ABSTRACT
Aerobic methanotrophic bacteria use methane as their sole source of carbon and energy and serve as a major sink for the potent greenhouse gas methane in freshwater ecosystems. Dissecting the molecular details of how these organisms interact in the environment may increase our understanding of how they perform this important ecological role. Many bacterial species use quorum sensing (QS) systems to regulate gene expression in a cell density-dependent manner. We have identified a QS system in the genome of Methylobacter tundripaludum, a dominant methane oxidizer in methane enrichments of sediment from Lake Washington (Seattle, WA). We determined that M. tundripaludum produces primarily N-3-hydroxydecanoyl-l-homoserine lactone (3-OH-C10-HSL) and that its production is governed by a positive feedback loop. We then further characterized this system by determining which genes are regulated by QS in this methane oxidizer using transcriptome sequencing (RNA-seq) and discovered that this system regulates the expression of a putative nonribosomal peptide synthetase biosynthetic gene cluster. Finally, we detected an extracellular factor that is produced by M. tundripaludum in a QS-dependent manner. These results identify and characterize a mode of cellular communication in an aerobic methane-oxidizing bacterium.
IMPORTANCE Aerobic methanotrophs are critical for sequestering carbon from the potent greenhouse gas methane in the environment, yet the mechanistic details of chemical interactions in methane-oxidizing bacterial communities are not well understood. Understanding these interactions is important in order to maintain, and potentially optimize, the functional potential of the bacteria that perform this vital ecosystem function. In this work, we identify a quorum sensing system in the aerobic methanotroph Methylobacter tundripaludum and use both chemical and genetic methods to characterize this system at the molecular level.
KEYWORDS: methane, methanotroph, quorum sensing, sociomicrobiology, acyl-homoserine lactone, biosynthetic gene cluster
INTRODUCTION
Aerobic methane-oxidizing bacteria (methanotrophs) are an important component of the carbon cycle that serve to sequester carbon from the potent greenhouse gas methane after it is produced by anaerobic microbial ecosystems (1, 2). Methanotrophs provide a carbon and energy source for communities of organisms that cannot oxidize methane themselves, thereby serving as key supporting species in the environment (3, 4). The methanotroph Methylobacter tundripaludum is a member of the Gammaproteobacteria (type I methanotroph), and it has been repeatedly identified as a dominant member of methane enrichment and stable isotope probing experiments of sediment from Lake Washington, Seattle, WA (3, 5–7). Methylobacter spp. have also been isolated from other geographically distinct regions, including the arctic (8), estuaries (9), and temperate wetlands (10), highlighting the diverse environments inhabited by this genus. We, therefore, use M. tundripaludum as part of a model system for understanding the function of aerobic methane-oxidizing bacterial communities.
Despite the critical ecosystem function performed by aerobic methanotrophs, little is known about the molecular mechanisms that mediate interactions within methane-oxidizing bacterial communities. Understanding these interactions is important in order to maintain, and potentially optimize, the functional potential of these groups. One well-studied bacterial interaction is quorum sensing (QS), which bacteria use to regulate gene expression in a density-dependent manner using diffusible signaling molecules (for review, see references 11 and 12).
In many species of Proteobacteria, QS systems use acyl-homoserine lactones (AHLs) produced by members of the LuxI family of AHL synthases. These signal molecules are bound by members of the LuxR family of transcription factors, which control gene expression in an AHL concentration-dependent manner. In many cases, AHLs control the production of extracellular factors, including proteases and antibiotics, which may allow resources to be dedicated to these metabolically expensive products solely under conditions in which the bacterial species has reached a sufficient density to impact its surrounding environment (13–15).
QS is thought to enable host-associated bacterial species to differentiate between low-density/free-living and high-density/host-associated states. The ecological function of QS systems that have been identified in non-host-associated species are not as well understood. Further analysis of the genes controlled by QS in these non-host-associated bacteria may help improve our understanding of how these species interact with other cells and their environment.
In this work, we identify and characterize a QS system in the methane-oxidizing bacterium M. tundripaludum strain 21/22. We identify the AHL signal as well as its cognate synthase and receptor. Furthermore, we determine that a secondary metabolite cluster is activated by QS in M. tundripaludum, and we identify an extracellular product controlled by QS. To our knowledge, a QS system has never before been characterized in a methanotroph. This characterization of the M. tundripaludum QS system, therefore, extends our knowledge of this widespread form of chemical communication.
RESULTS
Identification of quorum sensing genes flanking a putative nonribosomal peptide synthetase cluster in Methylobacter tundripaludum 21/22.
We identified QS genes in the recently sequenced genome of Methylobacter tundripaludum strain 21/22 (Fig. 1A) (5). This included a luxI family member, mbaI (T451DRAFT_0796), as well as a luxR family member, mbaR (T451DRAFT_0820). MbaI shares 68% amino acid sequence identity with the AHL synthase from the acidophilic bacterium Acidithiobacillus thiooxidans, strains of which have been reported to produce primarily N-3-oxo-octanoyl-l-homoserine lactone (3-oxo-C8-HSL) (16).
FIG 1.
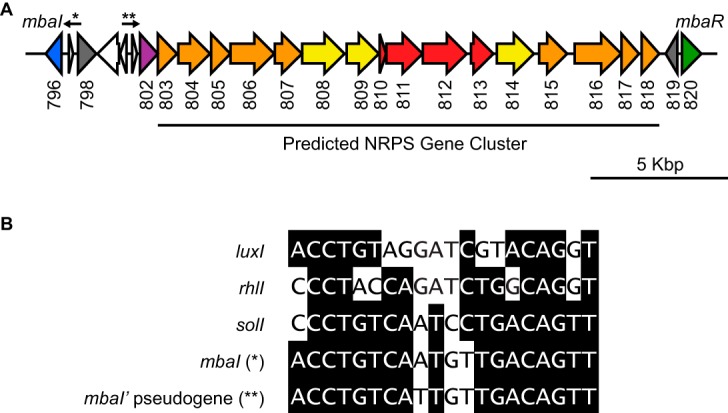
A quorum sensing system in Methylobacter tundripaludum. (A) The quorum sensing genes in Methylobacter tundripaludum flank a predicted nonribosomal peptide synthetase (NRPS) biosynthetic gene cluster. The arrows and asterisks identify putative MbaR-binding sites shown in Fig. 1B. The numbers below gene arrows are locus tags (T451DRAFT_0XXX) corresponding to the Joint Genome Institute Integrated Microbial Genomes (IMG) system (41). Due to space constraints, locus tags for transposase/integrase genes (white) have been omitted. Genes are colored as follows according to predicted function: luxI family AHL synthase gene mbaI, blue; luxR family transcription factor gene mbaR, green; antiSMASH-classified core NRPS genes, red (from left to right, acyl carrier protein, condensation domain, adenylation domain, and esterase genes); other biosynthetic genes, orange; efflux pump genes, yellow; transposase/integrase genes, white; mbaI' pseudogene, purple; and other genes, gray. (B) Comparison of putative MbaR-binding sites in front of the AHL synthase gene mbaI and the pseudogene mbaI' with known LuxR-type binding sites in the promoter sequences of Vibrio fischeri luxI, Pseudomonas aeruginosa rhlI, and Ralstonia solanacearum solI.
Interestingly, in M. tundripaludum, the QS genes flank a predicted nonribosomal peptide synthetase (NRPS) cluster (17), which we identified using the bioinformatics tool antiSMASH (antibiotics and secondary metabolite analysis shell) (Fig. 1A) (18). antiSMASH classifies several genes in this cluster as core NRPS genes, including genes encoding an acyl carrier protein, condensation and adenylation domains, and an esterase. These core genes are flanked by efflux pumps as well as additional biosynthetic genes. This cluster as well as the QS genes are also conserved in other M. tundripaludum isolates, including strains 31/32 (5) and SV96 (19) (see Fig. S1 in the supplemental material).
The putative NRPS cluster has a G+C content of 41%, while mbaI and mbaR more closely match the 49% G+C content of the rest of the genome (see Fig. S2 in the supplemental material). There are also several pseudogenes upstream of the cluster, including transposase fragments and an mbaI' pseudogene (T451DRAFT_0802) that contains several stop codons and does not contain all of the catalytic residues required for signal synthesis (Fig. 1A) (20). The genome of the related methanotroph Methylobacter whittenburyi also contains a QS system with mbaI and mbaR, and the products of these genes share high sequence identity with those in M. tundripaludum (79% and 69% amino acid identity, respectively) (21) (Fig. S1). However, in M. whittenburyi, this locus does not contain the predicted NRPS cluster, transposase fragments, or mbaI' pseudogene. These observations led us to speculate that this biosynthetic gene cluster is a potential genomic island (22) that was either inserted into the genome of M. tundripaludum or lost in other known members of the Methylobacter genus.
The Methylobacter tundripaludum quorum sensing signal is N-3-hydroxydecanoyl-l-homoserine lactone.
We sought to characterize the QS system in M. tundripaludum 21/22 by constructing a bioassay for QS signal detection. To do this, we took advantage of the fact that luxI-type synthase genes are often activated by QS, resulting in positive autoregulation of the system (12). We identified a candidate MbaR-binding site upstream of mbaI that conformed to the NNCTG-N10-CAGNN pattern and dyad symmetry typically recognized by LuxR-type transcription factors (Fig. 1B) (23). We then constructed a two-plasmid reporter system in Escherichia coli in which mbaR is expressed on one plasmid under its native promoter and the mbaI promoter is fused to gfp (PmbaI-gfp) on a separate vector.
We extracted the supernatant from a wild-type (WT) culture of M. tundripaludum 21/22 with ethyl acetate and added this extract to the PmbaI-gfp reporter strain, resulting in a 4-fold increase in green fluorescent protein (GFP) fluorescence compared to a solvent control (see Fig. S3 in the supplemental material). This suggests that an AHL signal is produced by this methane-oxidizing bacterium. Furthermore, the addition of this extract to a strain containing the PmbaI-gfp plasmid but not the mbaR-expressing plasmid resulted in no increase in GFP fluorescence, demonstrating that this process is MbaR dependent and that MbaR activates the expression of the synthase gene mbaI upon signal binding in a possible positive feedback loop (Fig. S3).
We separated the signal-containing extract by C18 reverse-phase high-performance liquid chromatography (HPLC) and assayed each fraction using the PmbaI-gfp reporter strain, resulting in a single peak of activity (Fig. 2A). This activity was not present in the supernatant extract from a ΔmbaI mutant, confirming that MbaI is the signal synthase and suggesting that the signal is an AHL (Fig. 2A). To determine the identity of the predicted AHL, we used tandem mass spectrometry to scan for parent ions in the ethyl acetate extract while monitoring for the production of the telltale lactone fragment ([M+H]+ = 102). We detected two peaks that produced this fragment (Fig. 3A). The parent [M+H]+ of the major peak is 272, which is consistent with the mass of N-3-hydroxydecanoyl-l-homoserine lactone (3-OH-C10-HSL) (Fig. 3B and D). The minor peak, which is approximately an order of magnitude smaller, has a parent [M+H]+ of 300, which is consistent with the mass of N-3-hydroxydodecanoyl-l-homoserine lactone (3-OH-C12-HSL) (Fig. 3C and E). The retention times and mass spectra of these purified compounds were indistinguishable from commercial standards (data not shown).
FIG 2.
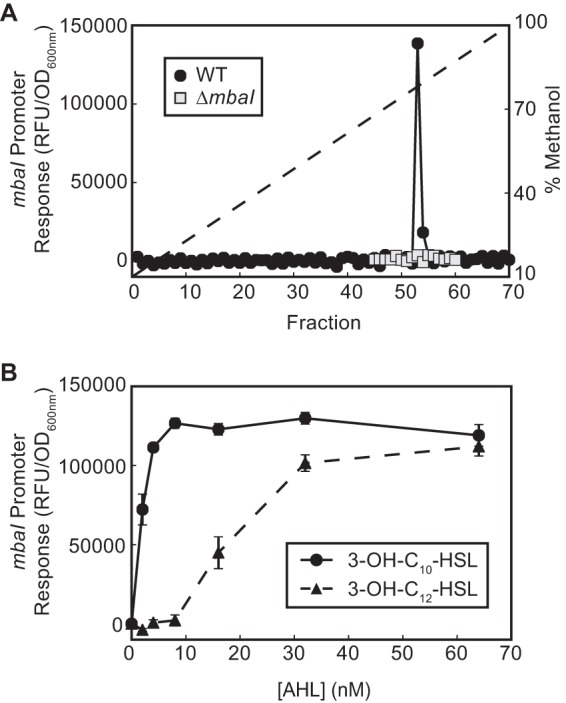
The M. tundripaludum quorum sensing system is active. (A) PmbaI-gfp activity of HPLC-fractionated culture supernatant extracts from wild-type and ΔmbaI M. tundripaludum strains. The dashed line shows the methanol gradient. (B) Dose response of the PmbaI-gfp E. coli reporter strain to 3-OH-C10-HSL and 3-OH-C12-HSL.
FIG 3.
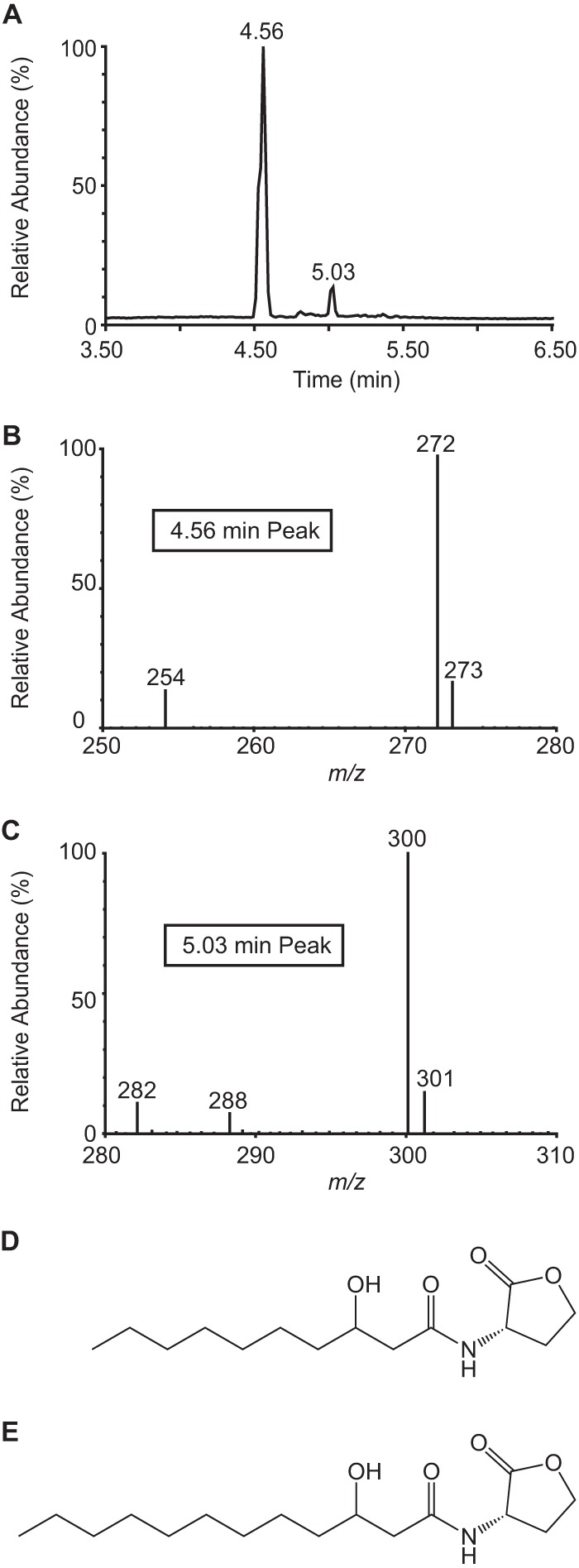
Tandem mass spectrometry reveals that M. tundripaludum produces primarily 3-OH-C10-HSL. (A) Scan for parent ions of homoserine lactone fragment ([M+H]+ = 102). (B, C) Parent mass spectrum for peaks at 4.56 min (B) and 5.03 min (C) in parent ion scan. (D, E) Structures of 3-OH-C10-HSL (D) and 3-OH-C12-HSL (E).
To determine whether these AHLs have biological activity, we tested commercial standards in the PmbaI-gfp reporter strain. MbaR is responsive to both 3-OH-C10-HSL and 3-OH-C12-HSL but is more sensitive to the former (half maximal activation with ∼3 nM and ∼25 nM signal, respectively) (Fig. 2B), which is consistent with this signal being the most abundant in the M. tundripaludum supernatant. Together, these results demonstrate that the QS system of the aerobic methanotroph M. tundripaludum 21/22 produces and responds primarily to the AHL 3-OH-C10-HSL (Fig. 3D).
Kinetics and magnitude of quorum sensing signal production.
In order to determine which genes are regulated by the M. tundripaludum QS system, we used the following approach shown to be successful in studies of other bacterial species: comparing the transcriptomes of an AHL synthase mutant grown in the presence and absence of added exogenous AHL (for example, see reference 24). To use this strategy with the M. tundripaludum ΔmbaI mutant, we determined how much 3-OH-C10-HSL to add and at which time points to harvest RNA for analysis.
Because the PmbaI-gfp E. coli reporter responds in a dose-dependent manner to the QS signal (Fig. 2B), we used it to quantify 3-OH-C10-HSL levels in the supernatant of a wild-type M. tundripaludum culture over the course of a growth curve (Fig. 4A). We saw maximum accumulation (approximately 330 nM) of the 3-OH-C10-HSL signal during stationary phase. Next, to determine the sensitivity of MbaR to 3-OH-C10-HSL in its native host, we constructed an M. tundripaludum reporter strain wherein the signal-responsive open reading frame (ORF) mbaI was replaced with the catechol dioxygenase reporter gene xylE (25). This mbaI::xylE reporter strain was responsive to as little as 3 nM 3-OH-C10-HSL (Fig. 4B).
FIG 4.
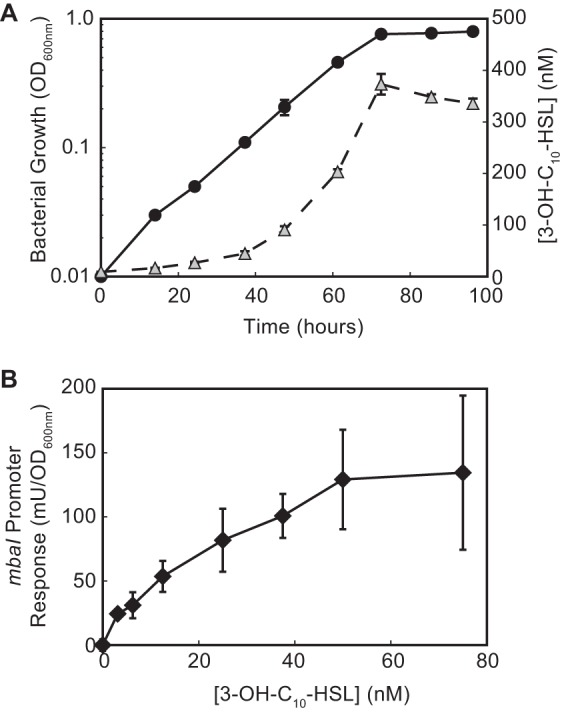
AHL production and detection in M. tundripaludum. (A) 3-OH-C10-HSL accumulation (triangles) during M. tundripaludum growth (circles). (B) M. tundripaludum mbaI::xylE strain response to 3-OH-C10-HSL. One unit (U) of catechol dioxygenase activity represents a change in absorbance (375 nm) per minute.
The predicted NRPS gene cluster is regulated by quorum sensing.
With these results in hand, we determined a minimal QS regulon of M. tundripaludum using transcriptome sequencing (RNA-seq) by analyzing differential gene expression in the ΔmbaI mutant in the presence or absence of 3-OH-C10-HSL. We chose to add 1 μM 3-OH-C10-HSL to ensure that the signal concentration would remain above the maximum concentration observed in a wild-type culture of ∼300 nM throughout the course of the experiment (Fig. 4A). Based on the mbaI::xylE reporter results (Fig. 4B), we extracted RNA from late-log-phase cultures (at an optical density at 600 nm [OD600] of ∼0.5) when the amount of signal produced by the wild-type strain had passed the threshold required to activate the mbaI promoter. We also examined differential gene expression in the early stationary phase (at an OD600 of ∼0.9), when the maximum AHL concentration occurs in a wild-type culture, to determine if additional genes are regulated at a later growth stage.
In the log-phase culture, the expression of 28 genes was ≥2.5-fold higher in the presence of the QS signal (Table 1). Many of these genes were grouped into clusters that may represent operons. The genes with the greatest increase in expression were located in the predicted NRPS gene cluster flanked by mbaI and mbaR. The expression of the first gene in the cluster, a haloacid dehalogenase (HAD) superfamily hydrolase gene (T451DRAFT_0803), was 28-fold higher in the presence of the signal. Interestingly, transcripts of the mbaI' pseudogene (T451DRAFT_0802) were also 4-fold higher in the presence of the signal (Table 1; see also Fig. S4 in the supplemental material). Reverse transcription-PCR confirmed that the mbaI' pseudogene and the hydrolase gene are transcribed together in the same operon (see Fig. S5 in the supplemental material).
TABLE 1.
Genes with increased expression in M. tundripaludum 21/22 ΔmbaI in the presence of 1 μM 3-OH-C10-HSL in late log phase
| Gene locus (T451DRAFT_) | Product | Fold change (+AHL/−AHL) | Padja |
|---|---|---|---|
| 0202 | Hypothetical protein | 10.2 | 2.6E−93 |
| 0203 | Transcriptional regulator, TetR family | 2.8 | 5.2E−22 |
| 0204 | Hypothetical protein | 3.4 | 4.7E−65 |
| 0796 | LuxI family acyl-homoserine lactone synthase MbaI | NAb | NA |
| 0802 | Product of mbaI' pseudogene | 4.2 | 5.9E−31 |
| 0803 | HAD superfamily subfamily IB hydrolase, TIGR01490 | 28.0 | 4.1E−239 |
| 0804 | Delta-aminolevulinic acid dehydratase | 18.6 | 2.0E−263 |
| 0805 | A-factor biosynthesis repeat protein | 11.8 | 2.8E−230 |
| 0806 | 2-Polyprenyl-6-methoxyphenol hydroxylase-like oxidoreductase | 7.0 | 7.7E−71 |
| 0807 | Predicted oxidoreductase, aryl-alcohol dehydrogenase-like protein | 4.3 | 8.6E−56 |
| 0808 | MFSc transporter, DHA2 family, multidrug resistance protein | 4.3 | 5.9E−62 |
| 0809 | Membrane fusion protein, multidrug efflux system | 3.5 | 2.0E−59 |
| 0810 | Acyl carrier protein | 3.4 | 2.8E−27 |
| 0811 | Putative polyketide synthase component | 3.2 | 9.3E−29 |
| 0812 | Long-chain acyl coenzyme A synthetase | 3.2 | 1.2E−35 |
| 0813 | Predicted hydrolase or acyltransferase of alpha/beta superfamily | 2.7 | 1.1E−17 |
| 0814 | Outer membrane protein | 2.9 | 1.2E−44 |
| 1642 | Cation/multidrug efflux pump | 3.0 | 4.4E−34 |
| 1643 | RND family efflux transporter, membrane fusion protein subunit | 3.6 | 6.1E−40 |
| 1644 | Efflux transporter, outer membrane factor lipoprotein, NodT family | 3.5 | 4.2E−35 |
| 1645 | Transcriptional regulator, TetR family | 3.6 | 2.2E−28 |
| 1678 | Hypothetical protein | 2.8 | 5.6E−27 |
| 1875 | RNA polymerase primary sigma factor | 3.0 | 1.1E−19 |
| 1876 | Membrane protease subunit HflK | 2.8 | 6.2E−16 |
| 1972 | Hypothetical protein | 9.5 | 8.6E−128 |
| 2702 | Outer membrane protein, multidrug efflux system | 4.5 | 2.1E−112 |
| 2703 | Multidrug efflux pump | 6.5 | 6.9E−111 |
| 2704 | Membrane fusion protein, multidrug efflux system | 6.9 | 3.7E−102 |
Padj, adjusted P value.
NA, not applicable. An accurate fold change cannot be computed for mbaI (T451DRAFT_0796) because these experiments were done in a ΔmbaI strain.
MFS, major facilitator superfamily.
We identified a potential MbaR-binding site upstream of the mbaI' pseudogene, centered approximately 50 bp before the transcription start site (Fig. 1B). This binding site contains only one mismatch compared to the one upstream of mbaI and has strong dyad symmetry, further supporting the idea that the mbaI' pseudogene, and consequently the predicted NRPS gene cluster, is regulated by QS. We also observed that the ΔmbaI mutant grows faster than the wild-type strain (9.7 ± 0.1 h versus 12.2 ± 0.5 h of generation time, respectively), suggesting that cell growth slows down after induction of these genes.
Other gene clusters with higher expression in the presence of the AHL signal included multiple efflux pump and hypothetical genes, as well as genes that encode transcriptional regulators (Table 1). However, while these gene clusters had reproducibly higher expression when signal was added, we were not able to identify putative MbaR-binding sites upstream of the approximate transcriptional start sites (see Materials and Methods for identification criteria).
When maximal signal production is reached in the early stationary phase, a subset of the same genes differentially expressed during log phase still had higher expression in the presence of the signal (see Table S3 in the supplemental material). The only additional genes with higher expression when signal was added coded for two hypothetical polypeptides, both of which narrowly missed the 2.5-fold cutoff in the log-phase analysis. Neither of these genes have a predicted MbaR-binding site. Twenty-two additional genes were downregulated at least 2.5-fold as well at this later time point, including multiple gene clusters related to redox reactions (see Table S4 in the supplemental material). Together, these results demonstrate a QS regulon in the aerobic methanotroph M. tundripaludum.
A product with strong UV absorbance is present in M. tundripaludum supernatants in a quorum-sensing-dependent manner.
In the process of separating the ethyl acetate extract of the M. tundripaludum supernatant over an HPLC gradient to identify the AHL signal (Fig. 2), we observed a product with a large absorbance maximum of 297 nm that eluted after 48 min (Fig. 5), prior to the AHL. This product was not detectable in the supernatant from the ΔmbaI strain but was present when the ΔmbaI strain was cultured with the addition of 3-OH-C10-HSL, suggesting that its production is QS dependent. The collected product had no discernible growth-inhibitory activity against E. coli MG1655 or Bacillus subtilis PY79. Together, these results suggest that the QS system in M. tundripaludum regulates the production of an extracellular factor with strong UV absorbance.
FIG 5.
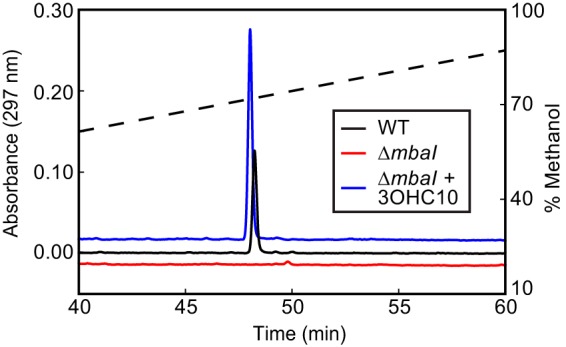
A product with strong UV absorbance is detected in M. tundripaludum culture supernatant in a quorum-sensing-dependent manner. UV absorbance (297 nm) of supernatant extract from cultures of M. tundripaludum wild-type (WT) and ΔmbaI strains and M. tundripaludum ΔmbaI with 1 μM 3-OH-C10-HSL separated by HPLC. The dashed line shows the methanol gradient. The absorbance traces are vertically offset for clarity.
DISCUSSION
Many species use QS systems to regulate gene expression in a cell density-dependent manner; however, this form of cellular communication has not been extensively studied in methanotrophs. Here, we describe an AHL-based QS system in M. tundripaludum. M. tundripaludum produces primarily 3-OH-C10-HSL, a signal that is also produced by environmental isolates of other bacterial species, including Burkholderia thailandensis (14) and Pseudomonas fluorescens (26). This opens up the possibility of cross talk between bacterial species in methane-oxidizing communities, as both Burkholderiales and pseudomonads have also been identified in enrichments from Lake Washington sediment (3, 7).
We discovered that in M. tundripaludum QS controls primarily the expression of a predicted NRPS gene cluster flanked by mbaI and mbaR in the genome. It is notable that the mbaI' pseudogene immediately upstream of this biosynthetic gene cluster is also transcribed and that there is a predicted MbaR-binding site upstream of this pseudogene that is nearly identical to the one upstream of the bona fide mbaI gene (Fig. 1B; see also Fig. S4 in the supplemental material). It is possible that mbaI and its upstream region were duplicated and that this led to the control of the biosynthetic gene cluster by QS.
Although the mbaI' pseudogene is transcribed, any resulting gene product does not appear to have AHL synthase activity, as there is no detectable AHL activity produced by the ΔmbaI mutant strain (Fig. 2A). This does not rule out another possible function for the product of mbaI'. In Methylobacterium extorquens AM1, which can utilize C1 compounds, including methanol and methylamine but not methane, the product of a truncated I gene termed tslI controls the expression of the AHL synthase gene msaI and, therefore, indirectly regulates QS in this bacterium (27).
The product of the predicted NRPS cluster and its function are unknown. We detected an ethyl acetate-extractable factor with strong UV absorbance in the M. tundripaludum supernatant that is produced in a QS-dependent manner. As the putative NRPS genes are upregulated in the presence of a QS signal and there are multiple genes encoding efflux pump systems in the cluster, these results are consistent with the extracellular factor being the product of the biosynthetic gene cluster. However, additional studies will be needed to confirm this link as well as to determine the structure and function of this factor.
QS systems often regulate the production of extracellular products (13), such as antibiotics (14, 15) and proteases (28). One hypothesis is that this regulation prevents these products from being produced at low cell densities, when the fitness cost for each individual bacterium may outweigh any benefit of the small amount of produced good (12). In M. tundripaludum, we observed a measurable fitness defect under laboratory conditions from the expression of QS-regulated clusters, as the AHL synthase ΔmbaI mutant grows approximately 20% faster than the wild type. Increased growth and respiration rates have been observed in QS-null mutants in Burkholderia spp. that may be explained by the abrogated production of QS-controlled goods or metabolic slowing under QS (29, 30). Increased growth rates have also been observed in Vibrio harveyi QS-null mutants that can be partially explained by alleviation of QS-controlled bioluminescence (31). The fact that the predicted NRPS gene cluster is conserved in M. tundripaludum strains isolated from such disparate geographic regions as Svalbard, Norway (strain SV96) (8), and Seattle, WA (strains 21/22 and 31/32) (5), suggests that there may be an ecological role for this gene cluster at high cell densities (see Fig. S1 in the supplemental material).
Several other gene clusters are more highly expressed in the presence of the QS signal; however, we were unable to identify strong candidates for MbaR-binding sites upstream of any of these clusters. One possible explanation for this is that we do not know all of the criteria for identifying sequence motifs that are bound by MbaR. However, another possibility is that these genes are indirectly regulated by QS, such as through activity of the putative NRPS gene cluster product. There are several examples of multiple layers of gene regulation via bioactive signaling molecules in one organism, such as in Pseudomonas and Burkholderia spp., which can have multitiered AHL-mediated QS systems that also regulate the production of signaling molecules such as 4-quinolones (32). Furthermore, the genes that we identified with lower expression in the presence of the signal in stationary phase may also be the result of the indirect effects of QS.
Identification of other genes with differential expression in the presence of 3-OH-C10-HSL may be due to technical aspects of the system we used to determine the M. tundripaludum QS regulon. For example, we cannot rule out that adding 1 μM exogenous 3-OH-C10-HSL signal to M. tundripaludum cultures triggered the activation of the clusters of efflux pump genes, such as T451DRAFT_1642 through T451DRAFT_1644 and T451DRAFT_2702 through T451DRAFT_2704, as a response to pump excess amounts of this molecule out of the cell. These genes would need to be tested individually before further conclusions can be made.
The predicted NRPS biosynthetic gene cluster may be the only set of genes activated directly by QS in M. tundripaludum. There is precedence for such a restricted QS regulon. Only a small subset of genes is activated in the photosynthetic bacterium Rhodopseudomonas palustris (33), as well as in the plant pathogen Pseudomonas syringae (34). This is in stark contrast to the QS systems of Pseudomonas aeruginosa, which regulate hundreds of genes (35, 36). This may reflect the difference in niches that can be occupied by these bacterial species. Pseudomonas aeruginosa is well known to occupy a diverse array of environmental niches, while aerobic methanotrophs, such as M. tundripaludum, are found mainly in the relatively restricted niche of opposing counter gradients of oxygen and methane in sediment (37). As these methanotrophs are generally not thought to be host associated, the difference in regulon size may also be the result of whether or not QS is needed to differentiate between a low-density planktonic environment and a high-density host-associated environment.
If there is indeed only one set of genes directly controlled by QS in M. tundripaludum, it brings up the question of what QS controls in other methanotrophs, such as M. whittenburyi, which has a QS system highly homologous to the one found in M. tundripaludum but does not have the predicted NRPS genes or virtually any other synteny at this locus. It is possible that there is no general function of QS in these methanotrophs but rather that this system represents a specific trait acquired for certain environmental niches. More studies will be needed to determine the role of QS in this ecologically significant group of bacteria.
Sequence data.
Normalized counts and computed ±3-OH-C10-HSL pairwise fold changes for the log- and stationary-phase RNA-seq experiments are available in Tables S5 and S6, respectively, in the supplemental material.
MATERIALS AND METHODS
Plasmid construction.
All plasmids were constructed using Gibson assembly (38), with the exception of the reporter vector pAWP112, which was constructed by inserting the 400 bp upstream of mbaI into the promoter probe plasmid pPROBE-GFP[LVA] (39) using the SacI and EcoRI restriction sites. Plasmids used in this study are listed in Table S1 in the supplemental material. Primers used in this study are listed in Table S2 in the supplemental material.
Strain growth and genetic manipulation.
Strains used in this study are listed in Table S1. E. coli and B. subtilis strains were grown in Luria-Bertani (LB) medium at 37°C. M. tundripaludum 21/22 was cultured in an atmosphere of 25% methane in air. Plates were incubated at room temperature in sealed jars (Oxoid Limited, Hampshire, United Kingdom), while liquid cultures were grown at 18°C in 250-ml glass serum bottles (Kimble Chase, Vineland, NJ) or 18- by 150-mm tubes (Bellco Glass, Vineland, NJ) sealed with rubber stoppers and aluminum seals (Wheaton, Millville, NJ) and shaken at 200 rpm. Cultures were grown in nitrate mineral salts (NMS) medium (10) containing 0.2 g/liter MgSO4·7H2O, 0.2 g/liter CaCl2·6H2O, 1 g/liter KNO3, and 30 μM LaCl as well as 1× trace elements; 500× trace elements contain 1.0 g/liter Na2-EDTA, 2.0 g/liter FeSO4·7H2O, 0.8 g/liter ZnSO4·7H2O, 0.03 g/liter MnCl2·4H2O, 0.03 g/liter H3BO3, 0.2 g/liter CoCl2·6H2O, 0.6 g/liter CuCl2·2H2O, 0.02 g/liter NiCl2·6H2O, and 0.05 g/liter Na2MoO·2H2O. A final concentration of 5.8 mM phosphate buffer (pH 6.8) was added immediately before use.
Plasmids were introduced via electroporation (for ΔmbaI) as previously described (40) or via conjugation using E. coli donor strain S17-1 (for mbaI::xylE). For conjugation, the M. tundripaludum biomass was spread on an NMS plate and grown under methane. After 2 days, an equal volume of donor biomass containing the plasmid of interest was added, and the mixture was grown under methane at room temperature for an additional 2 days. For both electroporation and mating, successful integrants (single crossovers) were selected on NMS plates containing kanamycin (50 μg/ml). Subsequent sucrose counterselection was performed as previously described (25).
antiSMASH analysis.
Putative biosynthetic gene clusters were identified using antiSMASH version 3.0.5 (18) with default settings using genomes downloaded from the Joint Genome Institute (JGI) Integrated Microbial Genomes (IMG) system (41).
Supernatant extraction and HPLC fractionation.
Two milliliters of M. tundripaludum culture supernatant was routinely extracted twice with an equal volume of ethyl acetate containing 0.01% acetic acid. For HPLC fractionation, dried extract from 15 ml of stationary-phase culture supernatant was resuspended in 150 μl of 1:1 methanol-water and separated using a Nucleosil C18 column (5 μm; 250 mm by 4.6 mm) (Supelco, Bellefonte, PA) at 0.75 ml/min using a linear gradient of 10% to 100% methanol in water over 70 min. A 5-μl aliquot of each 0.75-ml fraction was analyzed using the PmbaI-gfp assay described below.
AHL analysis using E. coli PmbaI-gfp reporter strain.
Acyl-homoserine lactone signals were detected as previously described (42). Briefly, an overnight culture of E. coli reporter strain containing plasmids pAWP112 and pAWP113 was subcultured to an OD of 0.1 in LB with kanamycin (50 μg/ml) and chloramphenicol (35 μg/ml). Subsequently, 500 μl was added to 1.5-ml tubes containing dried signal, and these tubes were shaken at 37°C for 4 h. Cultures were then pelleted and resuspended in 500 μl of 50 mM Tris, pH 7.5, and 100 μl was measured for GFP fluorescence (485-nm excitation, 510-nm emission) in a 96-well plate (Nunc black optical bottom) by using a plate reader (Tecan Infinite F500). Signal N-3-hydroxydecanoyl-l-homoserine lactone was purchased from Cayman Chemical (Ann Arbor, MI), and N-3-hydroxydodecanoyl-dl-homoserine lactone was purchased from Sigma-Aldrich (St. Louis, MO).
Catechol dioxygenase activity assay.
A log-phase M. tundripaludum mbaI::xylE culture was subcultured to an OD600 of 0.02, and 5-ml aliquots were added to tubes containing dried signal or supernatant extract. The cultures were grown to the early stationary phase (OD600 of ∼0.9), at which point catechol dioxygenase activity was assayed in whole cells by measuring the rate of change in absorbance at 375 nm in the presence of 1 mM catechol with a plate reader (Molecular Devices SpectraMax 190) as previously described (43).
Mass spectrometry analysis of AHL signal.
Extracted samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a Waters Xevo LC-MS system, consisting of an Acquity ultraperformance liquid chromatography (UPLC) system and a Xevo triple-quadrupole mass spectrometer (Waters, Milford, MA). An HSS T3 column (1.8 μm; 2.1 mm by 100 mm; Acquity UPLC) was used for liquid chromatography. The LC gradient started with 10% methanol in water and increased to 100% methanol at 5 min, with 50% methanol at 5.1 min, 10% methanol at 5.5 min, and a hold for 1 min at a flow rate of 0.45 ml/min.
AHLs were initially identified using a parent ion scan monitoring for the lactone fragment ([M+H]+ = 102). Multiple reaction monitoring (MRM) parameters for 3-OH-C10-HSL as well as 3-OH-C12-HSL were determined by tuning commercially available standards with the Xevo mass spectrum in positive mode. For biological samples, both MRM scan and MS scan (mass range from 250 to 350) were used.
RNA preparation, RNA-seq, and data analysis.
A log-phase M. tundripaludum ΔmbaI culture was subcultured to an OD600 of 0.01 and added to bottles containing dried 3-OH-C10-HSL (or solvent control), resulting in a final concentration of 1 μM signal. RNA was extracted at the desired time points as previously described (43). cDNA library preparation and RNA sequencing were performed by Genewiz (South Plainfield, NJ) using Illumina HiSeq 2500 1 × 50 (single-ended) reads. The raw reads from the sequencing facility were aligned to the M. tundripaludum strain 21/22 genome (genome identification number 2563366535) as downloaded from JGI's IMG system on 13 January 2016 (41). Alignment was performed using BWA version 0.7.12-r1044 with the BWA-MEM algorithm and default parameters (44). The alignments were postprocessed into sorted BAM files with SAMTools version 1.2-232-g87cdc4a (45). Reads were attributed to open reading frames (ORFs) as well as the mbaI' pseudogene T451DRAFT_0802 using the htseq-count tool from the HTSeq framework version 0.6.1p1 in the intersection-nonempty mode (46). Differential abundance analysis was performed with DESeq2 1.2.10 (47, 48) using R 3.3.0.
Genes were considered to be differentially expressed if there was an average change of ≥2.5-fold in comparisons of normalized counts (those in the presence versus the absence of 3-OH-C10-HSL), as well as an adjusted P value of less than 1E−05 (47).
MbaR-binding site identification.
To identify possible MbaR-binding sites upstream of differentially expressed gene clusters, approximate transcription start sites were first determined by visualizing the aligned RNA-seq reads using Integrative Genomics Viewer (IGV) (49). A sequence was considered a putative MbaR-binding site if (i) it was centered 40 to 80 bp upstream of the approximate transcription start site, (ii) it contained less than 10 mismatches compared to the MbaR-binding site upstream of mbaI (ACCTGTCAATGTTGACAGTT), (iii) it matched the general NNCTG-N10-CAGNN pattern with one mismatch or less, and (iv) eight or more of its base pairs had dyad symmetry (23).
Growth inhibition assay.
Ten milliliters of molten 1% LB agar was inoculated with 100 μl of stationary-phase E. coli MG1655 or B. subtilis PY79, and this mixture was overlaid onto spots of M. tundripaludum (pregrown for 7 days on an NMS1 agar plate) or onto a new 1.5% LB agar plate. In the latter case, 5 μl of the HPLC fraction containing the UV absorbent compound (containing compound from the equivalent of 65 μl of M. tundripaludum stationary-phase culture supernatant) was spotted onto the solidified overlay. The plates were then incubated at 37°C overnight and examined for zones of growth inhibition.
Accession number(s).
Normalized counts and computed ±3-OH-C10-HSL pairwise fold changes for the log- and stationary-phase RNA-seq experiments have been submitted to the Gene Expression Omnibus (GEO) database under accession number GSE85617.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Lidstrom and Greenberg labs for helpful discussions, M. Sadilek for help with LC-MS/MS instrumentation, and F. Chu for help with RNA-seq sample submission and for providing B. subtilis strain PY79.
This work was facilitated through the use of advanced computational storage and networking infrastructure provided by the Hyak supercomputer system supported in part by the University of Washington eScience Institute.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00773-16.
REFERENCES
- 1.Nisbet EG, Dlugokencky EJ, Bousquet P. 2014. Atmospheric science. Methane on the rise—again. Science 343:493–495. doi: 10.1126/science.1247828. [DOI] [PubMed] [Google Scholar]
- 2.Singh BK, Bardgett RD, Smith P, Reay DS. 2010. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 3.Beck DAC, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina Del Rio T, Ivanova N, Lidstrom ME, Chistoserdova L. 2013. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 1:e23. doi: 10.7717/peerj.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Ha D, Vanwonterghem I, Hoefman S, De Vos P, Boon N. 2013. Selection of associated heterotrophs by methane-oxidizing bacteria at different copper concentrations. Antonie Van Leeuwenhoek 103:527–537. doi: 10.1007/s10482-012-9835-7. [DOI] [PubMed] [Google Scholar]
- 5.Kalyuzhnaya MG, Lamb AE, McTaggart TL, Oshkin IY, Shapiro N, Woyke T, Chistoserdova L. 2015. Draft genome sequences of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc 3:e00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, Markowitz VM, Rigoutsos I, Tringe SG, Bruce DC, Richardson PM, Lidstrom ME, Chistoserdova L. 2008. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol 26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 7.Oshkin IY, Beck DA, Lamb AE, Tchesnokova V, Benuska G, McTaggart TL, Kalyuzhnaya MG, Dedysh SN, Lidstrom ME, Chistoserdova L. 2015. Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 9:1119–1129. doi: 10.1038/ismej.2014.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wartiainen I, Hestnes AG, McDonald IR, Svenning MM. 2006. Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard Islands, Norway (78 degrees N). Int J Syst Evol Microbiol 56:109–113. doi: 10.1099/ijs.0.63728-0. [DOI] [PubMed] [Google Scholar]
- 9.Smith KS, Costello AM, Lidstrom ME. 1997. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB51. Appl Environ Microbiol 63:4617–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittenbury R, Phillips KC, Wilkinson JF. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 11.Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 13.Bandara HMHN, Lam OLT, Jin LJ, Samaranayake L. 2012. Microbial chemical signaling: a current perspective. Crit Rev Microbiol 38:217–249. doi: 10.3109/1040841X.2011.652065. [DOI] [PubMed] [Google Scholar]
- 14.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill MEA, Parsek MR, Nierman WC, Greenberg EP. 2009. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol 191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoover SE, Perez AJ, Tsui H-CT, Sinha D, Smiley DL, DiMarchi RD, Winkler ME, Lazazzera BA. 2015. A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol Microbiol 97:229–243. doi: 10.1111/mmi.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz LM, Valenzuela S, Castro M, Gonzalez A, Frezza M, Soulère L, Rohwerder T, Queneau Y, Doutheau A, Sand W, Jerez CA, Guiliani N. 2008. AHL communication is a widespread phenomenon in biomining bacteria and seems to be involved in mineral-adhesion efficiency. Hydrometallurgy 94:133–137. doi: 10.1016/j.hydromet.2008.05.028. [DOI] [Google Scholar]
- 17.Miller BR, Gulick AM. 2016. Structural biology of nonribosomal peptide synthetases. Methods Mol Biol 1401:3–29. doi: 10.1007/978-1-4939-3375-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, Spang A, Bringel F, Vuilleumier S, Lajus A, Medigue C, Bruce DC, Cheng JF, Goodwin L, Ivanova N, Han J, Han CS, Hauser L, Held B, Land ML, Lapidus A, Lucas S, Nolan M, Pitluck S, Woyke T. 2011. Genome sequence of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol 193:6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton R, Kits KD, Ramonovskaya VA, Rozova ON, Yurimoto H, Iguchi H, Khmelenina VN, Sakai Y, Dunfield PF, Klotz MG, Knief C, Op den Camp HJ, Jetten MS, Bringel F, Vuilleumier S, Svenning MM, Shapiro N, Woyke T, Trotsenko YA, Stein LY, Kalyuzhnaya MG. 2015. Draft genomes of gammaproteobacterial methanotrophs isolated from terrestrial ecosystems. Genome Announc 3:e00515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzzini AC, Clardy J. 2016. Gene flow and molecular innovation in bacteria. Curr Biol 26:R859–R864. doi: 10.1016/j.cub.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antunes LCM, Ferreira RBR, Lostroh CP, Greenberg EP. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397. doi: 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. 2014. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol 196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puri AW, Owen S, Chu F, Chavkin T, Beck DA, Kalyuzhnaya MG, Lidstrom ME. 2015. Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol 81:1775–1781. doi: 10.1128/AEM.03795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact 11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 27.Penalver CGN, Cantet F, Morin D, Haras D, Vorholt JA. 2006. A plasmid-borne truncated luxI homolog controls quorum-sensing systems and extracellular carbohydrate production in Methylobacterium extorquens AM1. J Bacteriol 188:7321–7324. doi: 10.1128/JB.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An JH, Goo E, Kim H, Seo YS, Hwang I. 2014. Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci U S A 111:14912–14917. doi: 10.1073/pnas.1412431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol 191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nackerdien ZE, Keynan A, Bassler BL, Lederberg J, Thaler DS. 2008. Quorum sensing influences Vibrio harveyi growth rates in a manner not fully accounted for by the marker effect of bioluminescence. PLoS One 3:e1671. doi: 10.1371/journal.pone.0001671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5:e01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auman AJ, Stolyar S, Costello AM, Lidstrom ME. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol 66:5259–5266. doi: 10.1128/AEM.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 39.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 40.Yan X, Chu F, Puri AW, Fu Y, Lidstrom ME. 2016. Electroporation-based genetic manipulation in type I methanotrophs. Appl Environ Microbiol 82:2062–2069. doi: 10.1128/AEM.03724-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antunes LCM, Schaefer AL, Ferreira RBR, Qin N, Stevens AM, Ruby EG, Greenberg EP. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu F, Lidstrom ME. 2016. XoxF Acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J Bacteriol 198:1317–1325. doi: 10.1128/JB.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 49.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


