Abstract
Early childhood is a critical stage for the foundation and development of both the microbiome and host. Early-life antibiotic exposures, cesarean section, and formula feeding could disrupt microbiome establishment and adversely affect health later in life. We profiled microbial development during the first two years of life in a cohort of 43 US infants, and identify multiple disturbances associated with antibiotic exposures, cesarean section, and diet. Antibiotics delayed microbiome development and suppressed Clostridiales, including Lachnospiraceae. Cesarean section led to depleted Bacteroidetes populations, altering establishment of maternal bacteria. Formula-feeding was associated with age-dependent diversity deviations. These findings illustrate the complexity of early-life microbiome development, and microbiota disturbances with antibiotic use, cesarean section, and formula feeding that may contribute to obesity, asthma, and other disorders.
One Sentence Summary
Antibiotics, cesarean section, and infant formula alter patterns of microbial acquisition and succession during the first 2 years of childhood.
INTRODUCTION
The establishment of stable microbial communities within the gastrointestinal tract closely parallels host growth and immune development during early life (1). The intestinal microbiome helps regulate host metabolism (2) and immune function (3–4), and thus could play an important role in directing host development. Delayed or altered establishment of the intestinal microbiota in childhood, termed microbiota immaturity, has been associated with diarrhea and malnutrition in Bangladeshi children (5). The causes of these microbiota disturbances and their consequences in other populations have not been established, but they may be linked to host development.
Antibiotic use during childhood is prevalent in most parts of the world but the effects on microbiota maturation and human health are poorly characterized (6). The average US child receives about three antibiotic courses by the age of 2, and 10 courses by the age of 10 (7). Antibiotics directly perturb the intestinal microbiota, leading to altered compositional states in children and adults (6, 8), but the consequences of these changes on host physiology are not well-understood. Antibiotic exposure in children has been associated with increased risk of obesity (9), diabetes (10), inflammatory bowel disease (11), asthma (12), and allergies (13). We have shown previously that antibiotic exposure leads to increased adiposity in mice (14), that early-life exposures lead to prolonged effects on host metabolic characteristics (15, 16), and that the disturbed intestinal microbiota mediate these host effects (16).
Other disturbances, including birth mode and infant diet (17), also impact the intestinal microbiota during early life and are associated with later-in-life adiposity and other clinical effects. Cesarean delivery has been associated with asthma (18), allergies (19), type 1 diabetes (20), and obesity (21), possibly due to diminished exposure to maternal microbes during birth. Formula feeding similarly disrupts the intestinal microbiota (17) and may impair immune development (22) and normal metabolism (23).
While the impacts of antibiotic exposures on intestinal dysbiosis in adults are well-characterized, less attention has been given to their effects on microbiota development during early childhood (6). We hypothesized that antibiotics and other early disturbances may alter microbiome establishment during early life, potentially explaining associations with emerging health issues. We examined the intestinal microbiota to model its development over 2 years of life in a cohort of 43 healthy urban US infants. We then assessed the effects of birth mode, infant nutrition, and antibiotic exposures on intestinal microbiota development in this population. Antibiotic exposures, formula feeding, and cesarean section delayed and altered development of the gut microbial population, suggesting a possible link to the health risks associated with these procedures (18–21).
RESULTS
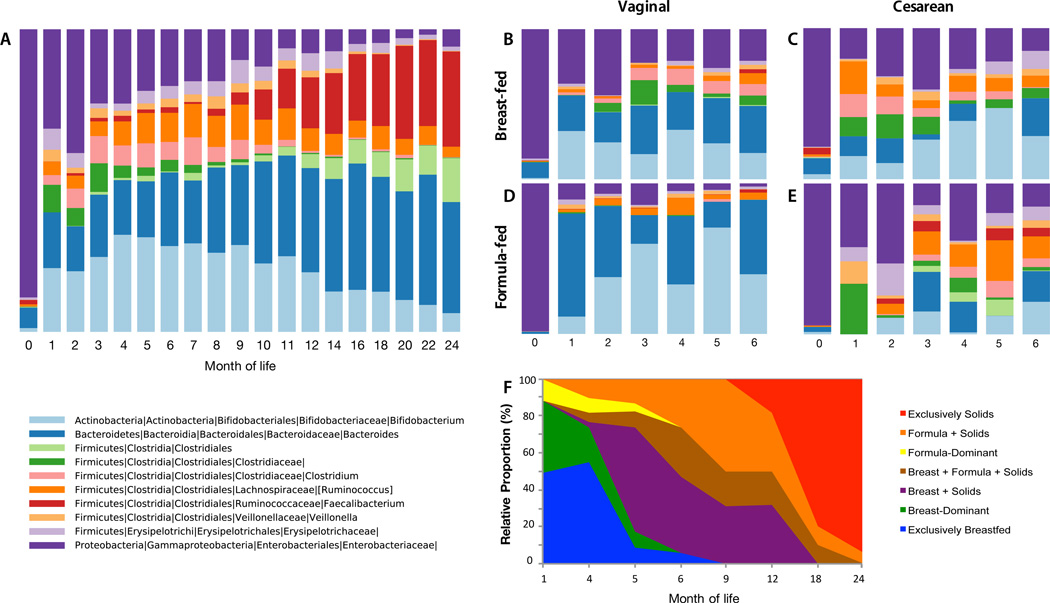
A total of 43 infants were enrolled for follow-up for up to the age of 2 years. Stool samples were collected from these infants and stool samples, vaginal swabs, and rectal swabs were collected from their mothers pre- and post-partum. Birth mode, feeding, and systemic antibiotic exposures in the infants are shown in Table 1. The succession of bacterial taxa during the first 2 years of life followed a predictable pattern (Fig. 1A), consistent with prior studies of the early-life microbiota (5, 24, 25). In the first month of life, stools were dominated by facultative aerobic Enterobacteriaceae before yielding to strict anaerobes — principally Bifidobacterium, Bacteroides, and Clostridium (Fig. 1A). These taxa were gradually displaced between months 6–24 by a diverse mixture of Clostridiales, roughly corresponding to the introduction and increased use of solid foods in these infants (Fig. 1G). However, even among infants who received no antibiotics in the first six months of life, those differing by birth mode and predominant diet showed substantial early differences (Fig. 1B–E; Table S6, Fig. S1). During this 2-year period, the microbiome was characterized by a period of community assembly, undergoing gradual succession of different taxa. While the microbiota begins to resemble an adult microbiota around 2 years of age (see below), it has not yet achieved an adult-like state, characterized by many different alternative states of semi-stable climax communities that exist in quasi-equilibrium (26). Hence, we focused on the trajectory of microbiota development in children in the context of early disturbances.
Table 1.
Characteristics of the 43 children in the study, by systemic antibiotic exposure, delivery mode, and diet
| Vaginal Delivery (N=24) | Cesarean Section (N=19) | |||
|---|---|---|---|---|
| Antibiotics + | Antibiotics − | Antibiotics + | Antibiotics − | |
| Breast milk-dominant | 10 (23%)* | 10 (23%) | 8 (19%) | 3 (7%) |
| Formula-dominant | 2 (5%) | 2 (5%) | 5 (12%) | 3 (7%) |
Values indicate number of children (% of total) categorized by delivery mode, whether they were exposed to antibiotics at any time during the study, and their predominant diet during the first 3 months of life.
Fig. 1. Microbial and dietary succession viewed over the first two years of life.
Mean relative abundance of fecal bacteria at the genus level at each month of life, for taxa with ≥1% mean relative abundance across all samples. Panel A. All subjects, first 2 years. Panels B–E: The first 6 months of life for the 32 subjects who were not antibiotic-exposed, organized by delivery mode [Vaginal (V), or Cesarean (C)], and predominant feeding mode [Breast (B), Formula (F). Group n’s are: V-B (15), C-B (7), V-F (3), C-F (7). Panel F: Dietary trends in all infants across the study period.
Antibiotic exposures alter bacterial diversity
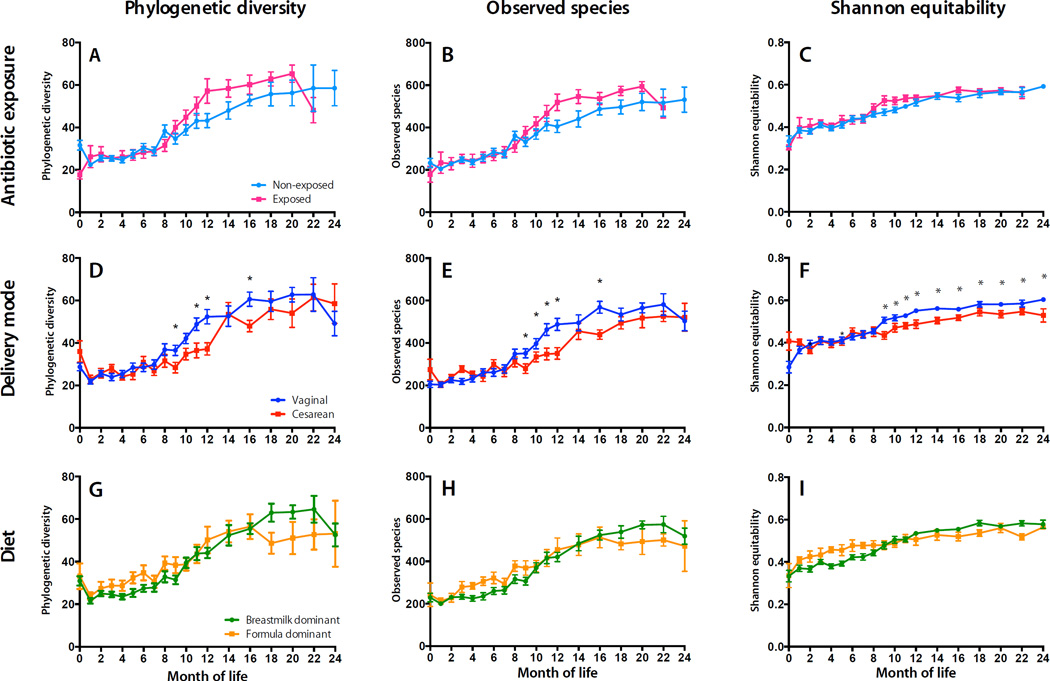
To assess the effects of disturbances on bacterial diversity, we measured bacterial richness, phylogenetic diversity, and evenness in each sample (Fig. 2). Antibiotic use significantly diminished phylogenetic diversity and richness immediately following birth (P < 0.0001), but accelerated their rates of increase during the first year of life (Fig. 2A–C, Table S6). However, α-Diversity was not significantly suppressed in individual children immediately following antibiotic administration (P > 0.05) (Fig. S2). Thus, antibiotic exposure altered the trajectory of α-diversity changes during the first 2 years of life, but transient effects were inconsistent.
Fig. 2. α-Diversity over the first two years of life in relation to early-life exposures.
Left column, Mean Phylogenetic Diversity (PD) ± SEM; second column, Mean observed OTUs ± SEM; third column, Mean Shannon Equitability (evenness) ± SEM. α-Diversity levels are shown for antibiotic use (Panels A-C), delivery mode (D-F), and diet (G-I). Asterisks and brackets indicate significant (LME P < 0.05) group differences at baseline or rate-of-change differences across age ranges.
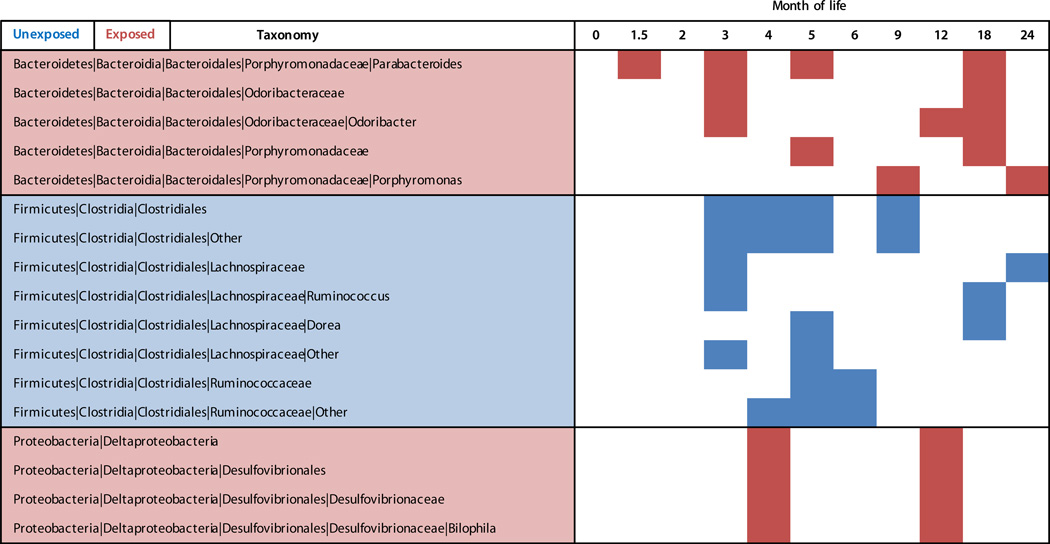
β-Diversity, measuring similarities between samples as a function of microbial composition, assesses how disruptions impact the composition and recovery of an entire microbial ecosystem. Infants who had not been exposed to antibiotics and those who had been exposed at any prior time differed significantly (Permutational MANOVA P < 0.001, R2 < 0.01) (Table S7). This observation provides evidence that antibiotics altered the intestinal microbiota in infants, but the effects were smaller than delivery mode (R2 = 0.02) and profoundly smaller than age (R2 = 0.14) (Table S7). Antibiotic exposure was associated with deficits in Clostridiales and Ruminococcus from 3 to 9 months of life, but with no consistent changes in other taxa (Fig. 3). These subjects were differentially exposed by age and by antibiotic class; thus, variation in alterations in most taxa is not surprising. Drug type, timing, route, duration, underlying conditions, number of exposures, and differences between individual subjects all may be confounding factors.
Fig. 3. Antibiotic exposure alters bacterial abundance.
Antibiotic exposure significantly altered abundance of diverse bacterial taxa over the first two years of life. Based on LefSe analysis, red-shaded taxa (rows) were significantly (P < 0.05) more abundant in antibiotic-exposed infants at the given time points (columns); blue shading indicates more abundant in unexposed infants.
Antibiotic exposures delay microbiota maturation
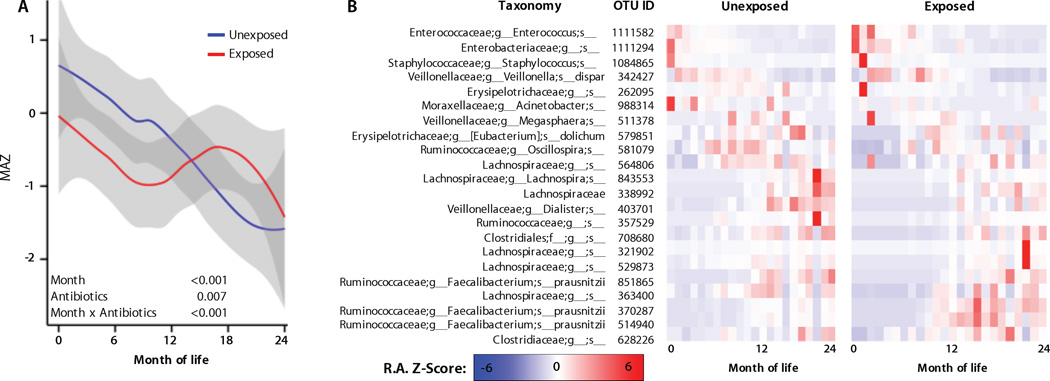
During the first 2–3 years of life, intestinal microbiota undergo a gradual succession, and while a large degree of inter-individual variation occurs, these age-dependent succession patterns share many features in different human populations (24). Microbiota maturation, then, is defined as the rate at which a child's microbiota develops, as measured by these age-dependent successional stages; a “mature” microbiota contains certain taxa that are biomarkers for that child's age group, while an “immature” or delayed microbiota resembles that of a younger child. Delayed microbiota maturation, as defined in healthy children, mirrors physiological disturbances in the host (5), and occurs in mice exposed to antibiotics (15). Thus, we examined whether antibiotic exposures and other disturbances similarly altered microbiota maturation in children, comparing relative maturation rates in a reference group (vaginally delivered, breast-fed, no pre-, peri-, or post-natal antibiotics), and antibiotic-exposed subjects using a Random Forests (27) regression model to predict a child's age as a function of their microbial composition, as reported (5). A defined microbiota maturation of the reference samples could be predicted using 22 key OTUs that explained the greatest degree of variation (pseudo R2 = 82.1) in the model and are thus biomarkers for normal development in this cohort (Fig. 4). Children exposed to antibiotics showed delayed microbiota maturation compared to those not exposed to antibiotics (Fig. 4A, Table S8). These effects were most pronounced during months 6–12, and thereafter no significant effect was observed. A delayed maturation pattern during early childhood was due to depletion of specific OTUs, including constituents of Enterobacteriaceae, Lachnospiraceae, and Erysipelotrichaceae (Fig. 4B). Within this cohort, intestinal bacterial communities followed a predictable pattern throughout the first two years (Fig. 1), but antimicrobials disturbed microbial succession at the OTU level, delaying microbial community development in the intestine relative to unexposed children (Fig. 4).
Fig. 4. Antibiotic exposure delays microbiota maturation during early life.
A, Microbiota-by-age Z-scores (MAZ) at each month of life between antibiotic-exposed and unexposed infants (infants never exposed to systemic pharmacologic antibiotic doses prior to the sampling time). MAZ scores indicate the number of standard deviations from the mean predicted age of age-matched control samples, as a function of microbiota maturation. Grey margins represent 95% confidence limits. Asterisks and brackets indicate significant (LME P < 0.05) group differences at baseline or rate-of-change differences across age ranges. The “unexposed” group contains both training set samples (from children who were never exposed to pre-, peri-, or post-natal systemic antibiotics; were vaginally delivered; and dominantly breast-fed), and all other samples from children who had not been previously exposed to systemic post-natal antibiotics. B, OTU abundance heat maps illustrate the relative abundance (RA) Z-scores of 22 maturity-marker OTUs in the antibiotic-exposed and unexposed groups throughout life. These OTUs were selected as those that best predict age of life in the control group, and hence can be used as markers of normal maturity. Substantial departures from the normal maturation profile alter predicted age of other samples. The color scale represents relative abundance (RA) Z-scores for each OTU, (i.e., the number of standard deviations from the mean RA of that OTU) across all samples at that age.
An important consequence of altered microbial patterns is changes to the functional gene repertoire present within the gut microbiome. We found substantial differences in the maturation of gene functions in the imputed metagenome (28) with relation to perturbations in exposures including antibiotics and predominant diet (Supplemental Materials, Fig. S3, S4, Table S9), but not delivery mode (Table S9).
Delivery Mode alters intestinal diversity
The first major microbial exposure for a vaginally born infant is in the birth canal, a potentially important event for establishing a healthy microbiome early in life. Cesarean section bypasses this exposure, altering the initial pool of microbes to which the neonate is exposed (29). We sought to investigate the impact of delivery mode on microbiota maturation and diversity during the first two years of life.
Cesarean-delivered infants had significantly altered intestinal bacterial richness and phylogenetic diversity (ANOVA P < 0.05), compared to vaginally born infants, throughout the first 2 years of life, and significantly diminished growth of evenness during the first year (Fig. 2D–F) (Table S6). These effects were not due to differences in absolute bacterial abundance (Fig. S5).
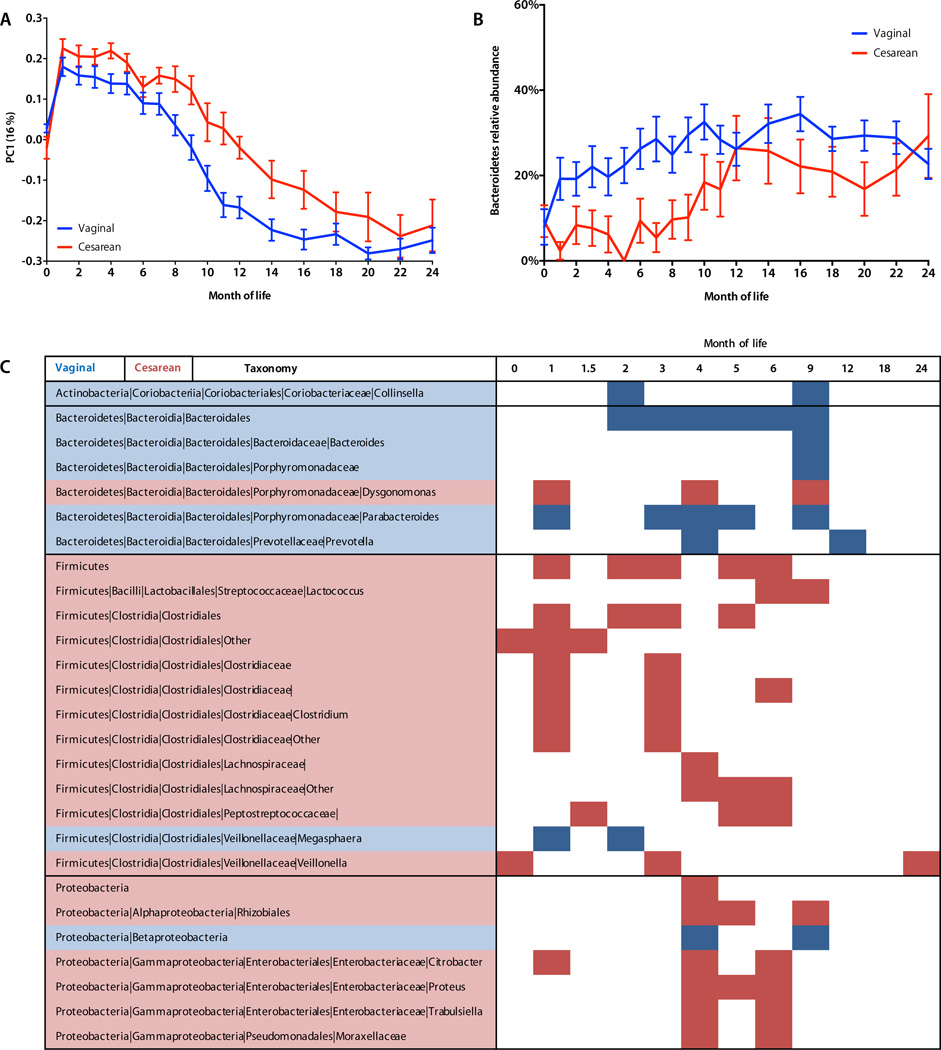
We also expected β-diversity to be altered in cesarean-delivered infants, reflecting both differing microbial exposures during birth and the observed altered α-diversity. For the babies’ first bowel movement (Mean ± SD 20.2 ± 18.3 hours of life), fecal β-diversity values were generally low, and not significantly different, suggesting that infants were colonized by microbiotas of similar complexity. Thereafter, cesarean section significantly altered microbial β-diversity compared to vaginally born children (unweighted UniFrac permutational MANOVA P < 0.001, R = 0.02) (Table S7, Fig. 5A). The abundances of several bacterial taxa were altered in cesarean-born infants, underlying the differences in α- and β-diversity (30) compared to those vaginally born (Table S6, S7). Most prominently, Bacteroides abundance was substantially and significantly lower in cesarean-delivered infants (Fig. 5B, C, Fig. S6) regardless of predominant feeding mode. By 12 months, the balance of Bacteroides, Bifidobacterium, and Enterobacteriaceae that dominated the first year in all infants was replaced by a mixture of Firmicutes, primarily Clostridiales (Fig. 1). Various Clostridiales and Enterobacteriaceae were significantly more abundant (LEfSe P < 0.05) in cesarean infants during the first year, filling the void left by Bacteroidales, but few taxa were significantly different during the second year of life (Fig. 5C). The increasing similarity between the intestinal microbial communities in children born vaginally or by Cesarean section after one year indicates that both undergo gradual maturation, eventually resembling the adult fecal microbiota (see below, Fig. 6). The taxa that dominated in the early months of life — whether or not disturbed by cesarean delivery or antibiotics — declined as later-life taxa replaced them.
Fig. 5. Delivery mode alters microbial diversity and composition.
A, Unweighted UniFrac principal coordinates analysis of the infant microbiome in relation to delivery mode over the first two years of life. Permutational MANOVA P < 0.05 (Table S7). B, Bacteroidetes relative abundance (Mean ± SEM) over time in relation to delivery mode. C, Cesarean section significantly alters abundance of diverse bacterial taxa over time. Red-shaded taxa (rows) were significantly more abundant (LEfSe P < 0.05) in cesarean-delivered infants at the given time points (columns); blue shading indicates more abundant in vaginally delivered infants
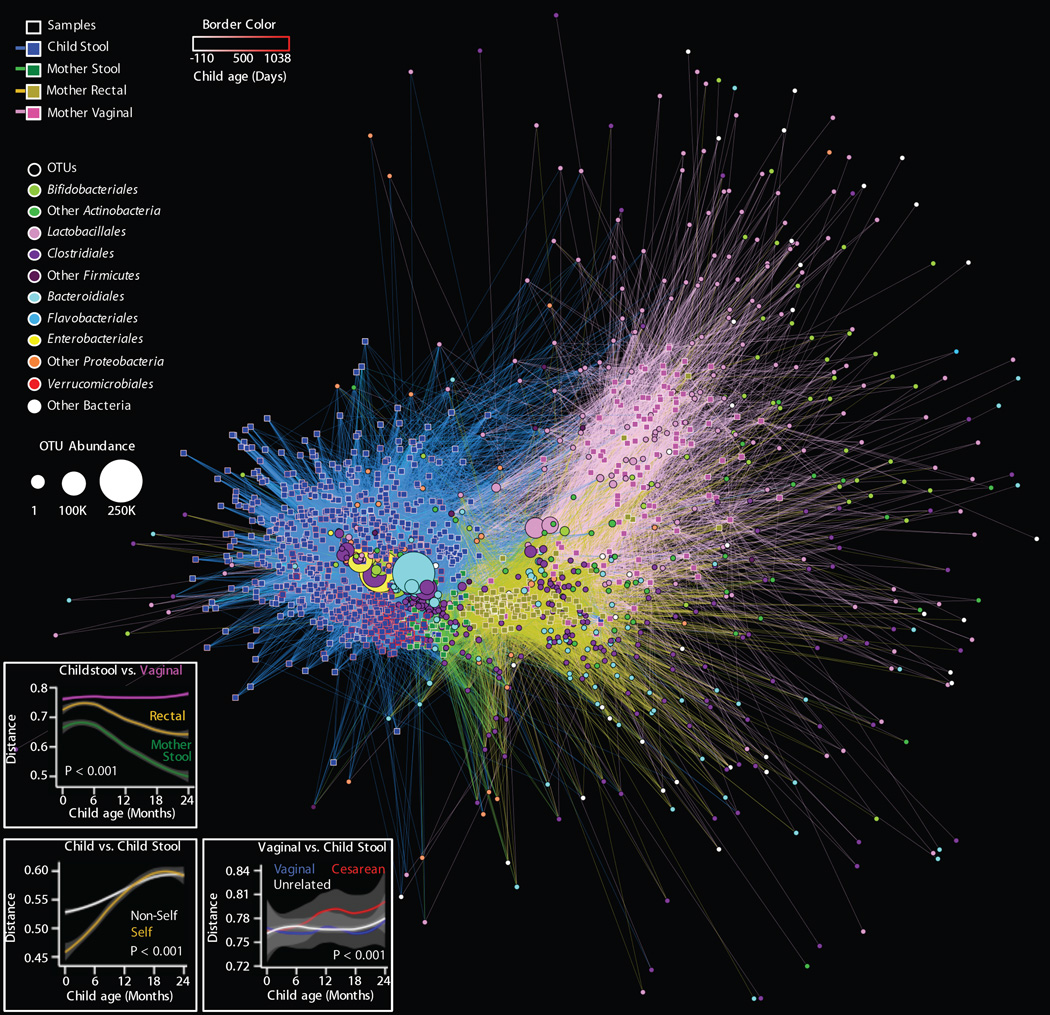
Fig. 6. Bipartite network comparing the relationships among all samples (squares) and OTUs (circles).
A, The distance between sample nodes and OTU nodes is a function of shared microbial composition. Samples with a large degree of OTU overlap (weighted by the number of observations of that OTU) form clusters. Edges connect a sample to each OTU detected in that sample, revealing shared OTUs between samples. Sample nodes and edges are colored by sample type; the border of sample nodes is a function of the age of the child, including pre-partum (negative) values for maternal samples (key at top-left). OTU nodes are colored by taxonomic family affiliation; the size of each OTU node is a function of that OTU's overall abundance, registered as OTU count in all samples (key at middle-left). See Fig 7 for specific analyses. B, Unweighted UniFrac distance between maternal vaginal, rectal, and stool microbiota and child stool microbiota as a function of child age. Shorter distance indicates greater similarity between microbial communities. C, Unweighted UniFrac distance between stool microbiota from the same child (self) and other children (non-self) as a function of the difference in age between sampling (Δ months). D, Unweighted UniFrac distance between maternal vaginal microbiota and stool microbiota of vaginally born dyads, unrelated children, or cesarean-delivered dyads as a function of child age. For panels B-D, lines indicate rolling-average mean values, grey shading = 95% CI. Grey shading = 95% CI. ANOVA P values are shown.
We hypothesized that the disruption of cesarean section could also alter microbiota maturation patterns in infants, similar to antibiotic exposure (Fig. 4). Using the microbiota maturation model described above, we found that cesarean- and vaginal-birth infants demonstrated similar degrees of microbiota maturation during the first 6 months of life. Subsequently, microbiota maturation stagnated in cesarean-delivered infants, with relative maturity dropping compared to vaginally born infants for the remainder of the study period (Fig. S7).
Infant Diet
Infant nutrition, surveyed routinely for the duration of the study, documented the extent of breast- or formula-feeding, and the timing of solid foods introduction (Fig. 1F). We compared two major dietary groups that best described dietary variation in this cohort: infants who were dominantly (> 50% of feedings) breast-fed or dominantly formula-fed for the first 3 months of life. Phylogenetic diversity and bacterial richness growth rates were significantly decreased in formula-fed children during 12–24 months of life (P < 0.05) (Fig. 2G–I, Table S6). Formula feeding also altered beta diversity (Table S7, Fig. S8A) and decreased microbiota maturation during 12–24 months of life (Fig. S8B). During this period, Lactobacillus, Staphylococcus, Megasphaera, and Actinobacteria were more abundant in breast-dominant children, and various genera of Clostridiales and Proteobacteria in formula-dominant children (Fig. S8C).
Maternal bacteria populate the infant gut during early life
The earliest source of microbial colonizers in the infant gut is the mother’s own microbiota, from passage through the birth canal to breastfeeding and skin contact. However, the extent to which a mother’s microbiota successfully establishes in her child, its dynamics over time, and the relative contributions of bacteria from different body sites have not been well-described. We characterized the microbiota of rectal swabs, vaginal swabs, and stool samples from mothers before and after birth (See Supplemental results, Fig. S9) to explore the microbial relationships shared between mother-infant dyads, as well as unrelated individuals, in the context of birth mode.
Network analysis explores the relationships between samples and their constituent OTUs, revealing that the infant microbiota matured from a neonatal state, associated with maternal OTUs from both vagina and rectum, to a post-infancy state resembling maternal stools (Fig. 6). In this analysis, sample nodes are connected by edges to all OTUs that they contain, indicating explicit connections between samples via shared OTUs. Edges are weighted based on OTU abundance, causing samples with similar OTU composition to cluster together along with their characteristic OTUs. These connections may be quantitatively measured directly as shared OTU counts between samples (Fig. 7, S10, see below) and indirectly as UniFrac distance (Fig. 6 panels B–D). Infant stool samples clustered together, associated strongly with several key Bacteroidales, Clostridiales, Enterobacteriales, and Bifidobacteriales OTUs (Fig. 6A). Infants cluster away from maternal stool samples, instead having more connections to maternal vaginal samples via robust links with several Lactobacillales and Bifidobacteriales OTUs (Fig. 6A). As the children aged beyond 12 months, their fecal microbiota came to resemble maternal stool and rectal samples more closely (Fig. 6B), indicating that their microbiota matured to a maternal-like configuration associated with numerous Clostridiales and Bacteroidales (Fig. 6A, Fig. S10). Stool microbiotas from the same child (“self”) were more similar to each other than those from unrelated children (“non-self”) (unweighted UniFrac ANOVA P < 0.001), and children’s stool microbiotas were most similar to those of the same age, highlighting that the succession of the intestinal microbiota occurs gradually (Fig. 6C). Conversely, at all ages, the children’s fecal microbiota was less similar to the mothers’ vaginal microbiota than to the mothers’ rectal and stool microbiotas (Fig. 6B). The dissimilarity between a mother’s vaginal microbiotas and her child’s fecal microbiotas was significantly more pronounced in cesarean-born children than in vaginally born mother-child pairs, whether dyads or unrelated (unweighted UniFrac ANOVA P < 0.001) (Fig. 6C). Considering two important genera, Bacteriodes and Bifidobacterium, there was a consistent diminution in the total OTU diversity acquired in cesarean-born infants, as well as shared OTUs; dyads shared significantly more of these taxa than did unrelated pairs (Fig. S10).
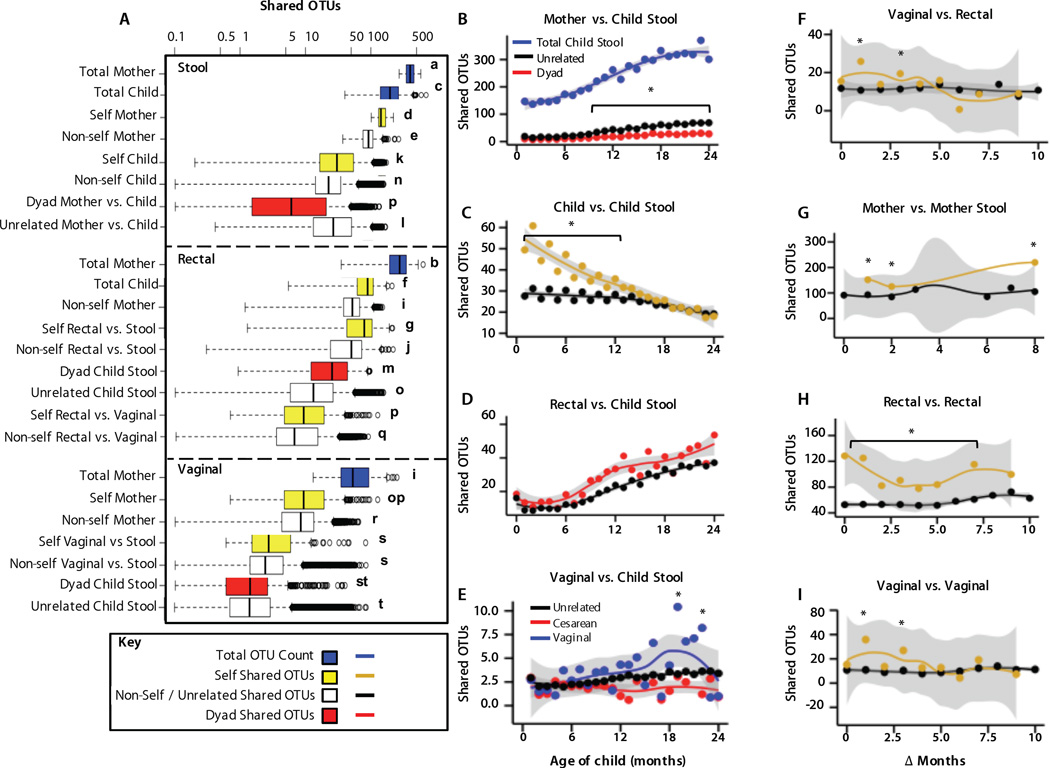
Fig. 7. Shared OTUs reveal microbial relatedness among mothers and children.
(A) Shared OTU counts (median ± quartiles) between individual stool samples (top), rectal swabs and stool samples (middle), and vaginal swabs and stool samples (bottom), represented in Fig. 6. Distributions represent the total number of OTUs within a single sample (blue) or shared OTUs between samples from the same individual (self, yellow), another individual (nonself, white/black), or a mother-infant dyad (red). Lowercase letters indicate significantly different shared OTU count distributions [one-way ANOVA, P < 0.0001, followed by false discovery rate (FDR)–corrected Fisher’s protected least significant difference (PLSD) test]. Key indicates coloring for box plots in (A) or line plots in (B) to (I). (B to I) Shared OTU counts over time between mothers and unrelated children, mother-infant dyads, and total OTUs in child stool samples. (C) Samples from the same child or unrelated children at different times (Δ months). (D) Mothers’ rectal swabs and stool samples from their own children (dyad) or unrelated children. (E) Mothers’ vaginal swabs and stool samples from unrelated children or dyads of children delivered vaginally or by cesarean section. (F) Vaginal and rectal swabs from the same mother or other mothers. (G) Stool samples from the same mother or other mothers. (H) Rectal swabs from the same mother or other mothers. (I) Vaginal swabs from the same mother or other mothers. (B), (D), and (E) compare mothers versus children, and x axes indicate the child’s age (months). For (C) and (F) to (I), x axes indicate the differences in child age (Δ months) between the times when these samples were obtained. Lines indicate rolling average mean values, and gray shading is equal to 95% CI. *P < 0.0001, ANOVA, followed by FDR-corrected Fisher’s PLSD test.
As microbial composition differs widely between adults and children, and between samples from different body sites, quantitation of shared OTUs is a metric to assess microbial transmission between these loci (Fig. 7). Samples with more shared OTUs are expected to have closer relationships, whether OTUs are transmitted directly between samples or both have a common external source. Maternal microbiota transmission implies that more maternal OTUs are expected in her child than in unrelated children, except when OTUs are highly dispersed among mothers. We examined the total number of OTUs in each sample, and the number shared between samples belonging to the same individual (“self”), other individuals (“non-self”), or between mother-infant dyads (Fig. 7). Results support expectations that OTUs are more shared between “self” samples and a mother’s rectal microbiota shares more OTUs with her own child’s stool than that of other children (Fig. 7A). Although vaginal swabs follow this trend, effects are not significant when examining samples from all time points and birth modes.
Child stool OTU counts gradually increased with time (Fig. 7B), approaching OTU counts in mothers by age 3 years (compare panels A and B). Unrelated mothers and children shared far fewer stool OTUs throughout this time, but surprisingly still shared more OTUs on average than mother-infant dyads (Fig. 7B). The shared OTU count increased as children grew older — for both dyads and unrelated mother-infant pairs — further evidence that children’s microbiotas gradually mature into an adult-like state. By 3 years of life, α-diversity (total OTU counts) approached adult levels (compare Fig. 7A and 7B), and β-diversity became lower between children and adults (Fig. 6B), but OTUs shared with adults remained low, indicating that different strains colonized children and adults even when similar bacterial taxa were present.
A child’s stool shared more OTUs with their other stools that were collected <14 months apart, compared to non-self stool samples; as time between sampling increased, shared OTUs decreased until self and non-self stools had similar shared OTU counts (occurring for samples >14 months apart) (Fig. 7C), indicating the dynamism of the microbiota during the first 2 years of life. As expected, the number of OTUs shared between self-stool samples gradually decreased as the time between sampling increased, indicating gradual succession of OTUs. As children aged, they also shared more OTUs with maternal rectal swabs whether or not they were related (Fig. 7D). Dyads shared more OTUs across all times (Fig. 7A), but were not significant at individual time points (Fig. 7D). Mothers shared significantly more vaginal OTUs with their children if they had been vaginally delivered, compared to both cesarean-delivered and unrelated infants (Fig. 7E), an effect only significant after 1 year of life, peaking between 18 and 24 months of life. Vaginal OTUs, relevant during early infancy, were detected less frequently in fecal specimens in later childhood, leading to the decline in shared OTUs. As expected, peri-partum maternal stool, vaginal, and rectal samples also showed fewer shared OTUs with samples from the same mother as the interval between sampling increased (Fig. 7F–I). As sampling interval grew, maternal samples shared as many OTUs with samples from other mothers as they did with self. Stool samples were the exception, with more shared self-OTUs across sampling than from other mothers (Fig. 7G).
DISCUSSION
The purpose of this study was to characterize early-life microbial development in the context of antibiotic use, cesarean section, and formula feeding. Each of these has been associated with conditions emerging later in life, including obesity (9), diabetes (10, 20), and allergies (13, 19). The cause for these relationships is unclear, but altered patterns of microbiota assembly during early life are plausible. We profiled microbiota development during the first two years of life, and document disturbances related to antibiotics, cesarean section, and diet.
Intestinal bacterial communities undergo a gradual succession during early life (Fig 1A), following a predictable, age-dependent pattern that is conserved between disparate human populations and stabilizes after age 3 years (5, 24, 25). These events may reflect a co-evolutionary relationship in which normal maturation of the gut microbiome during a critical window contributes to host development; disturbances may interrupt this delicate choreography, with consequences for long-term host health (16).
We observe three distinct phases in the childhood microbiome during the first years of life. During the first month of life, Enterobacteriaceae dominated the microbiota (Fig. 1), suggesting that these facultative anaerobes can exploit the naive conditions of the neonatal intestine. The intermittent stages of development, from approximately 1 to 24 months of life, were more dynamic and appeared to be sensitive to disturbances from birth mode, predominant nutrition, and antibiotic use. We speculate that even transient effects during this sensitive, developmental window could lead to long-lasting effects — as we have shown with short-term antibiotic exposures in mice (15, 16). Finally, as children reached 2 years of age the microbiota gradually stabilized toward an adult-like configuration (Fig. 6), characterized both by higher diversity and greater resilience to change. This period paralleled dietary transition from liquid to solid foods (Fig. 1F), an important catalyst for microbiota maturation in childhood (17), and a source of introduced microbes. Alternative hypotheses for the higher diversity include the diminution of the constraint imposed by C-section (Fig S9), and of breast-feeding with its strong selective effects (31). Higher immunologic tolerance during early life (32) may be ending, adding new constraints to acquiring non-founding microbes. These conserved transitions may be important, and suggest that disturbances during the first two years of life are likely to have strong effects on development of the microbiota and potentially for host health.
Microbial taxa and gene pathways that best define “microbial age” can be used as biomarkers to track the infant’s microbiota progress, paralleling how weight-for-height tracks a child’s development, and are similarly affected by disruptions to health (5, 15). The precise role that these bacteria play in development is unclear — we have yet to determine whether any maturity biomarkers are actually linked to host processes — but from a theoretical standpoint, this method allows identification of organisms that may play roles at key junctures during an infant’s development, and the results are striking. By suppressing early-life biomarkers including Lachnospiraceae, Enterobacteriaceae, Erysipelotrichaceae, and numerous predicted gene pathways, antibiotic exposures effectively stall the development of the intestinal microbiome, causing the intestinal communities in these infants to appear less developed (younger) than unexposed counterparts (Fig. 4).
Lachnospiraceae spp. and other Clostridiales were particularly sensitive, and were significantly depleted in antibiotic-treated infants often throughout early life (Fig. 3), in agreement with prior studies of adult human and mouse microbiotas (8, 33). Lachnospiraceae live almost exclusively in the mammalian gastrointestinal tract, often producing butyrate and other short-chain fatty acids (SCFA) (34) that regulate host immunity via epithelial cell signaling, colonic T regulatory cells (35), and macrophages (36). Butyrate synthesis could explain how Lachnospiraceae induce T regulatory cells, suppressing colitis and allergic diarrhea in mice (37), and are consistent with protective associations against type 1 diabetes development in infants (38). Butyrate, serving as an energy source for host epithelial cells, also regulates the cell growth- and differentiation-related AP-1 signaling pathway (39), potentially explaining links between butyrate producers (including Lachnospiraceae spp.) and body weight (40). Lachnospiraceae OTUs also were implicated as markers of intestinal microbiota maturation in Bangladeshi infants (5), suggesting that their blooms may be an important event in the developing infant gut across continents.
α-diversity, an important ecosystem characteristic, was disturbed by delivery mode, antibiotic use, and by diet (Fig. 2). During early life, overall diversity rapidly increases in the developing infant gut (24, 25) (Fig. 2) as children acquire bacterial strains encompassing greater phylogenetic diversity from diet, human contact, and environment. Since this process is occurring during a sensitive period when acquired intestinal bacteria train the nascent immune system (32), increasing diversity may be relevant to normal development (1). Disturbance from antibiotic use, formula feeding, or cesarean section, as well as the potential for cumulative damage from multiple disturbances, might affect intestinal homeostasis and long-term health. Decreased intestinal α-diversity during infancy precedes type 1 diabetes onset (38) and allergic manifestations (41), and has been reported in obese adults (40).
Whether or not intestinal α-diversity influences disease development or is only a marker, functional diversity differences could contribute (26). The substantial functional redundancy in the healthy human microbiome (26, 42) may provide insurance that the gut ecosystem can recover from temporary disturbances, maintaining productivity by minimizing risk of functional loss. In this model, a healthy microbiota with sufficiently high diversity can withstand normal environmental fluctuations, e.g., due to diet change or illness, and maintain key functions. However, disturbances during development may reduce diversity below thresholds sufficient to maintain an essential functional repertoire. Such lack of resilience may delay ecosystem recovery or lead to a new stable state (26); either event may promote later disease development. One challenge to the translation of this theory will be identifying microbial ecosystem functions important for critical developmental steps; as targets for future investigation, we identify several gene pathways impacted by early disturbances. (See Supplemental Results)
Another notable effect of cesarean section was a dramatic, persistent decrease in Bacteroides populations. Although several studies have noted decreased phylum Bacteroidetes in cesarean-delivered infants (43, 44), we now show that this effect persists throughout the first year of life and is associated with an altered metagenomic landscape in the gut. Since Bacteroides spp. regulate intestinal immunity (45), this long-lasting, large-scale disturbance could partially explain cesarean section-associated health consequences, including asthma, obesity, and allergies (18, 21). The deficiency of Bacteroides following cesarean section, with commensurate expansion of other taxa, may disrupt tolerogenic feedback loops (26), potentially contributing to development of inflammation and obesity. Disturbances to microbial metabolism and cell motility pathways in cesarean-born children suggest mechanisms by which microbial perturbation could influence hosts; targets for future investigation include the altered production of SCFA or other immunomodulatory metabolites (35).
The strong evidence for maternal transmission of early-life taxa in general, and specific dominant taxa in particular, provides support for the importance of the composition of the maternal microbiota for healthy development. Widespread practices that affect this reservoir include antibiotic therapies and prophylaxes, but also cleansings that were aimed for neonates in developing countries but have recently spread to more developed populations (46).
Some limitations are inherent to our study. Our sample size lacks sufficient power to account for complex interactions between many potential microbial disturbances during early life. Larger and longer studies may assess whether early-life microbiota disruptions are indeed cumulative, counteractive, or independent. Longer studies can determine how community assembly during childhood transitions to climax (e.g., adult) communities, characterized by diverse alternative stable states (47). Dissimilar trajectories of microbiota maturation during childhood may lead to different stable states in adulthood, without necessarily affecting the host. Microbial disruptions primarily delayed microbiota maturity during the first year, indicating transient effects rather than permanent alterations. Transient antibiotic disturbances during early life durably alter host development in mice (14–16), but our results do not necessarily imply that this effect extends to humans. An alternative possibility is that disruptions to microbiota composition and maturation are nullified if they are replaced by other functionally similar taxa. The disparate microbiota states observed in adults possess functional overlap despite compositional similarity, suggesting that community function may more relevant for predicting host interactions (26, 47). Our results suggest that community function and functional maturation are altered by antibiotics, cesarean section, and formula feeding (using PICRUSt-predicted metagenomes), but metagenomic and transcriptomic studies will be needed to assess whether early-life disruptions influence community function and behavior.
Our results identify multiple disturbances to microbial development during early life. These alterations were not linked to health outcomes, which we did not survey, but are potential targets for ongoing inquiry. The microbial populations and dynamics deserve further scrutiny for their role in host-microbial communication, host development, and health. Mounting evidence associates antibiotic use, cesarean section, and formula feeding with metabolic and immunologic disorders later in life, possibly as a consequence of disturbed microbiome development. Although these interventions often strongly benefit recipients, the hidden costs imply the need for careful consideration in children with less severe illnesses. Our findings warrant studies to establish whether and how early life microbial disturbances associated with antibiotic use, cesarean section, and formula feeding influence health outcomes; recent studies of Canadian children provide evidence that such early-life disturbances are important for asthma risk (12)
MATERIALS AND METHODS
An Institutional Review Board-approved study was conducted in healthy, pregnant NYC mothers in 2011–2014. Maternal vaginal, rectal, and fecal specimens, and fecal specimens from their infants were obtained from birth to the age of three years, chiefly in the first two years of life, and analyzed by microbiome sequencing. Information about home environment, delivery mode, infant diet, and antibiotic exposures was obtained for each mother-baby dyad. Complete details are provided in the Supplementary Materials.
Supplementary Material
Acknowledgments
Supported by R01 DK090989 from the National Institutes of Health, and by the Diane Belfer Program for Human Microbial Ecology, by the Daniel and Leslie Ziff Fund, and by an anonymous donor.
We thank Sukleen Bedi and Arielle Radin for clinical assistance.
Footnotes
List of Supplementary Materials:
FIGURES
Fig. S1. LEfSe analysis of taxon abundances in fecal samples in antibiotic-unexposed infants, combining delivery mode and predominant diet.
Fig. S2. Antibiotic exposures did not alter alpha diversity.
Fig. S3. Functional maturation of the microbiome is delayed by antibiotic exposure.
Fig. S4. Functional maturation of the microbiome is altered by formula feeding.
Fig. S5. Enumeration of 16S rRNA genes in fecal specimens in children at one and twelve months of age, by quantitative (q)PCR.
Fig. S6. Average relative abundance of Bacteroides in children in relation to feeding and delivery mode, in the first two years of life.
Fig. S7. Cesarean section exerts a modest impact on microbiota maturation.
Fig. S8. Infant diet alters microbial composition and maturity over the first two years of life.
Fig. S9. Relative abundance heat map of major taxa in 296 maternal samples.
Fig. S10. Development of diversity of Bifidobacterium and Bacteroides OTUs in children in early life.
TABLES
Table S1. Baseline characteristics of the 53 mothers in the study.
Table S2. Baseline characteristics of the 43 infants in the study.
Table S3. Prenatal antimicrobial use by class and purpose.
Table S4. Peri-natal antibiotic use by class and indication.
Table S5. Post-natal antimicrobial use by class and age of child
Table S6. Linear mixed effect model estimates of antibiotic, diet, and delivery effects on alpha diversity.
Table S7. Permutational MANOVA scores of antibiotic, diet, and delivery effects on UniFrac distance beta diversity.
Table S8. Linear mixed effect model estimates of antibiotic, diet, and delivery effects on microbiome maturation MAZ scores.
Table S9. Linear mixed effect model estimates of antibiotic, diet, and delivery effects on PICRUSt predicted metagenome maturation MAZ scores.
MJB, MJ, YC, GPP, and MGDB planned and designed experiments. NH and WS recruited study subjects. JC, XZ, and MC performed all laboratory experiments. NH, JC, and TB managed study data. NAB, TB, AL, FW, JC, and HL performed all analyses, and NAB and MJB wrote the manuscript. All authors contributed to and reviewed the final manuscript.
Competing interests: None
Data availability: All sequencing data are publicly available in QIITA database (http://qiita.microbio.me/) under study ID 10249.
References
- 1.Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nature Reviews Immunology. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrobial agents and chemotherapy. 2012;56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks LA, Taylor TH, Hunkler RJ. U.S. outpatient antibiotic prescribing, 2010. New England Journal of Medicine. 2013;368:1461–1462. doi: 10.1056/NEJMc1212055. [DOI] [PubMed] [Google Scholar]
- 8.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. PNAS. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. International journal of obesity. 2014 doi: 10.1038/ijo.2014.119. [DOI] [PubMed] [Google Scholar]
- 10.Kilkkinen A, Virtanen SM, Klaukka T, Kenward MG, Salkinoja-Salonen M, Gissler M, Kaila M, Reunanen A. Use of antimicrobials and risk of type 1 diabetes in a population-based mother-child cohort. Diabetologia. 2006;49:66–70. doi: 10.1007/s00125-005-0078-2. [DOI] [PubMed] [Google Scholar]
- 11.Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- 12.Arrieta M-C, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE, Finlay BB the CHILD Study Investigators. Early infancy microbial and metabolic alterations afect risk of childhood asthma. Science Translational Medicine. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 13.Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother’s and Offspring’s Use of Antibiotics and Infant Allergy to Cow’s Milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 14.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobel YR, Cox L, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nature Comm. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Jun W. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell host & microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2008;38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 19.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 20.Cardwell CR, Stene LC, Joner G, Cinek O, Svensson J, Goldacre MJ, Parslow RC, Pozzilli P, Brigis G, Stoyanov D, Urbonaite B, Sipetic S, Schober E, Ionescu-Tirgoviste C, Devoti G, de Beaufort CE, Buschard K, Patterson CC. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 21.Huh SY, Rifas-Shiman SL, Zera CA, Edwards JW, Oken E, Weiss ST, Gillman MW. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Archives of disease in childhood. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innis SM. Human milk: maternal dietary lipids and infant development. The Proceedings of the Nutrition Society. 2007;66:397–404. doi: 10.1017/S0029665107005666. [DOI] [PubMed] [Google Scholar]
- 23.Gale C, Logan KM, Santhakumaran S, Parkinson JR, Hyde MJ, Modi N. Effect of breastfeeding compared with formula feeding on infant body composition: a systematic review and meta-analysis. The American journal of clinical nutrition. 2012;95:656–669. doi: 10.3945/ajcn.111.027284. [DOI] [PubMed] [Google Scholar]
- 24.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 28.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone CA, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proceedings of National Academy of Sciences U.S.A. 2011;108(Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance, Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JE, Ballal SA, Morgan XC, Glickman JN, Gevers D, Huttenhower C, Garrett WS. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. The ISME journal. 2014;8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome biology and evolution. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 38.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, Lahdesmaki H, Franzosa EA, Vaarala O, de Goffau M, Harmsen H, Ilonen J, Virtanen SM, Clish CB, Oresic M, Huttenhower C, Knip M, Xavier RJ. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell host & microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nepelska M, Cultrone A, Beguet-Crespel F, Le Roux K, Dore J, Arulampalam V, Blottiere HM. Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells. PloS one. 2012;7:e52869. doi: 10.1371/journal.pone.0052869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 42.Huttenhower C, et al. Human Microbiome Project, Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ : Canadian Medical Association journal = journal de l'Association medicale canadienne. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 45.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. The Proceedings of the Nutrition Society. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 46.Mullany LC, Darmstadt GL, Tielsch JM. Safety and impact of chlorhexidene antisepsis interventions for improving neonatal health in developing countries. Pediatr Infect Dis J. 2006;25:665–675. doi: 10.1097/01.inf.0000223489.02791.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Relman DA. The human microbiome: ecosystem resilience and health. Nutrition reviews. 2012;70(Suppl 1):S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, Sears MR, Mandhane PJ, Turvey SE, Subbarao P, Becker AB, Scott JA, Kozyrskyi AL. CHILD Study Investigators, Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG: An International Journal of Obstetrics and Gynaecology. 2015 doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme Journal. 2012 doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aronesty E. Comparison of sequencing utility programs. Open Bioinform J. 2013;7:1–8. [Google Scholar]
- 51.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. Qiime allows analysis of high-throughput community sequence data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 54.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73 doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, Bittinger K, Bushman FD, DeSantis T, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla DM, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome research. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PloS one. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 60.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. Hoboken, NJ: John Wiley & Sons; 2004. [Google Scholar]
- 61.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. Journal of Clinical Microbiology. 2010;48:3575–3581. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.