.4×10−2 |
| Mouse AGO2 |
37 | (Salomon et al., 2015) | ≤3.6×10−3c | 3.6×10−2 | 0.15 |
Cleavage rate constant is shown for reference, as kcat can be limited by steps other than cleavage (such as product release). The values are based on single-turnover cleavage measurements (fly Ago2) or by global fitting of single-molecule cleavage data (mouse AGO2).
Dissociation measurements were carried out with targets containing 2’-O-methyl ribose flanked by phosphorothioate linkage at the scissile site to prevent cleavage.
The dissociation rate constant was measured for a target with seed complementarity only and represents an upper estimate of the dissociation rate constant from the fully complementary target.
Recent single-molecule measurements indicate that mouse AGO2 is also in the “sticky” enzyme kinetic regime for substrates that are fully complementary to the guide RNA, and the same likely holds for the 99% identical human AGO2 (Table 1, Salomon et al., 2015). Moreover, in vitro kinetic measurements of Streptococcus pyogenes Cas9 strongly suggest that the enzyme operates in a “sticky” regime: the observed cleavage rate constants are fast (reported values range from ~1 – 10 min−1 (Gasiunas et al., 2012; Jinek et al., 2012; Sternberg et al., 2014; Szczelkun et al., 2014; Sternberg et al., 2015), while dissociation from fully matched targets has been slower than observed time scales, estimated to occur over hours (Knight et al., 2015; Sternberg et al., 2014; Singh et al., 2016; Richardson et al., 2016).
Operating in the “sticky” enzyme regime may be of value to RNAi and CRISPR systems in vivo. For example, fly Ago2 has been implicated in siRNA-mediated silencing for defense against viral infection, whereby foreign RNA is processed by the RNAi machinery to generate RNA guides against additional copies of the intruder RNA (Wang et al., 2006); When under a viral onslaught, maximizing cleavage rates may be more important than maximizing specificity, and a certain degree of promiscuity may be beneficial in RNAi and CRISPR defense systems to accommodate rapidly mutating sequences of invading nucleic acids (Carroll, 2013; Paez-Espino et al., 2015; Sun et al., 2013). Furthermore, minimizing off-target effects may not be a major selective pressure for bacterial RGNs (such as CRISPR-Cas9), in part because of the small sizes of prokaryotic genomes (e.g., the genome of Streptococcus pyogenes, the source of the widely used Cas9 protein, is only ~1 Mb, 3000 fold smaller than the human genome).
Individual RNA-guided targeting systems evolved under different sets of selective pressures that presumably have optimized specificities and turnover rates for particular natural contexts. Consequently, it may not be surprising that initial efforts to apply these systems for laboratory applications revealed specificities that fall short of those required to target unique sites in mammalian transcriptomes or genomes (Jackson et al., 2003; Tsai and Joung, 2016). Developing RGN systems to be highly specific tools may therefore require modifying their inherent affinities and reaction rates by engineering or selection, and RGNs from certain biological contexts may be better starting points for these efforts.
Strategies for engineering RGNs to increase specificity
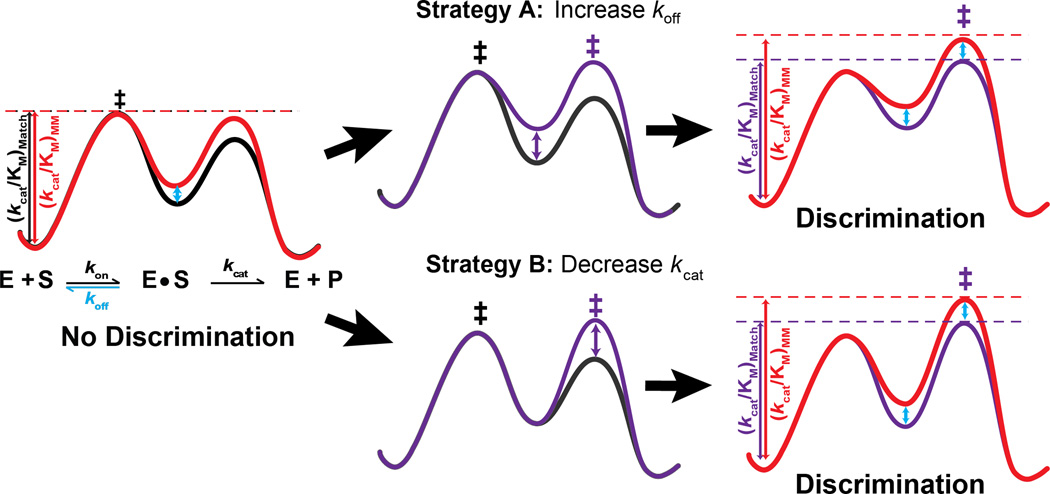
We present two general strategies for improving specificity based on the kinetic considerations laid out in the previous sections. The unifying premise is that the ability of an RGN to discriminate between correct and mismatched targets increases as dissociation becomes more favorable than cleavage. Thus, effective strategies to increase specificity will involve increasing koff or decreasing kcat, and the greatest discrimination will be achieved upon simultaneously changing both rate constants. Indeed, many of the successful approaches reported in the literature appear to follow these strategies (Table 2).
Table 2.
Strategies to improve specificity of RNA-guided targeting
| System | Modification | Hypothesized consequence |
Strategy A/Ba |
References |
|---|---|---|---|---|
| RNAi | Chemical modifications of guide RNA |
Weaker guide–target interactions/Perturbed active site |
A/B | (Jackson et al., 2006; Vaish et al., 2011) |
| Introduction of DNA nucleotides in the seed region |
Weaker guide–target interactions |
A | (Ui-Tei et al., 2008b) | |
| Introduction of G:U base- pairs in seed region |
Weaker guide–target interactions, weaker protein interactions with the guide–target helix |
A | (Ui-Tei et al., 2008a) | |
| Decreased GC content in guide RNA |
Weaker guide–target interactions |
A | (Gu et al., 2014) | |
| Introduction of seed mismatches to discriminate between alleles |
Weaker guide–target interactions |
A | (Dua et al., 2011; Ohnishi et al., 2008; Pfister et al., 2009) |
|
| Introduction of mismatches near the catalytic site |
Perturbed active site | B | (Pfister et al., 2009) | |
| CRISPR- Cas9 |
Truncated guide RNA |
Weaker guide–target interactions |
A | (Fu et al., 2014; Tsai et al., 2015) |
| Cas9 mutations neutralizing positive charge in the DNA binding grooves |
Weaker Cas9–target interactions |
A | (Kleinstiver et al., 2016; Slaymaker et al., 2016) |
|
| RNaseHb | Chemical modifications of guide DNA |
Perturbed active site | B | (Østergaard et al., 2013) |
Strategies A and B correspond to increasing koff and decreasing kcat, respectively, as described in the main text.
RNase H is technically a DNA-guided nuclease that cleaves the complementary RNA target.
We include it because it is conceptually similar to and has been utilized in similar fashion to the RGNs discussed herein.
Strategy A: Increasing koff
The RGN literature is rich in examples of increased specificity upon perturbing RGN-target interactions, and many of these examples could be explained by increased koff. The most straightforward strategy to weaken target-guide interactions involves shortening the guide to decrease the number of base-pairs formed with the target sequence. For example, truncated CRISPR guide RNAs showed reduced off-target activity, in some cases—with no observed reduction of cleavage of the intended target (Fu et al., 2014; Tsai et al., 2015). This observation is consistent with a switch from the “sticky” regime to a “rapid-equilibrium” regime (Figure 2, Strategy A), resulting in enhanced specificity. Although in practice RNA guides can only be shortened by a few bases—both to maintain sufficient number of contacts for robust on-target activity and because recognizing individual genomic sites imposes minimum length requirements on the guide—even truncating guide RNAs by 2–3 nucleotides provided substantial specificity enhancements in CRISPR-Cas9 targeting (Fu et al., 2014).
Figure 2. Strategies to increase specificity.
Left. Free energy profile representing the kinetics of a “sticky” enzyme for a fully complementary (black) and a mismatched target (red). The efficiency of cleavage, kcat/KM, is designated with vertical lines to the transition state (‡) of the reaction. Because dissociation is slow relative to cleavage, the efficiency of both reactions is the same and there is no specificity. The free energy diagram depicts a scenario in which the association rate (kon) and cleavage rate (kcat) are unchanged for the mismatched target, while the dissociation rate (koff) is faster for the mismatched versus matched target (blue arrow) (Salomon et al., 2015; Wee et al., 2012). Middle. Two strategies to increase discrimination, increasing koff and decreasing kcat, change the rate-limiting step from binding to cleavage (changes shown for the matched target only, but affect the off-target by the same amount). Reaction profiles are shown before (black) and after a change in the rate-limiting step (purple). Right. New free energy diagrams for the modified reactions of the matched (purple) and mismatched (red) target with koff increased (top) or kcat decreased (bottom). Modifications result in differences in kcat/KM (blue arrow) between the matched and mismatched substrates and thus enable discrimination.
Target binding can be weakened without sacrificing the number of residues recognized, by lowering the average base-pair stability. As was originally described in the context of ribozyme targeting (Herschlag, 1991), the decreased average stability of a base pair (e.g., in an AU-rich vs. a GC-rich sequence) leads to more discrimination in a random pool of target sequences, as more residues can be used in recognition before dissociation becomes slower than cleavage of mismatched targets. This concept can be extended to targeting by any RGN; for instance, specificity improvements in RNAi were observed after increasing the AU content in the guide (Gu et al., 2014), and the lower stability of DNA/RNA compared to RNA/RNA base pairs (Sugimoto et al., 1995) has been used to enhance specificity by replacing parts of the RNA guide with DNA (Ui-Tei et al., 2008b). It remains to be established to what extent the observed increases in specificity resulted from effects of these perturbations on koff rather than other steps in the maturation and loading of the RNA-induced silencing complex (Gu et al., 2014; Khvorova et al., 2003; Schwarz et al., 2003). In cases involving DNA/RNA base-pairs, the effects of increased AU content may be especially large because of the exceptionally low stability of dA-rU base-pairs (Martin and Tinoco, 1980). Certain chemical modifications of the guide could also destabilize target binding, by lowering base-pair stability and/or weakening contacts between target and the protein.
Target binding also can be weakened by introducing mutations into the nuclease itself. Significant improvements in CRISPR-Cas9 specificity have been achieved by substitutions of uncharged residues for positively charged ones in the DNA binding grooves of Cas9, hypothesized to weaken electrostatic interactions with the DNA backbone (Kleinstiver et al., 2016; Slaymaker et al., 2016). It remains to be established whether the effect is primarily on koff or other reaction steps.
Strategy B: Decreasing kcat
Decreasing the rate of target cleavage can increase the specificity of a “sticky” enzyme by promoting binding equilibration prior to cleavage (Figure 2, Strategy B). Although a decrease in on-target cleavage is generally viewed as a flaw and corresponding mutations and constructs are dismissed, it may be fruitful to reexamine nuclease and/or guide variants that give slower cleavage as they may be used to further optimize specificity.
How can kcat be manipulated? In vitro measurements have shown that mismatches in the miRNA-target duplex in proximity of the cleavage site reduce kcat (Wee et al., 2012), and the same likely applies to other RGNs, presumably as a result of active site perturbations. Chemical modifications of the guide have been reported to improve RNAi specificity, and some of these modifications may primarily affect kcat rather than the dissociation rate, or may affect both (Jackson et al., 2006; Vaish et al., 2011; Østergaard et al., 2013). Finally, nuclease domains with varying cleavage rates may be tethered to a catalytically inactive RGN to achieve a range of activities.
There will be a point at which these manipulations will make cleavage too slow to act on the required timescale, or dissociation will be too fast to efficiently target the desired molecules. Thus, there will be a kinetic “sweet spot” for a given RGN and application that can be identified through systematic studies. Moreover, some manipulations could potentially introduce detrimental effects on other steps (such as nuclease loading with the guide RNA), which will need to be accounted for. Given these complexities, one might be tempted to leave aside kinetic logic. We suggest instead, that conceptual strategies such as those presented herein are even more important in complex, less-intuitive systems and will be needed to guide rational re-engineering of RGNs, to identify the limits of improved RGN specificity, and to determine the in vivo factors that adjust RGN reaction properties.
A practical example: Discrimination between single-nucleotide polymorphisms
RGNs have long been recognized as promising therapeutics because of their potential to specifically target mutated genes associated with disease, especially autosomal dominant disorders, and several RNAi-based therapies are currently in clinical trials (Sullenger and Nair, 2016; Wittrup and Lieberman, 2015; Xiong et al., 2016). Among potential applications, targeting single-nucleotide polymorphisms (SNPs), which are linked to hereditary diseases, such as Huntington’s disease, hypertrophic cardiomyopathy, and amyotrophic lateral sclerosis, has proven to be a particularly challenging problem because the mutated target loci or associated SNPs differ from the wild-type allele by only a single nucleotide.
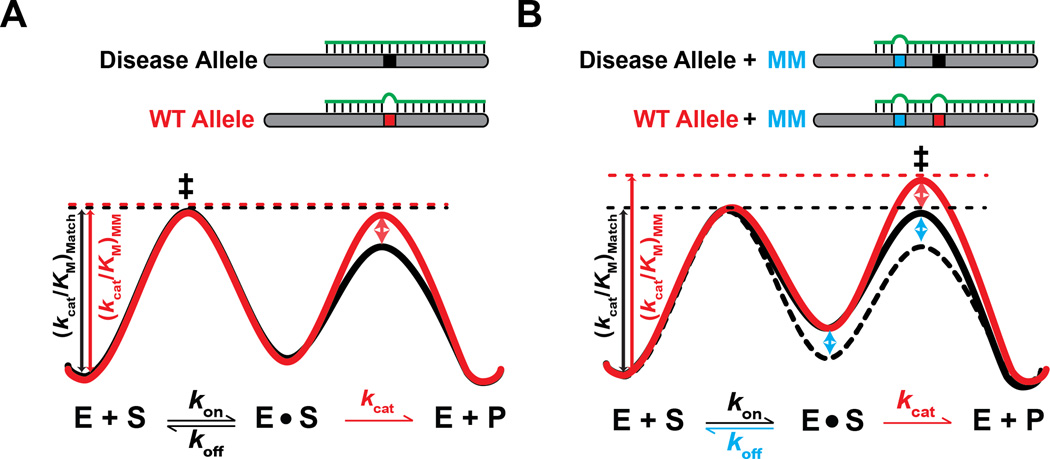
Early efforts toward therapeutic applications used short interfering RNA (siRNA) guides with full complementarity to the disease allele, and thus a single mismatch to the wild-type allele at the site of disease-associated polymorphism (Figure 3). However, cell culture studies indicated little or no discrimination in silencing between wild-type and disease alleles (e.g.,(Miller et al., 2003; Pfister et al., 2009; Schwarz et al., 2006; Yu et al., 2012)). Subsequently, much greater discrimination in siRNA-mediated silencing was found upon introduction of an additional mismatch against both the WT and disease allele (Dahlgren et al., 2008; Miller et al., 2003; Ohnishi et al., 2008; Pfister et al., 2009). This effect can be understood based on kinetic considerations. The first mismatch, against the wild-type allele only, is located in the central region of the guide RNA, and therefore likely reduces kcat (Ameres et al., 2007; Haley and Zamore, 2004; Wee et al., 2012). The second mismatch, against both the wild-type and the disease allele, is located in the seed region and is thus expected to increase koff (Salomon et al., 2015; Wee et al., 2012). Although neither individual mismatch is sufficient to allow discrimination (“sticky” regime is maintained), the two concomitant changes –slowing cleavage and speeding dissociation– together allow the system to cross a threshold from the “sticky” regime to the “rapid-equilibrium” regime, where binding equilibrates prior to cleavage and discrimination occurs (Figure 1). Here, we have rationalized results that were arrived at through trial and error; in the future, kinetic frameworks can be used to guide analogous successful engineering efforts.
Figure 3. Discrimination strategies for single-nucleotide polymorphisms (SNPs) in RNAi.
A. An siRNA that is fully complementary to the disease allele (black) contains a mismatch against the wild-type (WT) allele at the location of the SNP (red). This mismatch is often positioned in the central region of the siRNA (nucleotides 9–11) and is expected to affect the cleavage rate, kcat (Wee et al., 2012), as reflected in the free energy diagram (WT allele: red, disease allele: black). Because dissociation is still slower or comparable to cleavage, there is no (or little) discrimination in silencing (“sticky” regime, Box 1). B. Introduction of an additional seed mismatch (blue) destabilizes binding (blue arrow) of both the WT (red) and disease allele (black) targets relative to binding with only a single mismatch (dotted black line), and binding to the WT allele is further destabilized by the mismatch at the SNP location. This additional destabilization leads to a change in kcat/KM, and thus an increase in discrimination between the two alleles, by allowing the system to cross the threshold from a “sticky” kinetic regime to a “rapid-equilibrium” kinetic regime (Figure 1).
Conclusions
RGN systems hold tremendous technological and therapeutic promise, and recent advances in increasing their delivery, stability, and specificity have markedly lowered the barriers to wide application of these potent and versatile tools (Haussecker and Kay, 2015; Hendel et al., 2015; Hu et al., 2016; Sullenger and Nair, 2016; Wittrup and Lieberman, 2015; Xiong et al., 2016). Here we approached the widespread problem of off-target effects. We explain previous observations of RGN specificity by considering the kinetics of the targeting reaction, and we codify these observations in terms of a simple and generalizable model. This model leads to rational strategies for minimizing off-target activity and will help guide efforts to design more specific RGN systems.
The most obvious culprit in the widespread inability of RGN systems to efficiently discriminate between targets and very similar sequences is the inherent high stability of base-pairing interactions that underlie target recognition (Herschlag et al., 1991). Very slow dissociation of stable target-guide duplexes leads to “sticky” kinetics, where the RGN cleaves matched and partially mismatched targets before having an opportunity to discriminate between them. This limitation appears to be alleviated, in part, by RGN proteins that destabilize guide-target interactions by deforming the duplex and blocking helix propagation along the full length of the guide (Bartel, 2009; Elkayam et al., 2012; Nakanishi et al., 2013; 2012; Schirle and MacRae, 2012; Schirle et al., 2014), and by conformational changes that dissipate some of the binding energy (Alber, 1981; Herschlag, 1988; Jiang et al., 2015; Johnson, 2008; Schirle et al., 2014; Sternberg et al., 2014; Sternberg et al., 2015). Effective general strategies to improve RGN specificity are through further weakening of the RGN-target interactions and slowing of cleavage rates.
Our simple kinetic framework highlights fundamental commonalities between different RGN systems and we hope these similarities will generate cross-talk between RNAi, CRISPR and related RGN fields. Although it might appear that the complexities and idiosyncrasies of each RGN system render descriptions from the standpoint of basic principles hopelessly naïve, we believe just the opposite to be true. To understand and ultimately tame a complex system, a stepwise approach is needed. For each RGN, it will be important to build out from this simple, common framework to account for the individual reaction steps for that RGN, and the kinetics and thermodynamics of each step.
To our knowledge, cleavage and dissociation kinetics are not currently taken into account in any prediction algorithm for RGN target-sites and off-target effects. Quantitative studies of RGN systems (Chandradoss et al., 2015; De et al., 2013; Haley and Zamore, 2004; Jo et al., 2015; Salomon et al., 2015; Sternberg et al., 2014; Wee et al., 2012; Sternberg et al., 2015; Singh et al., 2016) inaugurate the types of analyses that will allow physical models to be developed and predictions to be made for specificity and efficacy. Careful, in-depth studies that build on these assays and results will lead to frameworks that provide quantitative predictions for each RGN system and identify steps whose modulation will improve specificity. With this knowledge, cellular factors that interact with these systems and alter their kinetic and thermodynamic properties can be identified. We hope that this perspective will stimulate these mechanistic studies of RGNs, which in turn will enable more accurate predictions and rational approaches to RGN re-engineering and application.
Box 2. Other strategies.
Other successful strategies for improving RGN specificity have been reported (Hu et al., 2016) and here we briefly note several of these approaches to place them in the context of basic kinetic and thermodynamic principles, analogous to the discussions above.
Cleavage guided by pairs of guide RNAs
Improvements in specificity have been observed with engineered Cas9 variants that only allow double strand cleavage upon two nearby binding events, i.e., by using a Cas9 nickase (Mali et al., 2013; Ran et al., 2013) or fusion of catalytically deactivated Cas9 with a monomer of the dimeric FokI nuclease (Guilinger et al., 2014; Tsai et al., 2014). In this approach, there are two binding and one or two cleavage events, which can be modeled by elaborating on the kinetic schemes presented herein and ultimately yield greater specificity than for each guide RNA alone.
Using low RGN concentrations
Improvements in specificity have been observed with lower RGN concentrations: i.e., with less production of the RGN complex or when RGN activity was controlled with small molecule activators and repressors (reviewed in Hu et al., 2016). At very high RGN concentrations, decrease in specificity can arise if the RGN concentration exceeds the amount of substrates and their respective KM values, such that targets and off-targets no longer need to compete for a common limiting pool of RGN complexes (Herschlag, 1988).
Limiting reaction time
Limiting the time of the RGN reaction has also been observed to increase specificity (Hu et al., 2016). In much of RGN literature ‘observed specificity’ corresponds to the relative amounts of target versus off-target cleaved at a given sampling time point, rather than relative rates of the reaction (Box 1). Indeed, the cleavage of all possible targets will go to completion at some point if a reaction is allowed to proceed long enough—i.e., the slower-cleaving off-targets will ‘catch up’ to targets and the apparent discrimination will lessen over time. This issue may account, in part, for the range of observed specificities for a particular RGN (Tsai and Joung, 2016); conversely, monitoring specificity over time will help in understanding the reason for a particular specificity outcome, and thereby help guide subsequent efforts to enhance specificity.
Acknowledgments
We thank Liang Wee, members of the Herschlag and Bartel labs, Matt Porteus, Andy Fire, Stanley Qi and Mark Kay for helpful discussions, comments, and for critical advice throughout this work. N.B. was supported by an NSF graduate fellowship, and this work was supported by a grant to D.H. from the National Institutes of Health [P01GM066275].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
N.B., I.J., and D.H. wrote the paper.
The authors declare no conflict or competing interests.
REFERENCES
- Alber TC. PhD thesis, Massachusetts Institute of Technology. 1981. Structural origins of the catalytic power of triose phosphate isomerase. [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GE, Haldane JB. A note on the kinetics of enzyme action. Biochem. J. 1925;19:338–339. doi: 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Staying on target with CRISPR-Cas. Nature Publishing Group. 2013;31:807–809. doi: 10.1038/nbt.2684. [DOI] [PubMed] [Google Scholar]
- Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. A dynamic search process underlies MicroRNA targeting. Cell. 2015;162:96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren C, Zhang H-Y, Du Q, Grahn M, Norstedt G, Wahlestedt C, Liang Z. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 2008;36:e53. doi: 10.1093/nar/gkn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De N, Young L, Lau P-W, Meisner N-C, Morrissey DV, MacRae IJ. Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol Cell. 2013;50:344–355. doi: 10.1016/j.molcel.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Zhang Y, Jin L, Huang Y, Zhang F, Bassik MC, Kampmann M, Kay MA. Weak base pairing in both seed and 3' regions reduces RNAi off-targets and enhances si/shRNA designs. Nucleic Acids Res. 2014;42:12169–12176. doi: 10.1093/nar/gku854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, Tsai SQ, Sander JD, Joung JK, Liu DR. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods. 2014a;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Publishing Group. 2014b;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Kay MA. RNA interference. Drugging RNAi. Science. 2015;347:1069–1070. doi: 10.1126/science.1252967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. The role of induced fit and conformational changes of enzymes in specificity and catalysis. Bioorg Chem. 1988:62–96. [Google Scholar]
- Herschlag D, Piccirilli JA, Cech TR. Ribozyme-catalyzed and nonenzymatic reactions of phosphate diesters: rate effects upon substitution of sulfur for a nonbridging phosphoryl oxygen atom. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- Hinz JM, Laughery MF, Wyrick JJ. Nucleosomes Inhibit Cas9 Endonuclease Activity in Vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, Torigoe SE, Tjian R, Weissman JS. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5 doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Davis KM, Liu DR. Chemical Biology Approaches to Genome Editing: Understanding, Controlling, and Delivering Programmable Nucleases. Cell Chemical Biology. 2016;23:57–73. doi: 10.1016/j.chembiol.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Isaac RS, Jiang F, Doudna JA, Lim WA, Narlikar GJ, Almeida R. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife. 2016;5 doi: 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo MH, Shin S, Jung S-R, Kim E, Song J-J, Hohng S. Human Argonaute 2 has diverse reaction pathways on target RNAs. Mol Cell. 2015;59:117–124. doi: 10.1016/j.molcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J Biol Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, Beheiry, El M, Masson J-B, Dahan M, Liu Z, Doudna JA, Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FH, Tinoco I. DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L, Menten ML. Die Kinetik der Invertinwirkung. Biochem. Z. 1913;49:333–369. [Google Scholar]
- Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. Biochemistry. 2011 doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013;3:1893–1900. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio. 2015;6 doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L, Hagedorn PH, Lindholm MW, Lindow M. A Kinetic Model Explains Why Shorter and Less Affine Enzyme-recruiting Oligonucleotides Can Be More Potent. Mol Ther Nucleic Acids. 2014;3:e149. doi: 10.1038/mtna.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel J-P, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nature Publishing Group. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Doudna JA, Ha T. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun. 2016;7:12778. doi: 10.1038/ncomms12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. Phage mutations in response to CRISPR diversification in a bacterial population. Environ. Microbiol. 2013;15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Publishing Group. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008a;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008b;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish N, Chen F, Seth S, Fosnaugh K, Liu Y, Adami R, Brown T, Chen Y, Harvie P, Johns R, Severson G, Granger B, Charmley P, Houston M, Templin MV, Polisky B. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute Divides Its RNA Guide into Domains with Distinct Functions and RNA-Binding Properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MJ, Guegler CK, Doudna JA, Greenleaf WJ. High-throughput biochemical profiling reveals Cas9 off-target binding and unbinding heterogeneity. bioRxiv. 2016 doi: 10.1073/pnas.1700557114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens N, Tsai SQ, Joung JK. Genome Editing in Human Cells Using CRISPR/Cas Nucleases. Curr Protoc Mol Biol. 2015;112:31.3.1–31.3.18. doi: 10.1002/0471142727.mb3103s112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Chen M, Lim WA, Zhao D, Qi LS. CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annu Rev Genomics Hum Genet. 2016;17:131–154. doi: 10.1146/annurev-genom-083115-022258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP, Corey DR. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard ME, Southwell AL, Kordasiewicz H, Watt AT, Skotte NH, Doty CN, Vaid K, Villanueva EB, Swayze EE, Bennett CF, Hayden MR, Seth PP. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]