Abstract
As a pathological condition, epilepsy is caused by abnormal neuronal discharge in brain which will temporarily disrupt the cerebral functions. Epilepsy is a chronic disease which occurs in all ages and would seriously affect patients' personal lives. Thus, it is highly required to develop effective medicines or instruments to treat the disease. Identifying epilepsy-related genes is essential in order to understand and treat the disease because the corresponding proteins encoded by the epilepsy-related genes are candidates of the potential drug targets. In this study, a pioneering computational workflow was proposed to predict novel epilepsy-related genes using the random walk with restart (RWR) algorithm. As reported in the literature RWR algorithm often produces a number of false positive genes, and in this study a permutation test and functional association tests were implemented to filter the genes identified by RWR algorithm, which greatly reduce the number of suspected genes and result in only thirty-three novel epilepsy genes. Finally, these novel genes were analyzed based upon some recently published literatures. Our findings implicate that all novel genes were closely related to epilepsy. It is believed that the proposed workflow can also be applied to identify genes related to other diseases and deepen our understanding of the mechanisms of these diseases.
1. Introduction
As a classical neurological condition that may suddenly interrupt normal life activities and result in physical injury, epilepsy has been widely used to describe a group of epileptic seizure associated diseases [1, 2]. Epileptic seizures are the typical symptoms of the disease, which is the consequence of a disruption of the electrical communications between neurons [3]. As a common neurological disease, epilepsies are found all over the world and affect people of all ages [4]. Only in America, more than one hundred thousand incident cases are diagnosed as epilepsy per year, seriously threatening their mental and physical health [5, 6]. Nowadays, the clinical study on epilepsy has been progressively deepening and typical diagnosis routines of epilepsy have been set up. Considering that epileptic seizures induced by disruption of electrical communications between neurons are the typical symptoms of epilepsy, the long-term paroxysmal epileptic seizures might suggest the initiation and progression of such neurological disease [7, 8]. Generally, it is quite necessary to turn to the doctor for help after someone has more than twice abnormal seizures excluding those with known medical conditions. Considering epilepsy has many subtypes induced by different pathogenic factors, resulting in different complications with similar early seizures symptoms, the diagnosis of epilepsy contributes not only to a confirmation of epilepsy but also to classification of the epilepsy from which the patients suffer into its subtype [9, 10]. The diagnosis of epilepsy can be divided into two main procedures: medical history taking and instrumental inspections. Previous experiments have confirmed that typical family history and multiple medical conditions may lead to neurological abnormalities, which may contribute to the initiation and progression of epilepsy [11–13]. Therefore, the first step of the diagnosis of epilepsy is to inquire about the medical history of the patients and their respective family. However, a definitive diagnosis of epilepsy is performed by the following instrumental inspections. As we have mentioned above, epilepsy is referred to as a group of neurological diseases induced by abnormal electrical communication between neurons [4]. Therefore, the measurement of electrical impulses in brain by an electroencephalogram (EEG) test has been regarded as one of the golden standards for epilepsy diagnosis [14, 15]. Apart from EGG, magnetic resonance spectroscopy (MRS), positron emission tomography (PET), and magnetic resonance imaging (MRI) have also been widely used to diagnose epilepsy [16–18].
Although great progresses have been made to diagnose epilepsy, the therapeutic methods to treat epilepsy are still quite limited and they mainly contribute to the symptomatic relief. There are two main functional methods to relieve the seizure symptoms of epilepsy: certain nutrient intakes and vagus nerve stimulations by surgery [19–22]. The therapeutic nutrients that have been confirmed to contribute to the relief of epilepsy include folic acid, melatonin, and vitamins (large doses) [19]. However, such a treatment cannot provide a permanent cure but tries to temporarily relieve the symptoms. The vagus nerve stimulation, as the most effective treatment for epilepsy, necessitates implantation of a pacemaker-like device in the patient's body to stimulate the vagus nerve, relieving the symptoms with few side-effects [23]. These treatments do not have an in-depth consideration of the pathogenic factors that cause the disease but mainly concentrate on the relief of the seizure symptoms. To develop more effective curing methods, it is quite fundamental to understand and reveal the pathogenesis of epilepsy.
As it is known, traditionally epilepsy is referred to a group of diseases characterized by similar symptoms (i.e., epileptic seizures), but not by its pathogenesis. And the underlying pathogenesis of epilepsy may be quite complicated. In the past, due to technological constraints, the pathogenesis of epilepsy is largely unknown. It has been reported that the occurrence of some epilepsy cases turned out to exhibit some degree of familial aggregation, not only implicating the significance of history taking, but also suggesting that the genetic background contributes to the disease [24, 25]. According to some clinical data, it is without any doubt that if a sibling suffers from epilepsy, the brothers and sisters who have similar genetic background inherited from their patients are at higher risk of epilepsy comparing to those who do not [26, 27]. However, the detailed pathogenesis cannot be clearly revealed. Recently, with the development of sequencing technologies, some epilepsy associated genes with either pathological mutations or copy number variants have been identified [28, 29]. Among these genes, a group of functional genes, the sodium channel protein family, encode the core sodium channel in the nerve system [30]; for example, genes like SCN1A and SCN8A encoding core component of the sodium channel have been confirmed to be associated with the progression of hereditary epilepsies [31–33]. Therefore, specific genes may play a definitive role during the initiation and progression of the epilepsy, as hereditary epilepsies are deemed to have a certain genetic background.
Although specific genes have been strongly suggested to be associated with epilepsy, however, it is quite hard to identify the core regulatory genes related to epilepsy by time-consuming experimental methods such as the western blot [34, 35]. Here, based on some known epilepsy-related genes, we presented a new computational workflow to search out potential genes of interest. The random walk with restart (RWR) algorithm was employed in our workflow to search possible novel genes in a protein-protein interaction (PPI) network. Compared to the network methods based on guilt-by-association [36] which only consider the neighbors of known genes [37–39], the RWR algorithm can inspect the whole network to make extensive decisions; that is, methods based on guilt-by-association used part of the network, while the RWR algorithm can utilize the whole network. The brief procedures of our workflow was described as follows. Firstly, the RWR algorithm was executed on a PPI network using validated epilepsy associated genes as seed nodes and genes receiving high probabilities were selected as possible candidate genes. Then, these possible genes were screened by a permutation test, followed by functional association tests, resulting in thirty-three novel epilepsy-related genes. Further analysis indicates that all genes obtained may directly or indirectly contribute to the initiation and progression of epilepsy. To the best of our knowledge, this is the first study attempting to identify core regulatory factors of epilepsy using computational methods. These newly found genes may reveal the underlying mechanisms of epilepsy, and the approach may be extended to solve the similar problems of other complex diseases.
2. Materials and Methods
2.1. Epilepsy Related Genes
499 genes related to epilepsy were retrieved from EpilepsyGene (http://61.152.91.49/EpilepsyGene/download.php) [40], a genetic resource for genes and their mutations related to epilepsy. The epilepsy genes in EpilepsyGene database were collected by searching the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed). Because our method was executed on a PPI network (referred to in Section 2.2), all 499 genes were linked to their Ensembl IDs. Those without Ensembl IDs or those Ensembl IDs do not occur in the PPI network were excluded. Finally, 470 genes with their Ensembl IDs were obtained for investigation in this study. The detailed information of these genes is provided in Supplementary Material S1 in Supplementary Material available online at https://doi.org/10.1155/2017/6132436.
2.2. PPI Network
The interactions between proteins within and outside the cells provide useful information about their activities, properties, and functions. Two proteins that can interact with each other produce a PPI (protein-protein interaction), which often share similar functions or involve in the same biological processes. The PPI network comprised of large amounts of PPIs representing proteins' complicated interaction relationships and remote functional relationships in signaling pathways, such as the proteins involved in regulation and catalysis activity in glycolysis and tricarboxylic acid cycle [41–43]. Some computational predictors and workflows employed PPIs to predict protein functions [44–46] and search for novel genes related to a variety of diseases [47–51]. Therefore, PPIs could be useful to infer novel epilepsy-related genes based on the validated 470 epilepsy genes mentioned in Section 2.1.
To obtain the PPI information and construct a PPI network, the human PPI information was retrieved from STRING (Version 10.0, http://string-db.org/) [52], a well-known public database collecting known and predicted protein-protein interactions. In the current version, it covers 9,643,763 proteins from 2,031 organisms. Interactions reported in STRING are derived from the following five sources: (I) Genomic Context Predictions; (II) High-throughput Lab Experiments; (III) (Conserved) Co-Expression; (IV) Automated Text mining; (V) Previous Knowledge in Databases. The human PPIs are collected in the file “9606.protein.links.v10.txt.gz” that can be accessed from the download page of STRING using “Homo sapiens” as a restriction to the data. Accordingly, we obtained 4,274,001 human PPIs covering 19,247 proteins. Because the 4,274,001 human PPIs include not only direct (physical) but also indirect (functional) interactions between proteins, these PPIs can offer relatively more information about the novel genes related to epilepsy.
For each PPI, there are two Ensembl IDs representing two proteins and a score ranging from 150 to 999 that indicates the strength of the interaction. A larger score assigned to a PPI indicates that the two proteins are more likely to interact with each other. For proteins pa and pb, their interaction score was denoted as S(pa, pb). In the network, the 19,247 proteins were denoted as the nodes and two proteins were connected by an edge if and only if they can form a PPI. Thus, there were 4,274,001 edges in the network, and each edge represented a PPI. In addition, the interaction score was added to the network as the weight of the corresponding edge. For convenience, the PPI network was denoted as G in the following sections.
2.3. RWR Algorithm
As a ranking algorithm, the RWR algorithm simulated a walker starting from a seed node or several seed nodes and randomly moved on the network G [53]. In this study, 470 Ensembl IDs of epilepsy genes were set as the seed nodes. Based on them, we aim to mine some potential genes functionally related to epilepsy. In the beginning of the algorithm, an initialization vector P0 was constructed with 19,247 components in it and each component was a score rating the probability of each node being a potential epilepsy-related gene. The probability scores of 470 Ensembl IDs that represented validated epilepsy genes in P0 were set to 1/470 (0.0021) and other components were set to zeros. If the vector Pi was the probability vector after the RWR algorithm was executed ith round, then the iteration equation can be formulated as follows:
| (1) |
where A was the column-wise normalized adjacency matrix and r was the probability that it returned to the start nodes, which was set to 0.8 in this study. When probability vector Pi+1 and Pi satisfy the inequality ‖Pi+1 − Pi‖L1 < 1E − 06, the iteration stopped and Pi+1 was output as the results of the RWR algorithm.
According to the probability vector yielded by RWR algorithm, each node (gene) in the network was given a number representing the probability of it being a novel epilepsy gene. Genes with larger values are more likely to be epilepsy-related genes. Threshold 1E − 05 was adopted in this study; that is, genes receiving scores larger than 1E − 05 were selected from the network G, because it filtered out a large portion of genes and there remained enough genes for further analysis. For convenience, the obtained genes were called RWR genes.
2.4. Filtering Methods
After RWR algorithm was executed on the network, many RWR genes could be selected. However, there are likely many false positive genes among them as elaborated in our previous study [44]. These genes are not special to the epilepsy and should be excluded. In this section, a two-step filtering method was proposed to screen out false positive genes.
2.4.1. Permutation Test
The structure of network G can influence the output of RWR algorithm, which may lead to the false selection of some RWR genes. For example, a node with a degree higher than average degree of the network G may receive a larger score by RWR algorithm even if it was not related to epilepsy. To mine this type of nodes in the network, a permutation test was applied on the network. Firstly, 1,000 Ensembl ID sets, denoted as S1, S2,…, S1000, were randomly produced and each set contained 470 random Ensembl IDs. Secondly, for each set, the RWR algorithm was executed on the network G using the 470 Ensembl IDs in this set as seed nodes, thereby yielding a probability for each RWR gene. Thirdly, for each RWR gene g, a measurement, namely, permutation FDR, was calculated based on the following equation:
| (2) |
where Θ was the number of randomly produced sets in which the score of gene g was larger than the score computed by the validated epilepsy related genes. According to (2), the higher permutation FDR an RWR gene had, the less possible the gene was an epilepsy related gene. Because 0.05 was widely used as a common cutoff in statistical test, it was also set to be the threshold of permutation FDR in this study. Therefore, the RWR genes with permutation FDRs less than 0.05 were selected and called candidate genes for further analysis.
2.4.2. Functional Association Test
Among the candidate genes, some of them were functionally highly associated with epilepsy while others weakly associated with it. To select essential genes among them, a functional association test that consisted of two selection schemes was proposed.
It is known that proteins in a PPI with a higher interaction score are more likely to share similar functions. Among the candidate genes, those having strong associations with validated epilepsy-related genes could be the most likely novel epilepsy genes. If a candidate gene has strong associations with exact one validated epilepsy-related gene and has weak or no associations with other epilepsy-related genes, it may still be a novel epilepsy-related gene. In view of this, we believe that using the associations between a candidate gene and its most related epilepsy-related gene is more proper to indicate its associations with epilepsy. Accordingly, an interaction measurement called maximum interaction score (MIS) was calculated for each candidate gene g, which can be defined as
| (3) |
Candidate genes with large MISs mean that it is highly possible that they can directly interact with at least one validated epilepsy gene and may cause the symptoms of epilepsy. In STRING, the value 900 is set to be the cutoff to achieve a highest confidence. Thus, it was also set to be the threshold of MIS; that is, candidate genes with MISs less than 900 were discarded.
Besides, genes related to epilepsy may share some common gene ontology (GO) [54] terms and often occurred in the same Kyoto Encyclopedia of Genes and Genomes (KEGG) [55] pathways. Thus, candidate genes sharing same or similar GO terms and KEGG pathways with validated epilepsy genes are more likely to be the genes related to epilepsy. The enrichment theory of GO terms (KEGG pathways) [56–58] was used to quantitatively measure the relationship between a gene and GO terms (KEGG pathways). For a gene g, let us denote the set containing g and its direct neighbor genes in the PPI network reported in STRING by H(g). Then, the relationship between g and one GO term or KEGG pathway can be encoded into a numeric value as follows:
| (4) |
where G represented a GO term or a KEGG pathway, N was the total number of genes in humans, M was the number of genes annotated to G, n was the number of genes in H(g), and m was the number of genes that are in H(g) and annotated by G. The values for all GO terms and KEGG pathways can be collected into a vector ES(g). The similarity of two genes g and g′ on GO terms and KEGG pathways can be measured by the proximity of the two vectors ES(g) and ES(g′) as follows:
| (5) |
It is clear that if the resultant number of (5) is large, g and g′ are similar on GO terms and KEGG pathways, implicating a strong relationship between them. With similar arguments on MIS, for each candidate gene g, the maximum function score (MFS) was calculated as follows:
| (6) |
A candidate gene receiving a high MFS means it shares relatively more GO terms and KEGG pathways with at least one validated epilepsy gene. In this study, we tried 0.9 as the threshold of MFS; that is, candidate genes with MFSs larger than 0.9 were selected.
In short, candidate genes resulting from permutation test with MISs larger than or equal to 900 and MFSs larger than 0.9 were selected. For convenience, they were named as core candidate genes.
3. Results
An outline for the procedure of the method, including RWR algorithm and filtering methods described in Sections 2.3 and 2.4, by a flowchart is illustrated in Figure 1. This section would show the detailed results yielded by the method.
Figure 1.
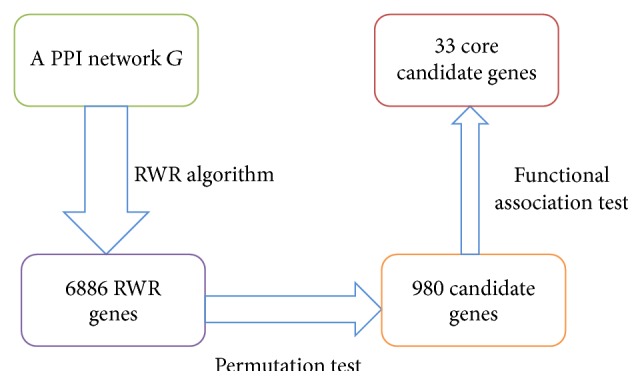
The flowchart of RWR algorithm and filtering methods for identifying core candidate genes.
As described in Section 2.3, the RWR algorithm was executed on the PPI network G, in which the 470 Ensembl IDs were used as seed nodes. A probability vector can be obtained, in which each composition represents the probability score of the corresponding node (gene) being a novel epilepsy-related gene. Genes with probabilities larger than 1E − 05 were selected, producing 6,886 RWR genes, which are listed in Supplementary Material S2.
For the 6,886 RWR genes derived from RWR algorithm, a permutation test was applied on them to screen out RWR genes that are not special for epilepsy. The permutation FDR was calculated for each RWR gene, which is provided in Supplementary Material S2. Value 0.05 was set to be the threshold of permutation FDR, thereby producing 980 candidate genes, which are listed in Supplementary Material S3.
To further select genes that are functionally related to epilepsy, a functional association test was applied to the 980 candidate genes. As described in Section 2.4.2, for each candidate gene, we calculated its MIS (cf. (3)) and MFS (cf. (6)). Values 900 and 0.9 were used as the threshold of MIS and MFS, respectively. And finally thirty-three core candidate genes were obtained. These genes were deemed to be closely related to epilepsy and are listed in Table 1. According to some recent published literature as discussed in Section 4, these core candidate genes, which had similar functions with the validated genes, are highly likely to be the novel epilepsy genes.
Table 1.
Thirty-three core candidate genes identified by our method.
| Gene symbol | Ensembl ID | Probability | Permutation FDR | MIS | MFS |
|---|---|---|---|---|---|
| ANK2 | ENSP00000349588 | 4.53E − 05 | <0.001 | 990 | 0.997 |
| ANK1 | ENSP00000265709 | 3.52E − 05 | 0.034 | 995 | 0.995 |
| EPHA7 | ENSP00000358309 | 3.28E − 05 | 0.025 | 906 | 0.988 |
| EPHA5 | ENSP00000273854 | 3.46E − 05 | 0.032 | 906 | 0.988 |
| PRIKCG | ENSP00000263431 | 4.75E − 05 | 0.025 | 905 | 0.987 |
| PTK7 | ENSP00000230419 | 3.10E − 05 | 0.023 | 943 | 0.987 |
| EPHA3 | ENSP00000337451 | 4.40E − 05 | 0.017 | 912 | 0.986 |
| PDE6C | ENSP00000360502 | 3.61E − 05 | 0.032 | 900 | 0.980 |
| EPHA4 | ENSP00000281821 | 4.16E − 05 | 0.008 | 990 | 0.980 |
| YWHAQ | ENSP00000238081 | 5.50E − 05 | 0.004 | 999 | 0.978 |
| GSK3A | ENSP00000222330 | 6.40E − 05 | 0.017 | 977 | 0.976 |
| CALM1 | ENSP00000349467 | 6.55E − 05 | 0.023 | 966 | 0.976 |
| EPHB2 | ENSP00000363763 | 4.39E − 05 | 0.026 | 908 | 0.975 |
| PRIKCA | ENSP00000408695 | 7.02E − 05 | 0.039 | 992 | 0.975 |
| CALM2 | ENSP00000272298 | 5.52E − 05 | 0.048 | 985 | 0.974 |
| YWHAE | ENSP00000264335 | 1.63E − 04 | 0.009 | 999 | 0.973 |
| YWHAB | ENSP00000300161 | 5.56E − 05 | 0.047 | 999 | 0.971 |
| ATP2A2 | ENSP00000440045 | 5.03E − 05 | 0.005 | 908 | 0.970 |
| YES1 | ENSP00000324740 | 6.54E − 05 | 0.002 | 967 | 0.969 |
| CALML3 | ENSP00000315299 | 3.96E − 05 | 0.027 | 906 | 0.968 |
| SGK1 | ENSP00000356832 | 4.90E − 05 | 0.034 | 999 | 0.966 |
| CALML6 | ENSP00000304643 | 4.44E − 05 | 0.01 | 909 | 0.965 |
| YWHAZ | ENSP00000309503 | 1.86E − 04 | 0.002 | 999 | 0.958 |
| MAPK7 | ENSP00000311005 | 7.15E − 05 | 0.031 | 999 | 0.956 |
| RRAS | ENSP00000246792 | 5.94E − 05 | 0.004 | 951 | 0.941 |
| RAP2A | ENSP00000245304 | 4.97E − 05 | 0.021 | 940 | 0.937 |
| RALA | ENSP00000005257 | 6.57E − 05 | 0.006 | 981 | 0.932 |
| RAP1B | ENSP00000250559 | 5.28E − 05 | 0.009 | 972 | 0.931 |
| MRAS | ENSP00000289104 | 4.70E − 05 | 0.022 | 932 | 0.931 |
| INSR | ENSP00000303830 | 7.27E − 05 | 0.024 | 996 | 0.927 |
| PAP1A | ENSP00000348786 | 6.49E − 05 | 0.014 | 995 | 0.925 |
| RND1 | ENSP00000308461 | 5.70E − 05 | 0.002 | 996 | 0.903 |
| FYN | ENSP00000346671 | 1.02E − 04 | <0.001 | 999 | 0.900 |
4. Discussions
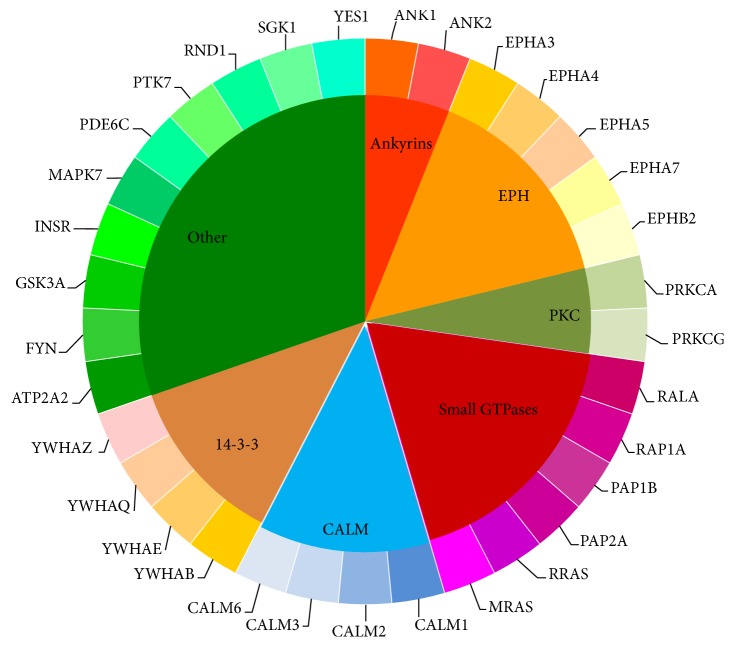
For a long time, epilepsy has been regarded as complicated neurological diseases with various pathogenesis. Based on clinical data, the occurrence of epilepsy has shown conspicuous familial aggregation characteristics, implying that genetic background features (such as mutations, copy number variants of genes) may play an irreplaceable role for epilepsy [59–61]. Recent publications have also confirmed such implication. Various epilepsy associated genes have been identified [62–66]. However, it is quite expensive and time-consuming to identify epilepsy associated genes with experiments. Based on our computational method, we identified thirty-three candidate epilepsy associated genes, listed in Table 1. According to some recent publications, all these core candidates show specific relationship with the initiation and progression of epilepsy, validating the effectiveness of our computational method. According to the gene families of these thirty-three core candidate genes, we classified them into seven clusters, shown in Figure 2, and analyzed them accordingly.
Figure 2.
The distribution of the thirty-three core candidate genes according to their protein families.
Ankyrins. In our prediction list, two of the candidate genes, ANK1 and ANK2, turned out to be the functional members of the ankyrins. As we all know, ankyrins are a group of connexin that link the integral membrane proteins to the cytoskeleton, which have been widely reported that they contribute to cell proliferation, motility, and the maintenance of specialized membrane domains [67–69]. In human bodies, ankyrins have been confirmed to bind to the voltage-gated potassium channel subunits KCNQ2 and KCNQ3, regulating their normal functions [70]. Considering that KCNQ2 and KCNQ3 turned out to directly contributing to the initiation and progression of epilepsy, our predicted genes ANK1 and ANK2 as the functional components of ankyrins may very probably be epilepsy associated genes, validating our prediction [71].
EPH Subfamily. Apart from the Ankyrin protein family, another group of proteins, the EPH subfamily, have also been identified to contribute to epilepsy. In our prediction list, five genes can be classified in such subfamily: EPHA3, EPHA4, EPHA5, EPHA7, and EPHB2. Such five genes all encode the receptors for the erythropoietin-producing hepatoma amplified sequences (EPH), acting as the tyrosine-protein kinase receptor [72, 73]. In mouse model, it has been confirmed that the activation of EPH receptor associated genes, like EPHB3, contributes to the onset of epilepsy [74]. Considering the functional similarity and underlying correlations, it is quite reasonable that our predicted genes of EPH receptor family may also contribute to such processes [75]. Apart from that, another publication confirmed that, by stimulating NMDA receptor activity, ERK activates the progression of epilepsy [76]. During the activation, various genes of our predicted EPH family have been identified to promote such biological processes, validating the crucial role of EPH family including our predicted genes EPHA3, EPHA4, EPHA5, EPHA7, and EPHB2 during epilepsy.
Protein Kinases. Two of our predicted candidates, PRKCA and PRKCG, can be clustered into another functional family, the family of serine- and threonine-specific protein kinases. Such two genes turn out to be functional components of the protein kinase C, a core member of the protein family we mentioned above [77–79]. The protein kinase C associated signaling pathway has been widely reported to be associated with epilepsy and may be a candidate therapeutic target for such disease [80]. As two major components for such pathway, our predicted genes PRKCA and PRKCG may definitely contribute to such disease. Apart from such evidence, a specific mutation of PRKCG (SCA-14) has been reported to be associated with a typical movement disorder, which can be called Ramsay Hunt phenotype [81]. Considering that such disorder has been widely identified in epilepsy patients, such mutation may be functionally related to the progression of epilepsy, validating our prediction [82, 83]. As for MAPK7, such gene has been widely regarded as a multifunctional gene that involves various biological processes including proliferation, differentiation, transcription regulation, and development [84]. MAPK7 has been reported to interact with a specific protein Aquaporin 4 (AQP4) in human beings [85]. Since AQP4 has been confirmed to accumulate in neuron cell during epilepsy and contribute to the pathological processes, it is quite reasonable to summarize that, as a functional related protein of AQP4, our predicted gene MAPK7 very probably contributes to epilepsy, validating the accuracy and efficacy of our prediction [85]. The next gene is also a crucial kinase for human beings, the PTK7. Although, different from other proteins from protein tyrosine kinase family, PTK7 lacks detectable catalytic tyrosine kinase activity, it has still been reported to contribute to the functional Wnt signaling pathway and regulate the cellular polarity and adhesion [86]. Though no direct relationship between PTK7 and epilepsy has been confirmed, recent publications reported that PTK7 may participate in the metabolism of antiepileptic drugs (AED), suggesting that there may remain uncovered interactions between PTK7 and epilepsy, validating our prediction [87]. GSK3A, as a multifunctional Ser/Thr protein kinase, has been reported to contribute to glycogen synthesis and transcriptional regulation [88, 89]. Such gene has been reported to be quite essential for the development and maturation of cortical neurons [90]. Considering that cortical neurons, especially the migration of neurons, are quite significant for epilepsy, our predicted gene GSK3A may very probably be epilepsy associated gene [91, 92].
Small GTPases. Apart from PKC associated genes, there are also six genes (RALA, RAP1A, RAP1B, RAP2A, RRAS, and MRAS) that can be clustered into the famous Ras family of small GTPases. Based on recent publications, various small GTPases have been identified to contribute to the progression of epilepsy, including Cdc42, RAB39B [93, 94]. As for our predicted candidates, it has been confirmed that, during the pathological processes of epilepsy, the normal function of RAP1A and its related Ras signaling pathway has been regulated and altered by microRNAs, implying the potential role of Ras signaling pathway during epilepsy [95]. As for RAP1B and RAP2A, RAP1B has been confirmed to be specifically activated in nerve system and contributes to the regeneration of neuronal connectivity, the dysfunction of which turns out to be one of the pathological factors for epilepsy, validating the regulatory role of RAP1B during such disease [96]. RAP2A has been validated and confirmed to contribute to the childhood absence epilepsy, a specific subtype of epilepsy, and may be related with a specific glioma inducing epilepsy associated symptoms, validating our prediction of epilepsy associated genes [97, 98]. As for RRAS and MRAS, considering the functional similarity of MRAS and MAPK, the detailed analysis of such genes can be seen below, while the inner relationship between RRAS and epilepsy has also been revealed in mouse model, validating our prediction [99, 100]. RALA, encoding a functional small GTPase belonging to Ras family, has been confirmed to mediate the transmembrane signaling by the occupancy of functional receptors [101]. Such gene has been definitely confirmed to be associated with epilepsy by regulating the drug resistance of such disease [102]. Another gene, RND1, encodes a small GTPase, which does not belong to Ras family but to Rho GTPase family. In response to various extracellular signaling, the protein encoded by such gene turns out to regulate the actin cytoskeleton [103, 104]. In intractable epilepsy, a clinical subtype of epilepsy which is hard to cure, recent publications confirmed the expression of such gene in the central nerve system of the patients, implying that such gene may definitely contribute to the progression and prognosis of such disease [105, 106]. MAPK7 and MRAS are two proliferation-associated genes in our candidate epilepsy associated gene list. Among them, MRAS turn out to contribute to Ras signaling pathway, which has been confirmed above to be associated with the progression of epilepsy [95]. What is more, as for MRAS itself, it has been reported that such gene may contribute to the development of brain in early stage and the abnormal activation of such gene may induce epilepsy-like syndrome, validating our prediction [107].
Calmodulin (CALM) Family. Apart from that, such genes may also contribute to the specific seizure like features during the progression of epilepsy, suggesting its core regulatory role [108]. Four genes (CALM1, CALM2, CALM3, and CALM6) of the functional calmodulin (CALM) family have also been predicted to contribute to epilepsy. Genes of calmodulin family mainly act as a calcium binding protein that participate in cell cycle and proliferation associated biological processes [109, 110]. Considering that epilepsy has been confirmed to be associated with abnormal calcium ion transportation, it is quite reasonable to speculate that our predicted genes of CALM family may contribute to epilepsy which has also been confirmed by recent publications [111, 112].
14-3-3 Family of Proteins. The remaining group of functional proteins, including YWHAB, YWHAE, YWHAQ, and YWHAZ, that contribute to epilepsy turn out to be encoded by the so-called 14-3-3 family of proteins. Such family of proteins contribute to the signaling transduction by binding to phosphoserine-containing proteins [113]. As we have mentioned above, epilepsy has been confirmed to be associated with the protein kinase C signaling pathway [81]. Recent publications identified that, during the progression of epilepsy, our predicted candidates, proteins of 14-3-3 family, may interact with protein kinase C and further promote the progression, implying the functional role of such genes [114]. Apart from that, such five genes that we sorted out have also been directly identified in epilepsy cases. Take YWHAB as an example. Such gene has been identified to contribute to the regeneration of neurons after physical or chemical injury. The dysfunction of such gene may be related to the initiation of epilepsy in certain pathological conditions [115]. Therefore, such four genes in our prediction list have all been confirmed to participate in epilepsy associated biological processes, validating the accuracy and efficacy of our prediction.
Other Crucial Genes. Apart from such genes, there still remain six genes with no clear family enrichment in our prediction list that may also contribute to epilepsy in their respective ways. Among such genes, ATP2A2 turns out to encode a significant intracellular pump located in the sarcoplasmic or endoplasmic reticula [116, 117]. Epilepsy has been confirmed to be a specific complication of various diseases, including Darier's disease [118, 119]. Our predicted gene ATP2A2 has been identified in clinical cases of Darier's disease and may directly contribute to epilepsy associated symptoms, validating the efficacy and accuracy of our prediction [120]. Another gene INSR turns out to be the functional receptor for a core endogenous hormone insulin, which further activates the downstream of insulin signaling pathway [121, 122]. As for the underlying relationship between INSR and epilepsy, it has been reported that a group of specific mutations in INSR: INSR H1085H C>T, G972R has been confirmed to specifically occur in the epilepsy patients in Han Chinese, validating the specific role of INSR during the progression of epilepsy [123]. Another gene FYN is also a candidate oncogene just like genes from Ras family as we have mentioned above [124, 125]. Recent publications reveal the potential relationship between our predicted gene FYN and amygdala kindling, a specific phenotype associated with epileptogenesis [126]. Participating in mTOR signaling pathway, though the detailed mechanism of the pathology of FYN initiated epilepsy has not been fully revealed, our predicted gene FYN may definitely be an epilepsy associated gene [126–128].
Phosphodiesterase 6C, encoded by our predicted gene PDE6C, has been confirmed to contribute to pyrimidine metabolism and phototransduction [129, 130]. Autosomal-dominant cerebellar ataxia is a specific symptom of epilepsy in human beings [131]. It has been reported that a specific mutation, p.Arg95His, of our predicted gene, PDE6C, may be associated with the autosomal-dominant cerebellar [132]. Considering the inner linkage between autosomal-dominant cerebellar ataxia and epilepsy in human beings, our predicted gene PDE6C may also contribute to the progression of epilepsy, validating our prediction [131]. As we have mentioned above, quite a lot of kinases have been reported to contribute to the abnormal biological process during epilepsy. The remaining two genes SGK1 and YES1 have also been suggested to contribute to the progression of epilepsy. SGK1 turns out to encode a serum/glucocorticoid regulated kinase, which further contributes to the regulation of cellular stress response [133, 134]. It has been confirmed that our predicted gene SGK1 is upregulated by a functional cellular component, aldosterone [134]. It has been confirmed that aldosterone has strong chemical and biological effects on epileptic seizures, implying that our predicted gene SGK1 may contribute to the regulation of the typical symptoms of epilepsy, the seizures [135]. Therefore, SGK1 very probably is a functional epilepsy associated gene. The last gene, YES1, also encodes a small GTPase that has been widely regarded as a tumor associated gene [136]. Although no direct report confirms the relationship between YES1 and epilepsy, considering the core regulatory role of small GTPases for epilepsy and that it has been confirmed that YES1 is expressed in brain and central nerve system, it is quite reasonable for us to believe that YES may be a functional epilepsy associated gene [137].
5. Conclusion
Based on our newly developed computational method, we have identified thirty-three novel genes that may contribute to the initiation and progression of epilepsy. According to the comprehensive analyses on these genes, they are strongly suspected to be either directly or indirectly related to epilepsy, validating the effectiveness of our method. In summary, this method can not only contribute to the identification of potential epilepsy associated genes but also provide a new tool to investigate the underlying mechanisms of the pathological processes of epilepsy.
Supplementary Material
The Supplementary Material consists of three files. In detail, Supplementary Material S1 lists 470 genes related to epilepsy with their Ensembl IDs and gene symbols; Supplementary Material S2 lists 6,886 RWR genes derived from RWR algorithm with probability larger than 1E-05; Supplementary Material S3 lists 980 candidate genes with permutation FDRs less than 0.05.
Acknowledgments
This paper is supported by Norman Bethune Program of Jilin University (2015218), Science and Technology Department of Jilin Province (20160414007GH, 20160414047GH), Education Department of Jilin Province (2015509, 2016449), and Development and Reform Commission of Jilin Province (2015Y032).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this article.
Authors' Contributions
Wei Guo and Dong-Mei Shang contributed equally to this work.
References
- 1.Wiebe S. Brain surgery for epilepsy. The Lancet. 2003;362:s48–s49. doi: 10.1016/s0140-6736(03)15075-1. [DOI] [PubMed] [Google Scholar]
- 2.Baxendale S. Epilepsy at the movies: possession to presidential assassination. Lancet Neurology. 2003;2(12):764–770. doi: 10.1016/s1474-4422(03)00589-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu F. M., Dai S., Napoli A., et al. Epileptic seizures are induced by intracerebral ablation of astrocytes in the brain, a novel model for dissecting the interaction of neurons with glial cells. Journal of NeuroVirology. 2015;21:S41–S42. [Google Scholar]
- 4.Sethi N. K. Psychogenic non-epileptic seizures—the age matters. Clinical Neurology and Neurosurgery. 2014;120:p. 142. doi: 10.1016/j.clineuro.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 5.Zepeda R., Gleason K. A., Bubrick E. J., Pallin D. J., Dworetzky B. A. Disparities of epilepsy care in the emergency department. Epilepsia. 2009;50:307–307. [Google Scholar]
- 6.Burneo J. G., Jette N., Theodore W., et al. Disparities in epilepsy: report of a systematic review by the North American Commission of the international league against epilepsy. Epilepsia. 2009;50(10):2285–2295. doi: 10.1111/j.1528-1167.2009.02282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugai K., Nakagawa E., Komaki H., Sakuma H., Saito Y., Sasaki M. Pharmacotherapy for childhood nonidio-pathic partial epilepsies based on seizure symptoms: retrospective and prospective studies. Epilepsia. 2009;50:113–113. [Google Scholar]
- 8.Choi H., Winawer M. R., Kalachikov S., Pedley T. A., Hauser W. A., Ottman R. Classification of partial seizure symptoms in genetic studies of the epilepsies. Neurology. 2006;66(11):1648–1653. doi: 10.1212/01.wnl.0000218302.03570.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aktekin B. Up-to-date critical review of the classification of epilepsies and epileptic seizures. Noropsikiyatri Arsivi. 2015;52(2):109–110. doi: 10.5152/npa.2015.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S.-H., Lim J. S., Kim J.-K., Yang J., Lee Y. Classification of normal and epileptic seizure EEG signals using wavelet transform, phase-space reconstruction, and Euclidean distance. Computer Methods and Programs in Biomedicine. 2014;116(1):10–25. doi: 10.1016/j.cmpb.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Jose M., Thomas S. V. Family history of congenital malformations does not increase the risk of fetal malformations in women with epilepsy. Epilepsia. 2015;56:31–31. [Google Scholar]
- 12.Alonso-Cerezo C., Herrera-Peco I., Fernández-Millares V., et al. Family history of epilepsy resistant to treatment. Revista de Neurologia. 2011;52(9):522–526. [PubMed] [Google Scholar]
- 13.Wieshmann U. C. Family history of epilepsy in epilepsy and other neurological conditions. Journal of Neurology, Neurosurgery & Psychiatry. 2004;75:799–800. [Google Scholar]
- 14.Bahbiti Y., Moutaouakil F., Ouichou A., El Hessni A., Benazzouz B., Mesfioui A. Epilepsy: electroencephalogram and brain maturation. Epilepsia. 2013;54:156–156. [Google Scholar]
- 15.Liao J., Song L., Chen Y. Seizures captured with video-electroencephalogram in infants with epilepsy. Epilepsia. 2012;53:p. 132. [Google Scholar]
- 16.Wang G. D., Dai Z. Y., Song W. G., et al. Grey matter anomalies in drug-naïve childhood absence epilepsy: a voxel-based morphometry study with MRI at 3.0 T. Epilepsy Research. 2016;124:63–66. doi: 10.1016/j.eplepsyres.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Lipatova L., Kapustina T. Functional neuroimaging using the method 1H MRS in epilepsy. European Journal of Neurology. 2015;22(supplement 1):p. 632. [Google Scholar]
- 18.Peter J., Houshmand S., Werner T. J., Rubello D., Alavi A. Novel assessment of global metabolism by 18F-FDG-PET for localizing affected lobe in temporal lobe epilepsy. Nuclear Medicine Communications. 2016;37(8):882–887. doi: 10.1097/mnm.0000000000000526. [DOI] [PubMed] [Google Scholar]
- 19.Kinney M., Morrow J. Vitamin K is important for epilepsy in pregnancy. British Medical Journal. 2016;354, article i3929 doi: 10.1136/bmj.i3929. [DOI] [PubMed] [Google Scholar]
- 20.Snoeijen-Schouwenaars F. M., Van Deursen K. C., Tan I. Y., Verschuure P., Majoie M. H. Vitamin D supplementation in children with epilepsy and intellectual disability. Pediatric Neurology. 2015;52(2):160–164. doi: 10.1016/j.pediatrneurol.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Bodin C., Aubert S., Daquin G., et al. Responders to vagus nerve stimulation (VNS) in refractory epilepsy have reduced interictal cortical synchronicity on scalp EEG. Epilepsy Research. 2015;113:98–103. doi: 10.1016/j.eplepsyres.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Mannino S., Colicchio G., Di Bonaventura R., et al. Patients/caregivers satisfaction following vagal nerve stimulation (Vns) for drug-resistant epilepsies. Epilepsia. 2014;55(1):103–104. doi: 10.1111/epi.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cukiert A., Burattini J., Cukiert C. Vagus nerve stimulation (Vns) in refractory epilepsy. Epilepsia. 2013;54:p. 84. [Google Scholar]
- 24.Fabera P., Krijtova H., Tomasek M., et al. Familial temporal lobe epilepsy due to focal cortical dysplasia type IIIa. Seizure. 2015;31:120–123. doi: 10.1016/j.seizure.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Chentouf A., Dahdouh A., Guipponi M., et al. Familial epilepsy in Algeria: clinical features and inheritance profiles. Seizure. 2015;31:12–18. doi: 10.1016/j.seizure.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Hames A., Appleton R. Living with a brother or sister with epilepsy: siblings' experiences. Seizure. 2009;18(10):699–701. doi: 10.1016/j.seizure.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Kurča E., Grofik M., Kučera P., Varsik P. Familial occurrence of adrenocortical insufficiency in two brothers with allgrove syndrome. A case report of 4A (Allgrove) syndrome with epilepsy and a new AAAs gene mutation. Neuroendocrinology Letters. 2005;26(5):499–502. [PubMed] [Google Scholar]
- 28.Purcell R. H., Papale L. A., Makinson C. D., et al. Effects of an epilepsy-causing mutation in the SCN1A sodium channel gene on cocaine-induced seizure susceptibility in mice. Psychopharmacology. 2013;228(2):263–270. doi: 10.1007/s00213-013-3034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escayg A., Goldin A. L. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51(9):1650–1658. doi: 10.1111/j.1528-1167.2010.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes T. H., Vanoye C. G., George A. L. Functional characterization of SCN1A sodium channel mutations associated with familial epilepsy. Biophysical Journal. 2005;88:p. 378a. [Google Scholar]
- 31.Baum L., Haerian B. S., Ng H.-K., et al. Case-control association study of polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, SCN3A, SCN1B, and SCN2B and epilepsy. Human Genetics. 2014;133(5):651–659. doi: 10.1007/s00439-013-1405-1. [DOI] [PubMed] [Google Scholar]
- 32.Barela A. J., Waddy S. P., Lickfett J. G., et al. An epilepsy mutation in the sodium channel SCN1A that decreases channel excitability. Journal of Neuroscience. 2006;26(10):2714–2723. doi: 10.1523/JNEUROSCI.2977-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lossin C., Rhodes T. H., Desai R. R., et al. Epilepsy-associated dysfunction in the voltage-gated neuronal sodium channel SCN1A. Journal of Neuroscience. 2003;23(36):11289–11295. doi: 10.1523/JNEUROSCI.23-36-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt A. B., Netoff T. I. Computational modeling of epilepsy for an experimental neurologist. Experimental Neurology. 2013;244:75–86. doi: 10.1016/j.expneurol.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefanescu R. A., Shivakeshavan R. G., Talathi S. S. Computational models of epilepsy. Seizure. 2012;21(10):748–759. doi: 10.1016/j.seizure.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Oliver S. Guilt-by-association goes global. Nature. 2000;403(6770):601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 37.Oti M., Snel B., Huynen M. A., Brunner H. G. Predicting disease genes using protein-protein interactions. Journal of Medical Genetics. 2006;43(8):691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krauthammer M., Kaufmann C. A., Gilliam T. C., Rzhetsky A. Molecular triangulation: bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(42):15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franke L., Van Bakel H., Fokkens L., De Jong E. D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. American Journal of Human Genetics. 2006;78(6):1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran X., Li J., Shao Q., et al. EpilepsyGene: a genetic resource for genes and mutations related to epilepsy. Nucleic Acids Research. 2015;43(1):D893–D899. doi: 10.1093/nar/gku943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Depré C., Rider M. H., Hue L. Mechanisms of control of heart glycolysis. European Journal of Biochemistry. 1998;258(2):277–290. doi: 10.1046/j.1432-1327.1998.2580277.x. [DOI] [PubMed] [Google Scholar]
- 42.Hue L., Rider M. H. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. The Biochemical Journal. 1987;245(2):313–324. doi: 10.1042/bj2450313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansford R. G., Zorov D. Role of mitochondrial calcium transport in the control of substrate oxidation. Molecular and Cellular Biochemistry. 1998;184(1-2):359–369. doi: 10.1023/A:1006893903113. [DOI] [PubMed] [Google Scholar]
- 44.Chen L., Zhang Y.-H., Huang T., Cai Y.-D. Identifying novel protein phenotype annotations by hybridizing protein-protein interactions and protein sequence similarities. Molecular genetics and genomics: MGG. 2016;291(2):913–934. doi: 10.1007/s00438-015-1157-9. [DOI] [PubMed] [Google Scholar]
- 45.Hu L., Huang T., Shi X., Lu W.-C., Cai Y.-D., Chou K.-C. Predicting functions of proteins in mouse based on weighted protein-protein interaction network and protein hybrid properties. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0014556.e14556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y.-F., Chen L., Cai Y.-D., Feng K.-Y., Huang T., Jiang Y. Predicting metabolic pathways of small molecules and enzymes based on interaction information of chemicals and proteins. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0045944.e45944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gui T., Dong X., Li R., Li Y., Wang Z. Identification of hepatocellular carcinoma-related genes with a machine learning and network analysis. Journal of Computational Biology. 2015;22(1):63–71. doi: 10.1089/cmb.2014.0122. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J., Yang J., Huang T., Shu Y., Chen L. Identification of novel proliferative diabetic retinopathy related genes on protein–protein interaction network. Neurocomputing. 2016;217:63–72. doi: 10.1016/j.neucom.2015.09.136. [DOI] [Google Scholar]
- 49.Chen L., Huang T., Zhang Y.-H., Jiang Y., Zheng M., Cai Y.-D. Identification of novel candidate drivers connecting different dysfunctional levels for lung adenocarcinoma using protein-protein interactions and a shortest path approach. Scientific Reports. 2016;6 doi: 10.1038/srep29849.29849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L., Yang J., Huang T., Kong X., Lu L., Cai Y.-D. Mining for novel tumor suppressor genes using a shortest path approach. Journal of Biomolecular Structure and Dynamics. 2016;34(3):664–675. doi: 10.1080/07391102.2015.1042915. [DOI] [PubMed] [Google Scholar]
- 51.Chen L., Xing Z., Huang T., Shu Y., Huang G., Li H.-P. Application of the shortest path algorithm for the discovery of breast cancer-related genes. Current Bioinformatics. 2016;11(1):51–58. doi: 10.2174/1574893611666151119220024. [DOI] [Google Scholar]
- 52.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43(1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Köhler S., Bauer S., Horn D., Robinson P. N. Walking the interactome for prioritization of candidate disease genes. The American Journal of Human Genetics. 2008;82(4):949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gene ontology consortium: going forward. Nucleic Acids Research. 2015;43(D1):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J., Chen L., Kong X., Huang T., Cai Y.-D. Analysis of tumor suppressor genes based on gene ontology and the KEGG pathway. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0107202.e107202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J., Xing Z., Ma M., et al. Gene ontology and KEGG enrichment analyses of genes related to age-related macular degeneration. BioMed Research International. 2014;2014:10. doi: 10.1155/2014/450386.450386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L., Zhang Y.-H., Zheng M., Huang T., Cai Y.-D. Identification of compound–protein interactions through the analysis of gene ontology, KEGG enrichment for proteins and molecular fragments of compounds. Molecular Genetics and Genomics. 2016;291(6):2065–2079. doi: 10.1007/s00438-016-1240-x. [DOI] [PubMed] [Google Scholar]
- 59.Fjaer R., Brodtkorb E., Øye A.-M., et al. Generalized epilepsy in a family with basal ganglia calcifications and mutations in SLC20A2 and CHRNB2. European Journal of Medical Genetics. 2015;58(11):624–628. doi: 10.1016/j.ejmg.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Partemi S., Cestèle S., Pezzella M., et al. Loss-of-function KCNH2 mutation in a family with long QT syndrome, epilepsy, and sudden death. Epilepsia. 2013;54(8):e112–e116. doi: 10.1111/epi.12259. [DOI] [PubMed] [Google Scholar]
- 61.Berghuis B., Brilstra E. H., Lindhout D., Baulac S., de Haan G. J., van Kempen M. Hyperactive behavior in a family with autosomal dominant lateral temporal lobe epilepsy caused by a mutation in the LGI1/epitempin gene. Epilepsy and Behavior. 2013;28(1):41–46. doi: 10.1016/j.yebeh.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 62.Bi W., Glass I. A., Muzny D. M., et al. Whole exome sequencing identifies the first STRADA point mutation in a patient with polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome (PMSE) American Journal of Medical Genetics A. 2016;170(8):2181–2185. doi: 10.1002/ajmg.a.37727. [DOI] [PubMed] [Google Scholar]
- 63.Gal M., Magen D., Zahran Y., et al. A novel homozygous splice site mutation in NALCN identified in siblings with cachexia, strabismus, severe intellectual disability, epilepsy and abnormal respiratory rhythm. European Journal of Medical Genetics. 2016;59(4):204–209. doi: 10.1016/j.ejmg.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 64.Li G., Shi R., Wu J., et al. Association of the hERG mutation with long-QT syndrome type 2, syncope and epilepsy. Molecular Medicine Reports. 2016;13(3):2467–2475. doi: 10.3892/mmr.2016.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sweeney M. G., Hammans S. R., Duchen L. W., et al. Mitochondrial DNA mutation underlying Leigh's syndrome: clinical, pathological, biochemical, and genetic studies of a patient presenting with progressive myoclonic epilepsy. Journal of the Neurological Sciences. 1994;121(1):57–65. doi: 10.1016/0022-510x(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 66.Hammans S. R., Sweeney M. G., Brockington M., et al. The mitochondrial-DNA transfer Rna(Lys) a-]G(8344) mutation and the syndrome of myoclonic epilepsy with ragged-red fibers (Merrf)—relationship of clinical phenotype to proportion of mutant mitochondrial-DNA. Brain. 1993;116:617–632. doi: 10.1093/brain/116.3.617. [DOI] [PubMed] [Google Scholar]
- 67.Chang K. J., Ho T. S., Susuki K., et al. Paranodal ankyrins: enigmatic glial anchors? Journal of Neurochemistry. 2013;125:p. 198. [Google Scholar]
- 68.Armani A., Giacomello E., Galli S., Rossi D., Sorrentino V. Muscle-specific ankyrins and the organization of the sarcoplasmic reticulum in striated muscle cells. Biophysical Journal 86(1): 222a. 2004;86(1):p. 222a. [Google Scholar]
- 69.Mohler P. J., Gramolini A. O., Bennett V. Ankyrins. Journal of Cell Science. 2002;115(8):1565–1566. doi: 10.1242/jcs.115.8.1565. [DOI] [PubMed] [Google Scholar]
- 70.Pan Z., Kao T., Horvath Z., et al. A common ankyrin-G-based mechanism retains KCNQ and Na V channels at electrically active domains of the axon. Journal of Neuroscience. 2006;26(10):2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cunha S. R., Mohler P. J. Ankyrin protein networks in membrane formation and stabilization. Journal of Cellular and Molecular Medicine. 2009;13(11-12):4364–4376. doi: 10.1111/j.1582-4934.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J., Song W., Amato K. Eph receptor tyrosine kinases in cancer stem cells. Cytokine & Growth Factor Reviews. 2015;26(1):1–6. doi: 10.1016/j.cytogfr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eriksson O., Ramström M., Hörnaeus K., Bergquist J., Mokhtari D., Siegbahn A. The Eph tyrosine kinase receptors EphB2 and EphA2 are novel proteolytic substrates of tissue factor/coagulation factor VIIa. Journal of Biological Chemistry. 2014;289(47):32379–32391. doi: 10.1074/jbc.M114.599332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang H., Li R. H., Yuan J. X., et al. Up-regulated ephrinB3/EphB3 expression in intractable temporal lobe epilepsy patients and pilocarpine induced experimental epilepsy rat model. Brain Research. 2016;1639:1–12. doi: 10.1016/j.brainres.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 75.Hock B., Böhme B., Karn T., et al. PDZ-domain-mediated interaction of the Eph-related receptor tyrosine kinase EphB3 and the ras-binding protein AF6 depends on the kinase activity of the receptor. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(17):9779–9784. doi: 10.1073/pnas.95.17.9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nateri A. S., Raivich G., Gebhardt C., et al. ERK activation causes epilepsy by stimulating NMDA receptor activity. EMBO Journal. 2007;26(23):4891–4901. doi: 10.1038/sj.emboj.7601911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sánchez-Fernández G., Cabezudo S., Caballero Á., et al. Protein kinase C ζ interacts with a novel binding region of Gαq to act as a functional effector. Journal of Biological Chemistry. 2016;291(18):9513–9525. doi: 10.1074/jbc.M115.684308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu A. M. F., Wong Y. H. G16-mediated activation of nuclear factor κB by the adenosine A1 receptor involves c-Src, protein kinase C, and ERK signaling. Journal of Biological Chemistry. 2004;279(51):53196–53204. doi: 10.1074/jbc.M410196200. [DOI] [PubMed] [Google Scholar]
- 79.Doering C. J., Kisilevsky A. E., Feng Z.-P., et al. A single Gβ subunit locus controls cross-talk between protein kinase C and G protein regulation of N-type calcium channels. The Journal of Biological Chemistry. 2004;279(28):29709–29717. doi: 10.1074/jbc.m308693200. [DOI] [PubMed] [Google Scholar]
- 80.Gajda Z., Török R., Horváth Z., et al. Protein kinase inhibitor as a potential candidate for epilepsy treatment. Epilepsia. 2011;52(3):579–588. doi: 10.1111/j.1528-1167.2011.02979.x. [DOI] [PubMed] [Google Scholar]
- 81.Visser J. E., Bloem B. R., Van De Warrenburg B. P. C. PRKCG mutation (SCA-14) causing a Ramsay Hunt phenotype. Movement Disorders. 2007;22(7):1024–1026. doi: 10.1002/mds.21414. [DOI] [PubMed] [Google Scholar]
- 82.Hsiao M.-C., Liu C.-Y., Yang Y.-Y., Lu C.-S., Yeh E.-K. Progressive myoclonic epilepsies syndrome (Ramsay Hunt syndrome) with mental disorder: report of two cases. Psychiatry and Clinical Neurosciences. 1999;53(5):575–578. doi: 10.1046/j.1440-1819.1999.00608.x. [DOI] [PubMed] [Google Scholar]
- 83.Bird T. D., Shaw C. M. Progressive myoclonus and epilepsy with dentatorubral degeneration: a clinicopathological study of the Ramsay Hunt syndrome. Journal of Neurology Neurosurgery and Psychiatry. 1978;41(2):140–149. doi: 10.1136/jnnp.41.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Javaid S., Zhang J., Smolen G. A., et al. MAPK7 regulates emt features and modulates the generation of CTCs. Molecular Cancer Research. 2015;13(5):934–943. doi: 10.1158/1541-7786.mcr-14-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee T. S., Eid T., Mane S., et al. Aquaporin-4 is increased in the sclerotic hippocampus in human temporal lobe epilepsy. Acta Neuropathologica. 2004;108(6):493–502. doi: 10.1007/s00401-004-0910-7. [DOI] [PubMed] [Google Scholar]
- 86.Golubkov V. S., Prigozhina N. L., Zhang Y., et al. Protein-Tyrosine Pseudokinase 7 (PTK7) directs cancer cell motility and metastasis. The Journal of Biological Chemistry. 2014;289(35):24238–24239. doi: 10.1074/jbc.m114.574459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ross M. E. Gene-environment interactions, folate metabolism and the embryonic nervous system. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(4):471–480. doi: 10.1002/wsbm.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bouskila M., Hirshman M. F., Jensen J., Goodyear L. J., Sakamoto K. Insulin promotes glycogen synthesis in the absence of GSK3 phosphorylation in skeletal muscle. American Journal of Physiology - Endocrinology and Metabolism. 2008;294(1):E28–E35. doi: 10.1152/ajpendo.00481.2007. [DOI] [PubMed] [Google Scholar]
- 89.Matys V., Kel-Margoulis O. V., Fricke E., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Research. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morgan-Smith M., Wu Y., Zhu X., Pringle J., Snider W. D. GSK-3 signaling in developing cortical neurons is essential for radial migration and dendritic orientation. eLife. 2014;3:p. e02663. doi: 10.7554/eLife.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sattarova D., Sigatullina M. I., Shamansurov S. S., Mirsaidova N. A. Outcome of epilepsy surgery in focal cortical dysplasia. European Journal of Neurology. 2012;19:p. 605. [Google Scholar]
- 92.Tanaka F., Otsubo H., Gaetz W. C., et al. FDG PET and MEG evaluation of focal cortical dysplasia: comparison with the results of intracranial invasive EEG and epilepsy surgery. Journal of Nuclear Medicine. 2000;41:p. 220. [Google Scholar]
- 93.Giannandrea M., Bianchi V., Mignogna M. L., et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. American Journal of Human Genetics. 2010;86(2):185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y., Liu J., Luan G., Wang X. Inhibition of the small GTPase Cdc42 in regulation of epileptic-seizure in rats. Neuroscience. 2015;289:381–391. doi: 10.1016/j.neuroscience.2014.12.059. [DOI] [PubMed] [Google Scholar]
- 95.Kretschmann A., Danis B., Andonovic L., et al. Different MicroRNA profiles in chronic epilepsy versus acute seizure mouse models. Journal of Molecular Neuroscience. 2015;55(2):466–479. doi: 10.1007/s12031-014-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakamura T., Yasuda S., Nagai H., et al. Longest neurite-specific activation of Rap1B in hippocampal neurons contributes to polarity formation through RalA and Nore1A in addition to PI3-kinase. Genes to Cells. 2013;18(11):1020–1031. doi: 10.1111/gtc.12097. [DOI] [PubMed] [Google Scholar]
- 97.Kano H., Takayama T., Midorikawa Y., Nagase H. Promoter hypomethylation of RAR-related orphan receptor α 1 is correlated with unfavorable clinicopathological features in patients with colorectal cancer. BioScience Trends. 2016;10(3):202–209. doi: 10.5582/bst.2016.01097. [DOI] [PubMed] [Google Scholar]
- 98.Wang L., Zhan W., Xie S., et al. Over-expression of Rap2a inhibits glioma migration and invasion by down-regulating p-AKT. Cell Biology International. 2014;38(3):326–334. doi: 10.1002/cbin.10213. [DOI] [PubMed] [Google Scholar]
- 99.Zhu Q., Wang L., Xiao Z., et al. Decreased expression of Ras-GRF1 in the brain tissue of the intractable epilepsy patients and experimental rats. Brain Research. 2013;1493:99–109. doi: 10.1016/j.brainres.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 100.Adachi M., Abe Y., Aoki Y., Matsubara Y. Epilepsy in RAS/MAPK syndrome: two cases of cardio-facio-cutaneous syndrome with epileptic encephalopathy and a literature review. Seizure. 2012;21(1):55–60. doi: 10.1016/j.seizure.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 101.Lu Z., Hornia A., Joseph T., et al. Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Molecular and Cellular Biology. 2000;20(2):462–467. doi: 10.1128/MCB.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Manguoglu E., Akdeniz S., Dündar N., et al. RLIP76 gene variants are not associated with drug response in turkish epilepsy patients. Balkan Journal of Medical Genetics. 2011;14(1):25–30. doi: 10.2478/v10034-011-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nobes C. D., Lauritzen I., Mattei M.-G., Paris S., Hall A., Chardin P. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. Journal of Cell Biology. 1998;141(1):187–197. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ishikawa Y., Katoh H., Negishi M. A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. Journal of Neuroscience. 2003;23(35):11065–11072. doi: 10.1523/JNEUROSCI.23-35-11065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wingard C. J., Chintalgattu V., Harris G., Nolan R., Narron J., Katwa L. C. Testosterone-dependent expression of RhoA, ROCK I, ROCK II and Rnd1 in rat corpus cavernosum. The FASEB Journal. 2005;19:p. A123. [Google Scholar]
- 106.Zanata S. M., Hovatta I., Rohm B., Püschel A. W. Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in semaphorin 3A-induced cytoskeletal collapse. Journal of Neuroscience. 2002;22(2):471–477. doi: 10.1523/JNEUROSCI.22-02-00471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Teixeira R. A., Zanardi V. A., Li L. M., Santos S. L. M., Cendes F. Epilepsy and destructive brain insults in early life: a topographical classification on the basis of MRI findings. Seizure. 2004;13(6):383–391. doi: 10.1016/j.seizure.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 108.Bae Y.-S., Chung W., Han K., et al. Down-regulation of RalBP1 expression reduces seizure threshold and synaptic inhibition in mice. Biochemical and Biophysical Research Communications. 2013;433(2):175–180. doi: 10.1016/j.bbrc.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 109.Lee M. C., Ban S. S., Woo Y.-J., Kim S. U. Calcium/calmodulin kinase II activity of hippocampus in kainate-induced epilepsy. Journal of Korean Medical Science. 2001;16(5):643–648. doi: 10.3346/jkms.2001.16.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Butler L. S., Silva A. J., Abeliovich A., Watanabe Y., Tonegawa S., Mcnamara J. O. Limbic epilepsy in transgenic mice carrying a Ca2+/calmodulin-dependent kinase II α-subunit mutation. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(15):6852–6855. doi: 10.1073/pnas.92.15.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ambrosino P., Alaimo A., Bartollino S., et al. Epilepsy-causing mutations in Kv7.2 C-terminus affect binding and functional modulation by calmodulin. Biochimica et Biophysica Acta—Molecular Basis of Disease. 2015;1852(9):1856–1866. doi: 10.1016/j.bbadis.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 112.Churn S. B., Kochan L. D., Delorenzo R. J. Chronic inhibition of Ca2+/calmodulin kinase II activity in the pilocarpine model of epilepsy. Brain Research. 2000;875(1-2):66–77. doi: 10.1016/S0006-8993(00)02623-8. [DOI] [PubMed] [Google Scholar]
- 113.Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84(6):889–897. doi: 10.1016/S0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 114.Kim Y. S., Choi M. Y., Kim Y. H., et al. Protein kinase Cdelta is associated with 14-3-3 phosphorylation in seizure-induced neuronal death. Epilepsy Research. 2010;92(1):30–40. doi: 10.1016/j.eplepsyres.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 115.Shinoda S., Skradski S. L., Araki T., et al. Formation of a tumour necrosis factor receptor 1 molecular scaffolding complex and activation of apoptosis signal-regulating kinase 1 during seizure-induced neuronal death. European Journal of Neuroscience. 2003;17(10):2065–2076. doi: 10.1046/j.1460-9568.2003.02655.x. [DOI] [PubMed] [Google Scholar]
- 116.Knopp E. A., Saraceni C., Moss J., McNiff J. M., Choate K. A. Somatic ATP2A2 mutation in a case of papular acantholytic dyskeratosis: mosaic Darier disease. Journal of Cutaneous Pathology. 2015;42:853–857. doi: 10.1111/cup.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dhitavat J., Dode L., Leslie N., Burge S., Hovnanian A. Effects of mutations in ATP2A2 on calcium transport across sarco/endoplasmic reticulum (ER) membrane. Journal of Investigative Dermatology. 2001;117:774–774. [Google Scholar]
- 118.Takagi A., Kamijo M., Ikeda S. Darier disease. Journal of Dermatology. 2016;43(3):275–279. doi: 10.1111/1346-8138.13230. [DOI] [PubMed] [Google Scholar]
- 119.Dodiuk-Gad R. P., Cohen-Barak E., Khayat M., et al. Darier disease in Israel: combined evaluation of genetic and neuropsychiatric aspects. British Journal of Dermatology. 2016;174(3):562–568. doi: 10.1111/bjd.14220. [DOI] [PubMed] [Google Scholar]
- 120.Jacobsen N. J. O., Lyons I., Hoogendoorn B., et al. ATP2A2 mutations in Darier's disease and their relationship to neuropsychiatric phenotypes. Human Molecular Genetics. 1999;8(9):1631–1636. doi: 10.1093/hmg/8.9.1631. [DOI] [PubMed] [Google Scholar]
- 121.Karimi K., Mahmoudi T., Karimi N., et al. Is there an association between variants in candidate insulin pathway genes IGF-I, IGFBP-3, INSR, and IRS2 and risk of colorectal cancer in the Iranian Population? Asian Pacific Journal of Cancer Prevention. 2013;14(9):5011–5016. doi: 10.7314/APJCP.2013.14.9.5011. [DOI] [PubMed] [Google Scholar]
- 122.Pechlivanis S., Pardini B., Bermejo J. L., et al. Insulin pathway related genes and risk of colorectal cancer: INSR promoter polymorphism shows a protective effect. Endocrine-Related Cancer. 2007;14(3):733–740. doi: 10.1677/ERC-07-0107. [DOI] [PubMed] [Google Scholar]
- 123.Che F., Fu Q., Li X., et al. Association of insulin receptor H1085H C>T, insulin receptor substrate 1 G972R and insulin receptor substrate 2 1057G/A polymorphisms with refractory temporal lobe epilepsy in Han Chinese. Seizure. 2015;25:178–180. doi: 10.1016/j.seizure.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 124.Wang Q., Qian J., Wang F., Ma Z. Cellular prion protein accelerates colorectal cancer metastasis via the Fyn-SP1-SATB1 axis. Oncology Reports. 2012;28(6):2029–2034. doi: 10.3892/or.2012.2025. [DOI] [PubMed] [Google Scholar]
- 125.Strom A., Diecke S., Hunsmann G., Stuke A. W. Cellular prion protein promotes glucose uptake through the Fyn-HIF-2 alpha-Glut1 pathway to support colorectal cancer cell survival. Cancer Science. 2011;103:606–606. doi: 10.1111/j.1349-7006.2010.01811.x. [DOI] [PubMed] [Google Scholar]
- 126.Cain D. P., Grant S. G. N., Saucier D., Hargreaves E. L., Kandel E. R. Fyn tyrosine kinase is required for normal amygdala kindling. Epilepsy Research. 1995;22(2):107–114. doi: 10.1016/0920-1211(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 127.Yang X., Marshall C., Dentchev T., et al. A topical PI3K/mTOR inhibitor induces regression of squamous cell carcinomas in K14-Fyn Y528F mice. Journal of Investigative Dermatology. 2012;132:p. S25. [Google Scholar]
- 128.Bermudez Y., Stratton S. P., Bowden G. T., et al. Abstract 3673: expression profile of phosphorylated proteins from the mTOR and Fyn/RSK2 signaling pathways in solar UV-induced skin carcinogenesis. Cancer Research. 2011;71(8 supplement):3673–3673. doi: 10.1158/1538-7445.am2011-3673. [DOI] [Google Scholar]
- 129.Chang B., Grau T., Dangel S., et al. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(46):19581–19586. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martinez S. E., Heikaus C. C., Klevit R. E., Beavo J. A. The structure of the GAF a domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. Journal of Biological Chemistry. 2008;283(38):25913–25919. doi: 10.1074/jbc.M802891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Filla A., De Michele G., Cocozza S., et al. Early onset autosomal dominant dementia with ataxia, extrapyramidal features, and epilepsy. Neurology. 2002;58(6):922–928. doi: 10.1212/WNL.58.6.922. [DOI] [PubMed] [Google Scholar]
- 132.Coutelier M., Blesneac I., Monteil A., et al. A recurrent mutation in CACNA1G alters Cav3.1 T-type calcium-channel conduction and causes autosomal-dominant cerebellar ataxia. American Journal of Human Genetics. 2015;97(5):726–737. doi: 10.1016/j.ajhg.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang H., Xu D., Toh M. F., Pao A. C., You G. Serum- and glucocorticoid-inducible kinase SGK2 regulates human organic anion transporters 4 via ubiquitin ligase Nedd4-2. Biochemical Pharmacology. 2016;102:120–129. doi: 10.1016/j.bcp.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Friedrich B., Feng Y., Cohen P., et al. The serine/threonine kinases SGK2 and SGK3 are potent stimulators of the epithelial Na+ channel α,β,γ-ENaC. Pflugers Archiv European Journal of Physiology. 2003;445(6):693–696. doi: 10.1007/s00424-002-0993-8. [DOI] [PubMed] [Google Scholar]
- 135.Seller M. J., Spector R. G. Effect of aldosterone and cortisol on leptazol-induced seizures in rats. British Journal of Pharmacology and Chemotherapy. 1962;19(2):271–273. doi: 10.1111/j.1476-5381.1962.tb01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tan W., Lim S.-G., Tan T. M. C. Up-regulation of microRNA-210 inhibits proliferation of hepatocellular carcinoma cells by targeting Yes1. World Journal of Gastroenterology. 2015;21(46):13030–13041. doi: 10.3748/wjg.v21.i46.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patel P. R., Sun H., Li S. Q., et al. Identification of potent Yes1 kinase inhibitors using a library screening approach. Bioorganic & Medicinal Chemistry Letters. 2013;23(15):4398–4403. doi: 10.1016/j.bmcl.2013.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Material consists of three files. In detail, Supplementary Material S1 lists 470 genes related to epilepsy with their Ensembl IDs and gene symbols; Supplementary Material S2 lists 6,886 RWR genes derived from RWR algorithm with probability larger than 1E-05; Supplementary Material S3 lists 980 candidate genes with permutation FDRs less than 0.05.