Abstract
Industrial glucose feedstock prepared by enzymatic digestion of starch typically contains significant amounts of disaccharides such as maltose and isomaltose and trisaccharides such as maltotriose and panose. Maltose and maltosaccharides can be utilized in Escherichia coli fermentation using industrial glucose feedstock because there is an intrinsic assimilation pathway for these sugars. However, saccharides that contain α-1,6 bonds, such as isomaltose and panose, are still present after fermentation because there is no metabolic pathway for these sugars. To facilitate more efficient utilization of glucose feedstock, we introduced glvA, which encodes phospho-α-glucosidase, and glvC, which encodes a subunit of the phosphoenolpyruvate-dependent maltose phosphotransferase system (PTS) of Bacillus subtilis, into E. coli. The heterologous expression of glvA and glvC conferred upon the recombinant the ability to assimilate isomaltose and panose. The recombinant E. coli assimilated not only other disaccharides but also trisaccharides, including alcohol forms of these saccharides, such as isomaltitol. To the best of our knowledge, this is the first report to show the involvement of the microbial PTS in the assimilation of trisaccharides. Furthermore, we demonstrated that an l-lysine-producing E. coli harboring glvA and glvC converted isomaltose and panose to l-lysine efficiently. These findings are expected to be beneficial for industrial fermentation.
Keywords: Escherichia coli, Maltose, Isomaltose, Panose, Assimilation, glvA, glvC, Bacillus subtilis, l-Lysine production
Introduction
In the industrial production of useful compounds by fermentation, glucose is one of the most frequently used carbon sources (Peters 2007). Industrial glucose feedstock is prepared from starch, a polysaccharide composed of glucose units linked together by α-1,4 and α-1,6 glycoside bonds, by means of enzymatic hydrolysis (Martin and Smith 1995; Hii et al. 2012). Complete hydrolysis of starch into glucose adds significant cost; therefore, most commercially available glucose feedstock is processed incompletely (Gokarn et al. 2014). Because of incomplete enzymatic hydrolysis and/or reverse reactions, the glucose feedstock contains significant amounts of maltose [4-O-α-D-glucopyranosyl-D-glucopyranose] (1–2%), isomaltose [6-O-α-D-glucopyranosyl-D-glucopyranose] (0.5–2%), and other oligosaccharides, such as panose [α-D-glucopyranosyl-(1->6)-α-D-glucopyranosyl-(1->4)-D-glucopyranose] (1% or less) (Hii et al. 2012; Gokarn et al. 2014; Hassan et al. 1998; Crabb and Shetty 1999; Sierkes and Svensson 1992; Takasaki 1988; Chaplin and Bucke 1994). If microorganisms used for fermentation cannot metabolize these saccharides, valuable carbohydrates would be wasted.
Escherichia coli is the most useful bacterial strain for the production of valuable compounds, such as amino acids and organic acids, because E. coli cells grow quickly, rapidly convert substrates to products, and are readily genetically engineered (Leuchtenberger et al. 2005; Wendisch et al. 2006). For example, l-lysine, which is used as a feed additive worldwide, is produced on the scale of approximately 1,500,000 t per year (Doi et al. 2014). However, saccharides that contain α-1,6 bonds, such as isomaltose and panose, are not used up during E. coli fermentation because E. coli cannot assimilate isomaltose and panose as carbon sources. Furthermore, these sugars, which contain reducing sugar moieties, can react with free amino groups of amino acids during the purification step (Smuda and Glomb 2011; Ledl and Schleicher 1990); this so-called Maillard reaction decreases the yield of the final product and contaminates the reaction mixture with undesirable compounds. These problems must be overcome in order to increase the yield and productivity of fermentation when using glucose feedstock as a carbon source.
The phosphotransferase system (PTS) is responsible for the transport and phosphorylation of sugars. The multi-component PTS comprises a phosphohistidine carrier protein (HPr), an enzyme I (EI) component, and a membrane-bound enzyme complex (EII). The HPr and EI components transfer a phosphoryl group of phosphoenolpyruvate (PEP) to the sugar-specific enzymes EIIA and EIIB. EIIC is an integral membrane protein permease that recognizes and transports the sugar, which is then phosphorylated by EIIB (Postma et al. 1993). There are 21 different EII complexes encoded in the E. coli chromosome; these complexes are involved in the transport of approximately 20 different carbohydrates (Escalante et al. 2012). Pikis et al. reported that the heterologous expression of Klebsiella pneumoniae aglA (a single-chain polypeptide of EIIC and EIIB that mediates the transport and phosphorylation of sucrose and various other α-linked glucosides) and aglB (a phospho-α-glucosidase) confers upon E. coli cells the ability to utilize isomaltose (Pikis et al. 2006; Thompson et al. 2001). Although E. coli K-12 strains have homologs of aglA and aglB (Thompson et al. 2001), these seemed to be cryptic or nonfunctional truncated proteins (Reizer 1994; Thompson et al. 1995). However, Bacillus subtilis strains, which are generally regarded as safe (GRAS) organisms by the Food and Drug Administration (FDA) (Harwood and Wipat 1996; Singh et al. 2009; Song et al. 2016; Zeigler et al. 2008), have glvA and glvC, functional homologs of aglB and aglA, respectively (Thompson et al. 2001). GlvA and GlvC are known to be involved in maltose assimilation in B. subtilis (Yamamoto et al. 2001; Thompson et al. 1998). Although a wide variety of phosphorylated α-linked aryl glucosides can be degraded by GlvA (Yip et al. 2007), there are no other reports describing its substrate specificity.
In this study, we found that the heterologous expression of glvA and glvC conferred upon E. coli cells the ability to assimilate isomaltose. Unexpectedly, the recombinant also assimilated trisaccharides containing α-1,6 bonds, such as panose, as well as the alcohol forms of these saccharides, such as isomaltitol. Our results may facilitate increased production yields using glucose feedstock in industrial-scale fermentation by E. coli.
Materials and methods
Bacterial strains, plasmids, and primers
All strains, plasmids, and primers used in this study are listed in Tables 1 and 2.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description, genotype, or sequence | Reference, source |
|---|---|---|
| Strains | ||
| E. coli K-12 MG1655 | F− l− ilvG rfb-50 rph-1 | CGSC Collection |
| E. coli WC196LC | W3110 NTG mutant (S-aminoethyl-L-cysteine-resistant mutant) Δldc ΔcadA | Kikuchi et al. (1997) |
| Bacillus subtilis 168 | trpC2 ypqP::SPβ | Zeigler et al. (2008) |
| MG1655 (empty vector) | E. coli K-12 MG1655 harboring pTWV229 and pMW219-Δplac | This study |
| MG1655 (glvAC) | E. coli K-12 MG1655 harboring pTWV229-self-glvA-Fw and pMW219-ΔPlac-*tac-glvC | This study |
| WC196LC (pCABD2) | E. coli WC196LC harboring pCABD2 | Kojima et al. (1994); Kikuchi et al. (1997); Doi et al. (2014) |
| Plasmids | ||
| pTWV229 | Cloning vector, Apr | Takara Bio Inc., (Japan) |
| pMW219 | Cloning vector, Kmr | Nippon Gene Co. Ltd. (Japan) |
| pMW219-ΔPlac | pMW219 derivative lacking the lac promoter region | This study |
| pMW219-ΔPlac-tac-glvC-R2 | pMW219-ΔPlac derivative harboring the Ptac-glvC gene | This study |
| pMW219-ΔPlac-Ptac4075-glvC-Rv | pMW219-ΔPlac derivative harboring the Ptac4075-glvC gene | This study |
| pTWV229-self-glvA-Fw | pTWV229 derivative harboring the Pself-glvA gene | This study |
| pMW219-ΔPlac-*tac-glvC | pMW219-ΔPlac-tac-glvC-R2 derivative harboring a mutation in the −10 region of the tac promoter (TATAAT to AATAAT) | This study |
| pCABD2 | pRSF1010 harboring mutated lysC, mutated dapA, mutated dapB, and C. glutamicum ddh | Kojima et al. (1994) |
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| pMW119-F-Hind3 | GCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGG |
| pMW119-R-Hind3 | CCCAAGCTTGCTAACTCACATTAATTGCGTTG |
| pTWV229-F-Hind3 | GCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGG |
| pTWV229-R-Hind3 | CCCAAGCTTCACATTACTTGGCAGAACATATCC |
| glvC-F-tac | CAATTTCACACAAGGAGACTGCCATGATGCAAAAAATTCAGCG |
| glvC-R2 | CCCAAGCTTCCCCTTTTTACTCGATTGTCTC |
| tac-promoter-glvC-1 | CGTATAATGTGTGGAATCGTGAGCGGATAACAATTTCACACAAGGAGACTGCCATGATGCAAAAAATTCAGCGCTTTGGA |
| Hind3-tac-promoter | CCCAAGCTTCCTGTTGACAATTAATCATCGGCTCGTATAATGTGTGGAATCGTGAGCGGATAACAATTTCACACAAGGAG |
| tac-promoter-glvC-2 | CGAATAATGTGTGGAATCGTGAGCGGATAACAATTTCACACAAGGAGACTGCCATGATGCAAAAAATTCAGCGCTTTGGA |
| glvA-self-Fw1 | AGAAATTTCCCGCTCTATGG |
| glvA-self-Rv1 | TGTAGTGCTGATTGATCAGTTC |
HindIII recognition site was underlined
Construction of vectors
The plasmid pMW219-ΔPlac was constructed by deleting the lac promoter from the vector plasmid pMW219 (Nippon Gene Co., Ltd., Tokyo, Japan) as follows. A DNA fragment was amplified using the primer set pMW119-F-Hind3 and pMW119-R-Hind3, and HindIII/PstI-digested pMW219 was used as a template. The polymerase chain reaction (PCR)-amplified fragment was digested by HindIII and subsequently self-ligated by DNA ligase. E. coli JM109 competent cells were transformed with the DNA, and transformants were selected on LB agar medium containing kanamycin.
Construction of glvC-expressing plasmids
A DNA fragment containing glvC was amplified by PCR with the primer set glvC-F-tac and glvC-R2 and with the B. subtilis 168 genome as a template. In order to add a promoter sequence upstream of glvC, the amplified DNA fragment containing glvC and synthetic single-strand DNA (tac-promoter-glvC-1) were mixed, and another PCR was then carried out using the primer set Hind3-tac-promoter, glvC-R2. The amplified DNA fragment and SmaI-digested pMW219-ΔPlac were ligated by DNA ligase. The plasmid pMW219-ΔPlac-glvC-R2 was extracted from transformants, and its structure was confirmed. The plasmid pMW219-ΔPlac-Ptac4075-glvC-Rv containing glvC under the control of the tac promoter variant Ptac4075, which was a weaker promoter than tac promoter because of a mutation in the consensus sequence, was constructed in a similar manner using primers Hind3-tac-promoter, glvC-R2, and synthetic DNA, tac-promoter-glvC-2.
Construction of glvA-expressing plasmid
A DNA fragment containing glvA and its upstream region containing a promoter sequence was amplified by PCR with the primer set glvA-self-Fw1 and glvA-self-Rv1 and with the B. subtilis 168 genome as a template. The PCR-amplified DNA and SmaI-digested pTWV229 were ligated by DNA ligase. In the resulting plasmid, pTWV229-self-glvA-Fw, glvA mRNA was transcribed via the lac promoter of pTWV229.
Assimilation test in M9 minimal medium
M9 liquid minimal medium (Miller 1992) supplemented with 2 g/L isomaltose or maltose was used for assimilation tests. E. coli strains were precultured overnight at 37 °C on LB medium. The cells were washed three times with cold saline and adjusted to an OD620 of 7.0. The cell suspension (70 μL) was added to 5 mL M9 minimal medium in an L-shaped test tube and cultured at 37 °C with shaking at 70 rpm using a Bio-Photorecorder (TN-1506; Advantec, Inc., Tokyo, Japan). In all experiments, appropriate antibiotics were added to the medium. M9 solid minimal medium (Miller 1992) supplemented with 2 g/L of various types of sugars and sugar alcohols was used for assimilation tests. E. coli strains were precultured overnight at 37 °C on LB medium. The cells were washed three times by cold saline and adjusted to OD620 of 5.0. The cell suspension (20 μL) was inoculated on M9 minimal medium plates containing various types of sugars and sugar alcohols and incubated at 37 °C for 48 h. Glucose and sucrose were purchased from Junsei Chemical Co., Ltd. (Tokyo, Japan). Maltose was purchased from Nacalai Tesque, Inc. (Kyoto, Japan). α-Methyl-glucoside was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Isomaltulose, maltotriitol, isomaltitol, lactitol, and erlose were purchased from Hayashibara Co., Ltd. (Okayama, Japan). Isomaltose, panose, isomaltotriose, maltitol, trehalose, turanose, maltulose, galactinol, cellobiose, gentiobiose, lactose, melibiose, lactulose, maltotriose, maltotetraose, maltopentaose, and raffinose were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). For solid medium, 15 g/L Bacto agar (Becton Dickinson and Company, USA) was added.
Assimilation tests for maltose and isomaltose in the presence of glucose
E. coli MG1655 harboring pTWV229-self-glvA-Fw and pMW219-ΔPlac-*tac-glvC (obtained by an unintended mutation, as described in the “Results” section) was inoculated into M9 minimal medium containing 1.0 g/L maltose or isomaltose combined with 1.0 g/L glucose. Cells were then cultured at 37 °C with shaking at 70 rpm using a Bio-Photorecorder (Advantec), and sugar concentrations were assayed.
l-Lysine production using glucose, maltose, isomaltose, and panose as carbon sources
The l-lysine-producing strain WC196LC harboring pCABD2 [encoding dapA24, lysC80, dapB, and ddh (Kojima et al. 1994; Kikuchi et al. 1997; Doi et al. 2014)] was transformed with pMW219-ΔPlac-Ptac4075-glvC and pTWV229-self-glvA-Fw. The transformant was inoculated on an LB plate containing 20 mg/L streptomycin, 100 mg/L ampicillin, and 50 mg/L kanamycin and incubated at 37 °C for 24 h. Colonies were scratched off, suspended in saline, and adjusted to an OD620 of 15. Next, 250 μL of the cell suspension was added to 5 mL L-lysine production medium containing 16 g/L glucose, 1.6 g/L maltose and/or isomaltose and/or 1.6 g/L panose, 1 g/L MgSO4 heptahydrate, 24 g/L (NH4)2SO4, 1 g/L KH2PO4, 2 g/L yeast extract, 0.1 g/L isoleucine, 12 mg/L FeSO4 heptahydrate, 9.6 mg/L MnSO4 pentahydrate, 30 g/L CaCO3 (as dry heat-sterilized powder), 20 mg/L streptomycin, 100 mg/L ampicillin, and 50 mg/L kanamycin (pH 7.0 with KOH). Cells were then cultivated at 37 °C for 41 h with reciprocal shaking at 120 rpm. Glucose and l-lysine were assayed by a biotech analyzer (AS310; Sakura Si Co., Ltd., Tokyo, Japan). Maltose, isomaltose, and panose were measured using an ICS-3000 Ion Chromatography System with a CarboPac PA1 column (Dionex, CA, USA).
Results
Evaluation of the functions of GlvA and GlvC from B. subtilis in E. coli
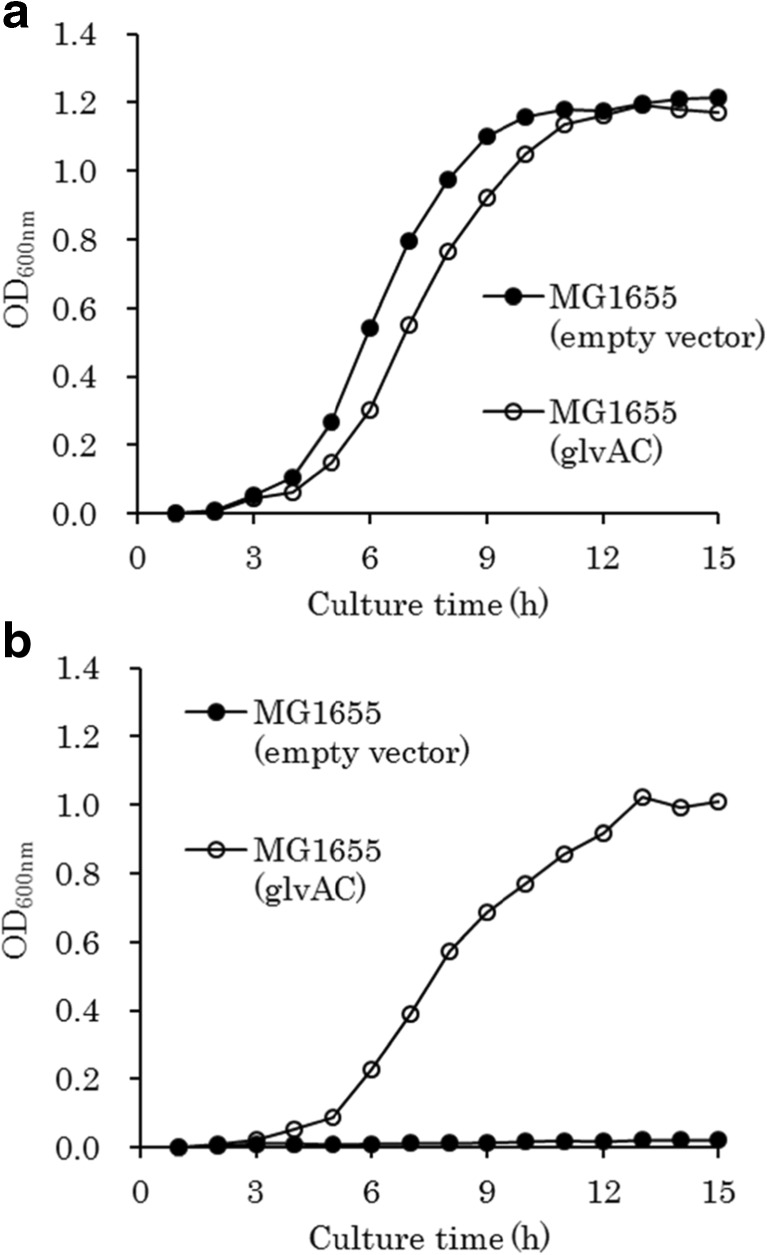
GlvA and GlvC have been reported to be involved in the assimilation of maltose in B. subtilis (Yamamoto et al. 2001; Thompson et al. 1998). We constructed the plasmids pTWV229-self-glvA-Fw and pMW219-ΔPlac-tac-glvC-R2 for the expression of glvA and glvC from B. subtilis in E. coli. The growth of the E. coli MG1655 recombinant harboring both of the plasmids on M9 medium containing isomaltose was severely limited (data not shown). However, after prolonged incubation (45 h) of the recombinant at 37 °C on the isomaltose medium, we found that some mutants started to form larger colonies. Because plasmids isolated from one of the mutants enabled E. coli to grow rapidly on medium containing isomaltose, we analyzed the DNA sequences of these plasmids. A mutation was found in the −10 region of the tac promoter (de Boer et al. 1983) (TATAAT to AATAAT) upstream of glvC. This mutation is expected to reduce the expression level of glvC by affecting the binding affinity of RNA polymerase. We speculated that strong expression of GlvC, a membrane permease, may be toxic in E. coli. The mutant plasmid was renamed pMW219-ΔPlac-*tac-glvC. Next, we tested the growth of E. coli MG1655 harboring pTWV229-self-glvA-Fw and pMW219-ΔPlac-*tac-glvC plasmids in M9 liquid medium containing isomaltose as a sole carbon source. The recombinant could grow on isomaltose efficiently, whereas E. coli harboring the empty vector plasmids could grow only on glucose (Fig. 1).
Fig. 1.
Growth curves of E. coli strains on M9-glucose (a) and M9-isomaltose (b). Each point in the curve represents the mean of two independent experiments
Simultaneous assimilation of glucose and isomaltose by glvA-expressing and glvC-expressing E. coli
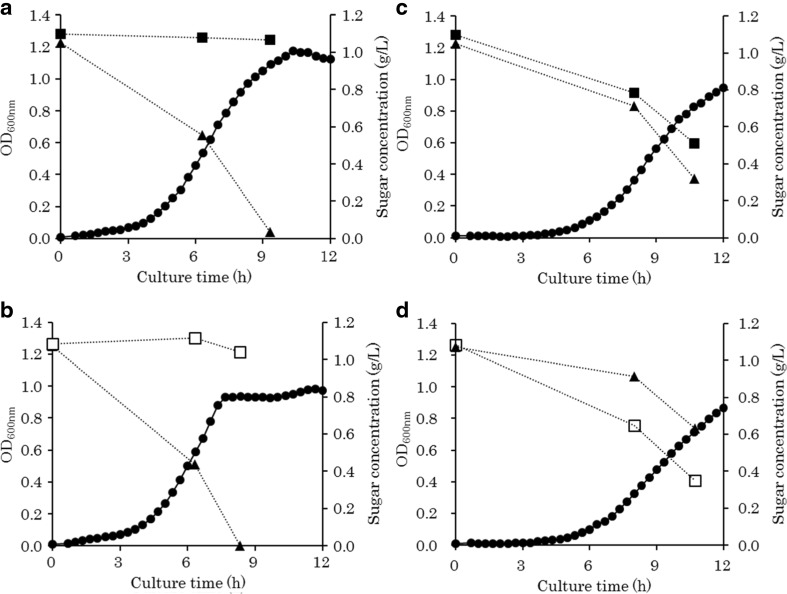
To test whether the E. coli recombinant could assimilate isomaltose without catabolite repression, we used M9 medium containing an excess amount (1.8 g/L) of isomaltose or maltose supplemented with a small amount (0.2 g/L) of glucose (Loomis and Magasanik 1967). In this medium, cell growth would stop temporarily at an OD600 of 0.1–0.2 if the strain consumed glucose first (showing diauxic growth). The E. coli recombinant showed relatively slow but smooth cell growth, without characteristics of diauxic growth (data not shown), suggesting that the E. coli recombinant could assimilate isomaltose without catabolite repression. Then, to demonstrate this phenomenon more clearly, we measured sugar concentrations during cultivation on M9 minimal medium containing 1.0 g/L maltose and glucose or 1.0 g/L isomaltose and glucose. The E. coli carrying empty vector did not assimilate maltose or isomaltose in the presence of glucose (Fig. 2a, b), because E. coli assimilates maltose under the control of catabolite repression (Dean et al. 1990; Boos and Shuman 1998) and cannot assimilate isomaltose. On the other hand, the recombinant assimilated maltose or isomaltose in the presence of glucose (Fig. 2c, d). Surprisingly, the recombinant preferentially assimilated isomaltose over glucose (Fig. 2d). These results indicated that the heterologous expression of glvA and glvC conferred upon the cells the ability to assimilate maltose and isomaltose even in the presence of glucose.
Fig. 2.
Growth curves of E. coli recombinants on M9-glucose-maltose or M9-glucose-isomaltose. a MG1655 (empty vector) on M9-glucose-maltose. b MG1655 (empty vector) on M9-glucose-isomaltose. c MG1655 (glvAC) on M9-glucose-maltose. d MG1655 (glvAC) on M9-glucose-isomaltose. Cell growth (closed circle), glucose concentration (closed triangle), maltose concentration (closed square), isomaltose concentration (open square)
Evaluation of the substrate specificity of GlvA and GlvC in E. coli
Pikis et al. reported that that expression of aglA and aglB from K. pneumoniae in E. coli allowed the cells to assimilate α-methyl-glucoside, isomaltose, trehalulose, turanose, maltulose, leucrose, and isomaltulose (Pikis et al. 2006). To investigate whether the heterologous expression of glvA and glvC enabled E. coli to assimilate other types of carbon sources, particularly panose, we performed growth tests using M9 solid medium supplemented with various types of sugars and sugar alcohols as a sole carbon source. We tested glucose, α-methyl-glucoside, sucrose, maltose, isomaltose, maltitol, trehalose, maltulose, isomaltulose, galactinol, cellobiose, gentiobiose, lactose, melibiose, lactulose, maltotriose, panose, isomaltotriose, maltotetraose, raffinose, maltotriitol, isomaltitol, lactitol, erlose, and maltopentaose (Table 3 and Fig. 3). Our results showed that the recombinant could assimilate many types of sugars and sugar alcohols, including α-methyl-glucoside, isomaltose, turanose, maltulose, and isomaltulose, which had been reported to be assimilated by AglA and AglB (Pikis et al. 2006). In contrast, galactinol, maltotriitol, and erlose could not be assimilated by the recombinant. Interestingly, the recombinant was able to assimilate trisaccharides, such as panose and isomaltotriose. Moreover, β-linked disaccharides such as sucrose and gentiobiose were also assimilated by the recombinant. It is an unexpected result because GlvA is classified as a 6-phospho-α-glucosidase (Yip et al. 2007).
Table 3.
Growth of E. coli recombinants on selected sugars
| Substrate | DP | Form | MG1655 (empty vector) | MG1655 (glvAC) | MG1655 (pAP2) (Pikis et al. 2006) |
|---|---|---|---|---|---|
| Glucose | 1 | Sugar | ++ | ++ | ++ |
| α-Methyl-glucoside | 1 | Sugar | NDG | ++ | ++ |
| Sucrose | 2 | Sugar | NDG | ++ | − |
| Maltose | 2 | Sugar | ++ | ++ | ++ |
| Isomaltose | 2 | Sugar | NDG | ++ | ++ |
| Maltitol | 2 | Sugar alcohol | NDG | ++ | ++ |
| Trehalose | 2 | Sugar | ++ | ++ | ++ |
| Turanose | 2 | Sugar | + | ++ | ++ |
| Maltulose | 2 | Sugar | NDG | ++ | ++ |
| Isomaltulose | 2 | Sugar | NDG | ++ | ++ |
| Galactinol | 2 | Sugar alcohol | NDG | NDG | No information |
| Cellobiose | 2 | Sugar | + | + | No information |
| Gentiobiose | 2 | Sugar | NDG | + | No information |
| Lactose | 2 | Sugar | ++ | ++ | No information |
| Melibiose | 2 | Sugar | ++ | ++ | No information |
| Lactulose | 2 | Sugar | ++ | ++ | No information |
| Maltotriose | 3 | Sugar | ++ | ++ | No information |
| Panose | 3 | Sugar | NDG | ++ | No information |
| Isomaltotriose | 3 | Sugar | NDG | ++ | No information |
| Maltotetraose | 4 | Sugar | ++ | ++ | No information |
| Raffinose | 3 | Sugar | + | ++ | No information |
| Maltotriitol | 3 | Sugar alcohol | NDG | NDG | No information |
| Isomaltitol | 3 | Sugar alcohol | NDG | ++ | No information |
| Lactitol | 2 | Sugar alcohol | ++ | ++ | No information |
| Erlose | 3 | Sugar | NDG | NDG | No information |
| Maltopentaose | 5 | Sugar | ++ | ++ | No information |
Data of MG1655 (pAP2) which expresses aglA and aglB from K. pneumoniae are described in the reference (Pikis et al. 2006) and listed in this table for comparison to show what kinds of sugars and sugar alcohols were newly assimilated by heterologous expression of glvA and glvC
DP degree of polymerization, NDG no detectable growth, − minimal growth, + slight growth, ++ clear growth
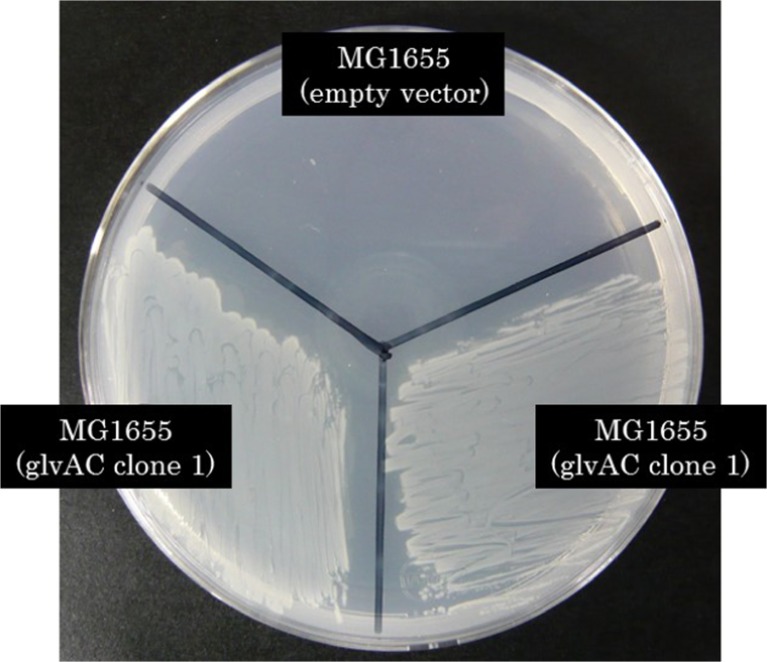
Fig. 3.
Growth of E. coli recombinants on M9 solid medium containing panose as a sole carbon source. Two clones were tested in the case of MG1655 (glvAC)
The expression of either glvA or glvC alone is not sufficient to confer upon the recombinant the ability to assimilate any of the tested sugars and sugar alcohols except α-methyl-glucoside. It is known that α-methyl-glucoside is transported by PtsG of E. coli with concomitant phosphorylation (Pikis et al. 2006). GlvA can hydrolyze the phosphorylated α-methyl-glucoside, conferring the ability to grow on the medium containing α-methyl-glucoside.
A common chemical structure among the substrates assimilated by the GlvA-expressing and GlvC-expressing recombinant was the presence of glucose at one terminal of the sugar or sugar alcohol. Therefore, GlvC appeared to recognize the glucose unit of the sugar or sugar alcohol and transport the unit with concomitant phosphorylation of the glucose terminal. Moreover, GlvA may hydrolyze the 6′-phospho-sugars and sugar alcohols to release glucose 6-phosphate. This is the first report to show expansion of the sugar substrates of E. coli to trisaccharides by heterologous expression of 6-phospho-α-glucosidase and PTS components.
Utilization of isomaltose and panose using GlvA and GlvC in an l-lysine-producing model strain
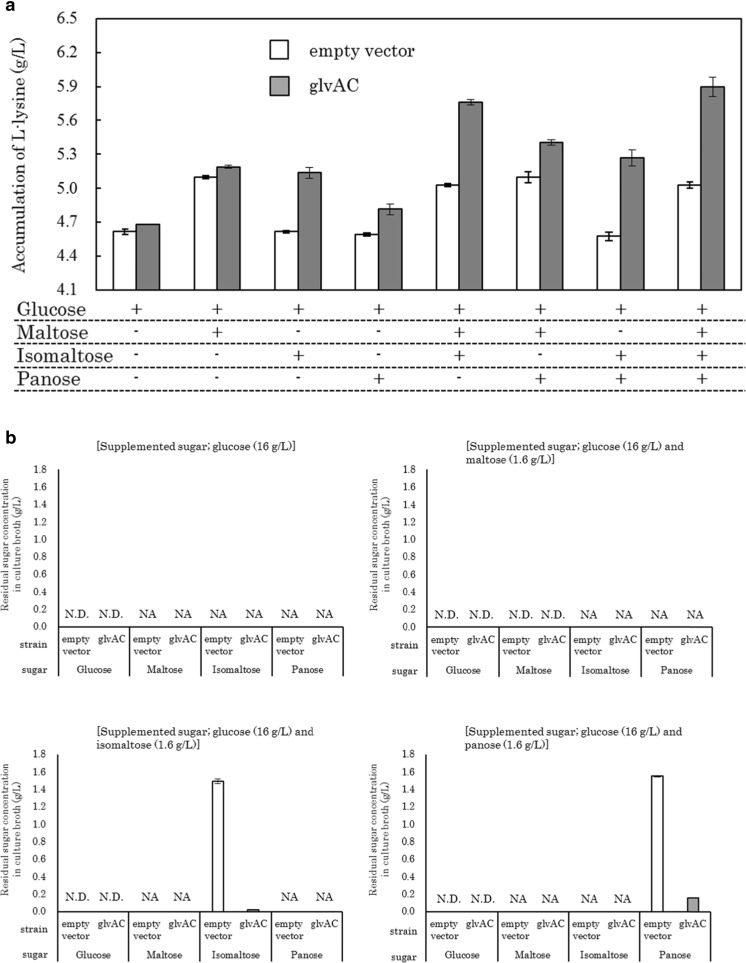
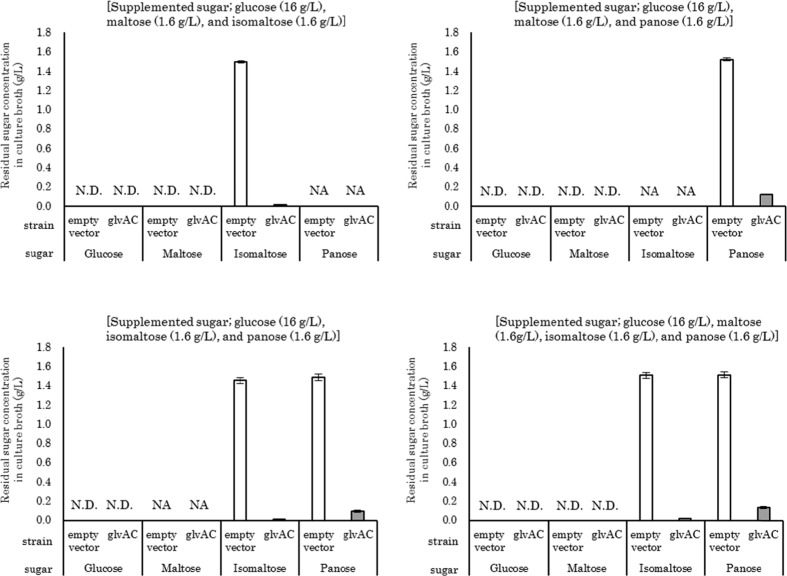
To evaluate the effects of isomaltose and panose utilization on fermentation efficiency, we introduced the plasmids to an l-lysine-producing E. coli strain WC196LC (pCABD2) (Kojima et al. 1994; Kikuchi et al. 1997; Doi et al. 2014). The recombinant was cultivated on l-lysine production medium supplemented with glucose or glucose combined with isomaltose, panose, or maltose (as a control). Additionally, media containing different combinations of the above saccharides were also prepared and used for l-lysine production tests. l-Lysine accumulation in the culture broth of E. coli WC196LC (pCABD2) harboring the empty vector plasmids was increased by approximately 0.5 g/L only when maltose, which can be assimilated intrinsically by E. coli, was contained in the medium in addition to glucose (Fig. 4a). In contrast, the l-lysine production by the recombinant was increased when maltose, isomaltose, and panose were all contained in the medium. In the case of isomaltose utilization by the recombinant, l-lysine accumulation in the culture broth was increased by approximately 0.5 g/L; in contrast, in the case of panose, it was increased by approximately 0.2 g/L, showing lower utilization efficiency compared with that for maltose and isomaltose (Fig. 4a). Residual sugar analysis (Fig. 4b) indicated that about 98% of supplemented isomaltose and 90% of panose were consumed by the recombinant. Although small amounts of isomaltose and panose remained in the culture broth, the assimilation of isomaltose and panose was clearly enhanced by the introduction of glvA and glvC. These results showed that isomaltose and panose could be utilized as carbon sources and converted to l-lysine, suggesting that the heterologous expression of glvA and glvC could increase the efficiency of glucose feedstock utilization.
Fig. 4.
Utilization of maltose, isomaltose, and panose in the l-lysine-producing model strain. a Accumulation of l-lysine in the l-lysine production medium supplemented with glucose or glucose combined with maltose, isomaltose, and panose at the end of fermentation. b Residual maltose, isomaltose, and panose in the culture broth at the end of fermentation. Values are the means of more than three independent samples. SE bars represent the standard error of the mean calculated with Excel software. WC196LC (pCABD2) harboring the empty vector plasmids, pTVW229 and pMW219-Δplac (empty vector); WC196LC (pCABD2) harboring the glvA-expressing and glvC-expressing plasmids, pTWV229-self-glvA-Fw and pMW219-ΔPlac-Ptac4075-glvC (glvAC); N.D. not detected, NA no addition
Discussion
The PTS is composed of the phosphohistidine carrier protein (HPr), the enzyme I (EI) component, and the enzymes EIIA, EIIB, and EIIC. Although heterologous expression of glvA (encoding phospho-α-glucosidase) and glvC (encoding EIICB) conferred upon E. coli the ability to assimilate isomaltose, panose, and various sugars and sugar alcohols, the combination of GlvA and GlvC did not provide all the components needed to produce PTS activity, functioning only as an EIICB enzyme. HPr and EI are not specific to particular sugars, and EIIAs also do not have strict substrate selectivity for each sugar and EIICB. For example, EIIAGlc can interact with glucose-PTS and trehalose-PTS (Postma et al. 1993). Pikis et al. reported that AglA, a homolog of GlvC, interacts with EIIAGlc, which is encoded by the endogenous crr gene in E. coli (Pikis et al. 2006). Therefore, GlvC is also likely to interact with EIIAGlc of E. coli. We disrupted the crr gene and tested whether the heterologous expression of glvA and glvC allowed the mutant to assimilate isomaltose. The crr mutant harboring the glvA and glvC plasmids could not grow on M9 medium containing isomaltose as a sole carbon source (data not shown). Our results suggested that GlvC (a single-chain polypeptide of EIIB and EIIC) derived from the gram-positive bacterium B. subtilis could associate with EIIAGlc of E. coli, similar to AglA of K. pneumoniae.
In industrial production of valuable compounds with E. coli, purified sugars are rarely used due to high cultivation cost, and hence, various sugar mixtures are used as carbon sources (Gokarn et al. 2014; Eiteman et al. 2008). However, assimilation of many sugars starts sequentially after consumption of glucose with lag phase, resulting in the extension of culture time and decrease of productivity (Eiteman et al. 2008; Aidelberg et al. 2014) due to carbon catabolite repression (Görke and Stülke 2008). In order to overcome this problem, several researchers have attempted to confer upon E. coli the ability to assimilate arabinose (Hernández-Montalvo et al. 2001), xylose (Dien et al. 2002; Hernández-Montalvo et al. 2001), and maltose (Tsujimoto et al. 2006) even in the presence of glucose. For example, Hernández-Montalvo used a mutant devoid of the phosphotransferase system to escape catabolite repression. In this study, we demonstrated that the heterologous expression of glvA and glvC under constitutive promoter allows E. coli to assimilate maltose and isomaltose in the presence of glucose. Surprisingly, the recombinant could also assimilate various other sugars and sugar alcohols, including several trisaccharides. This genetic engineering expanded the metabolizable sugars of E. coli and could increase product yield when using glucose feedstock. We demonstrated that an L-lysine-producing E. coli harboring glvA and glvC converted isomaltose and panose to l-lysine efficiently. This approach should increase the efficiency of industrial fermentation using E. coli and would facilitate full utilization of valuable carbohydrate resources.
Acknowledgements
We are grateful to Dr. Hisao Ito of Process Development Laboratories, Research Institute for Bioscience Products & Fine Chemicals, Ajinomoto Co., Inc. for providing technical advice.
Compliance with ethical standards
Funding
Nothing.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
References
- Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol. 2014;8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin MF, Bucke C (1994) Chapter 4: production of glucose syrup. In: Enzyme technology. Cambridge University Press
- Crabb WD, Shetty JK. Commodity scale production of sugars from starches. Curr Opin Microbiol. 1999;2:252–256. doi: 10.1016/S1369-5274(99)80044-7. [DOI] [PubMed] [Google Scholar]
- de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DA, Reizer J, Nikaido H, Saier MH., Jr Regulation of the maltose transport system of Escherichia coli by the glucose-specific enzyme III of the phosphoenolpyruvate-sugar phosphotransferase system. Characterization of inducer exclusion-resistant mutants and reconstitution of inducer exclusion in proteoliposomes. J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- Dien BS, Nichols NN, Bothast RJ. Fermentation of sugar mixtures using Escherichia coli catabolite repression mutants engineered for production of L-lactic acid. J Ind Microbiol Biotechnol. 2002;29:221–227. doi: 10.1038/sj.jim.7000299. [DOI] [PubMed] [Google Scholar]
- Doi H, Hoshino Y, Nakase K, Usuda Y. Reduction of hydrogen peroxide stress derived from fatty acid beta-oxidation improves fatty acid utilization in Escherichia coli. Appl Microbiol Biotechnol. 2014;98:629–639. doi: 10.1007/s00253-013-5327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, Lee SA, Altman E. A co-fermentation strategy to consume sugar mixtures effectively. J Biol Eng. 2008;2:3. doi: 10.1186/1754-1611-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A, Salinas Cervantes A, Gosset G, Bolívar F. Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: peculiarities of regulation and impact on growth and product formation. Appl Microbiol Biotechnol. 2012;94:1483–1494. doi: 10.1007/s00253-012-4101-5. [DOI] [PubMed] [Google Scholar]
- Gokarn R, Parsons M, Walcker D, Spencer J (2014) Batch feed process for fermenting sugars. Patent Cooperation Treaty No. WO2014093312
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Wipat A. Sequencing and functional analysis of the genome of Bacillus subtilis strain 168. FEBS Lett. 1996;389:84–87. doi: 10.1016/0014-5793(96)00524-8. [DOI] [PubMed] [Google Scholar]
- Hassan MA, Shirai Y, Kubota A, Abdul Karim MI, Nakanishi K, Hashimoto K. Effect of oligosaccharides on glucose consumption by Rhodobacter sphaeroides in polyhydroxyalkanoate production from enzymatically treated crude sago starch. J Ferment Bioeng. 1998;86:57–61. doi: 10.1016/S0922-338X(98)80034-2. [DOI] [Google Scholar]
- Hernández-Montalvo V, Valle F, Bolivar F, Gosset G. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotechnol. 2001;57:186–191. doi: 10.1007/s002530100752. [DOI] [PubMed] [Google Scholar]
- Hii SL, Tan JS, Ling TC, Ariff AB. Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012;2012:921362. doi: 10.1155/2012/921362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Kojima H, Tanaka T, Takatsuka Y, Kamio Y. Characterization of second lysine decarboxylase isolated from Escherichia coli. J Bacteriol. 1997;179:4486–4492. doi: 10.1128/jb.179.14.4486-4492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Ogawa Y, Kawamura K, Sano K (1994) Treaty patent WO95/16042. International Patent Cooperation.
- Ledl F, Schleicher E. New aspects of the Maillard reaction in foods and in the human body. Ang Chem. 1990;26:565–594. doi: 10.1002/anie.199005653. [DOI] [Google Scholar]
- Leuchtenberger W, Huthmacher K, Drauz K. Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol. 2005;69:1–8. doi: 10.1007/s00253-005-0155-y. [DOI] [PubMed] [Google Scholar]
- Loomis WF, Jr, Magasanik B. Glucose-lactose diauxie in Escherichia coli. J Bacteriol. 1967;93:1397–1401. doi: 10.1128/jb.93.4.1397-1401.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Smith AM. Starch biosynthesis. Plant Cell. 1995;7:971–985. doi: 10.1105/tpc.7.7.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1992) A short course in bacterial genetics. Cold Spring Harbor Laboratory Press
- Peters D. Raw materials. Adv Biochem Eng Biotechnol. 2007;105:1–30. doi: 10.1007/10_031. [DOI] [PubMed] [Google Scholar]
- Pikis A, Hess S, Arnold I, Erni B, Thompson J. Genetic requirements for growth of Escherichia coli K12 on methyl-α-D-glucopyranoside and the five α-D-glucosyl-D-fructose isomers of sucrose. J Biol Chem. 2006;281:17900–17908. doi: 10.1074/jbc.M601183200. [DOI] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J. Novel phosphotransferase system genes revealed by bacterial genome analysis: unique, putative fructose- and glucoside-specific systems. Protein Sci. 1994;3:440–450. doi: 10.1002/pro.5560030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierkes M, Svensson B (1992) Process for enzymatic hydrolysis of starch to glucose. United States Patent Number: 5,162,210
- Singh M, Patel SK, Kalia VC. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb Cell Factories. 2009;8:38. doi: 10.1186/1475-2859-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smuda M, Glomb MA. Novel insights into the Maillard catalyzed degradation of maltose. J Agric Food Chem. 2011;59:13254–13264. doi: 10.1021/jf203346b. [DOI] [PubMed] [Google Scholar]
- Song W, Nie Y, Mu XQ, Xu Y. Enhancement of extracellular expression of Bacillus naganoensis pullulanase from recombinant Bacillus subtilis: effects of promoter and host. Protein Expr Purif. 2016;124:23–31. doi: 10.1016/j.pep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Takasaki Y (1988) Method for production of glucose by use of transglucosidase. United States Patent Number: 4,855,232
- Thompson J, Gentry-Weeks CR, Nguyen NY, Folk JE, Robrish SA. Purification from Fusobacterium mortiferum ATCC 25557 of a 6-phosphoryl-O-α-D-glucopyranosyl:6-phosphoglucohydrolase that hydrolyzes maltose 6-phosphate and related phospho-α-D-glucosides. J Bacteriol. 1995;177:2505–2512. doi: 10.1128/jb.177.9.2505-2512.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Pikis A, Ruvinov SB, Henrissat B, Yamamoto H, Sekiguchi J. The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-α-glucosidase. Assignment to family 4 of the glycosylhydrolase superfamily. J Biol Chem. 1998;273:27347–27356. doi: 10.1074/jbc.273.42.27347. [DOI] [PubMed] [Google Scholar]
- Thompson J, Robrish SA, Immel S, Lichtenthaler FW, Hall BG, Pikis A. Metabolism of sucrose and its five linkage-isomeric α-D-glucosyl-D-fructoses by Klebsiella pneumoniae. Participation and properties of sucrose-6-phosphate hydrolase and phospho-α-glucosidase. J Biol Chem. 2001;276:37415–37425. doi: 10.1074/jbc.M106504200. [DOI] [PubMed] [Google Scholar]
- Tsujimoto N, Suzuki T, Ito H (2006) Method for producing target substance using microorganisms with reduced interactions between MalK and IIAGlc. United States Patent Number: 7,097,999
- Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Serizawa M, Thompson J, Sekiguchi J. Regulation of the glv operon in Bacillus subtilis: YfiA (GlvR) is a positive regulator of the operon that is repressed through CcpA and cre. J Bacteriol. 2001;183:5110–5121. doi: 10.1128/JB.183.17.5110-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip VL, Thompson J, Withers SG. Mechanism of GlvA from Bacillus subtilis: a detailed kinetic analysis of a 6-phospho-α-glucosidase from glycoside hydrolase family 4. Biochemistry. 2007;46:9840–9852. doi: 10.1021/bi700536p. [DOI] [PubMed] [Google Scholar]
- Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]