Abstract
Sulforaphane is a common antioxidant selectively abundant in cruciferous plants, which exhibits effective anti-cancer actions in control of tumorigenesis or progression of various cancers. A recent study has shown that sulforaphane attenuates the EGFR signaling pathway in non-small cell lung cancer (NSCLC), suggesting its potential anti-metastatic effects. In this study we assessed the involvement of sulforaphane and miR-616-5p in epithelial-mesenchymal transition (EMT) and NSCLC metastasis. Sulforaphane suppressed the cell proliferation in human NSCLC cell lines H1299, 95C and 95D with IC50 values of 9.52±1.23, 9.04±1.90 and 17.35±2.03 μmol/L, respectively. At low concentrations (1–5 μmol/L), sulforaphane dose-dependently inhibited the migration and invasion of 95D and H1299 cells with relatively high metastatic potential. The anti-metastatic action of sulforaphane was confirmed in 95D and H1299 cell xenografts in vivo. In fresh NSCLC tissue samples from 179 patients, miR-616-5p levels were upregulated in late-stage NSCLCs, and strongly correlated with risk of NSCLC recurrence and metastasis. Consistent with the clinic observation, miR-616-5p levels in the 3 NSCLC cell lines were correlated with their metastatic ability, and were decreased by sulforaphane treatment. Silencing miR-616-5p markedly suppressed the migration and invasion of 95D cells in vitro and NSCLC metastasis in vivo. Further studies revealed that miR-616-5p directly targeted GSK3β and decreased its expression, whereas sulforaphane decreased miR-616-5p levels by histone modification, and followed by inactivation of the GSK3β/β-catenin signaling pathway and inhibition of EMT, which was characterized by loss of epithelial markers and acquisition of a mesenchymal phenotype in NSCLC cells. Our findings suggest that sulforaphane is a potential adjuvant chemotherapeutic agent for the prevention of NSCLC recurrence and metastasis, and miR-616-5p can be clinically utilized as a biomarker or therapeutic target to inhibit metastasis.
Keywords: non-small cell lung cancer, metastasis, sulforaphane, microRNA, miR-616-5p, GSK3β, epithelial-mesenchymal transition (EMT)
Introduction
Sulforaphane is one of the most powerful and effective bioactive substances derived from plants in cancer research and is a common antioxidant that is selectively abundant in cruciferous plants. Many studies have examined the anti-cancer effects of sulforaphane in control of tumorigenesis or progression of lung cancer1, breast cancer2, prostate cancer3 and digestive system neoplasms4,5,6. However, the effect of sulforaphane on lung cancer metastasis, which limits the lifespan of lung cancer patients, has not been explored. The attenuation of the EGFR signal pathway by sulforaphane suggests a mechanism for the potential anti-metastatic effects of the drug in non-small cell lung cancers (NSCLCs)7.
MicroRNAs (miRNAs), which function as negative regulator of gene expression, are involved in various biological functions, including development and differentiation, metabolism, immune response, proliferation, apoptosis and metastasis. In recent years, many studies have revealed that miRNAs play an important physiological and pathological role in cancers; thus, therapies targeting miRNAs may be an effective strategy to block cancer oncogenesis and progression. Several miRNAs have previously been shown to be regulated by sulforaphane8,9,10. Sulforaphane can modify the activity of miR-200c/ZEB1 and the snail pathway to affect epithelial-mesenchymal transition (EMT)11, suggesting that sulforaphane may target specific miRNAs to promote its anti-metastatic effects in cancers. In EMT, the Wnt/β-catenin signal pathway plays an important role. Wnt ligands bind to Frizzled receptors, resulting in the inhibition of GSK3β. Consequently, β-catenin is prevented from being phosphorylated, ubiquitylated and degraded and translocates into the nucleus to bind transcription factors of the T cell factor/lymphoid enhancer binding factor (TCF/LEF) family; this process enables β-catenin to regulate gene expression12.
The detailed function of sulforaphane in NSCLC metastasis remains unknown. Here, we examined the effect of sulforaphane on NSCLC and the specific mechanisms underlying its anti-cancer ability and found that sulforaphane inhibited non-small cell lung cancer metastasis. Mechanistically, sulforaphane down-regulated miR-616-5p, which was associated with recurrence and metastasis in patients with NSCLC, to reverse EMT. The inactivation of the miR-616-5p/GSK3β/β-catenin signaling pathway may be a mechanism by which sulforaphane modulates NSCLC metastasis.
Materials and methods
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Cell culture
The human H1299, 95C, and 95D NSCLC cell lines and immortalized human bronchial epithelial cells (BEAS 2B) were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences and were propagated in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin, at 37 °C in a humidified atmosphere of 5% CO2 and 95% air in the incubator.
Chemicals
DL-Sulforaphane and 5-aza-2′-deoxycytidine (Decitabine) were purchased from LKT Laboratories (St Paul, MN, USA). Initially, 1 mol/L sulforaphane was prepared as a stock solution in dimethyl sulfoxide (DMSO) and stored at -20 °C in the dark. The stock sulforaphane solution was diluted to the final concentrations with phosphate buffered saline (PBS) just before use (final concentration of DMSO was 0.1%). The working solution of 5-aza-2′-deoxycytidine was prepared at a concentration of 0.1 μmol/L.
MTT assay
The exponentially growing cells were seeded into 96-well plates at a density of 8×103 cells/well in media and incubated overnight to allow for cell adherence. The cells were treated with various concentrations of sulforaphane for 48 h. The controls were incubated in DMSO vehicle at a concentration equal to that in the drug-treated cells. Cells viability was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays, as described previously13.
Migration and invasion assays
The in vitro migration and invasion were assessed using a 24-well 8.0 μm Transwell chamber (Corning Incorporated, Corning, NY, USA). For invasion assays, Transwells were coated with Matrigel (1 mg/mL, BD Biosciences). After treatment with sulforaphane for 48 h, 1×105 cells in 200 μL serum-free medium were harvested and seeded in the top chamber, and 600 μL 10% FBS culture medium was added to the lower chamber. Twenty-four hours later, the non-migrated cells were gently removed with cotton swabs, and the migrated cells on the bottom surface of the membrane were fixed with methanol for 30 min. The cells were then stained with crystal violet and photographed. Five randomly chosen fields were analyzed for each group.
In vivo tumor xenograft study
The protocols for the animal experiments were approved by the Institutional Animal Ethical Committee, Experimental Animal Center of Fujian Medical University, China. All surgery was performed under anesthesia, and all efforts were made to minimize animal suffering. Male BALB/c nude mice (SPF grade) at 4–5 weeks old (No SCXK 2012-002. SLAC Laboratory Animal Co Ltd, Shanghai, China) were used. To evaluate metastasis, cells were resuspended in 0.1 mL PBS and intravenously injected into the lateral tail vein at a density of 2×106 cells/mouse. One day after cell transplantation, the mice received the first intravenous injection of sulforaphane (25 mg/kg or 50 mg/kg) or PBS. The injections were repeated every three days, and mouse body weight was measured every three days until the termination of the experiment. The animals were sacrificed 4 weeks after the injection of tumor cells, and the harvested tissues from the nude mice were processed and conserved for subsequent experiments. Metastatic lung nodules were determined via Hematoxylin-Eosin H&E staining using a dissecting microscope.
Microarray analysis of miRNA expression
Based on the MTT assays, cells were treated with 5 μmol/L sulforaphane for 48 h. Total miRNA was isolated and labeled using a mirVANA™ miRNA isolation kit and labeling kit (Ambion, Austin, TX, USA). Samples, which were quantified by NanoDrop ND-2000 (Thermo Scientific) and labeled with Cy3/Cy5, were hybridized on an AgilentSure Print G3 Human Gene Expression Microarray. Each chip was scanned with an Agilent Scanner G2505C (Agilent Technologies). Data analyses were performed using Feature Extraction software (version 10.7.1.1, Agilent Technologies).
Real-time fluorescence quantitative PCR
Total RNA was extracted using TRIzol reagent (Invitrogen/Life Technologies, Grand Island, NY, USA) from cells or harvested tissues. cDNA synthesis of non-miRNA genes and miRNAs was performed using reverse transcription reagents (Thermo Scientific, Waltham, MA, USA). According to the manufacturer's instructions, real-time QPCR was performed on an ABI ViiA 7 detection system using a SYBR Premix Ex Taq™II kit (TaKaRa Bio, Inc, Shiga, Japan). The specific primer sets for PCR amplification of pri-miR-616-5p were as follows: forward 5′-AATATAAGTGCCACGGAG-3′ and reverse 5′-CTGCAGAAACTTTCTGAC-3′. The specific primer sets for PCR amplification of pre-miR-616-5p were as follows: forward 5′-TTAGGTAATTCCTCCTCTC-3′ and reverse 5′-CAGGTCATTCCTCTGCTCTTT-3′. The primer for mature miR-616-5p was 5′-CAAAACCCTTCAGTGACTT-3′. The non-miRNA and miRNA levels were normalized to GAPDH (forward 5′-CCATGGCACCGTCAAGGCTGA-3′ and reverse 5′-GGGCCATCCACAGTCTTCTGG-3′) and U6 (forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′), respectively. Data were analyzed using the comparative Ct method (2-ΔΔCt).
Bioinformatics analysis
MicroRNA expression profiles at various stages of non-small cell lung cancer were compared by analyzing miRNASeq data from the TCGA data set. A total of 370 samples were grouped according to the pathology tumor-node-metastasis (pTNM) system (AJCC), and 311 samples classified as Stage I and Stage II NSCLCs were defined as the early-stage NSCLC cohort, which was compared with the late-stage cohort (59 samples classified as Stage III and Stage IV NSCLCs). The differential expression profiles of microRNAs between early- (stage I and II) and late-stage (stage III and IV) NSCLC cohorts were analyzed using DESeq.
Patient tissues
One hundred and seventy-nine (179) primary fresh NSCLC tissues were collected from the First Hospital of Quanzhou Affiliated to Fujian Medical University, Quanzhou, China. All fresh samples were immediately preserved in liquid nitrogen. For the use of these clinical materials for research, prior approval from the Ethics Committee of the First Hospital of Quanzhou City was obtained. All specimens had confirmed pathological diagnoses and were staged according to the pathology tumor-node-metastasis (pTNM) system (AJCC). Total RNA was then isolated from the frozen samples using TRIzol reagent (Invitrogen/Life Technologies, Grand Island, NY, USA) for real-time fluorescence quantitative PCR.
Lentivirus production and infection
Based on the miR-616-5p precursor sequence, lentiviral particles (Neuron Biotech Co, Ltd, Shanghai, China) encoding the miR-616-5p precursor (miR-616-5p), shRNA targeting miR-616-5p (anti-miR-616-5p) and their control sequences (miR-NC and anti-miR-NC, respectively) were constructed. Then, 293T cells (Neuron Biotech Co, Ltd, Shanghai, China) were used to amplify the lentivirus. The harvested and titered lentivirus were transfected into cells using Polybrene reagent (Sigma-Aldrich, St Louis, MO, USA). For the following experiments, the stably transfected cells were incubated with puromycin, and the green fluorescence intensity was measured by fluorescence microscopy.
Western blot analysis
Cell lysates were prepared in lysis buffer consisting of 50 mmol/L Tris-HCl (pH 8.0), 150 mmol/L sodium chloride, 5 mmol/L ethylenediaminetetraacetic acid (EDTA), 1% NP-40, 0.02% NaN3, 50 mmol/L NaF and protease inhibitors (1 mmol/L phenylmethanesulfonyl fluoride (PMSF), 1 μg/mL aprotinin) on ice for 30 min. The protein concentration was measured with a BCA protein assay kit (Thermo Scientific, Waltham, MA, USA). Proteins were separated using 10% SDS–PAGE and transferred onto polyvinyldifluoride (PVDF) membranes (Millipore). The membranes were immunoprobed with the primary antibodies (Cell Signaling Technology, Inc, Danvers, MA, US) anti-GSK3β (12456), anti-β-catenin (8480), anti-E-cadherin (3195), anti-N-cadherin (13116), anti-Vimentin (5741) and anti-β-actin (4970) overnight at 4 ˚C and then probed for 1 h with the HRP-conjugated secondary antibody. The target proteins were detected on the membrane by an enhanced chemiluminescence reagent (Thermo Scientific, Waltham, MA, USA).
Immunofluorescence staining
Cells grown on glass culture slides were rinsed with phosphate-buffered saline (PBS) and fixed with cold 4% paraformaldehyde for 5 min at room temperature. Subsequently, the cells were blocked with 10% goat serum for 1 h and incubated with primary antibodies against E-cadherin (Cat No 3195, 1:200, CST), N-cadherin (Cat No 13116, 1:200, CST) and Vimentin (Cat No 5741, 1:100, CST) in PBS for 2 h at room temperature. After being washed three times in PBS, the cells were incubated for 1 h in the dark with Rhodamine Red or Alexa Fluor 488 goat anti-rabbit IgG (1:500, Bioworld Technology, Inc). After they were stained with 4-6-diamidino-2-phenylindole (DAPI), the slides were observed under a confocal microscope (Zeiss).
Luciferase reporter assay
GSK3β was predicted to be a direct target of miR-616-5p using miRWalk, RNAhybrid and TargetScan software. A fragment of the GSK3β 3′-UTR was amplified by RT-PCR and cloned into the pcDNA3.1 plasmid (named WT). A Site-Directed Mutagenesis kit (Invitrogen, Carlsbad, CA, USA) was used to generate point mutations in the miR-616-5p binding site of the GSK3β 3′-UTR (named Mut). For reporter assays, WT or Mut plasmid and the control pcDNA3.1 vector were transfected into H1299 cells with miR-616-5p inhibitor or mimics. Luciferase activity was detected at 48 h post-transfection based on the classical Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays (ChIP) were performed with a ChIP assay kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's protocol. For crosslinking, cells were incubated in 1% formaldehyde at room temperature for 10 min, and the reaction was stopped by adding glycine. The cells were lysed in lysis buffer and digested with Micrococcal Nuclease to generate DNA fragments. Anti-Histone H3 (acetyl K9) (ab10812) and anti-Histone H3 (tri-methyl K9) (ab8898) were used for immunoprecipitation. After elution and purification, the recovered DNA was analyzed by PCR. Primer sequences for the miR-616-5p promoter were forward 5′-TTCAAGCGATTCTTCTGC-3′ and reverse 5′-TAGTCGGTCGTGAGCCT-3′.
Statistical analysis
All data were analyzed by SPSS 19.0. The data are expressed as the mean±SD from at least three independent experiments. Statistical significance was determined using Student's two-tailed t test for two groups, one-way ANOVA for multiple groups and a parametric generalized linear model with random effects for MTT assays. Kaplan-Meier survival curves were generated for survival analysis. All statistical tests were two-sided, and a P value of <0.05 was considered statistically significant.
Results
Sulforaphane inhibited proliferation of non-small cell lung cancer cells
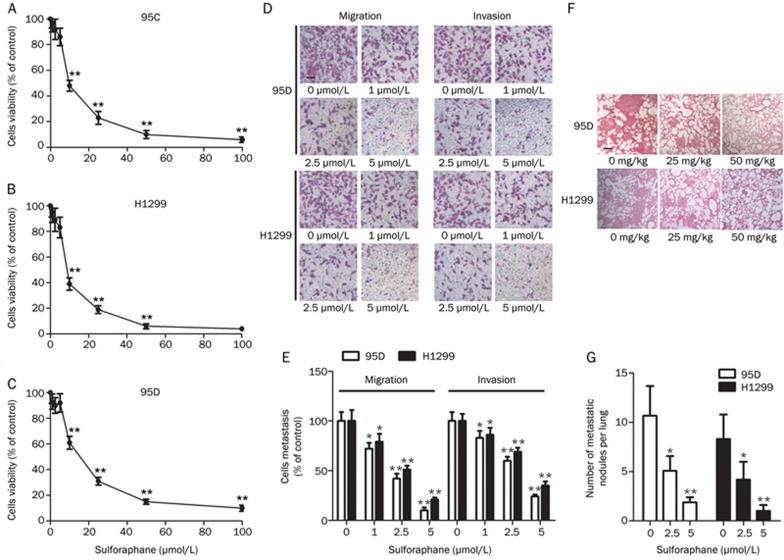
Sulforaphane was previously shown to inhibit proliferation in breast cancer cells14. The anti-proliferative effects of sulforaphane were evaluated in the human NSCLC cell lines H1299, 95C and 95D by MTT assays. Cells were treated with sulforaphane at concentrations of 0.5, 1, 2.5, 5, 10, 25, 50, and 100 μmol/L for 48 h. As shown in Figure 1A-1C, sulforaphane inhibited growth in a dose-dependent manner, with IC50 values of 9.04±1.90 μmol/L for 95C, 9.52±1.23 μmol/L for H1299 and 17.35±2.03 μmol/L for 95D.
Figure 1.
Sulforaphane inhibits the metastatic capacities of non-small cell lung cancer cells. (A–C) The viability of 95C, H1299 and 95D cells treated with different concentrations of sulforaphane for 48 h was measured by MTT assays. (D, E) Migration and invasion assays of 95D and H1299 cells treated with the different concentrations of sulforaphane (200×) (scale bar: 100 μm). (F, G) Microscopic pathology of lungs with H&E staining from the groups treated with the different concentrations of sulforaphane (100×). *P<0.05, **P<0.01 vs control.
Sulforaphane inhibited the migration and invasion of non-small cell lung cancer cells in vitro and in vivo
It is important to rule out any contribution by the anti-proliferation effect of sulforaphane on cell migration and invasion. No significant differences in the inhibitory ratio of the three cell lines treated with 5 μmol/L sulforaphane for 48 h were observed compared with that of the cells treated with 2.5 μmol/L sulforaphane (Figure 1A–1C). Therefore, the 95D cell line with high metastatic potential and the H1299 cell line were treated with 1, 2.5, and 5 μmol/L sulforaphane for 48 h, and the metastatic ability of the cell lines was evaluated. The 95D and H1299 cells treated with sulforaphane showed inhibited migration. After approximately 24 h, cells in the sulforaphane-treated group showed lower migration through the membranes than that of the control group. Furthermore, clear inhibition of cell invasion was observed in a dose-dependent manner. The invasive activity of the sulforaphane-treated cell lines was analyzed using Matrigel-coated membranes. Treatment with sulforaphane for 48 h resulted in a significant dose-response inhibition of cell invasion (Figure 1D, 1E).
To further confirm the anti-metastatic role of sulforaphane, 95D and H1299 cells were injected into nude mice through the lateral tail vein. Compared with the control group, histological analyses revealed that the incidence of lung metastasis was markedly decreased in mice treated with sulforaphane (Table 1), with the number of metastatic lung nodules decreased in the sulforaphane-treated groups (Figure 1F, 1G). However, the difference in body weight was not statistically significant (data not shown).
Table 1. Incidence of lung metastasis in mice.
| Group | Lung metastasis |
|---|---|
| Mice treated with 95D | 9/10 |
| Mice treated with 95D and 25 mg/kg sulforaphane | 5/10 |
| Mice treated with 95D and 50 mg/kg sulforaphane | 3/10 |
| Mice treated with H1299 | 7/10 |
| Mice treated with H1299 and 25 mg/kg sulforaphane | 4/10 |
| Mice treated with H1299 and 50 mg/kg sulforaphane | 2/10 |
miRNA-616-5p expression was increased in non-small cell lung cancer tissues
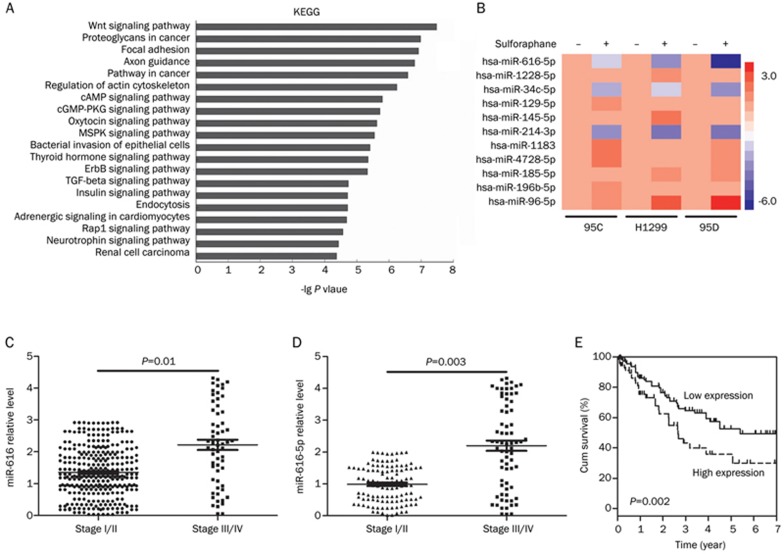
Based on the microarray data (Table 2 and Figure 2A, 2B), miRNA-616-5p was selected as a potential onco-miRNA. Differential expression analysis based on the TCGA data set identified miR-616 as a microRNA candidate that is upregulated in stage III/IV NSCLCs versus stage I/II NSCLCs (P=0.01) (Figure 2C). The mature miR-616 family has two isoforms, miR-616-5p and miR-616-3p. To validate the TCGA analysis, miR-616-5p and miR-616-3p expression levels in clinical samples (108 early-stage and 71 late-stage NSCLCs) were measured by qPCR analysis. miR-616-5p levels were decreased in early-stage NSCLCs compared to late-stage NSCLCs (P=0.003) (Figure 2D), which is consistent with the TCGA results. However, miR-616-3p levels were not statistically different between early- and late-stage NSCLC samples (Supplementary Figure S1). The clinical characteristics of the NSCLC patients are summarized in Table 3. miR-616-5p expression was positively correlated with the TNM stage in NSCLC patients (P<0.001), and patients with low miR-616-5p expression had longer survival times than those of patients with high miR-616-5p levels (log-rank test, P=0.002, Figure 2E).
Table 2. Top 10 differential expression microRNAs in 95D cells based on microarray data.
| Systematic_name | Fold change (sulforaphane-treated vs control) | Regulation (sulforaphane-treated vs control) | Active_sequence | Mirbase accession No |
|---|---|---|---|---|
| Hsa-mir-616-5p | −6.0681496 | Down | AAGTCACTGAAGGG | MIMAT0010357 |
| Hsa-mir-1228-5p | −5.106791 | Down | CACACACCTGCC | MIMAT0005582 |
| Hsa-mir-34c-5p | −4.9734235 | Down | GCAATCAGCTAACTACACTG | MIMAT0000686 |
| Hsa-mir-129-5p | −4.7998896 | Down | GCAAGCCCAGACCGC | MIMAT0000242 |
| Hsa-mir-145-5p | −4.7566605 | Down | AGGGATTCCTGGGAAAAC | MIMAT0000437 |
| Hsa-mir-214-3p | −4.5967555 | Down | ACTGCCTGTCTGT | MIMAT0000271 |
| Hsa-mir-1183 | 4.5329385 | Up | TGCCCACTCTCACCA | MIMAT0005828 |
| Hsa-mir-4728-5p | 4.6900268 | Up | TGCTTGCTGCCTCTC | MIMAT0019849 |
| Hsa-mir-185-5p | 4.7160745 | Up | TCAGGAACTGCCTTTCT | MIMAT0000455 |
| Hsa-mir-196b-5p | 4.8500404 | Up | CCCAACAACAGGAAACTACC | MIMAT0001080 |
| Hsa-mir-96-5p | 5.879141 | Up | AGCAAAAATGTGCTAGTGCCAA | MIMAT0000095 |
Figure 2.
miR-616 level is associated with NSCLC progression. (A) KEGG pathway analysis based on microarray data. (B) Heat map of miRNA expression in H1299, 95C, and 95D cells treated with 5 μmol/L sulforaphane or left untreated. (C) miRNASeq data based on TCGA between Stage I/II NSCLCs and Stage III/IV NSCLCs. (D) QPCR detections of miR-616-5p expression in Stage I/II NSCLCs and Stage III/IV NSCLCs. (E) Kaplan-Meier analysis of overall survival of NSCLC samples with low and high miR-616-5p levels.
Table 3. Correlation between the clinicopathologic characteristics and expression of miR-616-5P in NSCLCs.
| miR-616-5p expression |
||||
|---|---|---|---|---|
| Characteristics | n | Low | High | P value |
| Age (year) | ||||
| <50 | 81 | 28 (34.6%) | 53 (65.4%) | 0.787 |
| ≥50 | 98 | 32 (32.7%) | 66 (67.3%) | |
| Gender | ||||
| Male | 123 | 38 (30.9%) | 85 (69.1%) | 0.183 |
| Female | 56 | 23 (41.1%) | 33 (58.9%) | |
| Clinical stage | ||||
| I–II | 108 | 66 (61.1%) | 42 (38.9%) | <0.001 |
| III–IV | 71 | 16 (22.5%) | 55 (77.5%) | |
Down-regulation of miRNA-616-5p was critical for sulforaphane-mediated anti-metastatic effects in NSCLC cells
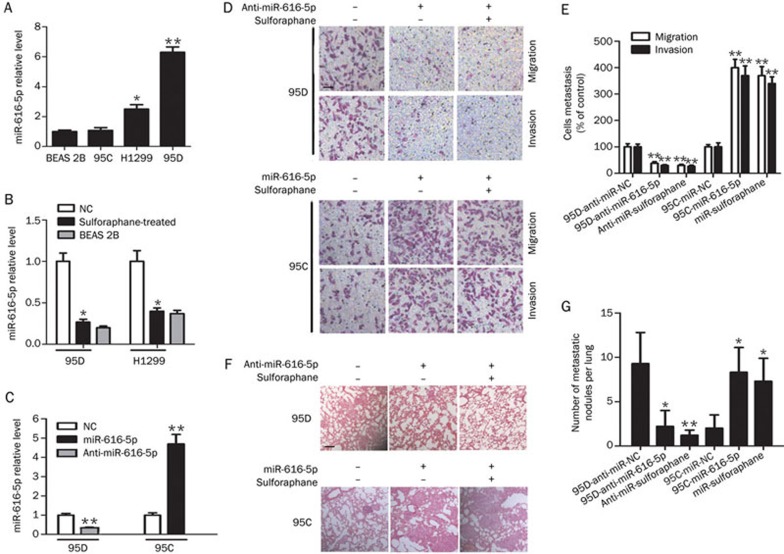
miR-616-5p expression in H1299, 95C, 95D and BEAS 2B cell lines was measured. As shown in Figure 3A, miR-616-5p levels were low in 95C cells with relatively low metastatic potential and were high in H1299 and 95D cells with relatively high metastatic potential. No significant differences were observed between 95C and BEAS 2B cells. Moreover, the expression of miR-616-5p was confirmed to be decreased by sulforaphane treatment in 95D and H1299 cell lines (Figure 3B). To further confirm the effect of miR-616-5p on NSCLC, we established stable miR-616-5p-silenced 95D cells and stable miR-616-5p-overexpressing 95C cells via lentiviral infection (Figure 3C). Up-regulation of miR-616-5p only slightly increased cell growth, and silencing miR-616-5p expression also had a small effect (Supplementary Figure S2). Then, the metastatic ability of the cell lines was analyzed in vitro and in vivo. We observed decreased cell migration and invasion in miR-616-5p-silenced 95D cells compared with anti-miR-NC-transfected cells and a significant increase in cell migration and invasion in miR-616-5p-transfected 95C cells compared with miR-NC-transfected cells (Figure 3D, 3E). Furthermore, in the intravenous injection animal model, the incidence of lung metastases and the number of metastatic lung nodules were markedly decreased in mice injected with 95D-anti-miR-616-5p cells compared with mice injected with 95D-anti-NC cells. In contrast, the incidence of lung metastasis and the number of metastatic lung nodules were increased in mice injected with 95C-miR-616-5p cells compared with mice injected with 95C-miR-NC cells. However, no significant changes in the metastatic ability were noted before and after sulforaphane treatment in both 95D-anti-miR-616-5p and 95C-miR-616-5p cells (Table 4 and Figure 3F, 3G).
Figure 3.
Sulforaphane suppresses miR-616-5p expression to inhibit NSCLC metastasis. (A) The endogenous expression levels of miR-616-5p in three lung cancer cell lines. The relative expression levels were normalized to those of BEAS 2B lung cells. (B) Decrease in miR-616-5p levels by sulforaphane treatment (5 μmol/L). (C) Relative miR-616-5p expression in cells after down- and up-regulation of miR-616-5p via lentiviral infection. (D, E) Migration and invasion assays of 95D-anti-NC, 95D-anti-miR-616-5p, 95C-miR-NC and 95C-miR-616-5p cells treated with 5 μmol/L sulforaphane or left untreated (200×). (F, G) Microscopic pathology of lungs with H&E staining from the groups treated with the different cells or 50 mg/kg sulforaphane (100×) (scale bar: 200 μm). *P<0.05, **P<0.01 vs control.
Table 4. Incidence of lung metastasis in mice.
| Group | Lung metastasis |
|---|---|
| Mice treated with 95D-anti-miR-NC | 8/10 |
| Mice treated with 95D-anti-miR-616-5p | 3/10 |
| Mice treated with 95D-anti-miR-616-5p and 50 mg/kg sulforaphane | 2/10 |
| Mice treated with 95C-miR-NC | 1/10 |
| Mice treated with 95C-miR-616-5p | 7/10 |
| Mice treated with 95C-miR-616-5p and 50 mg/kg sulforaphane | 6/10 |
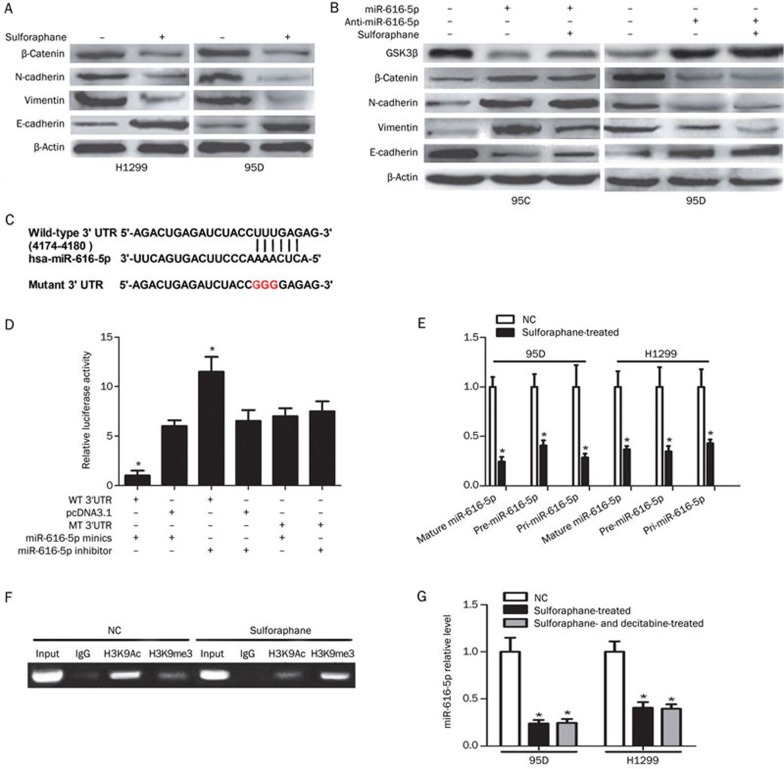
Sulforaphane suppressed EMT in non-small cell lung cancer cells
The expressions of β-catenin, E-cadherin, N-cadherin and Vimentin were measured to determine whether sulforaphane affected the EMT process. As shown in Figure 4A, sulforaphane treatment increased E-cadherin and decreased β-catenin, N-cadherin and Vimentin expression in H1299 and 95D cells. Using in vitro gain- and loss-of-function analyses, we observed that introduction of miR-616-5p decreased the expression of the epithelial marker E-cadherin and increased expression of β-catenin, N-cadherin and Vimentin in 95C-miR-616-5p cells. Furthermore, silencing of miR-616-5p reversed the EMT protein expression pattern in 95D-anti-miR-616-5p cells. However, additional treatment of sulforaphane had no effect on the expression of EMT markers in both 95C-miR-616-5p cells and 95D-anti-miR-616-5p cells (Figure 4B). Similar changes in E-cadherin, N-cadherin and Vimentin protein levels were observed by immunofluorescence staining (Supplementary Figure S3).
Figure 4.
Sulforaphane transcriptionally represses miR-616-5p expression, which directly targets GSK3β, to attenuate the EMT process in NSCLC cells. (A-B) After transfection with anti-miR-616-5p, miR-616-5p or treatment with sulforaphane, cells were analyzed by Western blotting for the expression levels of GSK3β, β-catenin and EMT-associated proteins. (C) miR-616-5p and its putative binding sequences in the 3′UTR of GSK3β, and a mutation was generated. (D) Luciferase reporter assays were performed to determine whether miR-616-5p directly targets the GSK3β 3′UTR. (E) After treatment with 5 μmol/L sulforaphane or no treatment, cells were analyzed by QPCR for the expression levels of mature miR-616-5p, precursor miR-616-5p and primary miR-616-5p. (F) ChIP analysis of histone H3K9Ac and H3K9me3 binding to the miR-616-5p promoter region in cells treated with 5 μmol/L sulforaphane or left untreated. (G) QPCR measurements of miR-616-5p expression in cells treated with sulforaphane or plus decitabine. *P<0.05 vs control.
miR-616-5p directly targeted GSK3β to down-regulate the Wnt/β-catenin signal pathway
GSK3β was predicted to be a direct target of miR-616-5p by three computational tools, miRWalk, RNAhybrid and TargetScan (Figure 4C). Silencing of miR-616-5p elevated GSK3β protein levels in 95D cells. Conversely, miR-616-5p overexpression impaired GSK3β protein levels in 95C cells (Figure 4B). GSK3β was further confirmed as a direct target of miR-616-5p by the luciferase reporter assay. Introduction of miR-616-5p mimics attenuated GSK3β luciferase activity; conversely, introduction of a miR-616-5p inhibitor increased the luciferase activity. As expected, these impacts on luciferase activity were abrogated when the cells were co-transfected with a mutated GSK3β reporter (Figure 4D).
Sulforaphane transcriptionally repressed the expression of miR-616-5p
To further elucidate the role of sulforaphane in the generation of miR-616-5p, we performed qPCR to measure primary miR-616-5p (pri-miR-616-5p), the precursor miR-616-5p (pre-miR-616-5p) and the mature miR-616-5p (miR-616-5p). Intriguingly, sulforaphane consistently down-regulated the expression of the three forms of miR-616-5p in 95D and H1299 cells (Figure 4E). Subsequently, ChIP assays were performed to confirm that sulforaphane could transcriptionally regulate the biogenesis of miR-616-5p. The level of H3K9Ac, a marker of activated chromatin, binding to the miR-616-5p promoter was reduced in sulforaphane-treated cells, and H3K9me3, a marker of inactivated chromatin, increased (Figure 4F). However, after treatment with 5-aza-2′-deoxycytidine (Decitabine) for 24 h, the suppressed expression of miR-616-5p by sulforaphane could not be rescued in 95D and H1299 cells (Figure 4G).
Discussion
Sulforaphane, which is extracted from broccoli sprouts, has been studied in many fields, including oncology. Its anti-cancer effects have attracted the attention of many researchers in recent years. In this study, we found that sulforaphane suppressed NSCLC metastasis without obvious toxicity and side effects. Mechanistically, sulforaphane exerted its anti-metastatic effects through down-regulation of miR-616-5p, which was identified as a marker associated with risk of relapse and metastasis in patients with NSCLC. The decreased expression of miR-616-5p, transcriptionally induced by sulforaphane treatment, contributed to the suppression of the EMT process in NSCLC cells.
It is well known that dramatic changes in cell proliferation can affect the accuracy in detecting cell metastatic ability. Researchers have shown that miR-616 can induce the growth of cancer cells15, and sulforaphane regulates the cell cycle in lung cancer16,17. Therefore, we ruled out the impact of miR-616-5p and sulforaphane on cell proliferation, and confirmed the pro-metastatic effect of miR-616-5p and the anti-metastatic function of sulforaphane in NSCLCs. Mechanically, microRNAs have been shown to be involved in the anti-cancer effect of natural plant extracts. miRNAs have various functions, such as induction of apoptosis18, anti-metastasis19, anti-angiogenesis20, proliferation inhibition21 and suppression of cancer stem-like cells22. Combining the microarray data with TCGA data, we identified miR-616-5p as a prominent regulator of NSCLC progression. Compared with control sera23, miR-616* (the previous name of miR-616-5p) was substantially increased in lung adenocarcinoma and was clinically related to TNM stage of NSCLCs in our study. These results also indicated the potential role of miR-616-5p in lung cancer oncogenesis or progression. The 95C and 95D cell lines with low and high metastatic potential, respectively, were subcloned from a poorly differentiated human large cell lung carcinoma cell line24. The expression pattern of miR-616-5p in NSCLC cells harboring different metastatic potentials further demonstrated that miR-616-5p, not miR-616-3p, played a crucial role in promoting metastasis of NSCLCs. Accordingly, as the pre-miRNA can generate two mature miRNAs25 (namely miRNA/miRNA* or miRNA-3p/miRNA-5p), the two miRNAs may exert different regulatory effects in specific cancers26. This hypothesis explains the distinct functions of miR-616-5p and miR-616-3p in the present work. The levels of miR-616-5p in 95C-miR-616-5p and 95D-anti-miR-616-5p cells were related to metastatic potential, which was not affected by sulforaphane, demonstrating that miR-616-5p is a downstream effector of sulforaphane-mediated signaling in NSCLCs.
MicroRNA-616 has been shown to promote metastasis and epithelial-mesenchymal transition (EMT) in HCC27. EMT is an important mechanism that leads to cancer metastasis. The loss of the epithelial marker E-cadherin and the gain of the mesenchymal marker N-cadherin are the characteristic features of EMT. β-catenin is the key molecule in Wnt signaling that promotes EMT and can transactivate Vimentin28. In addition, Wnt/β-catenin signaling phosphorylates Snail to increase the cytoplasmic retention and degradation of Snail. As a result, the expressions of E-cadherin and N-cadherin modulated by the transcription factor Snail were down-regulated and up-regulated during EMT, respectively29. Notably, our data showed that high expression of β-catenin, Vimentin and N-cadherin and low expression of E-cadherin in H1299 and 95D cells with relatively high levels of miR-616-5p could be reversed by sulforaphane, whereas sulforaphane only slightly influenced the EMT protein expression pattern in 95C-miR-616-5p and 95D-anti-miR-616-5p cells. However, GSK3β, a negative regulator of Wnt/β-catenin signaling, was negatively correlated with β-catenin in 95C and 95D cells. Collectively, these data indicated that sulforaphane might attenuate miR-616-5p expression to restrain EMT and lung cancer metastasis through the miR-616-5p/GSK3β/β-catenin signaling pathway. The regulation of miRNA expression is complicated. Before the transcription process, amplification, deletion or mutation of miRNA genes can alter the miRNA levels. During transcription, various transcription factors and epigenetic modifiers modulate the biogenesis of microRNAs. After transcription, the enzymes Drosha and Dicer process pri-miRNAs and pre-miRNAs to generate mature miRNAs30. Sulforaphane, as an effective cancer preventive factor, alters transcriptional activity of genes by histone acetylation and DNA methylation in cancers31. Additionally, histone demethylases have been shown to play critical roles in development, differentiation and diseases, such as cancer32. The present study first reported that sulforaphane transcriptionally suppressed the biogenesis of miR-616-5p through histone modification, including decreased histone acetylation and increased histone methylation, instead of DNA methylation in NSCLCs.
In summary, our findings suggest that sulforaphane inhibits cell migration and invasion through blockade of miR-616-5p expression and suppression of the EMT process in NSCLC cells, repressing metastasis of lung cancer. The miR-616-5p/GSK3β/β-catenin signaling pathway in the NSCLC cells might contribute to the underlying mechanism of the observed anti-metastatic ability of sulforaphane. Sulforaphane can be an effective agent, and miR-616-5p, which can serve as a potential biomarker for the clinical prognosis or diagnosis of NSCLCs, may be an effective anti-cancer target for the treatment of non-small cell lung cancer.
Author contribution
Zhi-qiang LIN and Da-xuan WANG designed the study; Yu-jiao ZOU wrote the paper; Xi-bin ZHUANG, Shu-xing CHEN, Yong LIN, Wen-lan LI, and Jun-jin LIN performed the experiments, data acquisition and analysis.
Acknowledgments
This study was supported by a grant (No 2015S0096) from the Fuzhou Science and Technology Plan Projects, a grant (No 2016D002) from the Science and Technology Plan Projects of Fujian Province and grants (No 2014-2-50 and 2015-1-73) from the Health and Family Planning Commission Foundation of Fujian Province, China.
Footnotes
Supplementary information is available at the Acta Pharmacologica Sinica website.
Supplementary Information
The relationship between miR-616 levels and pathological stage of NSCLC.
miR-616-5p slightly promotes proliferative capacities of non-small cell lung cancer cells.
The effect of sulforaphane and miR-616-5p on epithelial–mesenchymal transition (EMT).
References
- Jiang LL, Zhou SJ, Zhang XM, Chen HQ, Liu W. Sulforaphane suppresses in vitro and in vivo lung tumorigenesis through downregulation of HDAC activity. Biomed Pharmacother 2016; 78: 74–80. [DOI] [PubMed] [Google Scholar]
- Atwell LL, Beaver LM, Shannon J, Williams DE, Dashwood RH, Ho E. Epigenetic regulation by sulforaphane: opportunities for breast and prostate cancer chemoprevention. Curr Pharmacol Rep 2015; 1: 102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amjad AI, Parikh RA, Appleman LJ, Hahm ER, Singh K, Singh SV. Broccoli-derived sulforaphane and chemoprevention of prostate cancer: from bench to bedside. Curr Pharmacol Rep 2015; 1: 382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon YK, Yoo DR, Jang YH, Jang SY, Nam MJ. Sulforaphane induces apoptosis in human hepatic cancer cells through inhibition of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase4, mediated by hypoxia inducible factor-1-dependent pathway. Biochim Biophys Acta 2011; 1814: 1340–8. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung B, Kang YJ, Hwang SY, Kim MJ, Yoon JH, et al. Sulforaphane inhibits hypoxia-induced HIF-1alpha and VEGF expression and migration of human colon cancer cells. Int J Oncol 2015; 47: 2226–32. [DOI] [PubMed] [Google Scholar]
- Mondal A, Biswas R, Rhee YH, Kim J, Ahn JC. Sulforaphene promotes Bax/Bcl2, MAPK-dependent human gastric cancer AGS cells apoptosis and inhibits migration via EGFR, p-ERK1/2 down-regulation. Gen Physiol Biophys 2016; 35: 25–34. [DOI] [PubMed] [Google Scholar]
- Chen CY, Yu ZY, Chuang YS, Huang RM, Wang TC. Sulforaphane attenuates EGFR signaling in NSCLC cells. J Biomed Sci 2015; 22: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhauser C. Epigenetic impact of dietary isothiocyanates in cancer chemoprevention. Curr Opin Clin Nutr Metab Care 2013; 16: 405–10. [DOI] [PubMed] [Google Scholar]
- Lan F, Pan Q, Yu H, Yue X. Sulforaphane enhances temozolomide-induced apoptosis because of down-regulation of miR-21 via Wnt/beta-catenin signaling in glioblastoma. J Neurochem 2015; 134: 811–8. [DOI] [PubMed] [Google Scholar]
- Liu CM, Peng CY, Liao YW, Lu MY, Tsai ML, Yeh JC, et al. Sulforaphane targets cancer stemness and tumor initiating properties in oral squamous cell carcinomas via miR-200c induction. J Formos Med Assoc 2016. [DOI] [PubMed]
- Shan Y, Zhang L, Bao Y, Li B, He C, Gao M, et al. Epithelial-mesenchymal transition, a novel target of sulforaphane via COX-2/MMP2, 9/Snail, ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. J Nutr Biochem 2013; 24: 1062–9. [DOI] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 2012; 13: 767–79. [DOI] [PubMed] [Google Scholar]
- Lin X, Li HR, Lin XF, Yu ME, Tu XW, Hua ZD, et al. Silencing of Livin inhibits tumorigenesis and metastasis via VEGF and MMPs pathway in lung cancer. Int J Oncol 2015; 47: 657–67. [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Yoshizawa K, Uehara N, Miki H, Sasaki T, Kuro M, et al. Sulforaphane inhibits the growth of KPL-1 human breast cancer cells in vitro and suppresses the growth and metastasis of orthotopically transplanted KPL-1 cells in female athymic mice. Oncol Rep 2011; 26: 603–8. [DOI] [PubMed] [Google Scholar]
- Ma S, Chan YP, Kwan PS, Lee TK, Yan M, Tang KH, et al. MicroRNA-616 induces androgen-independent growth of prostate cancer cells by suppressing expression of tissue factor pathway inhibitor TFPI-2. Cancer Res 2011; 71: 583–92. [DOI] [PubMed] [Google Scholar]
- Liang H, Lai B, Yuan Q. Sulforaphane induces cell-cycle arrest and apoptosis in cultured human lung adenocarcinoma LTEP-A2 cells and retards growth of LTEP-A2 xenografts in vivo. J Nat Prod 2008; 71: 1911–4. [DOI] [PubMed] [Google Scholar]
- Zuryn A, Litwiniec A, Safiejko-Mroczka B, Klimaszewska-Wisniewska A, Gagat M, Krajewski A, et al. The effect of sulforaphane on the cell cycle, apoptosis and expression of cyclin D1 and p21 in the A549 non-small cell lung cancer cell line. Int J Oncol 2016; 48: 2521–33. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhao RH, Tseng KF, Li KP, Lu ZG, Liu Y, et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacol Sin 2016; 37: 519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Lee WJ, Tan P, Tang CH, Hsiao M, Hsieh FK, et al. Upregulation of miR-328 and inhibition of CREB-DNA-binding activity are critical for resveratrol-mediated suppression of matrix metalloproteinase-2 and subsequent metastatic ability in human osteosarcomas. Oncotarget 2015; 6: 2736–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wang X, Cui J, Wang P, Xiong M, Jia C, et al. Bidirectional regulation of angiogenesis and miR-18a expression by PNS in the mouse model of tumor complicated by myocardial ischemia. BMC Complement Altern Med 2014; 14: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhu M, Wang X, Tan HY, Tsao SW, Feng Y. Berberine-induced tumor suppressor p53 up-regulation gets involved in the regulatory network of miR-23a in hepatocellular carcinoma. Biochim Biophys Acta 2014; 1839: 849–57. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Song WJ, Zhang FQ, Liu WH, Dou KF. Differentiation-inducing activity of hydroxycamptothecin on cancer stem-like cells derived from hepatocellular carcinoma. Dig Dis Sci 2011; 56: 2473–81. [DOI] [PubMed] [Google Scholar]
- Rani S, Gately K, Crown J, O'Byrne K, O'Driscoll L. Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biol Ther 2013; 14: 1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Zhang J, Xu H, Wang Y, Chu Y, Liu R, et al. Differential expression of CXCR4 is associated with the metastatic potential of human non-small cell lung cancer cells. Clin Cancer Res 2005; 11: 8273–80. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–69. [DOI] [PubMed] [Google Scholar]
- Bu P, Wang L, Chen KY, Rakhilin N, Sun J, Closa A, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta. Nat Commun 2015; 6: 6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhou P, Wang W, Wang X, Li J, Sun X, et al. MicroRNA-616 promotes the migration, invasion and epithelial-mesenchymal transition of HCC by targeting PTEN. Oncol Rep 2016; 35: 366–74. [DOI] [PubMed] [Google Scholar]
- Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, et al. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res 2003; 63: 2658–64. [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J 2012; 18: 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman-Szymczyk A, Majewski G, Lubecka-Pietruszewska K, Fabianowska-Majewska K. The role of sulforaphane in epigenetic mechanisms, including interdependence between histone modification and dna methylation. Int J Mol Sci 2015; 16: 29732–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcipowski KM, Martinez CA, Ntziachristos P. Histone demethylases in physiology and cancer: a tale of two enzymes, JMJD3 and UTX. Curr Opin Genet Dev 2016; 36: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relationship between miR-616 levels and pathological stage of NSCLC.
miR-616-5p slightly promotes proliferative capacities of non-small cell lung cancer cells.
The effect of sulforaphane and miR-616-5p on epithelial–mesenchymal transition (EMT).