ABSTRACT
Cyclic di-GMP was the first cyclic dinucleotide second messenger described, presaging the discovery of additional cyclic dinucleotide messengers in bacteria and eukaryotes. The GGDEF diguanylate cyclase (DGC) and EAL and HD-GYP phosphodiesterase (PDE) domains conduct the turnover of cyclic di-GMP. These three unrelated domains belong to superfamilies that exhibit significant variations in function, and they include both enzymatically active and inactive members, with a subset involved in synthesis and degradation of other cyclic dinucleotides. Here, we summarize current knowledge of sequence and structural variations that underpin the functional diversification of cyclic di-GMP turnover proteins. Moreover, we highlight that superfamily diversification is not restricted to cyclic di-GMP signaling domains, as particular DHH/DHHA1 domain and HD domain proteins have been shown to act as cyclic di-AMP phosphodiesterases. We conclude with a consideration of the current limitations that such diversity of action places on bioinformatic prediction of the roles of GGDEF, EAL, and HD-GYP domain proteins.
KEYWORDS: cyclic dinucleotide second messenger, GGDEF domain, EAL domain, HD-GYP domain, DHH/DHHA1 domain, cyclic GAMP, cyclic di-AMP, cyclic di-GMP, second messenger
INTRODUCTION
The dinucleotide cyclic di-GMP is the most abundant second messenger in bacteria. It promotes the environmental lifestyle switch between sessility and motility, as well as the host-related lifestyle switch between acute and chronic/benign infection. A hallmark of the cyclic di-GMP signaling network is an apparent redundancy of cyclic di-GMP turnover proteins encoded in one genome. However, many of these proteins have distinct N-terminal sensing and signaling domains, suggesting that their activities in cyclic di-GMP turnover respond posttranslationally to various (and different) intra- and extracellular signals. In gross terms, the number of cyclic di-GMP turnover proteins is linearly correlated with genome size within the different bacterial phyla, with Thermotogae having one of the highest cyclic di-GMP-related “IQs,” the density of enzymes per megabase pair, with some species harboring over 100 cyclic di-GMP turnover proteins (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). As in other domain superfamilies, extensive sequence diversity exists. Here, we review the knowledge on the translation of sequence diversity of cyclic di-GMP turnover proteins into functional diversity. We conclude by discussing whether and how a unified nomenclature for cyclic di-GMP turnover proteins can be established.
FUNCTIONAL DIVERSIFICATION OF THE GGDEF DOMAIN
The approximately 180-amino-acid-long GGDEF domain catalyzes the synthesis of cyclic di-GMP from two molecules of GTP with the release of pyrophosphate (Fig. 1) (1, 2). So far, the GGDEF domain is the only identified protein domain to carry out this specific condensation reaction. Even before functional characterization, the GGDEF domain was recognized to be a structural homologue of the adenylate cyclase domain, both belonging to the RRM (ferredoxin) fold palm domain family, which includes other enzymes forming 3′-5′ phosphodiester bonds, such as reverse transcriptases, class A and B DNA polymerases, and RNA-dependent RNA polymerases (3, 4). In approximately 40% of proteins, the GGDEF domain is coupled not only to an N-terminal signaling domain but also to a C-terminal EAL domain. Standalone GGDEF domains are rare and have not been characterized extensively (5). The GGDEF domain frequently possesses suboptimal catalytic activity and requires dimerization for the condensation reaction to occur at the active half-sites of the two monomers. Dimerization can be further promoted by allosteric activation of the N-terminal sensor domain (6). Various mechanisms of activation are emerging, reflecting the diversity of cytoplasmic, transmembrane, and periplasmic signaling domains, as well as linker and signal transducing domains, which are potentially associated with sequence diversification of the turnover domain (1, 6–10). Notably, the DgcZ (YdeH) DGC is an active dimer, with Zn2+ ion binding to inhibit the catalytic activity (10). GGDEF domains can be differentiated into three major classes: enzymatically functional domains; enzymatically functional domains, linked to an EAL domain; and enzymatically nonfunctional domains (Fig. 2) (11). This classification is based on the homology of the entire domain in combination with the conservation of the extended signature motif (Fig. 2) (12, 13).
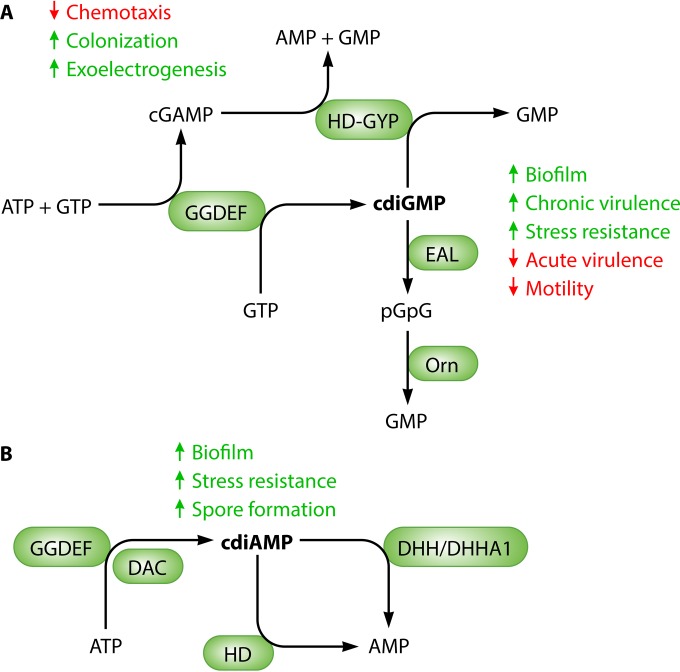
FIG 1.
Enzymes involved in the turnover of second messengers cyclic di-GMP (A) and cyclic di-AMP (B). GGDEF domain proteins are cyclic di-GMP synthases (2), but a few members can preferentially synthesize cyclic GMP-AMP (31). Cyclic di-GMP is degraded by EAL domain and HD-GYP domain phosphodiesterases into 5′-pGpG and GMP, respectively (66). 5′-pGpG is further hydrolyzed to GMP by the oligoribonuclease Orn. Cyclic di-AMP is synthesized by the DAC domain, and hydrolysis by HD and DHH/DHHA1 domain proteins has been demonstrated. Major phenotypes affected upon cyclic dinucleotide synthesis in many bacteria are indicated.
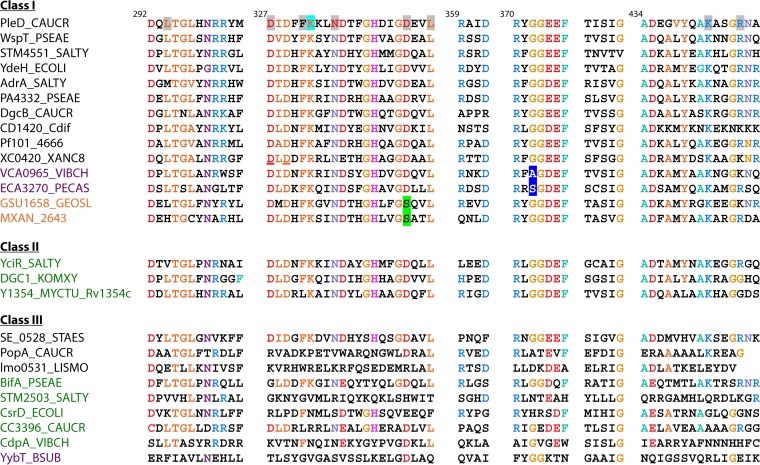
FIG 2.
Classification of GGDEF domains according to protein structure and conservation of signature motifs. Amino acids on a gray background interact with the substrate in the diguanylate cyclase PleD. K332, stabilizing the transition state, is on a cyan background. The RXXD I-site core motif is in blue. Unconventional amino acids still conferring enzymatic activity are on a blue background. Amino acids conferring cyclic GMP-AMP specificity are on a green background. Amino acids involved in protein-protein interactions are underlined. Conserved amino acids are color coded. GGDEF domain protein names are in black, and GGDEF-EAL proteins are in green. Unconventional GGDEF domain names are in violet, and cGMP-AMP-synthesizing proteins are in orange. Protein designations are given in the supplemental material. Modified from the work of Römling et al. (32).
KEY RESIDUES IN CATALYSIS AND ALLOSTERIC REGULATION
The GGDEF domain consists of the defining GG(D/E)EF sequence motif that includes the D/E catalytic base and other residues intimately involved in substrate binding and coordination of one of the two divalent cations (14). The position of the substrate GTP in the crystal structure(s) of GGDEF domain proteins indicates that the presence of the glycines provides space for the ribosyl sugar and phosphates, thus explaining the conservation of these residues (Fig. 3) (14, 15). In PleD, the most well-investigated diguanylate cyclase for which a crystal structure is available, the guanine base is bound in a pocket with N335 and D344 as key contact residues, curtailed by apolar side chains of L294, F331, and L247. D/E is the catalytic base, while K332 stabilizes the transition state. All those residues are well conserved in catalytically competent diguanylate cyclases (Fig. 2).
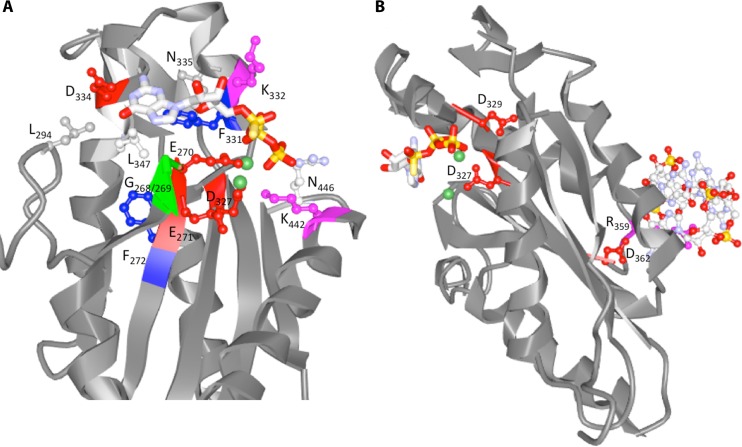
FIG 3.
Ribbon diagram of the GGDEF domain of PleD binding the substrate analog GTPαS (PDB code 2V0N). (A) Amino acids interacting with the substrate analog GTPαS (including Lys442 and Arg446, interacting with the phosphate group) and the Mg2+ ions (Asp327 and Glu370) (1, 6, 15) are indicated. Mg2+ ions are in green. (B) Amino acid motifs providing additional functionality to GGDEF domains. R359XXD362 is the core motif of the I-site. The XXD327XD329 motif was demonstrated to have a role in protein-protein interactions in GGDEF domain proteins others than PleD.
Nonfunctional GGDEF domains are usually characterized by a degenerate GGDEF motif, as any mutation within the GGDEF motif of catalytically active GGDEF domain proteins abolishes the catalytic activity, although there are exceptions. For example, the GGDEF domain protein of Staphylococcus aureus and Staphylococcus epidermidis with a well-conserved GGDEF motif has been experimentally proven to be nonfunctional (16). The structural basis of the nonfunctionality of staphylococcal GGDEF domains still remains an enigma. As to alteration in the signature motif, it is fairly common that GGDEF domains contain a degenerate GG(D/E)EF motif with the first G not conserved. Recent experimentally characterized proteins with a G→A or G→S substitution still exhibit significant functionality, demonstrating unexpected flexibility in the GGDEF containing active-site hairpin (Fig. 2) (17–19).
Besides the gross classification into catalytically active and nonactive GGDEF domains, the inhibitory site (I-site), designated by the central signature motif RXXD, is another functional feature which characterizes the activity profile (14, 20). The I-site, which is formed at an intra- or intermolecular interface bridged by a cyclic di-GMP dimer, variably extends beyond the central conserved RXXD cyclic di-GMP binding motif and mediates allosteric noncompetitive product inhibition, through feedback control of cyclic di-GMP synthesis (20, 21). The RXXD motif is absent in a proportion of GGDEF domains; alternative mechanisms to control cyclic di-GMP synthesis have been described for some of these proteins (6, 22, 23). A second recently discovered function of the I-site is the participation in protein-protein interaction with a cyclic di-GMP receptor, which ensures a stringent specificity of cyclic di-GMP signaling even in the presence of cyclic di-GMP production (21). In divergent GGDEF domain proteins (see below), a retained I-site in catalytically nonfunctional GGDEF domains converts these domains into cyclic di-GMP receptors (24–27). It should be noted that the enzymatic activity of the GGDEF domain can also be positively regulated by cooperative binding of the GTP substrate (19).
Some GGDEF domains have diverged to be enzymatically nonfunctional. These nonfunctional GGDEF domains can act as sensor domains that bind the substrate GTP, thereby allosterically regulating the enzymatic activity of a C-terminal EAL phosphodiesterase (28). In this way, the degenerate GGDEF motif is involved in allosteric control (20, 29). A surprisingly high catalytic plasticity has been demonstrated, as a highly degenerate GGDEF domain has been shown to display ATPase activity, albeit at suboptimal levels (30).
ALTERNATIVE CYCLIC DINUCLEOTIDES SYNTHESIZED BY GGDEF DOMAIN PROTEINS
A hallmark of binding of nucleotide and sugar derivatives to proteins is the low stringency of the specificity of the binding site. Accordingly, alteration of a few amino acids can alter the substrate specificity of nucleotides and sugars. Although it is the common perception that cyclic di-GMP synthases can be readily identified in bacterial genomes as being members of the GGDEF domain superfamily, GGDEF domain proteins that predominantly synthesize cyclic GMP-AMP, in parallel with cyclic di-GMP and cyclic di-AMP, have recently been identified (31). The relative specificity of cyclic GMP-AMP synthase activity as opposed to stringently using GTP as the substrate on this specific protein scaffold is determined by the amino acid serine, which has replaced aspartate at position 344 (designation according to PleD sequence), a key contact residue in the base binding pocket. As the exchange of aspartate for serine in an established diguanylate cyclase did not lead to the conversion into a cyclic GMP-AMP synthase, additional features of the protein scaffold must also contribute to substrate specificity.
SPECIFICITY IN REGULATORY ACTION
In general, GGDEF domains encoded by a single genome are functional paralogues, which have a low amino acid sequence identity/similarity, below 40%, while orthologues with identical domain structure and high sequence identity can be found even in distantly related species (32). One of the hallmarks of cyclic di-GMP signaling is a relative or absolute specificity of a phenotypic output of an individual chromosomally encoded GGDEF domain protein. This specificity is partly explained by the close proximity of signal production/degradation with receptor and/or effector proteins mediated through protein-protein interactions, a first example being the involvement of the I-site of a GGDEF domain in interaction with an EAL domain cyclic di-GMP receptor (21, 33). Interactions between the EAL domain protein YciR and diguanylate cyclase YdaM control a key step in E. coli biofilm formation through a suggested modulation of localized cyclic di-GMP levels (34). Functionality is also provided, however, by specific protein-protein interactions that are independent of the catalytic activity (19, 35). In this case, the XXDXDX motif, which is highly conserved in GGDEF domains, is required for the interaction with the HD-GYP domain. HD-GYP–GGDEF complex formation serves to control motility through recruitment of a PilZ domain protein and interaction with the pilus biogenesis machinery (35, 36). Overall, these data indicate that GGDEF domain proteins possess several protein interaction interfaces which participate in the formation of supramolecular complexes.
FUNCTIONAL DIVERSIFICATION OF THE EAL DOMAIN
The EAL domain was the first identified cyclic di-GMP-specific phosphodiesterase and remains the most well characterized (Fig. 4 and 5; see also Fig. S1 in the supplemental material) (2, 37, 38). The product of EAL phosphodiesterase activity is the dinucleotide 5′-pGpG, while hydrolysis of 5′-pGpG into GMP is considered to be too slow to be physiologically relevant. EAL phosphodiesterases require a divalent cation for enzymatic activity, which in most cases is a Mg2+ or Mn2+ ion, while Ca2+ and Zn2+ efficiently inhibit the enzymatic activity (39, 40). Catalytically active EAL domains usually have a high substrate affinity in the physiological nanomolar range, and cyclic di-GMP binding can increase the dimerization affinity (41). Although monomers can be catalytically active, dimerization substantially enhances protein stability and catalytic activity (37).
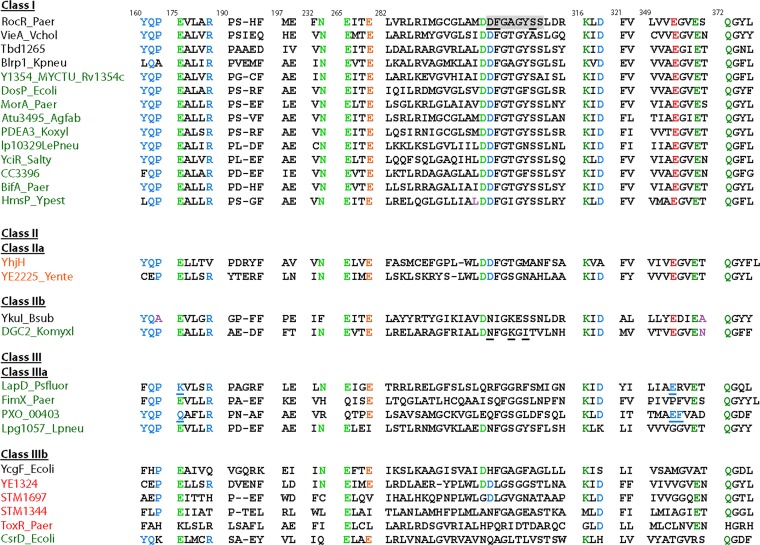
FIG 4.
Classification of EAL domains according to protein structure and conservation of signature motifs. The catalytic base glutamate is shown in red. Green, amino acids involved in Mg2+ binding; blue, amino acids involved in substrate binding. The glutamate-stabilizing loop 6 is shown in orange. Loop 6 amino acids are on a gray background. Mutated loop 6 amino acids in class I PleD, loop 6 amino acids determinative for lack of catalytic activity of the EAL domain in DGC2_Komyxl, and alternative amino acids involved in cyclic dinucleotide binding in class IIIA proteins are underlined. Names of EAL proteins are in black, EAL-only proteins are in red, and GGDEF-EAL proteins are in green. Protein designations are given in the supplemental material. Modified from the work of Römling et al. (32).
FIG 5.
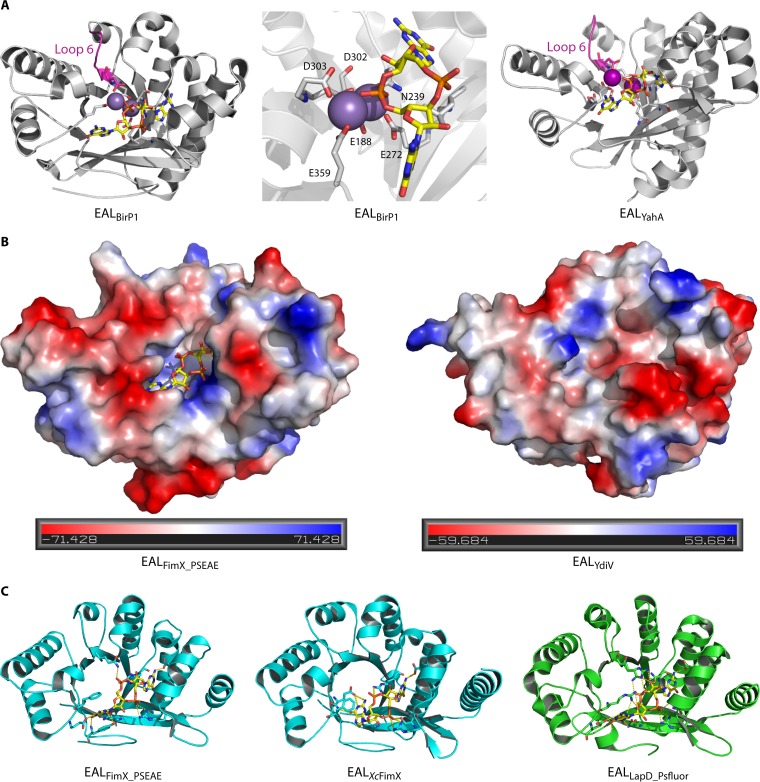
Substrate binding by EAL domains. (A) Ribbon diagram structure of the EAL domains of BlrP1, a fully functional class I PDE activated by light (44), and YahA (41) binding to the substrate cyclic di-GMP. In the middle is an enlarged view of the cyclic di-GMP binding site of BlrP1. Cations are shown in violet and pink. Cyclic di-GMP is shown as sticks with carbon atoms colored yellow. (B) Comparison of electrostatic surface representations of class III EAL domains FimX of P. aeruginosa and YdiV of E. coli. While the cyclic di-GMP binding site of class IIIa FimX is conserved (model is shown with cyclic di-GMP bound), the cyclic di-GMP binding pocket is not conserved in class IIIb member YdiV. The electrostatic surface potential shows highly electronegative (red) and electropositive (blue) patches of the two proteins. (C) Ribbon diagram structure of three class IIIa cyclic di-GMP binding EAL domains: EALFimX_PSEAE (Q9HUK6 of P. aeruginosa), EALXcFimX (A0A0H2X6E4 of Xanthomonas campestris pv. campestris), and EALLapD_Psfluor (Q3KK31 of Pseudomonas fluorescens Pf0-1). Note the different conformations and binding modes of cyclic di-GMP, which is displayed as sticks with carbon atoms in yellow, oxygen in red, phosphate in orange, and nitrogen in blue.
KEY RESIDUES FOR ACTIVITY
Systematic alanine substitutions of conserved signature amino acids have given insights into the catalytic mechanism, even before a crystal structure had become available (42, 43). That work showed that the EAL motif is part of a larger conserved signature motif that is required for catalytic activity, including amino acids required for binding of divalent cations, the substrate, and catalysis. In addition, a flexible loop (loop 6) extensively characterized in (β/α)8 barrel proteins (see below) mediates dimerization and controls substrate and cation binding, thus being required for catalytic activity (42, 44). The findings from this mutagenesis study enabled the differentiation of EAL domains in three classes, catalytically active, potentially catalytically active, and catalytically inactive EAL domains (32, 42), thus facilitating the prediction of the function of further EAL domains. Based on the functional characterization of additional EAL domains, further subclassifications can be made (Fig. 4).
The crystal structures of several EAL domain-containing proteins revealed that these proteins possess a protein fold variant of the (β/α)8 TIM-barrel structure, arranged as eight alternating alpha-helices and beta-strands (Fig. 5) (44). This arrangement of secondary structures is found in over 50 diverse protein superfamilies (45). The functionality of this highly conserved arrangement of secondary structures is highly flexible, as these protein families bind different substrates and catalyze different reactions. In case of the light-inducible phosphodiesterase Blrp1 of Klebsiella pneumoniae, interdomain interaction between the sensor domain and a nonconserved connector in the EAL domain of only four amino acids in length controls the catalytic activity in response to light (Fig. 5A) (44).
CLASSIFICATION OF DIVERGENT DOMAIN MEMBERS
As with the GGDEF domain, the EAL domain superfamily contains diverged members. Most EAL domains are class I EAL domains, which possess a N-terminal signaling domain and feature substantial, but still suboptimal, catalytic activity in the nonactivated state, requiring the correct positioning of conserved loop 6 (42, 44). Class II EAL domains potentially possess catalytic activity with deviations of some amino acids from the conserved signature motifs; they are most poorly characterized. Of note, catalytically active EAL-only domain proteins comprise a specific subgroup within the class II family. Class III EAL domains can already be recognized by bioinformatic analysis to be catalytically inactive, since class III domains possess deviations from the conserved signature motifs of active enzymes in several determinative positions. Nevertheless, some class III domains can still bind cyclic di-GMP, thus serving as cyclic di-GMP receptors (class IIIa), whereas others are unable to bind the dinucleotide (class IIIb) (Fig. 5B).
Cyclic di-GMP binding and nonbinding EAL domains cannot be distinguished with certainty (Fig. 4). However, in both cases, several conserved signature amino acids are missing, and loop 6 is not conserved. Binding of cyclic di-GMP to a receptor EAL domain allosterically controls subsequent events. In the conserved Lap system with the GGDEF-EAL receptor LapD, interactive inside-out/outside-in signals mediated by the HAMP domain couple cytoplasmic cyclic di-GMP binding to reinforcement of periplasmic protein-protein interactions controlling, e.g., periplasmic proteolysis of cell surface proteins (46, 47). Interestingly, homologous GGDEF-EAL receptors have variations in their cyclic di-GMP binding sites and bind cyclic di-GMP in different conformations, which reflects the structural polymorphism of this second messenger (48, 49), as well as binding site flexibility (Fig. 5C) (50). Such polymorphisms make it still challenging to predict cyclic di-GMP binding residues by bioinformatics.
Catalytically inactive noncyclic di-GMP binding EAL proteins function solely through protein-protein interactions. Several well-investigated class IIIb proteins of Escherichia coli and Salmonella enterica serovar Typhimurium, YdiV and Salmonella-specific STM1697, bind to the major flagellin regulator FlhDC with apparently similar but highly distinct interfaces (51–53). Furthermore, the class IIIb protein YdiV interacts in complex with FlhDC with the ClpXP protease, guiding FlhDC for degradation (54), and it regulates other physiological traits besides motility (55).
REGULATION OF DUAL-FUNCTION DIGUANYLATE CYCLASE PHOSPHODIESTERASES
Of particular complexity is the regulation of the activity of GGDEF-EAL domain proteins in cases where both domains are catalytically functional (56). Notably, the three DGCs and three PDEs of Komagataeibacter xylinus that affect cellulose production, the first biological function recognized to be affected by cyclic di-GMP signaling, are GGDEF-EAL domain proteins, and both domains are predicted to be functional by bioinformatics analysis (39). Differential regulation of the catalytic activity of these domains can include allosteric regulation by ligand binding, signal perception, or protein-protein interactions, which favor one catalytic activity over the other (7, 57–60), but could also include a combination of regulatory mechanisms, such as proteolytic cleavage in combination with signal perception (61). This points to a multifactorial regulation of catalytic activity in vivo. However, catalytically active domains can even predominantly affect certain aspects of physiology through protein-protein interactions. For example, the GGDEF-EAL phosphodiesterase YciR of E. coli affects the expression of csgD, a major biofilm regulator, through interaction with a DGC and a transcriptional regulator, which inhibits biofilm formation (34).
PHOSPHODIESTERASE INVOLVED IN pGpG DEGRADATION
The observation that the EAL domain hydrolyzes cyclic di-GMP into 5′-pGpG (Fig. 1) has raised the question of the possible cellular role and fate of this dinucleotide product (62). As an inhibitor of the enzymatic activity of particular EAL domain proteins, this molecule potentially impinges on cyclic di-GMP levels and signaling. Furthermore, it has been suggested that this nanoRNA (i.e., RNA oligonucleotide of ≤5 nucleotides) is a signaling molecule in its own right and is involved in the initiation of transcription by RNA polymerase (63). Two classes of enzymes are implicated in 5′-pGpG degradation: a subgroup of HD-GYP domain phosphodiesterases that can hydrolyze both cyclic di-GMP and 5′-pGpG (see below), and the oligoribonuclease Orn, recently identified as the primary degradative enzyme for 5′-pGpG in Pseudomonas aeruginosa (64, 65). Homologues of Orn are widely distributed in bacteria, although Cohen and colleagues (65) identified over 200 species that lack an Orn homolog but have EAL and HD-GYP domain proteins, as well as over 100 species that lack both an Orn homolog and EAL domain proteins but have HD-GYP domain proteins. Thus, in some bacteria, HD-GYP domain proteins may influence cyclic di-GMP levels both directly, by hydrolysis of the nucleotide, and indirectly, by preventing product inhibition of the activity of EAL domain enzymes.
FUNCTIONAL DIVERSIFICATION OF THE HD-GYP DOMAIN
There are fewer studies of HD-GYP domain proteins than those of the GGDEF and EAL domains. Although well-studied model organisms harbor mostly EAL domain phosphodiesterases, the HD-GYP domain is one-third as abundant throughout the phylogenetic tree (https://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). The prototype of an HD-GYP domain protein is the response regulator RpfG from Xanthomonas campestris (36, 66). This protein is part of a two-component system that affects the expression of multiple virulence functions in this plant pathogen (67, 68). In vitro, RpfG converts cyclic di-GMP to GMP via the intermediate 5′-pGpG dependent on Mn2+ (66, 69). An alanine substitution within the signature HD dyad leads to a loss of both enzyme activity and regulatory action (66). In contrast, although alanine substitutions in the signature GYP motif have little or no effect on enzyme activity, they do counteract the interaction of RpfG with particular GGDEF domain proteins to modulate a specific subset of RpfG-mediated phenotypes (35, 66, 70).
DIVERSITY IN METAL BINDING
The crystal structure of the enzymatically active HD-GYP phosphodiesterase from Persephonella marina EX-H1 (PmGH) unexpectedly showed a trinuclear Fe center with iron in two redox states, as Fe(II) and central Fe(III), buried at the bottom of the cavity forming the c-di-GMP binding site (Fig. 6) (71). In general, the HD domain superfamily of enzymes has been shown to catalyze phosphomonoesterase and phosphodiesterase reactions, depending on their catalytic metal center being mono- or binuclear, respectively. Variations in the metallic center of the HD-GYP domain were seen in the structure of the unconventional catalytically inactive Bd1817 from Bdellovibrio bacteriovorus (72), and PA4781, a two-component regulatory protein from Pseudomonas aeruginosa (73), which harbor binuclear metal centers, although of a distinct nature.
FIG 6.
Substrate binding by the HD-GYP domain of PmGH. (A) Surface representation of the PmGH HD-GYP domain monomer subunit showing the binding cavity for cyclic di-GMP, which is represented in stick mode and colored by atom type. (B) Superposition of the structures of PmGH bound to cyclic di-GMP and GMP. Both nucleotides are shown in stick mode. Bonding interactions are represented by dashed lines. The central metal iron has been labeled as the middle site (M) and the two flanking metal sites have been labeled H and G to reflect their proximity to the HD and GYP motifs, respectively. Residues that interact with cyclic di-GMP include Y285 of the GYP motif. Red spheres represent solvent and SIN-1, a succinate molecule (71).
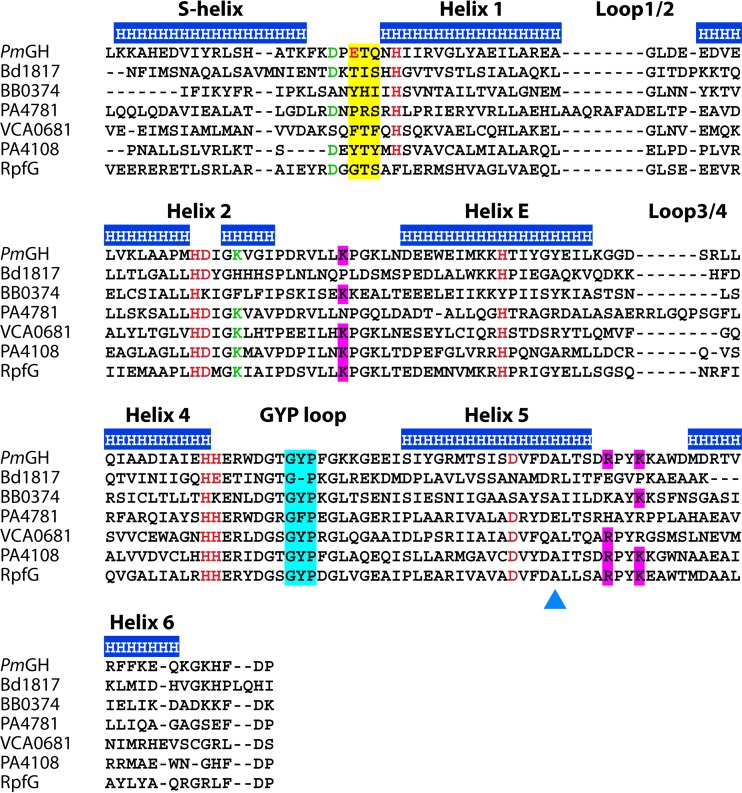
A phylogenetic comparison of HD-GYP domains showed a distinct separation into two evolutionary groups independent of the type of associated regulatory and/or sensory domains (71), with seven out of the eight PmGH metal ligand residues shared (Fig. 7) (71). The variable ligand which corresponds to E185 in PmGH is embedded in the signature motif E/D-T-G for the PmGH subfamily. E185 has been predicted to be determinative for a three-metal center valency (71, 74). Conversely, the other subfamily primarily presents a tyrosine or phenylalanine (Y/F) and lacks a unique signature. The separation of HD-GYP proteins into these two subfamilies is not entirely clear-cut, though (Fig. 7) (73, 75). For example, RpfG from X. campestris, despite phylogenetically clustering within the E/D-T-G subgroup, aligns a glycine in place of the E/D residue, as well as variation in an H-site metal ligand (Fig. 7). Thus, RpfG is more likely to possess a binuclear metal ion center.
FIG 7.
Primary sequence alignment of HD-GYP domains from proteins that have been characterized structurally and/or enzymatically reveals the diversity within the domain. Protein designations are given in the supplemental material. The top line indicates the helices in the structure of PmGH, with an annotation of the interhelix loops. Metal ligands are given in red, and proposed catalytic residues are given in green. The GYP motif is highlighted in cyan and the substrate binding ligands are in magenta. The region of the sequences with consensus motifs E(D)TG/YTY is highlighted in yellow. Note that these are not fully conserved. The triangle points to the E residue in PA4781 that may act in steric hindrance of cyclic di-GMP binding.
Recent work has provided evidence that the differences in the occupancy of the metal site and the redox status affect catalysis (74). The activity of VCA0681 requires Fe(II) at the bimetallic center, and derivatives with Fe(III) are inactive, suggesting that the activity of this protein is redox regulated (76). Also, isolated TM0186 from Thermotoga maritima with two Fe(III) atoms is inactive; reduction to Fe(II) enables the enzyme to generate 5′-pGpG but not GMP. Additional supplementation with either Mn(II) or Fe(II) leads to the production of GMP. The phylogenetic clustering of TM0186 within the E/D-T-G subgroup of HD-GYP domain proteins suggests that it has a trimetallic center. Furthermore, a variant protein with an alanine substitution of the glutamate generates only 5′-pGpG as a product. The findings point to the association of a trimetallic center with the ability to generate GMP from 5′-pGpG. Also, the action of HD-GYP domains in converting 5′-pGpG to GMP suggests regulation by the intracellular availability of metals and metal site occupancy. Finally, catalytically inactive SO2541 HnoD from Shewanella oneidensis and PA2572 from P. aeruginosa are variant at the HD dyad (SE and YN, respectively) and have only 1 conserved residue involved in metal chelation (77, 78); as a result, these proteins may exert their effect through protein interactions involving the GYP motif (77, 78).
DIVERSITY IN SUBSTRATE BINDING AND CATALYSIS
Determination of the structure of PmGH in complex with the substrate cyclic di-GMP and final reaction product, GMP, has revealed the mode of binding and shed light on the possible catalytic mechanism (71, 79). Adequate space is available for the substrate to bind and both hydrolysable phosphates to interact with the metal center to sequentially hydrolyze cyclic di-GMP to GMP. Cyclic di-GMP is bound in a cis conformation (71), in contrast to the more extended conformation observed when cyclic di-GMP is bound to EAL domain proteins (80) or predicted in binding to the HD-GYP domain protein PA4108 (81).
The structural analysis of the PmGH–cyclic di-GMP complex shows that the bound cyclic dinucleotide interacts with the central (M-site) Fe(III) and is involved in diverse hydrogen bonds and hydrophobic interactions (Fig. 6). As in RpfG, in PmGH alanine substitutions of six residues involved in metal binding in addition to the HD dyad (H221 and D222) (Fig. 7) essentially abolish or markedly reduce the phosphodiesterase activity. Alanine mutations of other conserved residues near the metal center (D183, D308, and K225) have a similar impact on activity (71). Alanine substitutions of residues implicated in cyclic di-GMP recognition do not, however, result in a substantial decrease in catalytic activity (71). The proposed enzymatic mechanism is that M-site Fe(III) directly interacts with a nonbridging oxygen of one of the scissile phosphate diesters of cyclic di-GMP to provide a strong Lewis acid catalyst, whereas a metal-activated bridging hydroxide ion of the M-H Fe pair is the likely nucleophile for the hydrolysis of the scissile bond (71). The occurrence of a hydroxide ion-bridging ligand is consistent with the metal-ligand bond lengths (72, 82). The structure does not reveal how the O3′ leaving group is protonated, however.
The structure of PA4781 reveals potential steric hindrance of cyclic di-GMP binding by a glutamate at position 314 (73). Accordingly, the purified enzyme has a relatively low affinity for cyclic di-GMP (Km, ∼120 μM) compared to 5′-pGpG (Km, ∼27 μM). In other enzymatically active HD-GYP domain proteins, position 314 is occupied by an alanine (Fig. 7), and an E314A variant of PA4781 shows substantially enhanced affinity for cyclic di-GMP (81). Detailed kinetic analyses indicate that PA4781 has low enzymatic activity but hydrolyzes 5′-pGpG more effectively than cyclic di-GMP (81). Although similar kinetic experiments on other HD-GYP domain proteins have not been reported, the available evidence suggests that differences in the relative activity against 5′-pGpG compared to cyclic di-GMP do occur (66, 69, 71, 76, 77, 83, 84).
STRUCTURAL INSIGHTS INTO THE MULTIFUNCTIONAL ROLES OF HD-GYP DOMAINS
A sequence-based analysis identified the GYP signature motif of HD-GYP proteins as part of a larger widely conserved motif, HHEXXDGXGYP (66). The PmGH structure suggests an extension of this consensus motif to HHEXXDGXGYPXXXXXXXI, to include a conserved isoleucine residue (I294 in PmGH) that stabilizes the structure of the loop by hydrophobic interactions with G284 from the GYP motif (71). The structural conservation of the GYP loop (Fig. 6) (73) between PmGH and PA4781 suggests that it is integral to the functions(s) of HD-GYP domain proteins. The GYP motif is critical for protein-protein interactions of RpfG with specific GGDEF domain proteins in X. campestris but is not necessary for the phosphodiesterase activity (66).
The available evidence suggests that the HD-GYP domain of RpfG can also interact with proteins of other classes, including the transcriptional regulator NtrC (36, 85). Furthermore, the enzymatically inactive HD-GYP domain response regulator HnoD can inhibit the activity of the EAL domain response regulator HnoB to regulate cyclic di-GMP levels in Shewanella oneidensis (77). The mechanistic basis of this inhibition is not known. Different HD-GYP domain proteins within the same organism may interact with different partners in vivo, although this remains to be tested experimentally.
The structure of the PmGH HD-GYP complex with cyclic di-GMP reveals that Y285 of the GYP motif is placed inside the substrate binding pocket, where it H-bonds to cyclic di-GMP (Fig. 6). This presents a conundrum for the action of RpfG. If GGDEF domains interact directly with Y285, they need to intercalate with the inner side of the HD-GYP nucleotide binding pocket. This would prevent cyclic di-GMP binding and phosphodiesterase activity, although such effects have not been observed in vitro (35). An intriguing alternative is that RpfG involvement in protein-protein complexes is determined not only by cyclic di-GMP binding but also by conformational alterations associated with cyclic di-GMP degradation, which would be “reported” via the GYP loop. In this way, RpfG would act as a trigger enzyme for protein complex formation and regulation, similar to what is suggested for the EAL domain protein YciR of Escherichia coli (34). However, mutation of the HD dyad of the HD-GYP domain of RpfG does not significantly affect its in vivo interaction with GGDEF domain proteins, as revealed by fluorescent resonance energy transfer (FRET) analysis (35). Only further work can reveal whether particular regulatory actions of HD-GYP domain proteins occur independently of their ability to bind or hydrolyze cyclic di-GMP or 5′-pGpG.
FURTHER SUBSTRATES FOR HD-GYP DOMAIN PROTEINS
In addition to cyclic di-GMP, bacteria have been shown to utilize cyclic di-AMP and, most recently, the dinucleotide 3′3′-cyclic GMP-AMP as intracellular signal molecules. The 3′3′-cyclic GMP-AMP molecule was discovered in Vibrio cholerae as a regulator of chemotaxis and of factors contributing to colonization of the intestine (86). A screen of potential phosphodiesterases for 3′3′-cyclic GMP-AMP from V. cholerae identified three HD-GYP domain proteins, VCA0210, VCA0681, and VCA0931, which were capable of hydrolysis of the cyclic dinucleotide into 5′-pApG, with VCA0681 having an additional 5′-nucleotidase activity to generate 5′-ApG (87). The nucleotidase and phosphodiesterase activities were associated with the HD and HD-GYP domains, respectively, which are present in tandem (87). All three proteins hydrolyze 3′3′-cyclic GMP-AMP specifically, with no activity against other cyclic GMP-AMP forms with different phosphodiester linkages, to include the mammalian innate immunity regulator 2′3′-cyclic GMP-AMP. Variant VCA0681 proteins with alanine substitutions in the signature HD dyad and GYP motif have no detectable activity (87), in contrast to the role of the GYP motif in PmGH and RpfG (70, 71).
FUNCTIONAL DIVERSIFICATION OF CYCLIC DI-AMP PHOSPHODIESTERASES
The functional diversification also extends to other cyclic dinucleotide signaling networks. As the currently most prominent example, DHH/DHHA1 proteins usually function as phosphatase or phosphodiesterases for hydrolyzing a wide variety of substrates that range from pyrophosphate to single-stranded DNA (ssDNA). The substrate specificity of DHH/DHHA1 enzymes is usually governed by the DHHA1 domain rather than the DHH domain. A bioinformatics search of potential phosphodiesterases for cyclic di-AMP, a universally essential cyclic dinucleotide second messenger in Gram-positive bacteria (88, 89), led to the discovery of a DHH domain protein (YybT or GdpP) from Bacillus subtilis as a cyclic di-AMP phosphodiesterase (30). GdpP is a metal ion-dependent phosphodiesterase that breaks down cyclic di-AMP into 5′-pApA at physiologically relevant substrate (micromolar) concentrations. In accordance with its specificity toward cyclic di-AMP, the DHHA1 domain of GdpP does not share significant sequence homology with the DHHA1 domains of other DHH/DHHA1 proteins. Importantly, a number of Arg residues critical for the binding of polyphosphate, RNA, or ssDNA in other DHHA1 domain proteins (e.g., RecJ and YtqI) are not conserved in YybT. Another DHH/DHHA1 protein (Pde2) that lacks the PAS and GGDEF domains of GdpP and degrades cyclic di-AMP into AMP was discovered in Streptococcus pneumoniae (90). Pde2 is an ortholog of B. subtilis YtqI (also named NrnA) that was claimed to be responsible for degrading nanoRNA (RNA oligonucleotides of ≤5 nucleotides) and dephosphorylating pAp to AMP (91, 92).
In addition to the DHH/DHHA1 proteins, a subfamily of HD domains possesses cyclic di-AMP phosphodiesterase activity. The first example is the Listeria monocytogenes protein PgpH (93). Biochemical and structural studies revealed binding of cyclic di-AMP with high affinity (Kd [dissociation constant], 0.3 to 0.4 μM) and hydrolysis to 5′-pApA in the presence of divalent metal ions, such as Mn2+ and Fe2+.
The discovery of the DHH/DHHA1- and HD domain-based phosphodiesterases for degrading cyclic di-AMP mirrors the converging evolution of the EAL and HD-GYP domains involved in cyclic di-GMP degradation. Although the structural basis for the recognition of cyclic di-AMP by the PDEs remains to be fully defined, the crystal structure of the standalone DHH/DHHA1 protein Rv2837c in complex with the hydrolytic intermediate 5-pApA suggests that a set of residues from both DHH and DHHA1 domains contribute to the binding of cyclic di-AMP (94). Even assuming that only two families of cyclic di-AMP phosphodiesterases are found in nature, identification of the members of the two families by bioinformatics should still proceed with caution, and experimental validation is necessary.
CONCLUDING REMARKS
As outlined above, diversity in the functions of the GGDEF, EAL, and HD-GYP domains is evident in terms of enzymatic activity, the ability to synthesize or degrade alternate dinucleotides, as well as in interactions with other proteins. This functional diversity certainly extends to other cyclic dinucleotide turnover proteins, such as the DHH/DHHA1 enzymes. Further biochemical and structural work is required to gain knowledge of the molecular bases for the substrate specificity or preference. Work on stringent cyclic mononucleotide synthases shows that quite limited variations give rise to different specificities; cyclic GMP synthases can be experimentally changed to cyclic AMP synthases, and vice versa, by just two or three amino acid exchanges (95, 96). On the other hand, relaxed enzymes can produce several different cyclic nucleotides (97). In addition, a three-amino-acid replacement in the human cyclic dinucleotide synthase cyclic GMP-AMP synthase (cGAS) changes the phosphodiester linkage specificity so that 3′3′ cyclic GMP-AMP rather than the noncanonical 2′3′ cyclic GMP-AMP is synthesized (98). The three new residues incorporated were the determinative amino acids in DncV, a bacterial homolog of cyclic GMP-AMP synthase (98). Indeed, ancient cGAS is a 3′3′ cyclic GMP-AMP synthase (99). As outlined above, distinct GGDEF domain proteins that have been shown to produce cyclic GMP-AMP (31) and some HD-GYP domain phosphodiesterases can have cyclic GMP-AMP hydrolytic activity (87). Changes in substrate specificity similar to those within the GGDEF and HD-GYP domain protein families could also occur within the EAL domain. In addition, novel enzymes with cyclic dinucleotide turnover activity might be recognized. Recently, CpdB, which displays a diffusion-limited reaction rate in 3′-AMP hydrolysis, was also shown to hydrolyze cyclic di-AMP with a reasonable turnover rate (100). With the current stage of knowledge, it thus appears difficult to assign substrate specificity and product outcome with certainty by bioinformatics. Thus, current species-specific nomenclatures might limit comparisons to distantly related species, which frequently harbor orthologous proteins, while functional paralogues of dinucleotide turnover proteins dominate within a species. The elucidation of the structures of cyclic di-GMP turnover domains in complex with other cyclic di-GMP turnover domains and other interacting proteins will also be necessary to provide a deeper understanding of the regulatory action of the diversity of these families of signaling proteins and to fully explore their true functions. This is certainly the case for those proteins that may be multifunctional and which may regulate different functions through protein-protein interactions and modulation of cyclic di-GMP levels.
Supplementary Material
ACKNOWLEDGMENTS
Cyclic di-GMP research of U.R. is funded by the Swedish Research Council for Natural Sciences and Engineering (grant 621-2013-4809). Research in Z.-X.L.'s laboratory is supported by a tier II ARC grant from MOE, Singapore. The work in the laboratory of J.M.D. has been supported in part by grants awarded by Science Foundation Ireland (SFI 07/IN.1/B955, SFI 07/IN.1/B955/IRPs, and SFI 11/TIDA/B2036) and the Wellcome Trust (project grant WT093314MA).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00790-16.
REFERENCES
- 1.Schirmer T. 2016. c-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol 428:3683–3701. doi: 10.1016/j.jmb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 2.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 3.Anantharaman V, Iyer LM, Aravind L. 2010. Presence of a classical RRM-fold palm domain in Thg1-type 3′-5′nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol Direct 5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei J, Grishin NV. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210–216. doi:. [DOI] [PubMed] [Google Scholar]
- 5.Deepthi A, Liew CW, Liang ZX, Swaminathan K, Lescar J. 2014. Structure of a diguanylate cyclase from Thermotoga maritima: insights into activation, feedback inhibition and thermostability. PLoS One 9:e110912. doi: 10.1371/journal.pone.0110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. 2007. Structure of BeF3−-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Mills E, Petersen E, Kulasekara BR, Miller SI. 2015. A direct screen for c-di-GMP modulators reveals a Salmonella Typhimurium periplasmic l-arginine-sensing pathway. Sci Signal 8:ra57. doi: 10.1126/scisignal.aaa1796. [DOI] [PubMed] [Google Scholar]
- 8.De N, Navarro MV, Raghavan RV, Sondermann H. 2009. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol 393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone JG, Jaeger T, Manfredi P, Dotsch A, Blanka A, Bos R, Cornelis GR, Haussler S, Jenal U. 2012. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog 8:e1002760. doi: 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21:1149–1157. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Seshasayee AS, Fraser GM, Luscombe NM. 2010. Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res 38:5970–5981. doi: 10.1093/nar/gkq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulesekara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Römling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci 62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland LM, O'Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O'Gara JP. 2008. A staphylococcal GGDEF domain protein regulates biofilm formation independently of c-di-GMP. J Bacteriol 190:5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter JL, Severin GB, Koestler BJ, Waters CM. 2014. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol 14:22. doi: 10.1186/1471-2180-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Mendoza D, Coulthurst SJ, Humphris S, Campbell E, Welch M, Toth IK, Salmond GP. 2011. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase. Mol Microbiol 82:719–733. doi: 10.1111/j.1365-2958.2011.07849.x. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira MC, Teixeira RD, Andrade MO, Pinheiro GM, Ramos CH, Farah CS. 2015. Cooperative substrate binding by a diguanylate cyclase. J Mol Biol 427:415–432. doi: 10.1016/j.jmb.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J Biol Chem 281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 21.Dahlstrom KM, Giglio KM, Sondermann H, O'Toole GA. 2016. The inhibitory site of a diguanylate cyclase is a necessary element for interaction and signaling with an effector protein. J Bacteriol 198:1595–1603. doi: 10.1128/JB.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang CY, Chin KH, Chuah ML, Liang ZX, Wang AH, Chou SH. 2011. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site. Acta Crystallogr D Biol Crystallogr 67:997–1008. doi: 10.1107/S090744491104039X. [DOI] [PubMed] [Google Scholar]
- 23.Rao F, Pasunooti S, Ng Y, Zhuo W, Lim L, Liu AW, Liang ZX. 2009. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal Biochem 389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Ozaki S, Schalch-Moser A, Zumthor L, Manfredi P, Ebbensgaard A, Schirmer T, Jenal U. 2014. Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol Microbiol 94:580–594. doi: 10.1111/mmi.12777. [DOI] [PubMed] [Google Scholar]
- 25.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koseoglu VK, Heiss C, Azadi P, Topchiy E, Guvener ZT, Lehmann TE, Miller KW, Gomelsky M. 2015. Listeria monocytogenes exopolysaccharide: origin, structure, biosynthetic machinery and c-di-GMP-dependent regulation. Mol Microbiol 96:728–743. doi: 10.1111/mmi.12966. [DOI] [PubMed] [Google Scholar]
- 27.Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 29.Tamayo R, Schild S, Pratt JT, Camilli A. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect Immun 76:1617–1627. doi: 10.1128/IAI.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallberg ZF, Wang XC, Wright TA, Nan B, Ad O, Yeo J, Hammond MC. 2016. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′,3′-cGAMP). Proc Natl Acad Sci U S A 113:1790–1795. doi: 10.1073/pnas.1515287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O'Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6(6):e01978-15. doi: 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan RP, McCarthy Y, Kiely PA, O'Connor R, Farah CS, Armitage JP, Dow JM. 2012. Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol Microbiol 86:557–567. doi: 10.1111/mmi.12000. [DOI] [PubMed] [Google Scholar]
- 36.Ryan RP, Dow JM. 2010. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence 1:404–408. doi: 10.4161/viru.1.5.12704. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. 2010. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol 402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. 2014. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J Biol Chem 289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. 2009. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol 191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 45.Toth-Petroczy A, Tawfik DS. 2014. The robustness and innovability of protein folds. Curr Opin Struct Biol 26:131–138. doi: 10.1016/j.sbi.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee D, Cooley RB, Boyd CD, Mehl RA, O'Toole GA, Sondermann H. 2014. Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP. eLife 3:e03650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chin KH, Kuo WT, Yu YJ, Liao YT, Yang MT, Chou SH. 2012. Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein. Acta Crystallogr D Biol Crystallogr 68:1380–1392. doi: 10.1107/S0907444912030594. [DOI] [PubMed] [Google Scholar]
- 49.Guzzo CR, Dunger G, Salinas RK, Farah CS. 2013. Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol 425:2174–2197. doi: 10.1016/j.jmb.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 50.Chou SH, Galperin MY. 2016. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang HW, Gu L. 2012. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res 40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, Anwar N, Chuah ML, Lunsdorf H, Frank R, Rhen M, Liang ZX, Lindqvist Y, Romling U. 2013. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella Typhimurium. Mol Microbiol 90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- 53.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J Bacteriol 193:1600–1611. doi: 10.1128/JB.01494-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol Microbiol 83:1268–1284. doi: 10.1111/j.1365-2958.2012.08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spurbeck RR, Alteri CJ, Himpsl SD, Mobley HL. 2013. The multifunctional protein YdiV represses P fimbria-mediated adherence in uropathogenic Escherichia coli. J Bacteriol 195:3156–3164. doi: 10.1128/JB.02254-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427. doi: 10.1099/mic.0.053892-0. [DOI] [PubMed] [Google Scholar]
- 57.Feirer N, Xu J, Allen KD, Koestler BJ, Bruger EL, Waters CM, White RH, Fuqua C. 2015. A pterin-dependent signaling pathway regulates a dual function diguanylate cyclase-phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens. mBio 6(4):e00156-15. doi: 10.1128/mBio.00156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber B, Riedel K, Kothe M, Givskov M, Molin S, Eberl L. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol Microbiol 46:411–426. doi: 10.1046/j.1365-2958.2002.03182.x. [DOI] [PubMed] [Google Scholar]
- 59.Qi Y, Rao F, Luo Z, Liang ZX. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 60.Lahiri T, Luan B, Raleigh DP, Boon EM. 2014. A structural basis for the regulation of an H-NOX-associated cyclic-di-GMP synthase/phosphodiesterase enzyme by nitric oxide-bound H-NOX. Biochemistry 53:2126–2135. doi: 10.1021/bi401597m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 62.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 63.Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. 2011. NanoRNAs prime transcription initiation in vivo. Mol Cell 42:817–825. doi: 10.1016/j.molcel.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, Lee VT. 2015. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38:986–1003. [DOI] [PubMed] [Google Scholar]
- 68.Ryan RP. 2013. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 159:1286–1297. doi: 10.1099/mic.0.068189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Wei C, Jiang W, Wang L, Li C, Wang Y, Dow JM, Sun W. 2013. The HD-GYP domain protein RpfG of Xanthomonas oryzae pv. oryzicola regulates synthesis of extracellular polysaccharides that contribute to biofilm formation and virulence on rice. PLoS One 8:e59428. doi: 10.1371/journal.pone.0059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. 2010. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc Natl Acad Sci U S A 107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellini D, Caly DL, McCarthy Y, Bumann M, An SQ, Dow JM, Ryan RP, Walsh MA. 2014. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol 91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. 2011. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio 2(5):e00163-11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinaldo S, Paiardini A, Stelitano V, Brunotti P, Cervoni L, Fernicola S, Protano C, Vitali M, Cutruzzola F, Giardina G. 2015. Structural basis of functional diversification of the HD-GYP domain revealed by the Pseudomonas aeruginosa PA4781 protein, which displays an unselective bimetallic binding site. J Bacteriol 197:1525–1535. doi: 10.1128/JB.02606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miner KD, Kurtz DM Jr. 2016. Active site metal occupancy and cyclic di-GMP phosphodiesterase activity of Thermotoga maritima HD-GYP. Biochemistry 55:970–979. doi: 10.1021/acs.biochem.5b01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun 79:3273–3283. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miner KD, Klose KE, Kurtz DM Jr. 2013. An HD-GYP cyclic di-guanosine monophosphate phosphodiesterase with a non-heme diiron-carboxylate active site. Biochemistry 52:5329–5331. doi: 10.1021/bi4009215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell 46:449–460. doi: 10.1016/j.molcel.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryan RP, Lucey J, O'Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. 2009. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 11:1126–1136. doi: 10.1111/j.1462-2920.2008.01842.x. [DOI] [PubMed] [Google Scholar]
- 79.Wigren E, Liang ZX, Römling U. 2014. Finally! The structural secrets of a HD-GYP phosphodiesterase revealed. Mol Microbiol 91:1–5. doi: 10.1111/mmi.12463. [DOI] [PubMed] [Google Scholar]
- 80.Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stelitano V, Giardina G, Paiardini A, Castiglione N, Cutruzzola F, Rinaldo S. 2013. c-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG. PLoS One 8:e74920. doi: 10.1371/journal.pone.0074920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown PM, Caradoc-Davies TT, Dickson JM, Cooper GJ, Loomes KM, Baker EN. 2006. Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism. Proc Natl Acad Sci U S A 103:15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKee RW, Kariisa A, Mudrak B, Whitaker C, Tamayo R. 2014. A systematic analysis of the in vitro and in vivo functions of the HD-GYP domain proteins of Vibrio cholerae. BMC Microbiol 14:272. doi: 10.1186/s12866-014-0272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rajeev L, Luning EG, Altenburg S, Zane GM, Baidoo EE, Catena M, Keasling JD, Wall JD, Fields MW, Mukhopadhyay A. 2014. Identification of a cyclic-di-GMP-modulating response regulator that impacts biofilm formation in a model sulfate reducing bacterium. Front Microbiol 5:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MC, Ramos CH, Farah CS. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv. citri. Mol Microbiol 62:537–551. doi: 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- 86.Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao J, Tao J, Liang W, Zhao M, Du X, Cui S, Duan H, Kan B, Su X, Jiang Z. 2015. Identification and characterization of phosphodiesterases that specifically degrade 3′3′-cyclic GMP-AMP. Cell Res 25:539–550. doi: 10.1038/cr.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis σM in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol 83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. 2013. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol 195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. 2009. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res 37:5114–5125. doi: 10.1093/nar/gkp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. 2007. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res 35:4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. 2015. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A 112:E747–756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He Q, Wang F, Liu S, Zhu D, Cong H, Gao F, Li B, Wang H, Lin Z, Liao J, Gu L. 2016. Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase. J Biol Chem 291:3668–3681. doi: 10.1074/jbc.M115.699801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. 2010. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem 285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tucker CL, Hurley JH, Miller TR, Hurley JB. 1998. Two amino acid substitutions convert a guanylyl cyclase, RetGC-1, into an adenylyl cyclase. Proc Natl Acad Sci U S A 95:5993–5997. doi: 10.1073/pnas.95.11.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Belyy A, Raoux-Barbot D, Saveanu C, Namane A, Ogryzko V, Worpenberg L, David V, Henriot V, Fellous S, Merrifield C, Assayag E, Ladant D, Renault L, Mechold U. 2016. Actin activates Pseudomonas aeruginosa ExoY nucleotidyl cyclase toxin and ExoY-like effector domains from MARTX toxins. Nat Commun 7:13582. doi: 10.1038/ncomms13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kranzusch PJ, Lee AS, Wilson SC, Solovykh MS, Vance RE, Berger JM, Doudna JA. 2014. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158:1011–1021. doi: 10.1016/j.cell.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, Vance RE. 2015. Ancient origin of cGAS-STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol Cell 59:891–903. doi: 10.1016/j.molcel.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.López-Villamizar I, Cabezas A, Pinto RM, Canales J, Ribeiro JM, Cameselle JC, Costas MJ. 2016. The characterization of Escherichia coli CpdB as a recombinant protein reveals that, besides having the expected 3′-nucleotidase and 2′,3′-cyclic mononucleotide phosphodiesterase activities, it is also active as cyclic dinucleotide phosphodiesterase. PLoS One 11:e0157308. doi: 10.1371/journal.pone.0157308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.