Abstract
Whole transcriptome analyses of next generation RNA sequencing (RNA-Seq) data from human cancer samples reveled thousands of uncharacterized non-coding RNAs including long non-coding RNA (lncRNA). Recent studies indicated that lncRNAs are emerging as crucial regulators in cancer processes and potentially useful as biomarkers for cancer diagnosis and prognosis. To delineate dysregulated lncRNAs in lung cancer, we analyzed RNA-Seq data from 461 lung adenocarcinomas (LUAD) and 156 normal lung tissues. FAM83H-AS1, one of the top dysregulated lncRNAs, was found to be overexpressed in tumors relative to normal lung and significantly associated with worse patient survival in LUAD. We verified this diagnostic/prognostic potential in an independent cohort of LUAD by qRT-PCR. Cell proliferation, migration and invasion were decreased after FAM83H-AS1 knockdown using siRNAs in lung cancer cells. Flow cytometry analysis indicated the cell cycle was arrested at the G2 phase after FAM83H-AS1 knockdown. Mechanistically, we found that MET/EGFR signaling was regulated by FAM83H-AS1. Our study indicated that FAM83H-AS1 plays an important role in lung tumor progression and may be potentially used as diagnostic/prognostic marker. Further characterization of this lncRNA may provide a novel therapeutic target impacting MET/EGFR signaling.
Lung cancer is the leading cause of cancer-related deaths globally and is histologically-defined as either small cell lung cancer (SCLC) or non-small cell lung cancer (NSCLC), with the latter accounting for 80% of all lung cancers1,2. NSCLC is a heterogeneous disease, with the most common subtypes being adenocarcinoma (LUAD) and squamous cell carcinoma (SCC). These subtypes represent distinct clinical entities, typically requiring different treatment options. Among these histological subtypes there exist cancers with diverse clinical outcomes, revealing heterogeneity in disease aggressiveness and underlying molecular alterations3,4,5. Indeed, the poor prognosis associated with lung cancer (15.7–18% 5-year survival) is related to the complex cellular, molecular and tumor microenvironment factors that impart a unique biological basis to an individual’s disease2. Discovery of oncogenic driver alterations have helped improve the outcomes in specific subtypes of patients with lung cancer, however the majority of the patients with lung cancer do not have an actionable molecular aberration6,7. Therefore, there is a vital need for new biomarkers and the identification of alternative treatments.
Long non-coding RNAs (lncRNAs) are RNA transcripts that are greater than 200 bp in length and lack an open reading frame encoding a protein8,9,10,11. LncRNA exhibit tissue or cancer specific expression patterns. In lung cancer, antisense RNA, long intergenic non-coding RNA (lincRNA), and processed transcripts are the most frequently expressed lncRNAs12. In the past few years, lncRNAs have emerged as novel mechanisms in mediating cancer biology13,14,15,16,17,18, although most lncRNAs remain undiscovered. LncRNAs appear to be involved in tumorigenesis, cell proliferation, differentiation, migration, immune response, apoptosis, and angiogenesis13,16,19,20,21. Several mechanisms associated with lncRNAs in tumor biological processes are remodeling of chromatin (HOTAIR, XIST, ANRIL), transcriptional co-activation or co-repression (H19, LincRNA-p21), protein inhibition (TERRA), post-transcriptional modifiers (MALT1) and decoy elements (PTENP1)22,23,24,25.
High-throughput RNA sequencing (RNA-Seq) of human cancer has shown remarkable potential to identify both novel markers of disease and uncharacterized aspects of tumor biology, particularly lncRNA species11,12,26,27. We recently generated transcriptome data using next-generation RNA-Seq to reveal both protein coding and lncRNA expression patterns in lung cancer7,11,12.
In the present study, we characterized a dysregulated lncRNA in lung cancer, FAM83H Antisense RNA1 (FAM83H-AS1), for its diagnostic/prognostic potential and role in cancer progression. We validated its differential expression in an independent LUAD cohort and established its diagnostic/prognostic importance. We next performed in vitro studies to delineate its oncogenic roles in cell proliferation, invasion and migration. Finally, we attempted to reveal which cancer related pathway was affected using a FAM83H-AS1 knockdown assay.
Results
FAM83H-AS1 expression is increased in lung adenocarcinomas and is associated with worse patient survival
In our previous study12, we analyzed 3 large RNA-Seq data sets representing independent tumor cohorts. These data sets are the University of Michigan (UM) cohort7 including 67 LUADs and 6 matched normal lung tissues, the Korean cohort (Seo)28 including 85 LUADs and 77 normal lung samples, and The Cancer Genome Atlas (TCGA) LUAD cohort29 including 309 LUADs and 73 normal lung samples. In order to identify lncRNAs whose expression patterns may have significant clinical utility, we performed a Receiver Operating Characteristic (ROC) curve analysis. The area under the curve (AUC) values was used to select the list of top differently expressed lncRNAs in LUAD. There were a total of 182 lncRNAs that had an AUC value greater than 0.7 and 99 lncRNAs that had an AUC value less than 0.25 in all 3 data sets12.
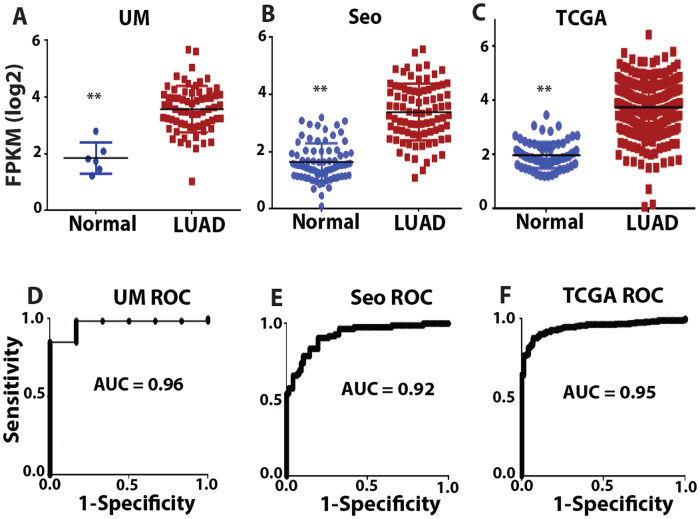
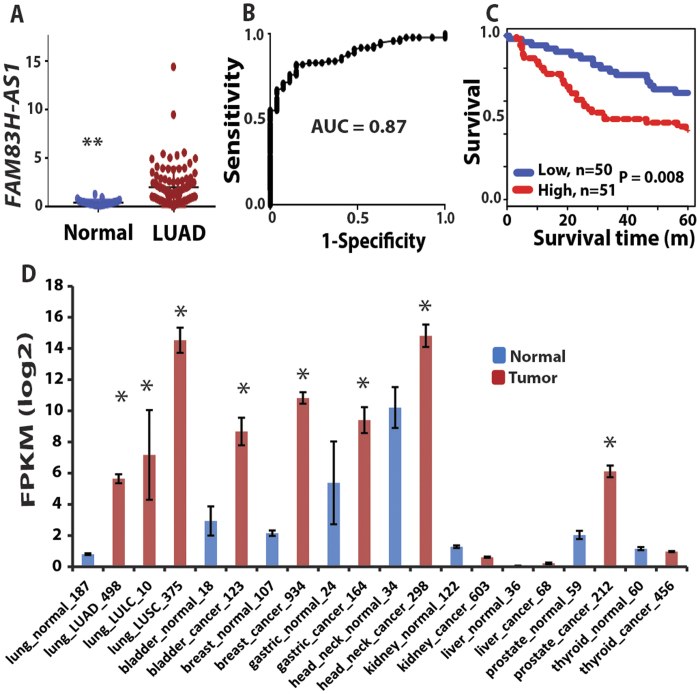
Among the most dysregulated lncRNAs, FAM83H-AS1 was found to be significantly increased in LUADs (Fig. 1A–C) and had AUC >0.9 in all 3 cohorts (Fig. 1D–F). Because FAM83H-AS1 is a novel lncRNA, there are no probe sets present on Affymetrix U133Plus2.0. There are FAM83H-AS1 probes on the Affymetrix exon array, but we did not find large number of lung tumor samples (or samples having survival information) using this platform on the GEO (Gene Expression Omnibus) database. In order to validate the expression of FAM83H-AS1 as well as evaluate its prognostic significance in lung cancer, we examined FAM83H-AS1 expression in an independent cohort of LUAD from UM including 101 lung ADs and 19 normal lung tissues using qRT-PCR. The boxplot shows that FAM83H-AS1 expression was significantly higher in cancer tissues as compared to normal lung tissues (p < 0.001) (Fig. 2A). The AUC = 0.87 indicates FAM83H-AS1 expression could significantly separate the tumors from normal lung tissues (Fig. 2B). We also found that FAM83H-AS1 expression was significantly associated with unfavorable survival in patients with lung cancer (Fig. 2C). We did not find that FAM83H-AS1 expression levels were associated with tumor stage, differentiation and other clinical variables in this validation set (Supplementary Table 1).
Figure 1. Increased FAM83H-AS1 expression in lung ADs.
(A–C) scatterplots of FAM83H-AS1 expression levels in LUAD and normal lung tissue samples in UM (67 LUAD vs. 6 Normal), Seo (85 LUAD vs. 77 Normal) and TCGA (309 LUAD vs. 73 Normal) RNA-Seq datasets (LUAD vs. Normal, p < 0.001 in all 3 data sets). (D–F) Receiver Operating Characteristic (ROC) curves of FAM83H-AS1 in UM, Seo and TCGA RNA-Seq data sets (LUAD vs. Normal). The area under the curve (AUC) >0.9 in all 3 data sets.
Figure 2. Validation of FAM83H-AS1 expression in an independent cohort of LUADs by qRT-PCR.
(A–C) validation of FAM83H-AS1 expression in an independent set of samples including 19 normal lung tissues and 101 LUAD tissues using qRT-PCR. FAM83H-AS1 expression was increased in tumors (vs. normal, **p < 0.001) (A), ROC curve of classifying the tumors from normal based on FAM83H-AS1 expression (AUC = 0.87) (B), and Kaplan-Meier survival curve indicated higher FAM83H-AS1 expression was unfavorable for patient survival (log-rank test, p = 0.008) (C). (D) Higher expression (FRKM log2 value > 2) of FAM83H-AS1 was also present in squamous cell (LUSC) and large cell lung cancer (LULC), as well as bladder, breast, gastric, head/neck, and prostate cancers, whereas lower expression was present in kidney, liver, and thyroid cancers (FRKM log2 value < 2) based on RNA-Seq expression data from MiTranscriptome. *Tumor vs. normal, p < 0.05.
In order to discover the FAM83H-AS1 expression status in other types of cancer, we examined RNA-Seq data based on 6,220 different types of cancer from one lncRNA database, MiTranscriptome26. The higher expression of FAM83H-AS1 (FRKM log2 value >2) was also found in squamous cell (LUSC) and large cell lung cancer (LULC) as well as bladder, breast, gastric, head/neck, prostate cancers. Lower expression was found in kidney, liver, and thyroid cancers (FRKM log2 value <2) (Fig. 2D). By comparing tumor vs. normal, we found that bladder, breast, gastric, head/neck, and prostate cancers were significantly increased. However, kidney, liver and thyroid cancer were not significantly changed between tumor and normal (Fig. 2D). Taken together, FAM83H-AS1 was highly expressed in not only lung cancer but also in other types of cancers and could be possibly used as a diagnostic or prognostic marker for various cancers.
Cell proliferation, invasion and migration were impaired after FAM83H-AS1 knockdown
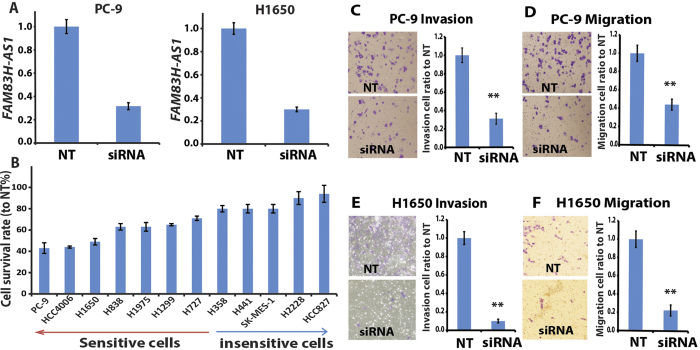
Since FAM83H-AS1 is overexpressed in most cancer tissues, we hypothesize that this lncRNA may play an oncogenic role in lung cancer progression. For this purpose, we examined cell proliferation, invasion and migration status after knockdown of FAM83H-AS1 by siRNA in lung cancer cells. The knockdown efficiency was 60–70%, measured by qRT-PCR, in 12 lung cancer cell lines (Fig. 3A and Supplementary Figure 1) with different genomic background (Supplementary Table 2) and different FAM83H-AS1 expression levels examined by RNA-Seq and RT-PCR (Supplementary Figure 2). Next, we tested the cell proliferation status after FAM83H-AS1 knockdown in these cell lines. We found that 7 out of 12 cell lines showed a significant decrease in cell proliferation (>30%). Surprisingly, we found that the cell growth was reduced in 4/5 of the tested EGFR mutated cells (Fig. 3B and Supplementary Table 2), including EGFR tyrosine kinase inhibitor (TKI) sensitive cells (PC-9) and resistant cells (HCC4006, H1975 and H1650). This result indicated that FAM83H-AS1 could affect cell proliferation even in EGFR TKI resistant cells. There were 5 cell lines not affected including two KRAS mutated cells (H358 and H441) and three TP53 mutated cells (H441, SKMES-1 and H2228, which also had a EML4-ALK fusion). HCC827 is an EGFR mutated and TKI sensitive cell as well as being EGFR amplified. We did not find a correlation between FAM83H-AS1 gene expression levels and a proliferation effect (Supplementary Figure 3). These results suggest that some cells demonstrate growth independent of FAM83H-AS1.
Figure 3. FAM83H-AS1 involved in tumor cell proliferation and invasion.
(A) The knockdown efficiency of FAM83H-AS1 siRNA in PC-9 and H1650 cells measured by qRT-PCR. The FAM83H-AS1 expression decreased more than 70% after FAM83H-AS1 siRNA (10 nM SMARTpool, at 48 h) transfection. (B) Cell proliferation was altered after FAM83H-AS1 siRNAs treatment (96–120 h) in 12 lung cancer cell lines. (C–F) cell invasion and migration decreased after FAM83H-AS1 knockdown by siRNA in both PC-9 and H1650 cell lines (20X) analyzed using Boyden chambers assay. siRNA = FAM83H-AS1 siRNA; NT = non-targeting control siRNA.
Since cell proliferation was affected in most EGFR mutated cells following FAM83H-AS1 knockdown, we selected the two most affected lung cancer cells, PC-9 (TKI sensitive) and H1650 (TKI resistant), for cell invasion and migration studies using Boyden chamber matrix assays. We found that cell invasion (with matrigel) was reduced by 70% versus control when FAM83H-AS1 was knocked down in both PC-9 and H1650 cell lines (Fig. 3C,E). Similarly, FAM83H-AS1 knockdown resulted in more than 60% decrease in cell migration (Fig. 3D,F). Colony formation was also decreased after FAM83H-AS1 knockdown (data not shown).
We found that FAM83H-AS1 could affect cell growth on one subset of lung cancer cells in vitro (Fig. 3B). Higher expression of this lncRNA is correlated with poor patient survival in clinical samples. To test if FAM83H-AS1 expression status is correlated with expression of the cell proliferation marker Ki-67 in clinical lung cancer samples, we performed Ki-67 immunohistochemistry (IHC) staining on 97 lung cancer tissues using tissue array. These 97 samples were also used for FAM83H-AS1 expression by RT-PCR in this study (Fig. 2C). Ki-67 staining was scored as 0 (negative), 1 (<10% positive cells), 2 (10–50% positive cells), or 3 (>50% positive cells) (Supplementary Figure 4). We did not find a significant correlation between FAM83H-AS1 expression and Ki-67 staining (Supplementary Figure 5).
Taken together, these results indicated that FAM83H-AS1 may not only be a potential biomarker for diagnosis and prognosis of lung cancer, but also play a critical role in lung cancer progression including cell proliferation, invasion, migration and colony formation.
FAM83H-AS1 knockdown causes cell cycle arrest at G2
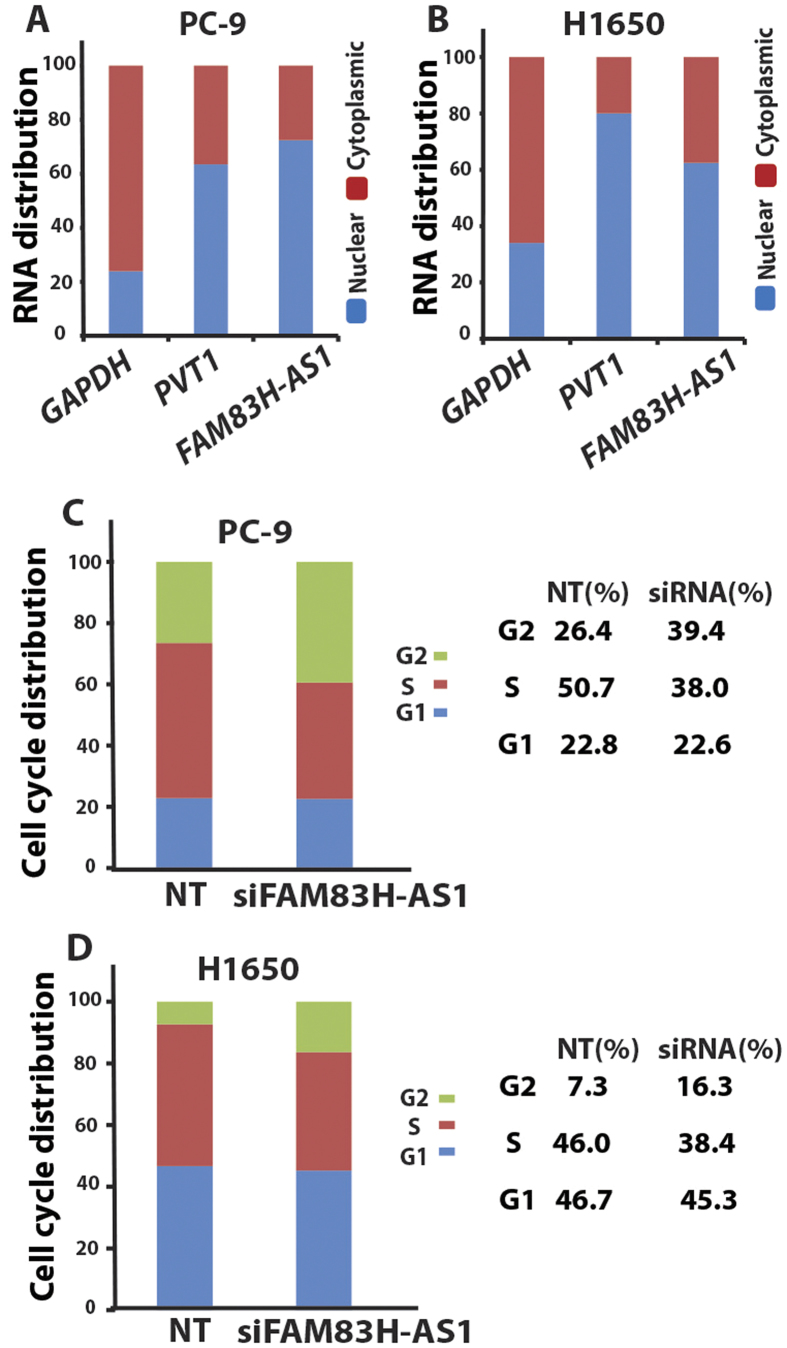
We first looked at the cellular localization of FAM83H-AS1 in PC-9 and H1650 cells. RNA was isolated from total, nuclear and cytoplasmic fractions of these two cell lines. Quantitative RT-PCR indicated FAM83H-AS1 expression was mainly in the nuclear fraction (Fig. 4A,B). Next, to determine whether FAM83H-AS1 affects cell cycle regulation, we performed flow cytometry to explore the effect on the cell cycle after FAM83H-AS1 knockdown. We found that FAM83H-AS1 knockdown by siRNA induced cell cycle arrest at the G2 phase in both PC-9 and H1650 cells (Fig. 4C,D). These results suggest that decreased tumor cell growth by FAM83H-AS1 knockdown might be through G2 arrest.
Figure 4. FAM83H-AS1 cell location and cell cycle regulation.
(A,B) FAM83H-AS1 is mainly located at nuclear measured by RT-PCR. GAPDH was used as cytoplasmic control and PVT1 as nuclear control. (C,D) FAM83H-AS1 knockdown using siRNA caused G2 arrested in both PC-9 (G2 cells increased from 26.4% to 39.4% after siRNA treatment) and H1650 (G2 cells increased from 7.3% to 16.3% after siRNA treatment) lung cancer cell lines. Right panel is the actual number of cell cycles calculated from left panel.
FAM83H-AS1 regulates MET/EGFR signaling
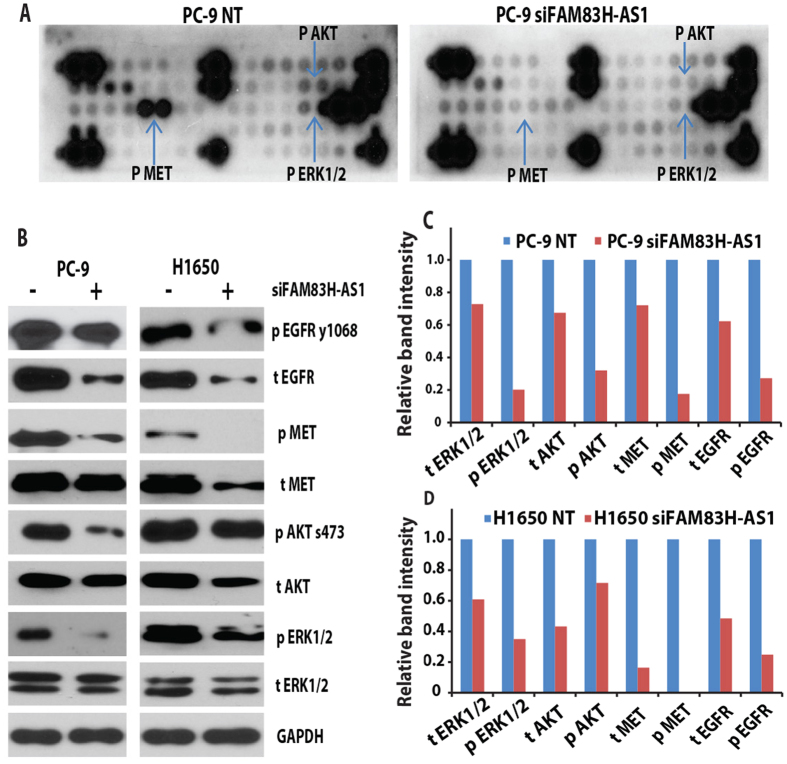
Since cell proliferation was affected in most EGFR mutated cells, we hypothesized that FAM83H-AS1 affects tumor progression through the EGFR pathway. We first performed receptor tyrosine kinase phosphorylation antibody array analysis, which included 49 different phosphorylated proteins covering most cancer-related pathways. We found that MET was the most affected protein after FAM83H-AS1 knockdown by siRNA in the EGFR mutated cell PC-9 (Fig. 5A). We next performed Western blot to confirm this results in PC-9 and H1650 cells and found that EGFR, AKT and ERK1/2 were also affected after FAM83H-AS1 knockdown (Fig. 5B–D). As indicated in Fig. 5, EGFR, MET and their downstream proteins AKT and ERK1/2 were significantly decreased after FAM83H-AS1 knockdown in both PC-9 and H1650 cells. Identification of the FAM83H-AS1 direct binding proteins using RNA pull-down assay or identification of the functional domains of FAM83H-AS1 using domain-specific chromatin isolation by RNA purification (dChIRP)30 are currently being investigated.
Figure 5. FAM83H-AS1 regulated MET/EGFR signaling.
(A) Receptor tyrosine kinase phosphorylation antibody array indicated phosphor-MET is the most significantly decreased protein after FAM83H-AS1 knockdown by siRNA (10 nM at 96 h) as compared to NT in PC-9 cell. Phosphor-AKT and ERK1/2 were also decreased. (B) Western blot showing MET, EGFR, AKT and ERK1/2 were decreased after FAM83H-AS1 siRNA treatment on PC-9 and H1650 lung cancer cell lines. (C,D) Quantitative analysis of image in B using Image J software. GAPDH was used as protein loading control (NT = 1). Abbreviation: for protein labeling, p = phosphor protein, t = total protein.
In order to explore the gene expression pathways regulated by FAM83H-AS1, we performed RT-PCR on 20 genes which are involved in pathways of MET/EGFR, cell cycle, EMT, etc. We found MET mRNA was decreased by 40%. Other genes were not affected after FAM83H-AS1 siRNA treatment on PC-9 cell (Supplementary Figure 6), indicating MET may be regulated at the transcription level and the others at the translational level.
Discussion
Genome-wide analyses indicate that lncRNA expression can confer clinical information about patient outcome and may have utility in diagnostic tests22,31,32,33. For example, MALAT1 not only has a crucial significance in predicting metastasis risk, but could also be a diagnostic biomarker detectable in the blood screening for lung cancer34,35. Prostate cancer gene 3 (PCA3) has been used as a biomarker in clinical practice for predicting prostate cancer volume36. H19 expression was found to be higher in gastric cancer (GC) tissues37, and highly expressed in the plasma of patients with GC38. HULC is reportedly detectable in peripheral blood cells of HCC patients39. Patients with high expression of HOTAIR are more likely to have tumor recurrence and poor prognosis40. HOTAIR might also be a biomarker for lymph node metastasis in HCC41. The characterization of the RNA species, their functions, and their clinical application has therefore become an area of biological and clinical importance in cancer research. In our previous study, we identified lncRNAs differently expressed in lung cancer as compared to normal lung using three large RNA-Seq data sets. Some of the lncRNAs were also related to patient outcome in lung cancer. These lncRNAs could potentially serve as diagnostic or prognostic markers in lung cancer. In this study, we have validated the differently expressed lncRNA, FAM83H-AS1, in an independent set of LUADs using RT-PCR and found that FAM83H-AS1 was significantly increased in cancer and correlated with worse patient survival. We also found that FAM83H-AS1 was increased in squamous cell and large cell lung cancer, as well as bladder, breast, gastric, head/neck, and prostate cancers. Increased FAM83H-AS1 expression in many types of cancers supports the hypothesis that this lncRNA plays a role in cancer progression and has potential use as a cancer biomarker. We found that knockdown of FAM83H-AS1 by siRNA impairs tumor cell proliferation, migration and invasion. These effects may be through cell cycle regulation by inducing G2 arrest.
LncRNAs may represent good candidates as therapeutic targets42,43. HOTAIR can reduce the sensitivity of lung adenocarcinoma cells to chemotherapeutic drugs such as cisplatin43. Down-regulation of MALAT1 expression reduced tumor growth in vivo35. EGFR is a major therapeutic target in EGFR mutated lung cancers. So far, all anti-EGFR therapies are targeted towards inhibition of its kinase activity, either by using small molecule tyrosine kinase inhibitors (erlotinib and gefitinib) or by monoclonal antibodies (cetuximab)44. However, clinical responses to such therapies have improved the outcome for only a subset of patients. Furthermore, erlotinib-sensitive patients often develop resistance due to acquisition of a T790M EGFR mutation45. Thus, there is an urgent need to identify additional strategies capable of targeting mutant EGFR. In this study, we found that MET/EGFR and their downstream signaling ERK1/2 and AKT may be the targets of FAM83H-AS1 in lung cancer, indicating that FAM83H-AS1 may be a potential therapeutic target. Further characterization of the FAM83H-AS1 direct binding molecules is ongoing.
Gene expression profiling after FAM83H-AS1 knockdown would provide more unbiased information regarding pathways affected at the transcriptional levels. In this study, we have only measured 20 genes involved in MET/EGFR, cell cycle, EMT, and other pathways. We found MET mRNA was decreased by 40%, but other genes were not affected after FAM83H-AS1 knockdown indicating MET may be regulated at the transcriptional level and others at the translational levels.
We did not find a significant correlation between FAM83H-AS1 expression and Ki-67 staining (Supplementary Figure 5). The cell growth in some cells (H358, H441 and HCC827) were not affected by FAM83H-AS1 siRNA knockdown, although these cells have relatively higher levels of FAM83H-AS1 expression, indicating these cells grow independent of FAM83H-AS1 levels. Cell growth in other cells (H838, H1299 and H1975) were affected by FAM83H-AS1 siRNA knockdown, although these cells have relatively low levels of FAM83H-AS1 expression, indicating that cell growth is affected if FAM83H-AS1 expression reaches a certain level (Supplementary Figure 3). This could partially explain why the correlation between FAM83H-AS1 expression and Ki-67 expression was not significant. Another reason may be that Ki-67 also represents a subset of tumors with higher proliferation rate. This was supported by proliferation-related gene cluster analysis (Supplementary Figure 7). Finally, some other unknown genomic events may be involved.
FAM83H-AS1 is a novel lncRNA with little known about its roles in primary tumors. Interestingly, using DAVID Gene Ontology analysis, we uncovered that FAM83H-AS1 may be involved in the inhibition of immune response in lung cancer (data not shown). Further validation and characterization of this mechanism may support a role for this lncRNA in tumor immune regulation.
In summary, this study provides a comprehensive analysis of newly characterized lncRNA FAM83H-AS1 in lung cancer. Future experiments including RNA pull-down, gene overexpression, and gene profiling affected by FAM83H-AS1 are warranted in order to establish the roles of FAM83H-AS1 in cancer progression and its potential use as a diagnostic or prognostic marker. Our findings also warrant further exploration of this lncRNA as a potential therapeutic target for patients with cancer.
Methods
Lung cancer/normal specimen
For qRT-PCR validation, 101 lung cancer and 19 paired non-tumor lung tissues were obtained from patients undergoing cancer surgery during the period from 1991 to 2012 at the University of Michigan Health System. All the patients provided informed consent, and all experimental protocols were approved by the University of Michigan Institutional Review Board and Ethics Committee. The methods were carried out in accordance with approved guidelines. Resected specimens were stored at −80 °C until use. Frozen tissues for regions containing a minimum of 70% tumor cellularity, defined by cryostat sectioning, were used for RNA isolation. None of the patients received any preoperative radiation or chemotherapy. The clinical information is shown in Supplementary Table 1. The median follow-up time was 8.1 years among the patients that remained alive.
Lung cancer cell lines
All 12 lung cancer cell lines used in this study were obtained from the American Type Culture Collection and maintained in RPMI 1640, supplemented with 10% FBS and 1% antibiotic-antimycotic. Cells were cultured at 37 °C in a 5% CO2 cell culture incubator. Genotyping for identity of all cell lines was performed at the University of Michigan Sequencing Core, and testing of mycoplasma contamination was routinely examined.
RNA isolation and quantitative RT-PCR
Total RNA isolation, cDNA synthesis, and qRT-PCR from tissue samples and cell lines were performed as described previously12. The oligonucleotide primers for FAM83H-AS1 were forward: 5′-TAGGAAACGAGCGAGCCC, and reverse: 5′-GCTTTGGGTCTCCCCTTCTT. The housekeeping genes GAPDH and ACTB were used as loading controls. Fold-changes were calculated relative to control siRNA or the median value of normal tissue samples.
Western Blot analysis
Lung cancer cell lysates were boiled in sample buffer, separated by polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride membranes. After blocking for 1 h with 5% non-fat milk or BSA, the membranes were incubated with primary antibodies against human MET (1:1000 dilution), EGFR (1:1000 dilution), AKT (1:1000 dilution), ERK1/2 (1:1000 dilution), and GAPDH (1:10,000 dilution) overnight at 4 °C. After incubation with HRP-conjugated secondary antibody (at a 1:10,000 dilution) for 1 h at room temperature, the membranes were developed using ECL and exposed to X-ray film.
siRNA knockdown assay
Cells were plated at a desired cell number and 24 h after plating, target siRNAs and non-targeting controls (scribe siRNA #1 Dharmacon, USA) were transfected at a final concentration of 10 nM. Lipofectamine RNAiMax Reagent and OptiMEM medium were used for knockdown experiments according to the manufacturer’s instructions (Invitrogen, USA). SMARTpool of FAM83-AS1 siRNA (Dharmacon) was used in this study. Knockdown efficiency was measured by qRT-PCR.
Cell proliferation, invasion and migration assays
Detailed experiments of these assays were described previously12. In short, for cell proliferation, cells were plated in 96-well plates at a desired concentration and transfected FAM83H-AS1 siRNA or non-targeting controls at 24 h after plating. At 96 h or 120 h after transfection with siRNAs, the proliferation rates were measured by Cell Proliferation Reagent (WST-1) (Roche) according to manufacturer’s instructions. For invasion and migration assays, cells were treated with the indicated siRNAs. After 48 h siRNA transfection, cells were seeded onto Boyden chambers (8 mm pore size, BD) present in the insert of a 24-well culture plate (Matrigel was used for invasion, purchased from BD). After 48 h, invasive cells located on the lower side of the chamber were stained with Diff-QuikTM Stain Set (SIEMENS).
Flow cytometry for cell cycle analysis
PC-9 and H1650 cells were collected after 48 h treatment with FAM83-AS1 siRNA and control siRNA. The cells were fixed with 70% ice-cold ethanol and washed with PBS. Cells were re-suspended in 1 ml of propidium iodide (PI) staining solution ((0.1% (v/v) Triton X-100, 10 μg/mL PI, and 100 μg/mL DNase-free RNase A in PBS), and then incubated for 30 minutes at room temperature in the dark. Cell cycle was measured in the University of Michigan flow cytometry core.
Statistical analysis
Data were analyzed using GraphPad Prism 6 (GraphPad software) and R software. The Receiver Operating Characteristic (ROC) curve analysis was used for diagnostic accuracy measured by the area under the curve (AUC). The Kaplan-Meier Survival curves and the log-rank test were used for survival analysis. Other data, such as proliferation, invasion, and migration, were evaluated by unpaired Student’s t-test. A two-tailed p value < 0.05 was considered significant. The correlation of proliferation related genes was analyzed with hierarchical cluster and heat map was visualized using Tree View software. To determine potential underlying biological processes associated with FAM83H-AS1 correlated (Pearson correlation) or regulated genes, Gene Ontology enrichment analysis was performed based on significantly correlated genes using the DAVID bioinformatics website46.
Additional Information
How to cite this article: Zhang, J. et al. Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci. Rep. 7, 42819; doi: 10.1038/srep42819 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (grant R01CA154365 to D.G.B. and A.M.C.; grant U01CA157715 to D.G.B.; grant R21CA205414 to G.C.), the University of Michigan’s Cancer Center Support Grant (P30 CA46592), University of Michigan’s Cancer Center Thoracic Oncology Program Research Grant (G.C.), a University of Michigan Department of Surgery RAC grant (G.C.), and the National Natural Science Foundation of China (NSFC) (81660324 to S.F. and 81360409 to S.B.).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.Z. performed the major experiments. J.Z., D.B., Y.G. and G.C. designed the study and wrote the manuscript. S.F., W.S., S.B., L.X., L.W. and D.T. provided technical help. G.C. performed statistical analysis. J.L., R.R., P.C., W.L. and A.C. provided tumor samples and scientific discussion. All authors contributed to the discussion and review of the manuscript.
References
- Siegel R. L., Miller K. D. & Jemal A. Cancer statistics, 2015. CA Cancer J Clin 65, 5–29, doi: 10.3322/caac.21254 (2015). [DOI] [PubMed] [Google Scholar]
- Herbst R. S., Heymach J. V. & Lippman S. M. Lung cancer. N Engl J Med 359, 1367–1380, doi: 10.1056/NEJMra0802714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoi N. et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol 32, 810–827, doi: 10.1097/PAS.0b013e31815cb162 (2008). [DOI] [PubMed] [Google Scholar]
- He Y. et al. MAP3K3 expression in tumor cells and tumor-infiltrating lymphocytes is correlated with favorable patient survival in lung cancer. Scientific reports 5, 11471, doi: 10.1038/srep11471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal E. et al. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Scientific reports 5, 12464, doi: 10.1038/srep12464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075, doi: 10.1038/nature07423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran S. M. et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nature communications 5, 5893, doi: 10.1038/ncomms6893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227, doi: 10.1038/nature07672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R. et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29, 742–749, doi: 10.1038/nbt.1914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J. T., Colognori D. & Lee J. T. Long noncoding RNAs: past, present, and future. Genetics 193, 651–669, doi: 10.1534/genetics.112.146704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbin O. A. et al. The landscape of antisense gene expression in human cancers. Genome Res 25, 1068–1079, doi: 10.1101/gr.180596.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 7, 11487–11499, doi: 10.18632/oncotarget.7203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792, doi: 10.1126/science.1240925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. Regulatory long non-coding RNA and its functions. J Physiol Biochem 68, 611–618, doi: 10.1007/s13105-012-0166-y (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular cell 49, 1083–1096, doi: 10.1016/j.molcel.2013.01.010 (2013). [DOI] [PubMed] [Google Scholar]
- Yang L. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500, 598–602, doi: 10.1038/nature12451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12, 433–446, doi: 10.1038/nrd4018 (2013). [DOI] [PubMed] [Google Scholar]
- Batista P. J. & Chang H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307, doi: 10.1016/j.cell.2013.02.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS genetics 9, e1003368, doi: 10.1371/journal.pgen.1003368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300, doi: 10.1038/nature10398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419, doi: 10.1016/j.cell.2010.06.040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076, doi: 10.1038/nature08975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E. et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742, doi: 10.1016/j.cell.2013.01.034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038, doi: 10.1038/nature09144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell 39, 925–938, doi: 10.1016/j.molcel.2010.08.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47, 199–208, doi: 10.1038/ng.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 34, 3076–3084, doi: 10.1038/onc.2014.236 (2015). [DOI] [PubMed] [Google Scholar]
- Seo J. S. et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 22, 2109–2119, doi: 10.1101/gr.145144.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550, doi: 10.1038/nature13385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. J. et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol 32, 933–940, doi: 10.1038/nbt.2943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok J. B. et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res 62, 2695–2698 (2002). [PubMed] [Google Scholar]
- Meseure D., Drak Alsibai K., Nicolas A., Bieche I. & Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. BioMed research international 2015, 320214, doi: 10.1155/2015/320214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Gao G. & Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers 2016, 9085195, doi: 10.1155/2016/9085195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F. et al. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem Biophys Res Commun 465, 293–298, doi: 10.1016/j.bbrc.2015.08.027 (2015). [DOI] [PubMed] [Google Scholar]
- Schmidt L. H. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 6, 1984–1992, doi: 10.1097/JTO.0b013e3182307eac (2011). [DOI] [PubMed] [Google Scholar]
- Roobol M. J. et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. European urology 58, 475–481, doi: 10.1016/j.eururo.2010.06.039 (2010). [DOI] [PubMed] [Google Scholar]
- Song H. et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 11, 225, doi: 10.1186/1479-5876-11-225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Yin C., Dang Y., Ye F. & Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Scientific reports 5, 11516, doi: 10.1038/srep11516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132, 330–342, doi: 10.1053/j.gastro.2006.08.026 (2007). [DOI] [PubMed] [Google Scholar]
- Kogo R. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71, 6320–6326, doi: 10.1158/0008-5472.CAN-11-1021 (2011). [DOI] [PubMed] [Google Scholar]
- Geng Y. J., Xie S. L., Li Q., Ma J. & Wang G. Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of international medical research 39, 2119–2128 (2011). [DOI] [PubMed] [Google Scholar]
- Gutschner T. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73, 1180–1189, doi: 10.1158/0008-5472.can-12-2850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PloS one 8, e77293, doi: 10.1371/journal.pone.0077293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardiello F. & Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 358, 1160–1174, doi: 10.1056/NEJMra0707704 (2008). [DOI] [PubMed] [Google Scholar]
- Riely G. J. et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 12, 839–844, doi: 10.1158/1078-0432.CCR-05-1846 (2006). [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.