Abstract
Thioredoxin reductase (TrxR) is a component of several redox-sensitive signaling cascades that mediate important biological processes such as cell survival, maturation, growth, migration and inhibition of apoptosis. The expression levels of TrxR1 in some human carcinoma cell lines are nearly 10 times higher than those in normal cells. Ethaselen is a novel antitumor candidate that exerts potent inhibition on non-small cell lung cancer (NSCLC) by targeting TrxR. In this study we explored the relationship between the ethaselen dose and TrxR activity level and the relationship between TrxR degradation and tumor apoptosis in a human lung carcinoma A549 xenograft model. BALB/c nude mice implanted with human NSCLC cell line A54 were administered ethaselen (36, 72, 108 mg·kg−1·d−1, ig) or vehicle for 10 d. The tumor size and TrxR activity levels in tumor tissues were daily recorded and detected. Based on the experimental data, NONMEM 7.2 was used to develop an integrated dose-biomarker-response model for describing the quantitative relationship between ethaselen dose and tumor eradication effects. The time course of TrxR activity levels was modeled using an indirect response model (IDR model), in which the influence of the tumor growth rates on Kin with the linear correction factor γ1 (0.021 d/mm). The drug binding-inhibition effects on Kout was described using a sigmoidal Emax model with Smax (5.95), SC50 (136 mg/kg) and Hill's coefficient γ2 (2.29). The influence of TrxR activity inhibition on tumor eradication was characterized by an Emax model with an Emax (130 mm3/d) and EC50 (0.0676). This model was further validated using a visual predictive check (VPC) and was used to predict the efficacy of different doses. In conclusion, the properties and characteristics of ethaselen acting on TrxR degradation and subsequently resulting in tumor apoptosis are characterized by the IDR model and integrated dose-biomarker-response model with high goodness-of-fit and great predicative ability. This approach shed new light on the detailed processes and mechanism of ethaselen action and may offer a valuable reference for an appropriate dosing regimen for use in further clinical applications.
Keywords: non-small cell lung cancer (NSCLC), thioredoxin reductase, anticancer drug, ethaselen, dose-biomarker-response model, NONMEM
Introduction
Mammalian thioredoxin reductase (TrxR) is a component of several redox-sensitive signaling cascades that mediate some physiological processes, especially cell survival, maturation, growth, migration and inhibition of apoptosis1,2,3,4,5,6. The TrxR1 expression levels in some human carcinoma cell lines are approximately 10 times higher than those in normal cells7, indicating their close relationships with cell proliferation and progression. In addition, it has been reported that the TrxR activity level is closely associated with the malignancy grade8. That is to say that malignant tumors tend to have high activity levels compared to benign hyperplasia. Recent studies have shown that TrxR is a promising target for cancer therapy in a wide range of tumors9,10.
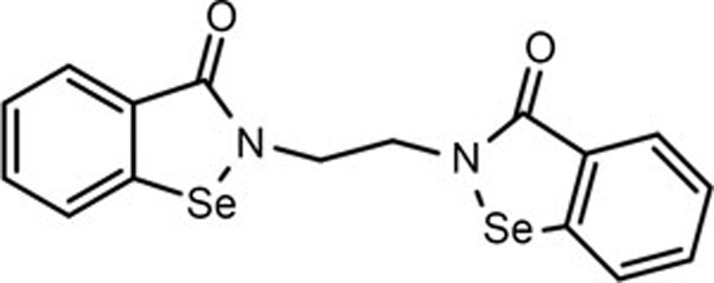
Ethaselen, as shown in Figure 1, was first synthesized in our group and has now entered a clinical Phase II trial as a novel antitumor candidate that targets TrxR, exhibiting good antitumor effects in many cancer models11. Ethaselen was designed to inhibit mammalian TrxR activity by selectively targeting SeCys498/Cys497, the C-terminal active sites of mammalian TrxR, with high binding potential12,13,14. Ethaselen can inhibit TrxR activity in several human carcinoma cell lines and tumor-bearing mouse models11,12,13,14,15,16; in one example, the antitumor effects on A549, a human non-small cell lung cancer cell line, appeared to be excellent. In addition, previous studies have demonstrated that TrxR inactivation correlates with cell death/apoptosis in the A549 cell line, and the mechanism is related to inducing mitochondria-dependent apoptosis17.
Figure 1.
The chemical structure of Ethaselen.
A dose-response model is usually an excellent option for characterizing the relationship between drug effects and doses; this relationship is crucial for adequate dose selection18,19,20. For Ethaselen, to make use of the available pharmacokinetic (PK) measurements in blood samples, complicated extraction methods and large systems of differential equations are needed to characterize the Ethaselen dynamics21. Taking the difficulty and complexity of PK measurements into account, we chose a simpler dose-biomarker-response model that is based on a pharmacokinetic-pharmacodynamic (PK-PD) model22, where the PK component in the unobserved effect compartment is treated as a latent variable23.
The dynamic levels of a biomarker are a key indicator revealing the details and processes of the drug action24. Furthermore, by exploring the quantitative relationship between changes in the biomarker level and in the tumor apoptosis effects, direct evidence for and detailed information on the drug actions were obtained, such as the pharmacological response of the drug, the degree of effects and the time course of response changes25. Previous studies have demonstrated that the TrxR activity level, which can be specifically inhibited by Ethaselen, is a potent, effective biomarker for the initiation and development of cancer26. As a result, TrxR activity was adopted as the biomarker for this pharmacodynamic study.
It has been reported that TrxR is involved in many aspects of tumor physiology, such as proliferation, apoptosis and metastasis1,2,3,4,5. In recent years, an increasing number of TrxR inhibitors have received attention and have eventually been demonstrated as effective7,27,28,29; Ethaselen is one promising, potent candidate26. However, to the best of our knowledge, no studies have investigated the quantitative relationship between Ethaselen administration and tumor eradication, let alone possible models based on the complicated process and mechanism of Ethaselen action and the TrxR signaling pathway. In this study, we explored the relationship between Ethaselen dose and the level of TrxR activity and explored the relationship between TrxR degradation and tumor apoptosis effects in a human lung carcinoma A549 xenograft model. This is the first time that an integrated dose-biomarker-response model has been established to quantitatively describe the relationship between Ethaselen dose, TrxR activity levels and tumor volume in BALB/c nude mice xenografted with A549 (a non-small cell lung cancer). Through data-based development and model-based analysis, this integrated dose-biomarker-response model sheds new light on the detailed processes and complicated mechanism of Ethaselen action in vivo and allows verification of the appropriate dose for clinical treatments.
Materials and methods
Drugs and reagents
Ethaselen (PCT: CN02-00412, purity >99%) was designed and synthesized in our laboratory (State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing, China). Other chemicals were purchased from Sigma-Aldrich unless otherwise specified.
Cell culture
The human non-small cell lung cancer cell A549 was obtained from the Cell Center of the Chinese Academy of Medical Sciences (Beijing, China) and cultured in DMEM (Macgene, Beijing, China) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Cells in the exponential growth phase were used for the experiments.
Animals
Five-week-old female BALB/c nude mice were purchased from the Animal Center of Peking University Health Science Center. All animal studies and experiments were approved and conducted strictly under the ethical guidelines issued by Ethical Committee of Peking University. Animals were housed under controlled conditions (25±2 °C, with 12 h light/dark cycle). Tumors were established by injecting the right armpits of mice with human non-small cell lung cancer cells from the A549 cell line (5×106/0.2 mL per mouse).
TrxR activity assay
TrxR activity was determined by the 5,5′-Dithiobis-(2-nitrobenzoic acid) (DTNB)-reducing method30,31 and expressed as an increase in the absorbance at 412 nm, which is proportional to the TrxR-dependent reduction of DTNB into TNB−. After tissue homogenization, the same amount of protein was diluted with an equal volume of 0.1 mol/L PBS and mixed with the enzymatic reaction solution (100 mmol/L KH2PO4/K2HPO4 (pH 7.0), 5 mmol/L DNTB, 0.2 mmol/L NADPH, 10 mmol/L EDTA, and 0.2 mg/mL BSA). Absorbance was measured at 412 nm every 5 s for 400 s, and the reaction temperature was sustained at 37 °C.
TrxR activity was calculated with the following formula:
 |
The TrxR activity unit was defined as the nmol value of the reduced DTNB in units of time. The linear range of this method was 0–1000 U/mL, and all data detected in the study were within the linear range.
Pharmacodynamics study based on TrxR activity levels in tumor tissues
A TrxR activity assay was used to investigate the inhibitory effect of Ethaselen on the TrxR activity levels in the tumors of A549 xenograft mice. When the tumor volume reached approximately 100 mm3, 132 mice were randomly divided into the following four groups: control group (receiving 0.5% CMC-Na alone, pH 7.4); low-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 36 mg/kg, ig, QD, ×10 d); middle-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 72 mg/kg, ig, QD, ×10 d); and high-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 108 mg/kg, ig, QD, ×10 d). Each group consisted of 33 mice. In each group, before daily dosing (with the first drug administration set as time 0 d), the subcutaneous tumor volume was measured by Vernier calipers and recorded as the pharmacodynamics data. Then, the tumor tissue was resected after decapitating the mice (n=3 per day for each group) and stored at -20 °C until further TrxR activity analysis by the DTNB-reducing assays, as described above.
Pharmacodynamics study based on tumor volume
Until the tumor size reached approximately 100 mm3, 28 mice were randomly divided into the following four groups: control group (receiving 0.5% CMC-Na alone, pH 7.4); low-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 36 mg/kg, ig, QD, ×10 d); middle-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 72 mg/kg, ig, QD, ×10 d); and high-dose group (receiving Ethaselen dissolved in 0.5% CMC-Na at a dose of 108 mg/kg, ig, QD, ×10 d). Each group consisted of 7 mice. Ethaselen was dissolved in 0.5% CMC-Na (pH 7.4). The subcutaneous tumor volume was measured using Vernier calipers daily and calculated as shown in Eq 2. Combined with the tumor volume data recorded from the above assays of TrxR activity, the whole tumor volume data set was adopted as the pharmacodynamics data for the dose-response modeling.
Dose-biomarker-response model
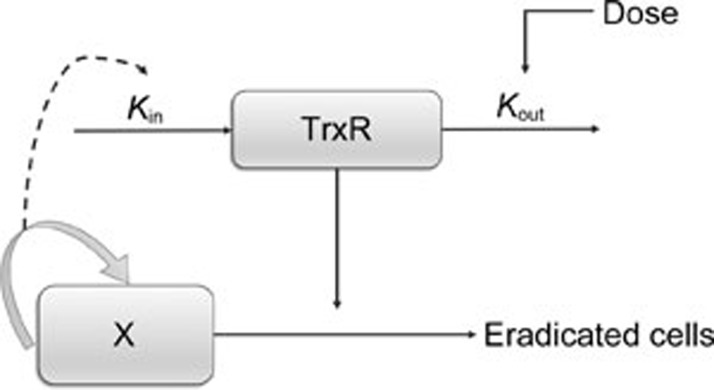
Previous studies have shown that the mechanism of Ethaselen action, including specific binding to the TrxR enzyme, inhibits its activities and subsequently induces tumor apoptosis17. In addition, it has been reported that the TrxR activity level is closely associated with the malignancy grade8. As for the human non-small cell lung cancer xenograft mouse model, the malignancy grade can be simplified and represented by the degree of tumor proliferation. Therefore, it was assumed that the TrxR activity levels might be quantitatively related to the tumor growth rates. Finally, an indirect response model and an integrated dose-biomarker-response model, as shown in Figure 2, were established to reveal some aspects of the pharmacodynamics properties of Ethaselen.
Figure 2.
Schematic of the integrated model, including the IDR model for TrxR inhibition. Kin and Kout, TrxR generation and degradation rate constants. X, tumor compartement.
Dose-biomarker-response modeling based on TrxR activity levels
The TrxR activity levels were modeled using an indirect response model (IDR model)32. In this model, Kin was influenced by tumor growth rates with a linear relationship, and Kout was affected by Ethaselen binding-inhibition after dosing. The latter was characterized by a sigmoidal Emax model using an Smax value (ie, the maximal TrxR inhibition) and an SC50 value (ie, the dose of Ethaselen causing one-half maximal TrxR inhibition). The inhibition effects are described in the block diagram shown in Figure 2 and can be calculated using Eqs 3–4.
In Eq 3, TrxR represents the TrxR activity level. Kin is the zero order rate constant of the generation of TrxR, and Kout is the first order rate constant of the degradation process of TrxR. Here, the Gompertz model was used, with the exponential and linear phases describing the natural tumor growth rates; the total TrxR generation rates were affected by the natural growth rates of the tumor. Additionally, γ1 is the linear correction factor, and γ2 is the Hill's coefficient.
Dose-biomarker-response modeling based on tumor volume
The Gompertz model was adopted to apply to the vehicle control group (natural tumor growth)33. In this model, the following two different phases were suggested: initial exponential growth in the early phase and a subsequent phase of linear growth until reaching a plateau34. However, because the plateau phase was not achieved over the experimental period, an alternative and simplified model was applied, placing more emphasis on the exponential and linear phases. The tumor natural growth equations (Eqs 5–6) were as follows:
Where X represents the tumor size and W represents the initial tumor volume just before dosing. Additionally, in this model, λ0 and λ1 represent the exponential and linear growth rate constants, respectively.
Here, the ratio of TrxR inhibition, P, was used as the variable representing the relative TrxR activity inhibition levels. The tumor apoptosis effects caused by TrxR degradation were characterized by a sigmoidal Emax model with zero order elimination. The Emax value is the maximal tumor eradication effect, and the EC50 value is the ratio of TrxR inhibition P causing one-half of the maximal tumor eradication effects that represent the tumor apoptosis effects throughout TrxR degradation. In this study, transit compartments were not used based on the goodness-of-fit with the data. The equations (Eqs 7,8,9) were as follows:
The tumor size data of the vehicle control group, 36 mg/kg Ethaselen group, 72 mg/kg Ethaselen group and 108 mg/kg Ethaselen group were applied to construct the integrated dose-biomarker-response model.
Modeling construction, evaluation, and simulation
All modeling and simulations were performed on NONMEM 7.1.2 (ICON Development Solutions, Ellicott City, MD, USA), Pirana 2.8.0 and Perl-speaks-NONMEM (PsN, Version 3.5.3) with the FOCEI algorithm. All graphs were drawn with the R software. The inter-individual and residual variabilities were estimated and used for evaluating the precision and reliability of the parameters. Selection of the models was based on the change in the objective function value (OFV), value of estimate, relative standard error (RSE), and exploratory analysis of the goodness-of-fit plots. Data for the vehicle control and different treatment groups were used for model development. For model validations, the visual predictive check (VPC) was conducted with 1000 simulations, and the 10th, 50th, and 90th percentiles of the simulations were compared with the observed value. The simulations for the different doses (36, 50, 72, 90, and 108 mg/kg Ethaselen) were performed by fixing all parameters obtained from the integrated model above.
Results
Pharmacodynamic studies
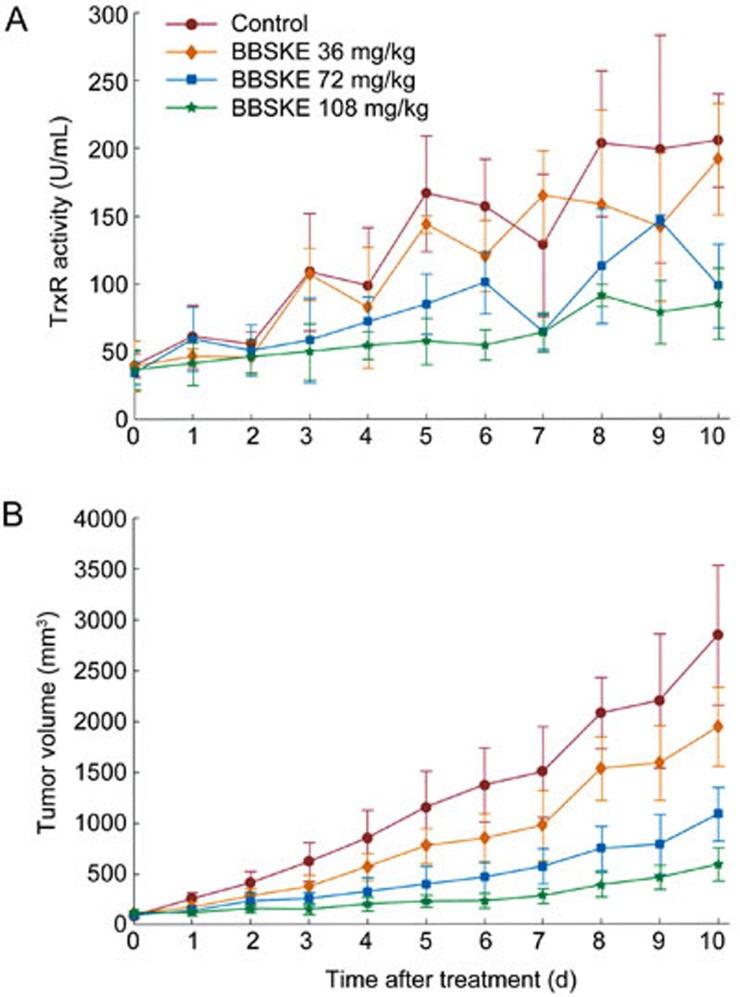
TrxR activities at various time points are shown in Figure 3A. The TrxR activities increase as the tumor grows. After Ethaselen administration, an evident decrease in the TrxR activity levels was observed compared to the control group at each time point. This type of inhibition increased in conjunction with the dose. Compared with the control group, the TrxR activity levels of the high dose group (108 mg/kg) decreased more than the middle dose group (72 mg/kg) and low dose group (36 mg/kg). These results indicated that TrxR inhibition was dose-dependent.
Figure 3.
(A) The mean±SD (n=3) data of TrxR activity levels in tumor tissue of the A549 xenografted mice after treatment with ethaselen at 0 (control), 36, 72, and 108 mg/kg dosage. (B) The mean±SD (n=10) data of tumor volume in the A549 xenografted mice after treatment with ethaselen at 0 (control), 36, 72, and 108 mg/kg daily.
Figure 3B shows the inhibitory effects of Ethaselen (at various doses) on tumor growth in A549 xenograft mice. Compared with the control group, the treatment groups (36, 72, and 108 mg/kg Ethaselen) showed increased inhibition of tumor growth, and the inhibition levels increased with the dose. The tumor volume-time profiles of the different doses were applied to a dose-time-response model as pharmacodynamic data.
Modeling
Modeling based on TrxR activity levels
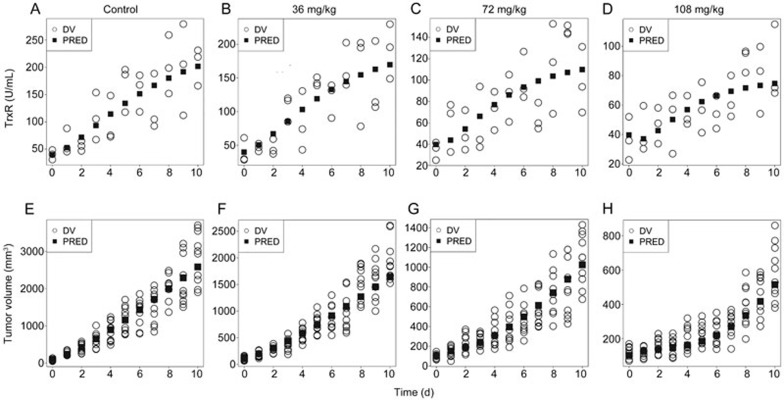
Time courses of the levels of TrxR activity for all the groups were described by the indirect response model. The observed and model-predicted TrxR activities for each group are graphically presented in Figure 4A–4D, and the estimated parameters are shown in Table 1.
Figure 4.
(A–D) The observed and model-predicted TrxR activity levels in tumor tissue of the A549 xenografted mice after treatment with ethaselen at 0 mg/kg as control (A), 36 mg/kg (B), 72 mg/kg (C), and 108 (D) mg/kg. (E–H) The observed and model-predicted tumor volume-time profiles of the control group (E), and groups treated with 36 mg/kg (F), 72 mg/kg (G), and 108 mg/kg (H) of ethaselen daily. DV represented the observed value.
Table 1. Estimates of parameters for the integrated dose-biomarker-response model.
| Parameter | Definition | Estimates | RSE (%)1 | IIV (CV%)2 |
|---|---|---|---|---|
| Kin (U·mL−1·d−1) | TrxR production rate | 8.27 | 42.4 | 12.4 |
| Base (U/mL) | Initial TrxR activity | 39.7 | 8.60 | 9.6 |
| γ1 (d/mm) | Linear correction factor | 0.021 | 16.5 | NE3 |
| γ2 (−) | Hill's coefficient | 2.29 | 17.3 | NE |
| Smax (−) | Maximal TrxR inhibition | 5.95 | 31.9 | NE |
| SC50 (mg/kg) | Dose of ethaselen causing one-half maximal TrxR inhibition | 136 | 25.2 | NE |
| λ0 (d−1) | Exponential growth rate | 0.704 | 31.4 | NE |
| λ1 (mm3/d) | Linear growth rate | 321 | 11.5 | 32.7 |
| W (mm3) | Initial tumor volume | 103 | 3.9 | 14.2 |
| Emax (mm3/d) | Maximal tumor eradication effects | 130 | 4.8 | NE |
| EC50 (−) | Ratio of TrxR inhibition P causing one-half maximal tumor eradication effects | 0.0676 | 23.1 | 19.3 |
| Errpro | Proportional error | 20.22% | 13.6 | |
| Erradd (mm3) | Additive error | 141 | 78 |
1 Percentage relative standard error.
2 Percentage coefficient of inter-individual variability of the estimate.
3 Inter-individual variability was fixed to zero when CV<0.1%.
Modeling based on tumor volume
The integrated model, which included the indirect response model for TrxR inhibition, was used to describe the pharmacodynamic profiles of the tumor growth data. The results of the observed and predicted tumor volumes are shown in Figure 4E–4H, and the estimated parameters of the integrated model are listed in Table 1.
Model evaluation and simulation
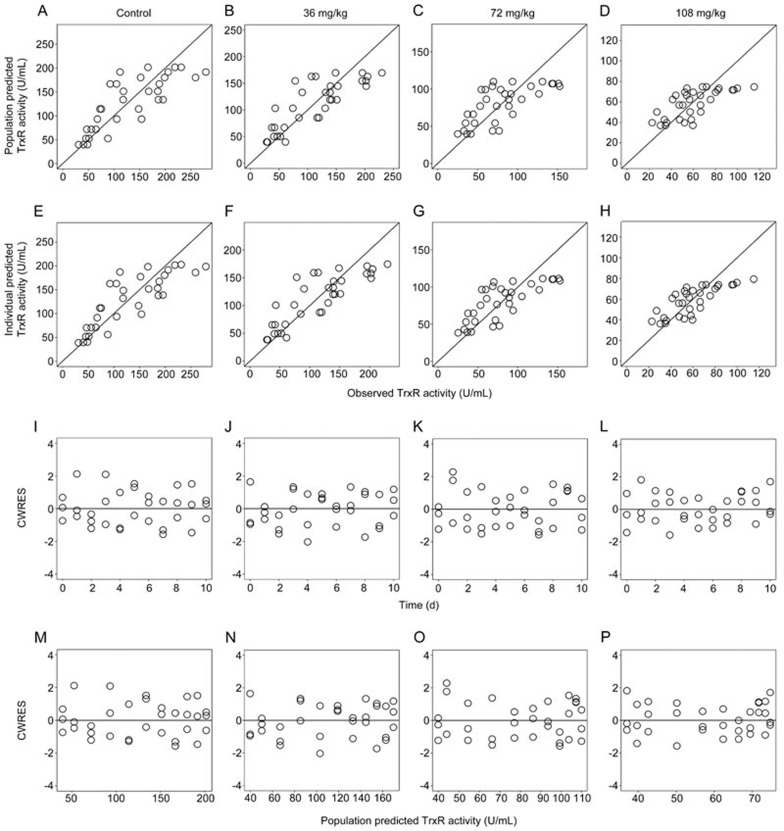
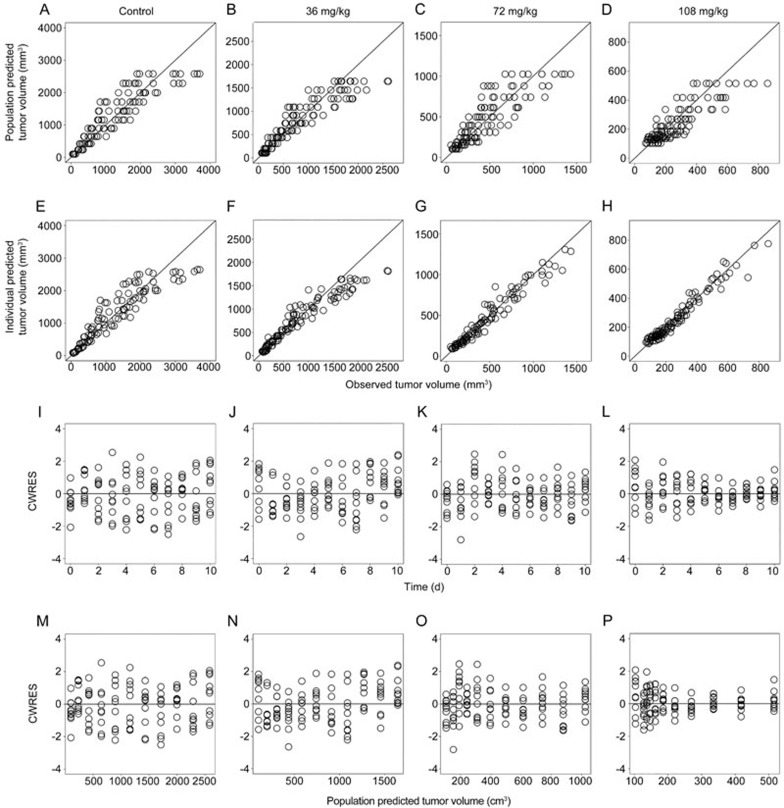
For the IDR and integrated models, the goodness-of-fit plots are shown in Figure 5 (the IDR model) and Figure 6 (the integrated dose-biomarker-response model), respectively. The dots of the population predicted value (PRED) or individual predicted value (IPRE) versus observed data (OBS) were randomly distributed around y=x. Additionally, the conditional weighted residuals (CWRES) were homogenously distributed around the x axis. The time profiles at the different doses tested by the model were a reasonable fit and the parameters were estimated with good precision.
Figure 5.
The goodness-of-fit plots of the IDR model: Relationship between population predicted values versus observed TrxR activities in the control group (A), and groups treated with 36 mg/kg (B), 72 mg/kg (C), and 108 mg/kg (D) of ethaselen daily; Relationship between individual predicted values versus observed TrxR activities in the control group (E), and groups treated with 36 mg/kg (F), 72 mg/kg (G), and 108 mg/kg (H) of ethaselen daily; Conditional weighted residuals at different time points of the control group (I), and groups treated with 36 mg/kg (J), 72 mg/kg (K), and 108 mg/kg (L) of ethaselen daily; Conditional weighted residuals at different predicted values of the control group (M), and groups treated with 36 mg/kg (N), 72 mg/kg (O), and 108 mg/kg (P) of ethaselen daily.
Figure 6.
The goodness-of-fit plots of the integrated dose-biomarker-response model: Relationship between population predicted values versus observed tumor volumes of the control group (A), and groups treated with 36 mg/kg (B), 72 mg/kg (C), and 108 mg/kg (D) of ethaselen daily; Relationship between individual predicted values versus observed tumor volumes of the control group (E), and groups treated with 36 mg/kg (F), 72 mg/kg (G), and 108 mg/kg (H) of ethaselen daily; Conditional weighted residuals at different time points of the control group (I), and groups treated with 36 mg/kg (J), 72 mg/kg (K), and 108 mg/kg (L) of ethaselen daily; Conditional weighted residuals at different predicted values of the control group (M), and groups treated with 36 mg/kg (N), 72 mg/kg (O), and 108 mg/kg (P) of ethaselen daily.
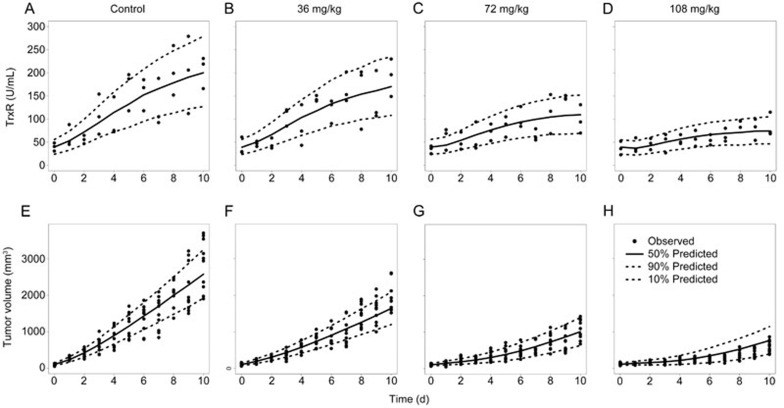
The VPC results of the IDR and integrated models for the different treatment groups (0, 36, 72, and 108 mg/kg Ethaselen) are shown in Figure 7. The 50th percentiles of the predictions nearly pass through the middle of the observed values, and most observed values were within an 80% confidence interval of the predictions, suggesting that the model accurately fits the current data of TrxR activity and tumor volume with a high predictability.
Figure 7.
(A–D) The VPC results of the IDR model in BALB/c nude mice bearing an A549 xenograft using a simulation that was conducted 1000 times: (A) Control group, (B) 36 mg/kg Ethaselen, (C) 72 mg/kg ethaselen, (D) 108 mg/kg ethaselen. (E–H) The VPC results of the integrated dose-biomarker-response models in BALB/c nude mice bearing an A549 xenograft using a simulation that was conducted 1000 times: (E) Control group, (F) 36 mg/kg ethaselen, (G) 72 mg/kg ethaselen, (H) 108 mg/kg ethaselen. The range between the dashed lines represents the 80% confidence intervals, the solid lines are the medians of the simulated data, and the solid dots are the observed data.
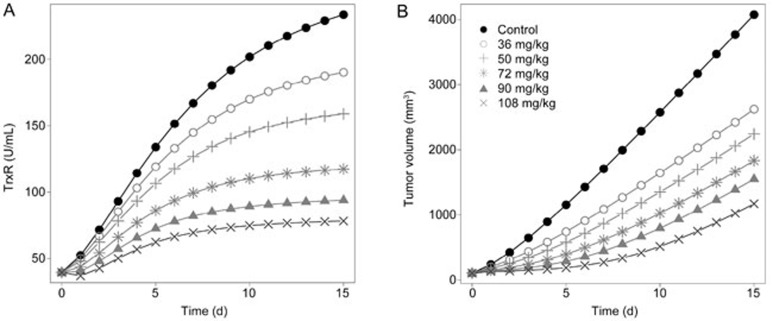
To better understand the drug action and optimize the therapeutic regimen of Ethaselen treatment, simulations of the TrxR activity-time and tumor volume-time profiles for a series of different dose regimens (0, 36, 50, 72, 90, and 108 mg/kg) were performed by fixing the parameters. The dose-dependent efficacy in both TrxR activity inhibition and tumor eradication effects was observed, as shown in Figure 8.
Figure 8.
The simulation results of different dose regimens of ethaselen. (A) The predicted TrxR activity levels in the A549 xenografted nude mice at the dosages of 36, 50, 72, 90, and 108 mg/kg ethaselen. (B) The predicted tumor volumes in the A549 engrafted nude mice at the dosages of 36, 50, 72, 90, and 108 mg/kg ethaselen.
Discussion
The present research sheds new light on the dynamic dose-biomarker-response relationships of Ethaselen, a TrxR-specific inhibitor, in a human lung carcinoma A549 xenograft model. Previous studies have demonstrated that many tumor processes are closely related to TrxR, including growth, metastasis and inhibition of apoptosis1,2,3,4,5,17. Ethaselen is a small molecule that is specifically combined with TrxR and reduces the TrxR activity levels, exerting good inhibitory effects on several human carcinoma cell lines11,12,13,14,15,26. However, to the best of our knowledge, no studies have detected TrxR activities after Ethaselen administration, and the dynamic relationship between the degree of TrxR degradation and tumor apoptosis effects remains unclear. The TrxR activity detection results showed that, without drug treatment, TrxR increased with tumor growth in a positive correlation. After the administration of Ethaselen, the TrxR activity levels in the tumor tissues of A549 xenograft mice were inhibited in a dose-dependent manner.
As previously reported and demonstrated, carcinoma cell proliferation relies on ribonucleotide reductase (RNR) for dNTP biosynthesis, and the reduction of RNR is regulated by TrxR35,36,37. Therefore, the high tumor growth and proliferation rates depend on the increased replication rates and accelerated metabolism of nucleic acid, resulting in elevated TrxR expression, which is a characteristic of many cancers38,39. Clinical observations8,40,41 also showed that the TrxR activity level is closely associated with the malignancy grade. In this way, malignant tumors with high proliferation rates tend to have higher activity levels than benign hyperplasia. Both the molecular mechanism and clinical observations suggest that the TrxR activities may be proportional to the tumor growth rates. To investigate the changing TrxR activity levels in the tumor, an IDR model was adopted based on the assumption that the rates of the onset and offset of TrxR activity are regulated by an indirect mechanism of action. In addition, from the point of view of goodness-of-fit terms, we explored the importance of the tumor growth rate on the Kin, the zero order rate constant of the generation of TrxR, in the IDR model equation. The IDR model allowed for better adherence to the observed data when considering the influence of the tumor growth rates on the Kin. The model not only reasonably described the dose-time-biomarker relationships between TrxR inhibition and Ethaselen dose, but it also provided the best fit to the observed biomarker data.
TrxR inhibition, tumor eradication effects and their dynamic relationships were illuminated by an integrated dose-biomarker-response model. Two parameters (λ0 and λ1), representing the exponential and linear phases, characterized the properties of natural tumor growth in the vehicle control group. The outcomes of model fitting showed that the tumor growth model that lacks transit compartments has higher goodness-of-fit with the data compared to adding transit compartments or non-proliferating compartments in the model. This may be due to Ethaselen-induced rapid apoptosis (within 1 h) in A549 cells17. The Emax model was used to describe the drug-elicited effects in the tumor eradication process, which was fit by the zero order elimination beyond the classical first-order elimination based on the goodness-of-fit for the observed data. This model adequately described the antitumor effects of Ethaselen based on the action of Ethaselen, which involves the inhibition of TrxR followed by induction of the mitochondrial apoptosis pathway and eventually results in tumor death17. The dose-biomarker-response model reflected the quantitative relationship between TrxR degradation and tumor apoptosis effects. Focusing on the biomarker, the integrated dose-biomarker-response model demonstrated that TrxR activity is an important index for drug action and a key bridge for the dynamic linkage between drug administration and tumor eradication effects, which improves our knowledge of the Ethaselen mechanism of action and will inform future related clinical applications.
The simulation results of the tumor volume in groups after the administration of 50 and 90 mg/kg Ethaselen were conducted well using the integrated dose-biomarker-response model. According to the simulation results, the excellent prediction abilities of the dose-biomarker-response model were confirmed, which may provide an easy and effective way to predict tumor apoptosis effects without conducting tumor growth studies and may optimize the experimental design in preclinical and clinical studies.
Although previous studies have demonstrated that several tumor types respond to Ethaselen exposure and TrxR inhibition, this is the first time that the dose-biomarker-response model was constructed in A549 xenograft nude mice with Ethaselen administration. Therefore, the value of TrxR activity as a biomarker must be assessed in more detail in future studies. One consideration is that the TrxR inhibitor Ethaselen might induce tumor apoptosis through some other biomarker responses and signaling pathways. To better understand the dynamic dose-biomarker-response relationship, more mechanistic models need to be constructed and evaluated. Another consideration is that because of the temporary absence of effective Ethaselen plasma concentration measurements, more studies should be performed to explore an effective approach for detecting the Ethaselen plasma concentration and to construct a pharmacokinetic–pharmacodynamic (PK–PD) model, which is a typical and widely applied modeling strategy for acquiring comprehensive, quantitative knowledge of the Ethaselen plasma concentration, TrxR degradation and tumor apoptosis effects in a xenograft model. However, to the best of our knowledge, the analysis presented here is currently the most informative available for developing Ethaselen as a promising, potent anticancer drug.
In conclusion, the relationship between the Ethaselen dose and TrxR activity levels in tumor tissue and the relationship between TrxR activity levels and tumor growth in A549 xenograft mice were appropriately characterized by an integrated dose-biomarker-response model that was reliable and predictable. The outcomes of this study confirmed the mechanism of Ethaselen action and corresponding therapeutic responses, which could improve experimental designs in future preclinical and clinical studies.
Author contribution
Suo-fu YE, Jian LI, Shuang-min JI, Wei LU, and Hui-hui ZENG designed the research; Suo-fu YE performed the research; Jian LI, Shuang-min JI, and Suo-fu YE analyzed the data; and Suo-fu YE wrote the manuscript.
Acknowledgments
This work was supported in part by the National Natural Science Foundation (81372266) and the National Science and Technology Major Project, China (2011zx09101-001-03).
References
- Powis G, Mustacich D, Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic Biol Med 2000; 29: 312–22. [DOI] [PubMed] [Google Scholar]
- Mustacich D, Powis G. Thioredoxin reductase. Biochem J 2000; 346: 1–8. [PMC free article] [PubMed] [Google Scholar]
- Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, et al. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol 2005; 25: 1980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, et al. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol 2004; 24: 9414–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol 2006; 16: 420–6. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryotic Cell 2004; 3: 1476–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Lu J, Holmgren A. Thioredoxin reductase is irreversibly modified by curcumin: a novel molecular mechanism for its anticancer activity. J Biol Chem 2005; 280: 25284–90. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zheng X, Wang X. Oxidative stress measured by thioredoxin reductase level as potential biomarker for prostate cancer. Am J Cancer Res 2015; 5: 2788–98. [PMC free article] [PubMed] [Google Scholar]
- Yan C, Shieh B, Reigan P, Zhang Z, Colucci MA, Chilloux A, et al. Potent activity of indolequinones against human pancreatic cancer: identification of thioredoxin reductase as a potential target. Mol Pharmacol 2009; 76: 163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Awwad RT, Smart DD, Spitz DR, Gius D. Thioredoxin reductase as a novel molecular target for cancer therapy. Cancer Lett 2006; 236: 164–74. [DOI] [PubMed] [Google Scholar]
- Wang L, Yang Z, Fu J, Yin H, Xiong K, Tan Q, et al. Ethaselen: a potent mammalian thioredoxin reductase 1 inhibitor and novel organoselenium anticancer agent. Free Radic Biol Med 2012; 52: 898–908. [DOI] [PubMed] [Google Scholar]
- Xing FX, Li SL, Ge XY, Wang CY, Zeng HH, Li D, et al. The inhibitory effect of a novel organoselenium compound BBSKE on the tongue cancer Tca8113 in vitro and in vivo. Oral Oncol 2008; 44: 963–69. [DOI] [PubMed] [Google Scholar]
- Shi C, Yu L, Yang F, Yan J, Zeng H. A novel organoselenium compound induces cell cycle arrest and apoptosis in prostate cancer cell lines. Biochem Biophys Res Commun 2003; 309: 578–83. [DOI] [PubMed] [Google Scholar]
- Zhao F, Yan J, Deng SJ, Lan LX, He F, Kuang B, et al. A thioredoxin reductase inhibitor induces growth inhibition and apoptosis in five cultured human carcinoma cell lines. Cancer Lett 2006; 236: 46–53. [DOI] [PubMed] [Google Scholar]
- Wang L, Fu JN, Wang JY, Jin CJ, Ren XY, Tan Q, et al. Selenium-containing thioredoxin reductase inhibitor ethaselen sensitizes non-small cell lung cancer to radiotherapy. Anticancer Drugs 2011; 22: 732–40. [DOI] [PubMed] [Google Scholar]
- Fu JN, Li J, Tan Q, Yin HW, Xiong K, Wang TY, et al. Thioredoxin reductase inhibitor ethaselen increases the drug sensitivity of the colon cancer cell line LoVo towards cisplatin via regulation of G1 phase and reversal of G2/M phase arrest. Invest New Drugs 2011; 29: 627–36. [DOI] [PubMed] [Google Scholar]
- Xiong K, Xu W, Zeng H. Reactive oxygen species mediate ethaseleninduced rapid apoptosis in A549 cells. J Chin Pharm Sci 2014; 23: 54–9. [Google Scholar]
- Bretz F, Pinheiro JC, Branson M. Combining multiple comparisons and modeling techniques in dose-response studies. Biometrics 2005; 61: 738–48. [DOI] [PubMed] [Google Scholar]
- Bornkamp B, Bretz F, Dmitrienko A, Enas G, Gaydos B, Hsu CH, et al. Innovative approaches for designing and analyzing adaptive dose-ranging trials. J Biopharm Stat 2007; 17: 965–95. [DOI] [PubMed] [Google Scholar]
- Tan H, Gruben D, French J, Thomas N. A case study of model-based Bayesian dose response estimation. Stat Med 2011; 30: 2622–33. [DOI] [PubMed] [Google Scholar]
- Lowe PJ. Applying physiological and biochemical concepts to optimize biological drug development. Clin Pharmacol Ther 2010; 87: 492–6. [DOI] [PubMed] [Google Scholar]
- Gabrielsson J, Weiner D. Pharmacokinetic and pharmacodynamic data analysis: concepts and applications. Stockholm Swedish Pharmaceutical Press. 2007.
- Jacobs T, Straetemans R, Molenberghs G, Adriaan Bouwknecht J, Bijnens L. A latent pharmacokinetic time profile to model dose-response survival data. J Biopharm Stat 2010; 20: 759–67. [DOI] [PubMed] [Google Scholar]
- Jackson RC. Pharmacodynamic modelling of biomarker data in oncology. ISRN pharmacol 2012; 2012: 590626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res 2007; 96: 213–68. [DOI] [PubMed] [Google Scholar]
- He J, Li DD, Xiong K, Ge YJ, Jin HW, Zhang GZ, et al. Inhibition of thioredoxin reductase by a novel series of bis-1,2-benzisoselenazol-3(2H)-ones: organoselenium compounds for cancer therapy. Bioorg Med Chem 2012; 20: 3816–27. [DOI] [PubMed] [Google Scholar]
- Urig S, Becker K. On the potential of thioredoxin reductase inhibitors for cancer therapy. Semin Cancer Biol 2006; 16: 452–65. [DOI] [PubMed] [Google Scholar]
- Zeng HH, Wang LH. Targeting thioredoxin reductase: anticancer agents and chemopreventive compounds. Med Chem 2010; 6: 286–97. [DOI] [PubMed] [Google Scholar]
- Hashemy SI, Ungerstedt JS, Zahedi Avval F, Holmgren A. Motexafin gadolinium, a tumor-selective drug targeting thioredoxin reductase and ribonucleotide reductase. J Biol Chem 2006; 281: 10691–7. [DOI] [PubMed] [Google Scholar]
- Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Meth Enzymol 1995; 252: 199–208. [DOI] [PubMed] [Google Scholar]
- Smith AD, Guidry CA, Morris VC, Levander OA. Aurothioglucose inhibits murine thioredoxin reductase activity in vivo. J Nutr 1999; 129: 194–8. [DOI] [PubMed] [Google Scholar]
- Jusko WJ, Ko HC. Physiologic indirect response models characterize diverse types of pharmacodynamic effects. Clin Pharmacol Ther 1994; 56: 406–19. [DOI] [PubMed] [Google Scholar]
- Norton L, Simon R. Growth curve of an experimental solid tumor following radiotherapy. J Natl Cancer Inst 1977; 58: 1735–41. [DOI] [PubMed] [Google Scholar]
- Bissery MC, Vrignaud P, Lavelle F, Chabot GG. Experimental antitumor activity and pharmacokinetics of the camptothecin analog irinotecan (CPT-11) in mice. Anticancer Drugs 1996; 7: 437–60. [DOI] [PubMed] [Google Scholar]
- Zimanyi CM, Chen PY, Kang G, Funk MA, Drennan CL. Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli. Elife 2016; 5: e07141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye Y, Li M, Long MJC, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 2015; 34: 2011–21. [DOI] [PubMed] [Google Scholar]
- Sengupta R, Holmgren A. Thioredoxin and glutaredoxin-mediated redox regulation of ribonucleotide reductase. World J Biol Chem 2014; 5: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Gong C, Liu H, Zhang X, Mei L, Song M, et al. E2F1 promote the aggressiveness of human colorectal cancer by activating the ribonucleotide reductase small subunit M2. Biochem Biophys Res Commun 2015; 464: 407–15. [DOI] [PubMed] [Google Scholar]
- Xu CW, Wang G, Wang WL, Gao WB, Han CJ, Gao JS, et al. Association between epidermal growth factor receptor mutations and the expression of excision repair cross-complementing protein 1 and ribonucleotide reductase subunit M1 mRNA in patients with non-small cell lung cancer. Exp Ther Med 2015; 9: 880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yang N, Yueqin L, Zhao Y, Ye S, Liu L, et al. Evaluation of thioredoxin reductase as a novel biomarker in the diagnosis and treatment of breast cancer. J Chin Pharm Sci 2014; 23: 711–15. [Google Scholar]
- Ye S, Yang N, Ma W, Fu Y, Wu L, Li Y, et al. Prospective clinical application of thioredoxin reductase as a novel diagnostic tumor marker. J Biosci Med 2014; 2: 44–53. [Google Scholar]