Abstract
Purpose
Insufficient sensitivity and specificity prevent the use of most existing biomarkers for early detection of breast cancer. Recently, it was reported that serum microRNAs (miRNAs) may be potential biomarkers in many cancer diseases. In this study, we investigated whether serum levels of 5 miRNAs including miR-21, miR-125b, miR-145, miR-155, and miR-365 could discriminate breast cancer patients and healthy controls.
Methods
Serum levels of miRNAs were measured by using quantitative real-time polymerase chain reaction in 99 breast cancer patients and 21 healthy controls. The abundance change of serum miRNAs were also evaluated following surgical resection in 20 breast cancer patients. Receiver operating characteristic (ROC) curve analysis was performed to assess the sensitivity and specificity of miRNAs as diagnostic biomarkers.
Results
Serum levels of miR-21 and miR-155 was significantly higher, while miR-365 was significantly lower in breast cancer as compared with healthy controls. The serum levels of miR-21 and miR-155 significantly decreased following surgical resection. Additionally, the serum level of miR-155 at stages I and II was significantly higher compared to stage III. The serum miR-145 level was remarkably higher in progesterone receptor (PR)-positive patients than PR-negative. The positivity of miR-21, miR-155, and miR-365 was high compared to CA 153 and CEA in breast cancer. ROC curve analyses of a combination of miR-21, miR-155, and miR-365 yielded much higher area under curve and enhanced sensitivity and specificity in comparison to each miRNA alone.
Conclusion
The combination of serum miR-21/miR-155/miR-365 may potentially serve as a sensitive and specific biomarker that enables differentiation of breast cancer from healthy controls.
Keywords: Serum, MicroRNAs, miRNA-21, miRNA-155, miRNA-365, Breast neoplasms
INTRODUCTION
Breast cancer, one of the most common types of female cancers, is a highly heterogeneous disease that has multiple subtypes with distinct clinical outcomes and a high fatality rate globally [1]. In clinic, the status/expression level of hormone receptor including estrogen receptor (ER), progesterone receptor (PR), and human EGF-like receptor 2 (HER2) as well as tumor grade are often used for classification and target therapy indicators of breast cancers [1,2]. Although current early detection and target therapies based on the measurement of hormone receptors have remarkably reduced the rate of mortality from breast cancer, their application is still limited due to insufficient sensitivity and specificity as well as the invasive, unpleasant and inconvenient nature of diagnostic procedures. To identify unique therapeutic targets, it is necessary to develop predictive and prognostic biomarkers that can be conveniently and reliably used in clinic.
MicroRNAs (miRNAs), a class of naturally occurring, small and noncoding RNA molecules with a length of 19–25 nucleotides, can specifically regulate gene mRNA expressions at posttranscriptional levels [3]. The dysregulation of miRNAs may affect some crucial biological processes of cells leading to tumor development by increasing proliferation, decreasing apoptosis, and enhancing the metastatic potential [3,4]. Recently, it has been reported that miRNAs expression level/profile is altered in various cancers exhibiting great potential in improving the diagnosis and prognosis of cancer [5]. Notably, it has been documented that certain secreted miRNAs originating from cancer tissues are protected from endogenous RNase by some unknown mechanisms [6], and thus can be detected in blood and other body fluids [7,8]. In breast cancer tissues, miRNAs have been shown to function as either oncogenes or tumor suppressors [9], and circulating miRNAs may correlate with disease progression, therapeutic responses and patient survival [10,11], suggesting that the evaluation of miRNAs levels in the blood, either serum or plasma, may be used as noninvasive blood-based biomarkers. Even though many studies have compared changes of miRNAs levels in blood between breast cancer patients and healthy controls in order to map the profiling of miRNAs and identify some specific miRNAs as potential biomarkers in breast cancer [12], the results are not always consistent due to the differences in study design, such as sample size, patient source and characteristics, and RNA preparation, as well as detection methods or profiling platforms that were used.
In our study, the serum levels of 5 miRNAs including miR-21, miR-155, miR-125b, miR-145, and miR-365 that have been indicated as a recurrent presence in breast cancer patients [12,13], were compared between breast cancer patients and healthy controls. We found that the serum level of miR-21 was significantly higher, while miR-155 and miR-365 was significantly lower in breast cancer than healthy control, and receiver operating curve (ROC) analyses showed that the combination of miR-21/miR-155/miR-365 led to higher sensitivity and specificity in distinguishing breast cancer from healthy controls, indicating a potential as biomarkers for the diagnosis and monitoring of breast cancer.
METHODS
Patients
Blood samples were collected from 99 patients with breast cancer (range, 31–77 years; mean, 48.95 years) and 21 age-matched healthy female volunteers (range, 35–59 years; mean, 45.38 years; without current or previous malignancy or inflammatory condition). In addition, the paired blood samples were collected from a 20-patient subset both before and 3 weeks after breast cancer surgery. All participants had signed an informed consent to participate in this study. The study was approved by the Ethics Review Board of Harbin Medical University.
All patients' breast cancer was histologically confirmed. The surgical patients' clinicopathological and relevant demographic characteristics were documented in our prospectively maintained breast cancer database. The clinical stage of breast cancer of all patients was classified according to the TNM classification system of the American Joint Committee on Cancer. The number of various groups' breast cancer patients is summarized in Table 1.
Table 1. The number of patients with breast cancer in various groups (n = 99).
Serum preparation
A 5-µL sample of the whole blood was collected in a Vacutainer Serum Separator Tube (Becton Dickinson, Franklin Lakes, NJ, USA). The blood was left to clot at room temperature for 30 minutes, and then centrifuged at 2,000 rpm for 10 minutes at 4℃. The resulting serum was collected, aliquoted, and stored at –80℃.
RNA extraction
Total RNA was extracted from 300 µL of serum using the mirVana PARIS Kit (Ambion, Carlsbad, CA, USA), and finally eluted into 100 µL of preheated (95℃) Elution Solution according to the manufacturer's protocol. The eluate was then collected and stored at –20℃. RNA concentration and integrity were determined using NanoDrop spectrophotometry (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE, USA). The RNA concentration (µg/µL) was calculated by the value of OD260 (OD260 × dilution factor × 40), and adjusted to 0.002 µg/µL. The total RNA was stored at –80℃.
RT and real-time qPCR
The level of miRNAs was quantified in duplicate using quantitative reverse transcription polymerase chain reaction (RT-PCR) and human TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). The RT reaction was carried out in a 15-µL TaqMan MicroRNA Reverse Transcription System containing 5 µL of RNA extract, 0.15 µL of 100mM dNTPs, 1 µL of Multiscribe Reverse Transcriptase (50 U/µL), 1.5 µL of 10 × RT buffer, 0.19 µL of RNase inhibitor (20 U/µL), 1 µL of genespecific primer and 4.16 µL of nuclease-free water. For cDNA synthesis, the above mixtures were incubated at 16℃ for 30 minutes, 42℃ for 30 minutes, and 85℃ for 5 minutes.
A 4-µL cDNA solution was amplified using 10 µL of TaqMan Gene Expression Master Mix, 1 µL of gene-specific primers/probe and 5 µL of nuclease-free water in a final volume of 20 µL. Quantitative PCR was performed on a 7000 Real-Time PCR system (Applied Biosystems) by incubation at 95℃ for 10 minutes, and 40 cycles of 95℃ for 15 seconds and 60℃ for 1 minute. The values of cycle threshold (Ct) were calculated with SDS 1.4 software (Applied Biosystems).
Measurement of hormone status, p53, CA 153, and CEA
The levels of CA 153 and CEA were measured through electrochemiluminescence assays, and the level of hormone status including ER, PR, HER2, and p53 was measured by immunohistochemistry assay in the Laboratory Department of the Third Affiliated Hospital of Harbin Medical University.
Statistical analysis
The unpaired t-test, a two-tailed Mann-Whitney test, was used to compare the differential expression of serum miRNAs between breast cancer and normal samples, between family history positive and negative patients, and between the patients with ER (or PR, HER2, p53) positive and negative. The paired t-test was used for comparison of serum miRNAs level between pre- and postoperative samples in breast cancer. The one-way analysis of variance was used to compare the differential serum miRNAs level between normal and breast cancer patients at different TNM stages. ROCs were generated using logistic regression models, to evaluate the diagnostic performance of various miRNAs and a combination of miRNAs. Area under curve (AUC) was used as the evaluation criteria; the higher AUC, the better diagnostic performance.
All analysis was performed using SAS 9.5 (SAS Institute Inc., Cary, NC, USA) and GraphPad Prism ver. 6 (GraphPad Software Inc., La Jolla, CA, USA). The P < 0.05 was considered as significant difference.
RESULTS
Serum miRNAs levels in breast cancer and healthy samples
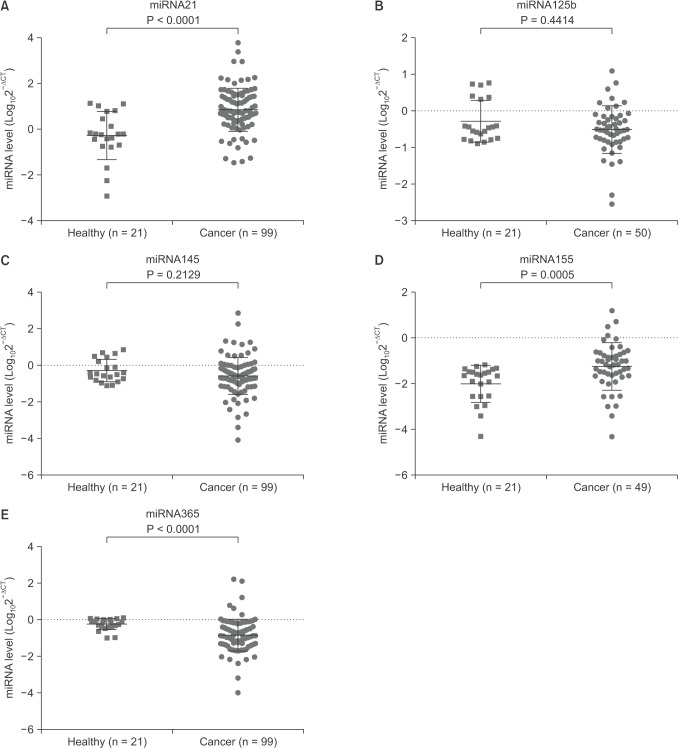
It is difficult to extract RNA from serum due to its low abundance. In our experiments, the average concentration of the total RNA from 300 µL of serum was 0.118 mg/µL ranging from 0.042 to 0.208 mg/µL. As compared with normal samples, the level of miR-21 and miR-155 was significantly higher (miR-21: 0.86 ± 0.941 vs. –2.74 ± 1.055, P <0.0001; miR-155: –1.24 ± 1.022 vs. –2.01 ± 0.808, P = 0.0005), while miR-365 was significantly lower (–0.84 ± 0.873 vs. –0.24 ± 0.317, P < 0.0001) in breast cancer patients' serum. The level of serum miR-125b and miR-145 showed no significant difference between breast cancer and normal samples (Fig. 1).
Fig. 1. The serum abundance of microRNAs (miRNAs) in breast cancer patients and healthy controls. Total RNA was extracted from serum and quantitative polymerase chain reaction was performed for evaluation of miRNA abundance. As compared with healthy controls, the serum levels of miR-21 (A) and miR-155 (D) were significantly higher, while miR-365 (E) was significantly lower in breast cancer patients. The serum concentrations of miR-125b (B) and miR-365 (C) showed no difference between breast cancer patients and healthy controls. Data are expressed as mean ± standard deviation.
Effects of family history and TNM stage on serum miRNAs level in breast cancer patients
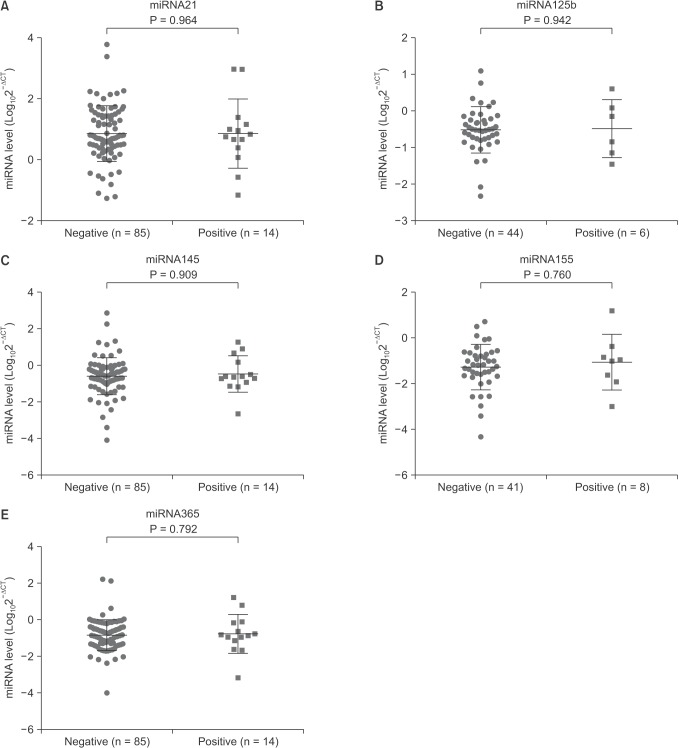
We compared the serum level of the five miRNAs in breast cancer patients with or without family history. The five miRNAs levels were not significantly different between family history positive and negative patients with breast cancer (Fig. 2).
Fig. 2. Effects of family history on the serum microRNAs (miRNAs) level in breast cancer patients. The serum level of miRNAs was assessed in family history positive and negative breast cancer patients. Family history showed no effects on serum miRNAs level in breast cancer patients. Data are expressed as mean ± standard deviation.
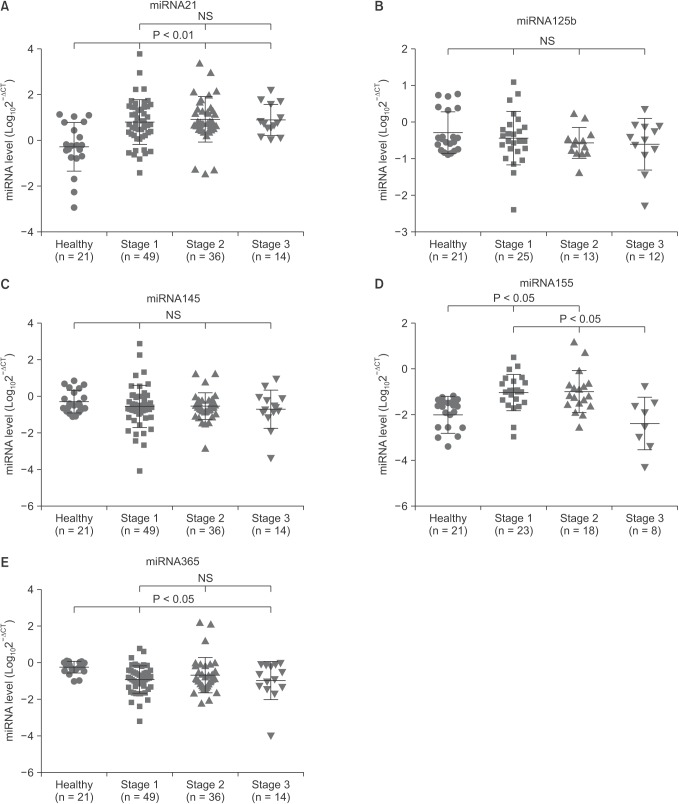
The serum levels of miR-21, miR-125b, miR-145, miR155, and miR-365 in breast cancer patients at different TNM stages were also evaluated to determine if the serum miRNAs could be detected in early-stage breast cancer (Fig. 3). Across 3 stages, the serum levels of miR-21, miR-125b, miR-145, and miR-365 showed no marked difference. In comparison to stages I and II, the serum miR-155 level was remarkably lower (stage III: –2.40 ± 1.151 vs. stage I: –1.03 ± 0.790, stage II: –1.00 ± 0.919, P < 0.05) in breast cancer patients at stage III. Notably, compared to normal samples, the serum level of miR-21 was significantly higher (stage I: 0.80 ± 0.980, stage II: 0.92 ± 0.994, stage III: 0.87 ± 0.674 vs. healthy controls: –0.27 ± 1.055; P < 0.01) in breast cancer at any TNM stage. Nevertheless, in comparison with normal controls, the serum level of miR-365 was significantly lower (stage I: –0.92 ± 0.753, stage III: –0.97 ± 1.043 vs. healthy control: –0.24 ± 0.317; P < 0.05) at both stages I and III.
Fig. 3. Comparisons of the serum microRNAs (miRNAs) level in breast cancer patients across different TNM stages. The serum levels of miR-21 (A), miR-125b (B), miR-145 (C), and miR-365 (E) showed no difference across different TNM stages. Compared to stages I and II, the serum miR-155 level was lower in breast cancer patients at stage III (D). In comparison with healthy controls, the miR-21 level was remarkably higher in breast cancer patients at any TNM stage (A), miR-155 was significantly higher at stages I and II (D), whereas miR-365 was significantly lower at stages I and III (E). Data are expressed as mean ± standard deviation. NS, not significant.
Comparison of serum miRNAs levels in various groups of breast cancer patients
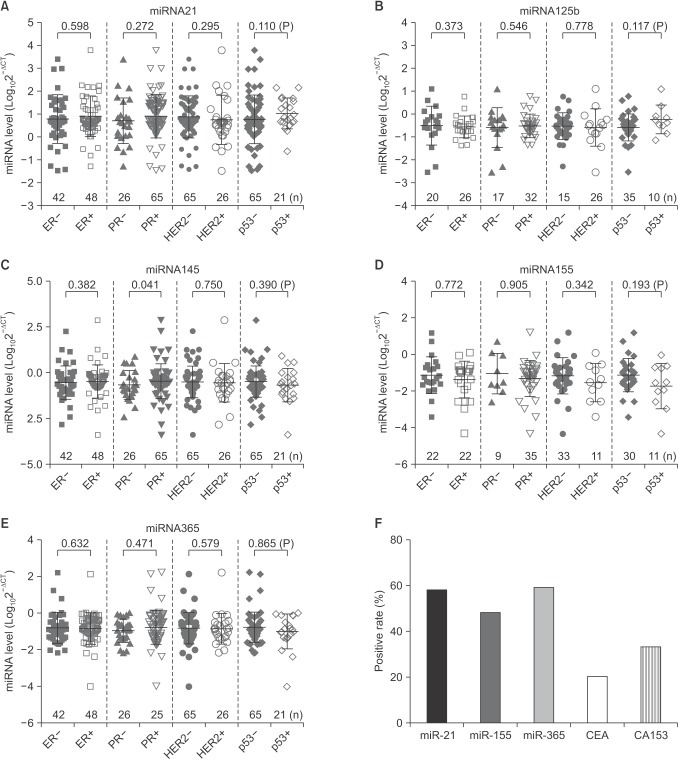
We compared the serum levels of miR-21, miR-125b, miR-145, miR-155, and miR-365 in various groups of breast cancer patients classified by hormone status including ER, PR, and HER2 as well as p53 (Fig. 4). We only detected that the serum level of miR-145 was significantly higher (–0.46 ± 0.953 vs. –0.66 ± 0.777, P = 0.041) in PR-positive patients as compared with PR negative (Fig. 4C). Additionally, in breast cancer patients, the positivity of miR-21, miR-155 and miR-365 was 58%, 48%, and 59%, respectively. Nevertheless, the positivity of CA 153 and CEA was 33% and 20%, respectively (Fig. 4F).
Fig. 4. Comparisons of the serum miRNAs levels in breast cancer patients with different hormone status. (A-E) The serum levels of miRNAs were compared in various groups of breast cancer patients classified by hormone status including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) as well as p53. The serum level of miR-145 was significantly higher in PR-positive patients as compared with PR negative. The number for each group was indicated in the images. Data are expressed as mean ± standard deviation. (F) The positive rates of miR-21, miR-155, and miR-365 as well as CEA and CA153 in breast cancer patients. Positivity of the three miRNAs was determined based on the confidence intervals (CI). The value of miR-21 and miR-155 level ≥ the CI, and the value of miR-365 level ≤ the CI, was considered positive.
Comparison of serum miRNAs levels before and after surgery in breast cancer patients
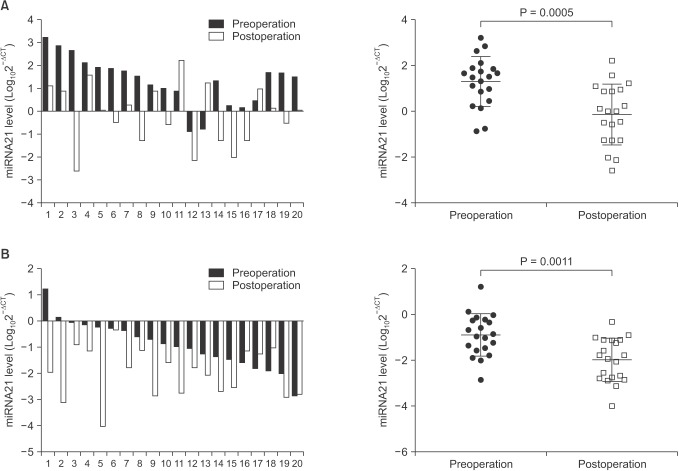
The serum level of miRNAs was analyzed in the paired pre- and postoperative samples from 20 patients with breast cancer. As compared with the preoperative samples, the level of miR-21 and miR-155 decreased significantly (miR-21: –0.01 ± 1.318 vs. 1.31 ± 1.082, P = 0.0005; miR-155: –1.98 ± 0.944 vs. –0.89 ± 0.921, P = 0.0011) in the post-operative samples (Fig. 5).
Fig. 5. Alteration of the serum miR-21 and miR-155 levels in breast cancer patients receiving surgery. Total serum RNA was extracted preoperation and postoperation in 20 breast cancer patients, and quantitative polymerase chain reaction was performed for evaluation of changes of miRNAs levels. In comparison with preoperation, the serum levels of miR-21 (A) and miR-155 (B) were significantly reduced 3 weeks postsurgery in breast cancers. Data are expressed as mean ± standard deviation.
ROC curve analysis
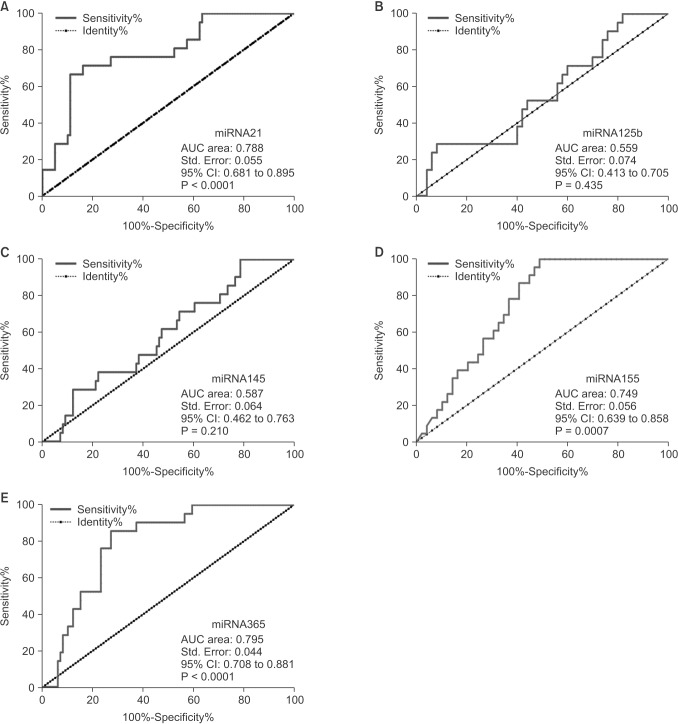
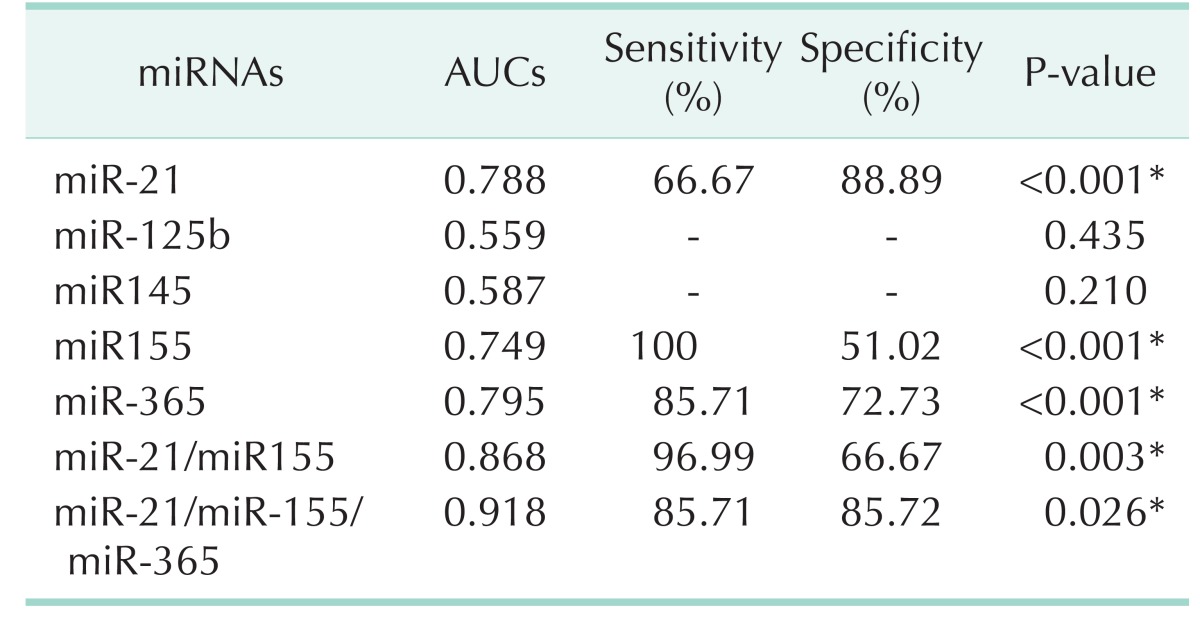
ROC curve analysis was performed to evaluate the diagnostic value for the five miRNAs. The AUCs closer to 1 reflect more substantial differences between breast cancer and normal samples. ROC curve analysis revealed that the three miRNAs, miR-21, miR-155, and miR-365, had significantly higher AUCs with values of 0.788, 0.749, and 0.795, respectively (Fig. 6, Table 2). At the optimal cutoff value of –0.089, with the values of sensitivity plus specificity considered to be maximal for miR-21, the sensitivity and specificity were 66.67% and 88.89%, respectively. At the optimal cutoff value of –1.171 for miR-155, the sensitivity and specificity were 100% and 51.02%, respectively. At the optimal cutoff value of –0.4722 for miR-365, the sensitivity and specificity were 85.71% and 72.73%, respectively.
Fig. 6. Area under curve (AUCs) of receiver operating characteristic (ROC) for serum miRNAs levels. The three miRNAs including miR-21 (A), miR-155 (D), and miR-365 (E) show higher sensitivity and specificity than miR-125b (B) and miR-145 (C). The AUC area, standard (Std.) error, 95% confidence interval (CI), and the P-values are indicated in the images.
Table 2. The area under curve, sensitivity and specificity of miRNAs.
AUC, area under curve; miRNA, microRNA.
*P < 0.05, statistically significant.
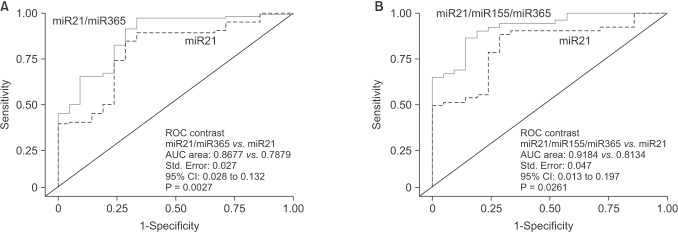
We further performed ROC curve analyses for combinations of miR-21, miR-155, and miR-365. Compared to miR-21 alone, the combination of miR-21/miR-365 yielded higher AUC (0.8677 vs. 0.7879, P = 0.0027), and better sensitivity (96.99%) and specificity (66.67%) (Fig. 7A). A combination of the three miRNs also created higher AUC (0.9184 vs. 0.8134; P = 0.0261), and better sensitivity (85.71%) and specificity (85.72%) (Fig. 7B).
Fig. 7. Area under curve (AUCs) of receiver operating characteristic (ROC) for combination of microRNAs (miRNAs). The combination of miR-21 and miR-365, as well as miR-21, miR-155 and miR-365, shows higher sensitivity and specificity than each miRNA alone, e.g. miR-21. The AUC area, standard (Std.) error, 95% confidence interval (CI), and the P-values are indicated in the images.
DISCUSSION
Endocrine treatment is conventionally used for ER+ patients, and trastuzumab for HER2+ patients [14,15]. Although these biomarkers are commonly used in clinic for target therapy and management of breast cancer, there is still room for improvement, such as early diagnosis of tumor lesions, identification of high-risk patients, development of predictive biomarkers for monitoring progress and therapy effects, etc. By mapping the circulating miRNAs signature and profiling, some miRNAs have been found differentially expressed in breast cancer and normal tissues [16]. Although the clinical application of serum miRNAs as a noninvasive diagnostic strategy is promising, the miRNA signatures should be further investigated and validated for different subtypes of breast cancers. In the current study, we analyzed the serum level of five miRNAs including miR-21, miR-125b, miR-145, miR-155, and miR-365 in breast cancers and healthy controls, followed by construction of ROC curves to determine the sensitivity and specificity of circulating miRNAs as potential biomarkers for breast cancer.
Our results showed that the serum concentrations of miR-21 and miR-155 were significantly elevated, while miR-365 was significantly downregulated in patients with breast cancer compared with healthy controls (Fig. 1). MiR-21 is involved in regulating the expression of multiple tumor suppressor genes, and plays a crucial role in the occurrence and development of a variety of cancer diseases [17]. In breast cancer, several studies have shown that up-regulation of miR-21 was detected in both serum and tissues [18], and that serum miR-21 could be used as an non-invasive biomarker for diagnosis and prognosis of breast cancer as well as an indicator for the invasiveness of the tumor [19]. MiR-155 plays a role in various physiological and pathological processes, and oversilencing by miR-155 may result in apoptotic resistance and thus triggering oncogenic cascades. It has been reported that the amount of both circulating miR-155 in serum and non-circulating miR-155 in tissue were elevated in patients with breast cancer [20,21]. Therefore, miR-21 and miR-155 may act as oncogenic factors, and thus may be potential targets for breast cancer therapy. Additionally, we found that miR-155 level was remarkably higher at stages I and II compared to stage III in breast cancer patients (Fig. 3). Similarly, it has been shown that miR-7 was associated with breast cancer grades [22]. Reduction of tissue miR-365 has been reported in different types of cancers such as hepatocellular carcinoma (HCC) [23] and lung cancer [24]. An in vitro experiment showed that overexpression of miR-365 remarkably suppressed proliferation and migration capacities of HCC cell [23]. Kodahl et al. [25] also showed that the serum miR-365 was downregulated in early stage breast cancer patients. Family history is an important predisposing factor in breast cancer. However, we did not find differential expression of the five miRNAs between family history positive and negative breast cancer patients (Fig. 2).
In this study, the serum abundance of miR-125b and miR-145 displayed no remarked difference between breast cancer and healthy control (Fig. 1). Nevertheless, by comparing the miRNAs profile between 61 breast cancers and 10 healthy controls in the Mexican population, it was found that the serum levels of miR-145 and miR-125b were significantly higher in patients with breast cancer than healthy controls [26]. The current analysis just found that serum miR-145 abundance was significantly higher in PR-positive breast cancer patients compared to the PR-negative (Fig. 4C), indicating that serum miR-145 level may separate the PR-positive subtypes of breast cancer. A previous study also examined the association of miRNAs expression in serum with different tumor hormone status, and they found 7 miRNAs with differential expression for women whose breast cancer differed by HER2 expression [27]. Notably, it was reported that the level of miR-145 was downregulated both in the in vitro cultured breast cancer cell lines [28] and in the serum from early stage breast cancer patients [25]. CA153 and CEA are the most widely used circulating biomarkers in monitoring patients with breast cancer. Nevertheless, we found that the positivity of miR-21, miR-155 and miR-365 was higher in comparison with CEA and CA 153 in breast cancer patients (Fig. 4F). Similarly, serum miR-21 and miR-30a has a higher sensitivity in diagnosis of breast cancer compared with CEA and CA153 [29,30].
The serum level of miR-21 and miR-155 was further analyzed in 20 patients with breast cancer before surgery and 3 weeks after tumor removal. The abundance of serum miR-21 and miR-155 was significantly reduced in the postoperative samples as compared with the paired pre-operative samples (Fig. 5). Consistently, Sochor et al. [20] found that early breast cancer patients significantly overexpressed several oncogenic miRNAs such as miR-155, miR-181b, miR-19a, and miR-24, which dramatically decreased following surgical resection. Kodahl et al. [25] reported that circulating miR-338-3p, miR-223, and miR-148a exhibited lower, and miR-107 exhibited higher levels postoperatively than the preoperative samples from 24 postmenopausal women with ER+ early-stage breast cancer. These findings suggest that serum oncogenic miRNAs may be potentially used for diagnostic purpose and relapse judgement.
To determine the diagnostic performance of miRNAs for breast cancer, we performed ROC curve analysis, and the AUC was used as the evaluation criteria; the higher AUC, the better diagnostic performance. Our data revealed that the 3 miRNAs miR-21, miR-155, and miR-365 had significantly higher AUC with the values of 0.788, 0.749, and 0.795, respectively (Fig. 6), suggesting that we were able to discriminate breast cancer from healthy controls. The sensitivity for the three miRNAs (miR-21, miR-155, and miR-365) was 66.7%, 100%, and 85.7%, respectively, and the specificity was 88.9%, 51.02%, and 72.7%, respectively (Fig. 6, Table 2). It has been reported that a miRNA panel could accurately distinguish cancers from normal subjects [4,7,8,11,12]. A combination of ROC curve analyses of miR-145, miR-155, and miR-382 exhibited much better sensitivity and specificity than each miRNA alone [30]. In this study, ROC curve was analyzed in a panel of 3 miRNAs (miR-21, miR-155, and miR-365). Our data demonstrate that a combination of miR-21 and miR-365 yielded a significantly high AUC (0.868), sensitivity (96.97%), and specificity (66.67%). The combination of miR-21, miR-155, and miR-365 further generated much higher AUC (0.918), accompanied by relatively higher sensitivity (85.71) and specificity (85.72) (Fig. 7, Table 2). Accordingly, our results implied that a panel of miRNAs (e.g., a combination of miR-21/miR-155/miR-365) remarkably enhanced the diagnostic performance with high sensitivity and specificity, and thus may provide an improved indicator for breast cancer diagnosis and screening.
Taken together, our study provides evidence that evaluation of serum miRNAs level can be used as biomarkers for diagnosis of breast cancer, and that a combination of miR-21/miR-155/miR-365 may potentially serve as a sensitive and specific biomarker that enables the differentiation of breast cancer from healthy controls.
Footnotes
CONFLICTS OF INTEREST: No potential conflict of interest relevant to this article was reported.
References
- 1.Schettini F, Buono G, Cardalesi C, Desideri I, De Placido S, Del Mastro L. Hormone Receptor/Human Epidermal Growth Factor Receptor 2-positive breast cancer: Where we are now and where we are going. Cancer Treat Rev. 2016;46:20–26. doi: 10.1016/j.ctrv.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Duffy MJ. Role of tumor markers in patients with solid cancers: a critical review. Eur J Intern Med. 2007;18:175–184. doi: 10.1016/j.ejim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Pileczki V, Cojocneanu-Petric R, Maralani M, Neagoe IB, Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89:50–55. doi: 10.15386/cjmed-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomides CC, Evans BJ, Navenot JM, Vadigepalli R, Peiper SC, Wang ZX. MicroRNA profiling in lung cancer reveals new molecular markers for diagnosis. Acta Cytol. 2012;56:645–654. doi: 10.1159/000343473. [DOI] [PubMed] [Google Scholar]
- 5.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 8.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chavez J, Trevino V, Rodriguez-Padilla C, et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–59. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 9.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 11.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravelli A, Reuben JM, Lanza F, Anfossi S, Cappelletti MR, Zanotti L, et al. Breast cancer circulating biomarkers: advantages, drawbacks, and new insights. Tumour Biol. 2015;36:6653–6665. doi: 10.1007/s13277-015-3944-7. [DOI] [PubMed] [Google Scholar]
- 13.Al-Khanbashi M, Al-Moundhri M. Microribonucleic acid and carcinogenesis: breast cancer as an example. Oncol Rev. 2015;9:279. doi: 10.4081/oncol.2015.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fatima S, Faridi N, Gill S. Breast cancer: steroid receptors and other prognostic indicators. J Coll Physicians Surg Pak. 2005;15:230–233. [PubMed] [Google Scholar]
- 15.Stark AT, Claud S, Kapke A, Lu M, Linden M, Griggs J. Race modifies the association between breast carcinoma pathologic prognostic indicators and the positive status for HER-2/neu. Cancer. 2005;104:2189–2196. doi: 10.1002/cncr.21463. [DOI] [PubMed] [Google Scholar]
- 16.Graveel CR, Calderone HM, Westerhuis JJ, Winn ME, Sempere LF. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer (Dove Med Press) 2015;7:59–79. doi: 10.2147/BCTT.S43799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB, Cheng XC. MicroRNA-21 gene and cancer. Med Oncol. 2013;30:376. doi: 10.1007/s12032-012-0376-8. [DOI] [PubMed] [Google Scholar]
- 18.Savad S, Mehdipour P, Miryounesi M, Shirkoohi R, Fereidooni F, Mansouri F, et al. Expression analysis of MiR-21, MiR-205, and MiR-342 in breast cancer in Iran. Asian Pac J Cancer Prev. 2012;13:873–877. doi: 10.7314/apjcp.2012.13.3.873. [DOI] [PubMed] [Google Scholar]
- 19.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 20.Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, et al. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236–1243. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 22.Lyng MB, Lænkholm AV, Sokilde R, Gravgaard KH, Litman T, Ditzel HJ. Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant tamoxifen monotherapy: a DBCG study. PLoS One. 2012;7:e36170. doi: 10.1371/journal.pone.0036170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Huang Z, Ye Q, Ming Y, Zhang S, Zhao Y, et al. Prognostic significance and anti-proliferation effect of microRNA-365 in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- 24.Kang SM, Lee HJ, Cho JY. MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 2013;335:487–494. doi: 10.1016/j.canlet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ERpositive early-stage breast cancer: a case control study. Mol Oncol. 2014;8:874–883. doi: 10.1016/j.molonc.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godfrey AC, Xu Z, Weinberg CR, Getts RC, Wade PA, DeRoo LA, et al. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:R42. doi: 10.1186/bcr3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012;109:14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP, et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013;30:477. doi: 10.1007/s12032-013-0477-z. [DOI] [PubMed] [Google Scholar]
- 30.Gao J, Zhang Q, Xu J, Guo L, Li X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res. 2013;25:743–748. doi: 10.3978/j.issn.1000-9604.2013.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]