ABSTRACT
Rhodobacter sphaeroides is an alphaproteobacterium that has two complete sets of flagellar genes. The fla1 set was acquired by horizontal transfer from an ancestral gammaproteobacterium and is the only set of flagellar genes that is expressed during growth under standard laboratory conditions. The products of these genes assemble a single, subpolar flagellum. In the absence of the Fla1 flagellum, a gain-of-function mutation in the histidine kinase CckA turns on the expression of the fla2 flagellar genes through the response regulator CtrA. The rotation of the Fla1 and Fla2 flagella is controlled by different sets of chemotaxis proteins. Here, we show that the expression of the chemotaxis proteins that control Fla2, along with the expression of the fla2 genes, is coordinated by CtrA, whereas the expression of the chemotaxis genes that control Fla1 is mediated by the master regulators of the Fla1 system. The coordinated expression of the chemosensory proteins with their cognate flagellar genes highlights the relevance of integrating the expression of the horizontally acquired fla1 genes with a chemosensory system independently of the regulatory proteins responsible for the expression of fla2 and its cognate chemosensory system.
IMPORTANCE Gene acquisition via horizontal transfer represents a challenge to the recipient organism to adjust its metabolic and genetic networks to incorporate the new material in a way that represents an adaptive advantage. In the case of Rhodobacter sphaeroides, a complete set of flagellar genes was acquired and successfully coordinated with the native flagellar system. Here we show that the expression of the chemosensory proteins that control flagellar rotation is dependent on the master regulators of their corresponding flagellar system, minimizing the use of transcription factors required to express the native and horizontally acquired genes along with their chemotaxis proteins.
KEYWORDS: Rhodobacter sphaeroides, chemotaxis, bacterial flagellum, horizontal transfer, FleQ, CtrA, RpoN, sigma-54
INTRODUCTION
Chemotaxis is a complex response that enables bacteria to detect and swim toward or away from positive or negative stimuli in the surrounding environment. To achieve chemotaxis, a signal transduction system senses and controls the flagellar motor. Signals are sensed by specialized receptors that are assembled together with the chemotaxis proteins CheW and CheA to form complex arrays. In response to receptor occupancy, CheA phosphorylates the response regulator CheY, which in turn binds to the flagellar switch complex to control flagellar rotation. The response is terminated by lowering the levels of CheY-P, through the action of the phosphatase CheZ. The chemotaxis proteins CheB and CheR play a key role in adaptation by chemically modifying the chemotaxis receptors. The methylesterase CheB is stimulated upon phosphorylation by CheA, and CheR is a constitutive methyltransferase. The activity of these proteins resets the signaling state of the receptor (for reviews, see references 1–5).
This basic chemotaxis system is found in many species other than Escherichia coli and Salmonella enterica. However, many species have multiple copies of the chemotaxis genes. The products of these additional copies frequently are involved in chemotaxis, but in some species these copies have evolved to control other cellular functions such as development, biofilm formation, and twitching motility mediated by type IV pili (6, 7).
In several bacterial species, the chemotaxis genes are expressed coordinately with the flagellar genes. Therefore, the cytoplasmic components that control flagellar rotation are present only when the flagellum is formed. In the paradigmatic bacteria E. coli and S. enterica, the chemotaxis genes are expressed when the cytoplasmic and membrane components of the flagella are already formed (8–10). A similar situation has been observed in Pseudomonas aeruginosa and Xanthomonas campestris (11, 12). In the alphaproteobacteria Caulobacter crescentus and Sinorhizobium meliloti, chemotaxis genes are expressed under the control of the master activator protein of the flagellar genes (13, 14). Therefore, the chemotaxis genes are expressed early during flagellar biogenesis.
Rhodobacter sphaeroides is an alphaproteobacterium with two different flagellar systems, multiple copies of the chemotaxis genes, and four different rpoN genes, encoding functionally different σ54 factors. σ54-2 has been shown to be required to transcribe one of the flagellar sets (15–17).
This bacterium assembles a single, subpolar flagellum under standard laboratory growth conditions (18), and the products of the fla1 genes are responsible for the formation and functioning of this structure. The expression of these genes follows a hierarchical pattern. At the top of the hierarchy, the master protein FleQ, together with the RNA polymerase core (E) associated with the σ54-2 factor, promotes the expression of the class II flagellar operon fleT-fliEFGHI. Subsequently, the hetero-oligomeric complex FleQ/FleT and Eσ54-2 bring about the transcription of genes encoding the proteins that form the hook and the basal body (HBB), fliA (which encodes σ28) and flgM (encoding the anti-sigma factor of σ28). Once the HBB has been assembled, FlgM is exported out of the cell, and σ28 is free to associate with E to transcribe the late flagellar genes, such as those encoding flagellin (FliC) and its scaffold protein, FliD (19).
The other flagellar system present in R. sphaeroides, Fla2, enables the formation of several polar flagella (16, 20). The fla2 genes are not expressed under standard laboratory growth conditions. So far, the Fla2 flagellar system has been studied in mutant strains that are unable to form Fla1 due to a mutation in a regulatory or structural gene and importantly contain a gain-of-function mutation in the histidine kinase CckA. In these strains, it has been determined that the expression of fla2 is dependent on the two-component system formed by the histidine kinase CckA, the phosphotransferase protein ChpT, and the response regulator CtrA (21).
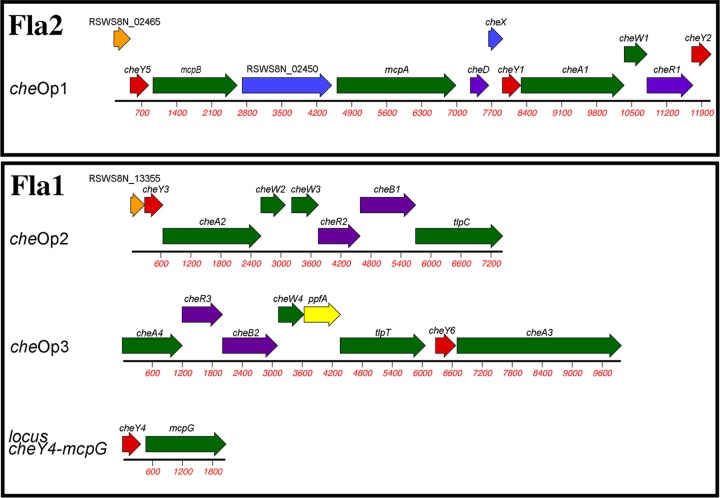
Phylogenetic studies have shown that the fla2 genes constitute the native flagellar genes of R. sphaeroides, whereas the fla1 set was acquired by horizontal transfer from an ancestral gammaproteobacterium (16). In R. sphaeroides, the multiple copies of the chemotaxis genes are organized in four different operons (15) (Fig. 1).
FIG 1.
Schematic representation of the organization of the chemotaxis genes in R. sphaeroides. The multiple cheY genes are in red. Black outlines group the chemotaxis operons that control Fla1 or Fla2.
The control of Fla1 was shown to be mediated by the products of che operon 2 (cheOp2) and cheOp3 (15, 22–24), whereas the control of Fla2 is mediated by the chemosensory proteins encoded in cheOp1 (25, 26). Specifically, the chemotaxis response dependent on Fla1 is mainly controlled by CheY6 (encoded in cheOp3), as well as either CheY3 (encoded in cheOp2) or CheY4 (24). Fla2 is controlled by CheY2 and CheY5 (both encoded by cheOp1) (25).
Regarding the expression of these genes, cheOp3 expression reportedly depends on the σ54-3 factor (27). This is unexpected, given that the expression of flagellar and chemotaxis genes in bacteria is usually coordinated and that σ54-3 has not been implicated in the transcription of any fla1 gene. The expression of cheOp1 has yet to be studied. The other chemotaxis operon whose expression has already been analyzed is cheOp2, which is dependent on σ70 and σ28 (FliA) (27). Therefore, whether cheY4 and cheOp1 are expressed concurrently with fla1 and fla2, respectively, remains to be determined.
Another important aspect that remains to be determined is the identity of the activator protein that is responsible for interacting with Eσ54-3 to promote cheOp3 expression; since Eσ54 is unable to form open complex by itself, the presence of an activator protein of the bacterial enhancer-binding protein (bEBP) family is compulsory (28). In this context, it should be stressed that most bacteria have a single σ54 factor (encoded by rpoN) that interacts with multiple bEBPs that recognize specific binding sites to achieve the transcription of a specific set of genes (29). In contrast, R. sphaeroides has four σ54 factors, two of which, σ54-1 and σ54-2, are activated by specific bEBPs (i.e., NifA and FleQ/FleT, respectively) (17, 30). Potential bEBP-encoding genes were previously identified in silico in the genome of R. sphaeroides, revealing that aside from the bEBPs for σ54-1 and σ54-2, only two loci remain that encode proteins with bEBP family signatures that could be the activators for σ54-3 and σ54-4 (19).
In this work, we have determined that the expression of cheOp3 is dependent on σ54-2 and not on σ54-3, as was previously reported (27), and that the bEBP of the fla1 system (FleQ) is the activator protein for this operon. In addition, we found out that cheY4 expression is dependent on σ28 (FliA) and, consequently, is also a part of the flagellar transcription hierarchy. Our results also show that cheOp1 expression is controlled by CtrA. Together, these results establish that the expression of the chemotaxis genes in R. sphaeroides is coordinated with the expression of their cognate flagellar system.
RESULTS
In this work, we used derivatives of the WS8N and AM1 strains to study the regulation of the flagellar and chemotaxis genes. WS8N is the wild-type strain that constitutively expresses the Fla1 flagellum but is unable to express the fla2 genes given that the two-component system CckA/ChpT/CtrA is inactive. The AM1 strain is a WS8N derivative that is unable to express the fla1 genes, given that it carries a mutation in the flagellar master activator fleQ and also carries a point mutation in cckA that activates the CckA/ChpT/CtrA two-component system (21, 25); therefore, this strain has a Fla1− Fla2+ phenotype.
cheOp1 expression is dependent on the master regulator of the fla2 genes.
The expression of three chemotaxis proteins encoded in cheOp1 (Fig. 1) was evaluated by Western blotting using total cell extracts obtained from AM1 (25) and its derivative, EA1 (ΔctrA::aadA) (21). The proteins CheY2 and CheY5 were detected in AM1 but not in the absence of ctrA (Fig. 2A), indicating that cheOp1 expression is dependent on the presence of CtrA. To test for another protein encoded in cheOp1, we isolated a chromosomal fusion of mcpA with the Flag epitope in the AM1 strain. The resulting strain, JV1 (mcpA-Flag) and its derivative, JV2 (mcpA-Flag ΔctrA::hyg), were tested by immunoblotting using an anti-Flag antibody (Fig. 2A). Consistent with the hypothesis that CtrA is required to transcribe cheOp1, McpA-Flag was detected in total cell extracts of JV1 but not of JV2.
FIG 2.
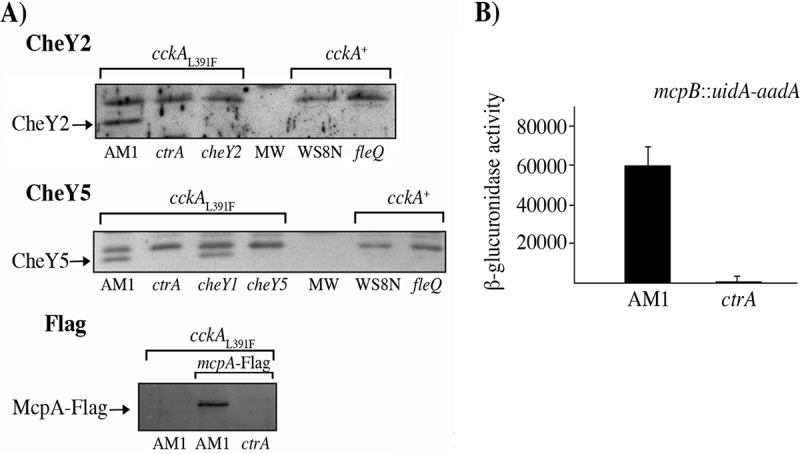
Determination of the presence of proteins encoded in cheOp1 by immunoblotting and measuring the transcriptional activity of cheOp1 using a reporter gene. (A) Total cell extracts were obtained from cultures grown in Sistrom's minimal medium supplemented with 80 μM succinic acid and harvested at an OD600 of 0.3. The notation at the top of the Western blots regarding the status of the cckA allele indicates that the WS8N and ΔfleQ (SP13) strains do not carry the gain-of-function mutation in CckA that enables the expression of the fla2 genes whereas the AM1 strain and its derivatives carry the mutations ΔfleQ and, importantly, cckAL391F, which activate the CckA/ChpT/CtrA two-component system. Each lane is labeled according to the relevant mutation present in the strain. The antibody used for each blot is indicated at the top. MW indicates the lane where the molecular weight marker was loaded. Anti-CheY2 and anti-CheY5 antibodies were used at a 1:3,000 dilution, and anti-Flag antibody (Sigma-Aldrich) was used at a 1:10,000 dilution. (B) β-Glucuronidase activity promoted by the chromosomal fusion mcpB::uidA-aadA. AM1 and its ΔctrA derivative (LC7 strain) carrying the transcriptional fusion mcpB::uidA-aadA were grown in 80 μM succinic acid, and total cell extracts were used for the enzymatic reaction. Activity is reported in picomoles of 4-methylumbelliferone produced per minute per milligram of protein. The mean values and standard deviations of the results of three independent determinations are shown.
To rule out the possibility that an undetected mutation other than ΔctrA was responsible for the lack of CheY2, CheY5, and McpA-Flag, we introduced a plasmid that expresses CtrA (pRK_ctrA) into the EA1 and JV2 strains. In these complemented strains, the expression of CheY2, CheY5, and McpA-Flag was restored (see Fig. S1 in the supplemental material).
Consistent with the proposed model of activation of the CckA/ChpT/CtrA system (21), we did not observe the presence of either CheY2 or CheY5 in the wild-type strain WS8N or in the SP13 (ΔfleQ::kan) mutant (Fig. 2A), indicating that cheOp1 is expressed only when the CckA/ChpT/CtrA two-component system is active. To test the hypothesis that cheOp1 expression could be transcriptionally controlled by CtrA, a transcriptional fusion of mcpB to the promoterless uidA gene was made. This fusion was used to replace the mcpB+ gene in AM1 and ΔctrA::hyg (LC7) strains. uidA expression was recorded as the amount of β-glucuronidase present in total cell extracts. We noticed that the enzymatic activity was severely reduced in the absence of CtrA (Fig. 2B), implying that transcription of cheOp1 is directly or indirectly controlled by CtrA.
The control region of cheOp1 has a potential CtrA binding site relevant for its expression.
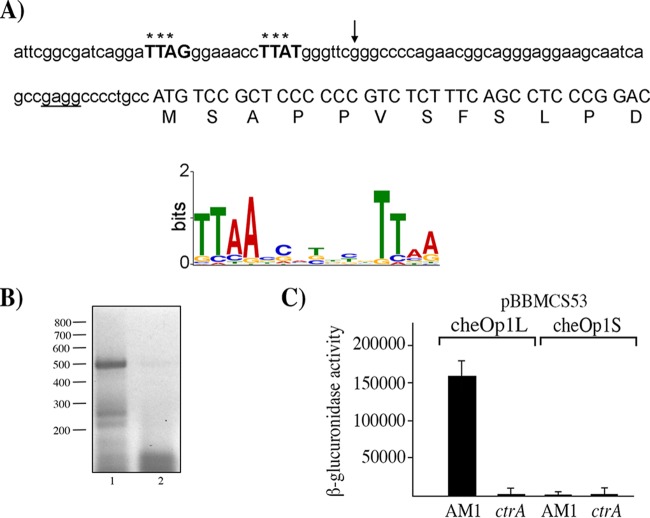
Given that the expression of several of the proteins encoded in cheOp1 was dependent on CtrA, we searched for potential CtrA binding sites upstream of the annotated genes in chromosome 1 using a CtrA position weight matrix (PWM) built from the regulatory regions of 54 genes identified to be controlled by CtrA in Caulobacter crescentus (31). From this analysis, the sequence TTAGN7TTAT was found 67 bp upstream of the translational start site of RSWS8N_02465, the gene located upstream of cheY5 (Fig. 1 and 3A). The identified sequence is highly similar to the CtrA consensus binding sequence (TTAAN7TTAA) previously characterized in C. crescentus and other species (32), suggesting that cheOp1 transcription may be activated by CtrA from this site. This finding implies that RSWS8N_02465 might be part of cheOp1. To evaluate this prediction, we performed a reverse transcription (RT)-PCR experiment using specific primers for RSWS8N_02465 and cheY5. Using total RNA from AM1, the main product amplified from this reaction had the expected size for a DNA fragment that expands from RSWS8N_02465 to cheY5 (Fig. 3B). Further supporting the idea that RSWS8N_02465 is part of cheOp1, it should be noted that the stop codon of RSWS8N_02465 overlaps the start codon of cheY5. RSWS8N_02465 encodes a 109-amino-acid protein with a STAS (sulfate transporter and anti-sigma factor antagonist) domain that has not been implicated in chemotaxis so far.
FIG 3.
Nucleotide sequence upstream of cheOp1, activity of the reporter gene uidA fused to different fragments from the regulatory region of cheOp1 in the pBBMCS53 plasmid, and RT-PCR data for cheY5 and its upstream gene. (A) Nucleotide sequence upstream of RSWS8N_02465. The first residues of the protein encoded by this gene are shown below the corresponding codons. The putative Shine-Dalgarno sequence is underlined, and nucleotides similar to the consensus binding site of CtrA, identified using a previously reported position weight matrix (31), are shown in bold and capital letters. The LOGO sequence of the CtrA-binding site is shown below; it was reported as part of a genomic analysis performed in C. crescentus (32). Asterisks above the sequence indicate nucleotides that match with the most frequent base in the LOGO sequence. An arrow indicates the 5′ end of the fragment cloned into pBBMCS53_cheOp1S. The 5′ end of the fragment cloned into pBBMCS53_cheOp1L includes 25 bp upstream of the sequence shown. (B) Agarose gel showing the RT-PCR products of the primer pair that amplifies the region encompassing CheY5 and RSWS8_02465. A 469-bp product was expected for this reaction. Lane 1, 5 μl from the RT-PCR; lane 2, 5 μl from the control reaction carried out in the absence of reverse transcriptase. The relevant molecular sizes from a 100-bp DNA ladder are shown on the left. (C) β-Glucuronidase activity of cell extracts from the AM1 and EA1 (ΔctrA::aadA) strains carrying the indicated plasmids. Cell cultures were grown in Sistrom's minimal medium supplemented with 80 μM succinic acid. Activity is reported in picomoles of 4-methylumbelliferone produced per minute per milligram of protein. The mean values and standard deviations (SD) of the results of three independent determinations are shown.
To evaluate if the upstream region of RSWS8N_02465 exhibits CtrA-dependent promoter activity, we cloned this region upstream of the reporter gene uidA in the pBBMCS53 plasmid. The resulting construct (pBBMCS53_cheOp1L) promoted high β-glucuronidase expression in the AM1 strain but not in EA1 (ΔctrA::aadA) (Fig. 3C). In addition, we cloned a shorter fragment from the upstream region of RSWS8N_02465, which excludes the putative CtrA binding site, into the pBBMCS53 plasmid. The resulting plasmid (pBBMCS53_cheOp1S) did not promote β-glucuronidase expression in either of the two strains (Fig. 3C). The lack of expression of the reporter gene in this construction could be explained by the loss of either the CtrA binding site or the −35 promoter region. In this context, it should be stressed that most of the CtrA-activated promoters in C. crescentus show overlapping of these two sequence motifs (32, 33).
cheOp3 expression is dependent on σ54-2 and the master regulator of the fla1 genes.
CheY6 and either CheY3 or CheY4 are needed to control the chemotaxis response of the Fla1 flagellum (24, 34). CheY6 is encoded in cheOp3, which is located within the 56-kb fla1 gene cluster, and CheY3 is encoded in cheOp2, located elsewhere on chromosome I, whereas CheY4 is located on chromosome II (34) (Fig. 1).
cheOp3 expression was previously shown to depend on σ54-3 (27); however, fla1 gene expression is dependent on σ54-2, raising the possibility that the expression of cheOp3 is unlinked from the expression of Fla1. Given that Eσ54 is unable to form open complex by itself, we decided to identify the bEBP responsible for activating Eσ54-3. The in silico identification of bEBP-encoding genes in the R. sphaeroides genome revealed only five genes that encode proteins with the characteristic signatures of the σ54 transcriptional activators (19): nifA, fleQ, fleT, RSWS8N_RS13335, and RSWS8N_RS04480. We previously reported the isolation of strains with mutations in the genes nifA, fleQ, and fleT (19, 30); therefore, we isolated strains with mutations in RSWS8N_RS13335 and RSWS8N_RS04480.
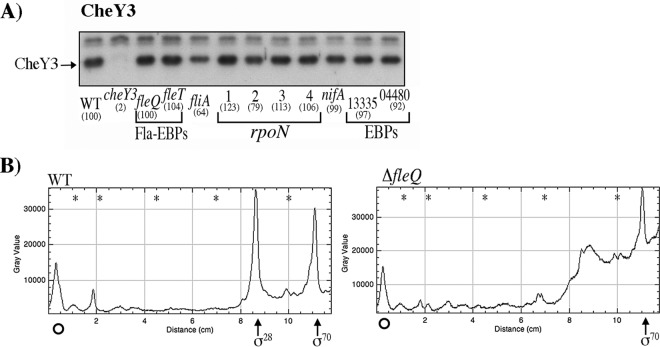
The expression of CheY6 (encoded in cheOp3) was tested in total cell extracts obtained from the wild-type strain, along with cell extracts from the strains with mutations in each of the five bEBP-encoding genes. As shown in Fig. 4A, CheY6 expression was abolished in the strain carrying the ΔfleQ::kan allele and was not affected by the absence of RSWS8N_RS13335, RSWS8N_RS04480, or nifA, indicating that CheY6 is expressed as a part of the fla1 transcriptional hierarchy.
FIG 4.
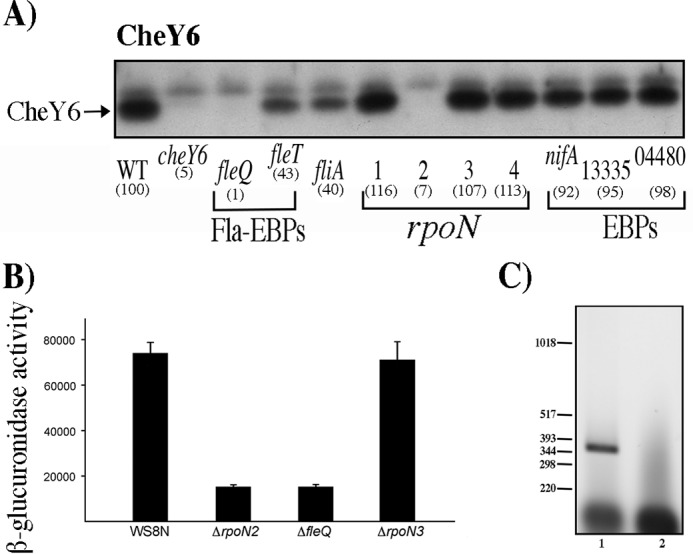
Determination of the presence of CheY6 in different strains, activity of the reporter gene uidA fused to the regulatory region of cheOp3 in the pBBMCS53 plasmid, and RT-PCR of cheY6 and its upstream gene. Total cell extracts obtained from cell cultures grown in Sistrom's minimal medium and harvested at an OD600 of 0.6 were tested as described in Materials and Methods. (A) Total cell extracts were separated by gel electrophoresis, transferred to nitrocellulose, and probed with anti-CheY6 antibodies (1:3,000). WT, WS8N cell extract. Each lane is labeled according to the relevant mutation present in the strain. Square brackets group the samples according to their function. Fla-EBPs indicate the two enhancer binding proteins required for activation of the fla1 genes, rpoN indicates the genes encoding σ54-1, -2, -3, or -4, and EBPs indicate the genes encoding proteins that belong to the enhancer binding protein family in the R. sphaeroides genome sequence. The signal identified as CheY was quantified by densitometry using ImageJ; the resulting value was normalized using an unspecific band on the blot. The values shown in parentheses are the average results of three independent experiments and are expressed as a percentage of the WT level. Figure S3 shows the graph of these values with SD. (B) β-Glucuronidase activity of the wild-type and mutant strains carrying the plasmid pBBMCS53_cheA4. Activity is reported in picomoles of 4-methylumbelliferone produced per minute per milligram of protein. The mean values and standard deviations of the results of three independent determinations are shown. (C) Agarose gel of the RT-PCR products of the primer pair that amplifies the region encompassing cheY6 and its upstream gene tlpT. A 391-bp product was expected for these primers. Lane 1, 5 μl from the RT-PCR; lane 2, 5 μl from the control reaction carried out in the absence of reverse transcriptase. The relevant sizes in base pairs of the DNA molecular size marker X (Roche Life Science) are shown on the left.
An approximately 60% reduction in CheY6 levels was noticed in the absence of FleT and FliA. In both of these strains, the FleQ activator is present and the σ28 (FliA) factor is absent, suggesting that wild-type CheY6 levels could arise from the combined activity of a class I FleQ-activated promoter and a class IV σ28-dependent promoter.
In contrast to the proposal that cheOp3 expression is dependent on σ54-3 (27), we observed that the expression of CheY6 (encoded in cheOp3) was dependent only on σ54-2 and that the absence of σ54-3 did not affect CheY6 expression levels (Fig. 4A). Given that cheY6 and the upstream gene tlpT are located 204 bp apart, cheY6 may be transcribed independently of the upstream chemotaxis genes (Fig. 1). Therefore, to determine if the expression of the genes located upstream of cheY6 was also dependent on FleQ and σ54-2, we tested the expression of a reporter gene under the control of the region located upstream of cheA4 which has been reported to carry the cheOp3 promoter (27). To this end, an 854-bp fragment of the upstream region of cheA4, starting from codon 48 of cheA4, was cloned upstream of the reporter gene uidA in pBBMCS53 (35). The resulting plasmid was introduced into the WS8N, ΔrpoN2::kan, and ΔfleQ::kan strains. As a control, this plasmid was also introduced into the ΔrpoN3 strain.
Total cell extracts of these strains were assayed for β-glucuronidase activity. A strong reduction in the amount of β-glucuronidase was detected for the ΔrpoN2::kan and ΔfleQ::kan mutants compared with that in the wild-type WS8N or ΔrpoN3::kan strains (Fig. 4B), suggesting that the expression of cheA4 is also dependent on σ54-2 and FleQ. In addition, RT-PCR revealed that cheY6 is cotranscribed with tlpT, suggesting that these genes could form a single operon (Fig. 4C).
σ28 is involved in the expression of cheY4.
The expression of CheY3 and CheY4 was analyzed in each of the mutant strains lacking either one of the bEBP or σ54 factors. We observed that the expression of CheY4 was severely reduced in ΔfleQ::kan (SP13), ΔfleT::aadA (SP12), ΔrpoN2::kan (SP7), and ΔfliA::kan (SP15) strains (Fig. 5A). Since it has been shown that the expression of fliA is dependent on FleQ, FleT, and σ54-2 (19), these results suggest that cheY4 expression is dependent on σ28 (fliA) and that the absence of CheY4 in the ΔfleQ, ΔfleT, and ΔrpoN2 strains is the expected consequence of this regulation.
FIG 5.
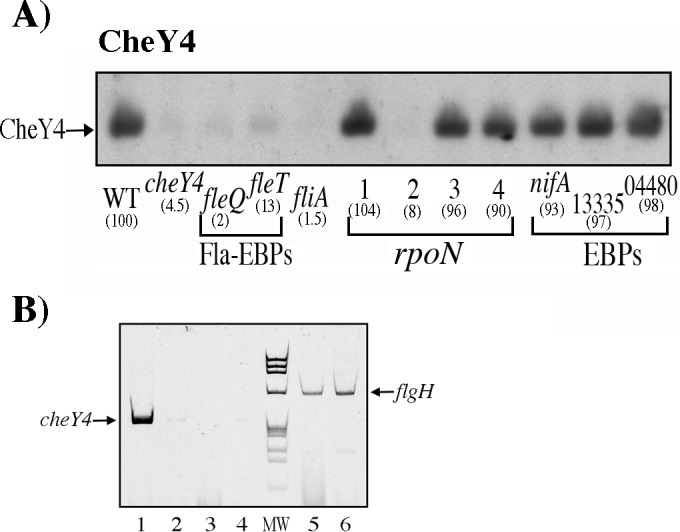
Determination of the presence of CheY4 in different mutant strains by immunoblotting. Total cell extracts obtained from cell cultures grown in Sistrom's minimal medium and harvested at an OD600 of 0.6 were tested as described in Materials and Methods. Anti-CheY4 antibodies were used at 1:3,000. WT denotes the WS8N cell extract. Each lane is labeled according to the relevant mutation present in the strain. Other labels are the same as described in the legend for Fig. 4A. (B) Acrylamide gel electrophoresis of the RT-PCR products corresponding to the primer pair that amplifies cheY4 (lanes 1 to 4) and the primer pair that amplifies flgH (lanes 5 and 6). Lanes: 1, 3, and 5, total RNA from WS8N; 2, 4, and 6, total RNA from the fliA::kan mutant strain. Control reactions were carried out in the absence of reverse transcriptase (lanes 3 and 4). Lane MW corresponds to PhiX174 DNA digested with HaeIII and was used as a molecular size marker (fragment sizes, 1,353, 1,078, 872, 603, 310, 281, 271, 234, 194, and 118 bp). The expected sizes for the cheY4 and flgH products are 361 and 639 bp, respectively).
To verify that the expression of cheY4 was dependent on the presence of σ28, we carried out an RT-PCR assay using specific oligonucleotides for cheY4. The expected product of 361 bp was detected when the total RNA from WS8N was included in the reaction, whereas a faint product was detected when RNA from the fliA::kan strain was used (Fig. 5B, lanes 1 and 2, respectively). It should be noted that for this RNA, a similar signal was detected when no reverse transcriptase was included in the reaction mixture (Fig. 5B, lane 4), indicating that this slight signal is the product of a small amount of DNA in the sample. To be certain that both samples of RNA had the appropriate quality to yield an RT-PCR product, we performed a reaction using a pair of oligonucleotides that amplified a region of the flgH gene (Fig. 5B, lanes 5 and 6).
Given that our results suggest that CheY4 is expressed from a σ28-dependent promoter, we searched upstream of cheY4 for a sequence similar to the σ28 consensus promoter. We analyzed a 159-bp region from the stop codon of RSWS8N_17744 (RSP_3301) to the start codon of cheY4 by using a PWM built from 24 σ28-dependent promoters (36). This analysis revealed a possible σ28-promoter located 104 bp upstream of the start codon of cheY4 (Fig. S2). The sequence of this putative promoter is similar to that of previously reported σ28-dependent promoters in R. sphaeroides (19).
A more complex situation was observed for CheY3, where we detected a reduction in the amount of CheY3 in the ΔfliA mutant strain but no reduction in the absence of FleQ or FleT (Fig. 6A); this result was unexpected given that the expression of fliA depends on FleQ and FleT (19). It has been shown that cheOp2 contains a regulatory region with two promoters, one dependent on σ70 and the other dependent on σ28 (27), which explains the reduction of CheY3 in the absence of σ28 (FliA). However, the CheY3 expression level in the absence of FleQ and FleT (in these strains, fliA is not expressed [19]) is similar to that observed in the wild-type strain, suggesting that FleQ and FleT may have a negative effect on CheY3 expression.
FIG 6.
Determination of the presence of CheY3 in different strains by immunoblotting and plot profile of the primer extension experiment for cheOp2. (A) Total cell extracts obtained from cell cultures grown in Sistrom's minimal medium and harvested at an OD600 of 0.6 were tested as described in Materials and Methods. Anti-CheY3 antibodies were used at 1:3,000. WT, WS8N cell extract. Each lane is labeled according to the relevant mutation present in the strain. Other labels are the same as described in the legend for Fig. 4A. (B) Total RNA was isolated from cell cultures of WS8N and SP13 (ΔfleQ::kan) strains grown in Sistrom's minimal medium. After hybridization of the RNA samples with the 5′-end-labeled oligonucleotide, cDNA synthesis was performed using AMV reverse transcriptase. The reactions were subjected to gel electrophoresis, and the images were visualized using a Typhoon scanner (GE Healthcare Life Science) and quantified using ImageJ. The plot profiles initiate immediately below the largest transcript detected (position −155 from the translational start site of RSWS8N_13355), denoted by an open circle. The transcripts that originated from the σ70 and σ28 promoters are indicated by vertical arrows. The asterisks correspond to the migration of the 151-, 140-, 118-, 100-, and 82-bp (from left to right) DNA fragments of PhiX174 that were obtained after digestion with HinfI (Promega) and 5′ end labeled.
To evaluate this possibility, we carried out a primer extension experiment using the oligonucleotide cheOp2B, which targets the first gene of cheOp2 (i.e., RSWS8N_13355). The reaction mixtures were subjected to gel electrophoresis, and the products were analyzed using ImageJ. The plot profile of the sample from the wild-type strain revealed the previously reported σ28 and σ70 promoters (27) (Fig. 6B). From two independent experiments, a 26% average increase was observed in the amount of the σ70-dependent transcript in the ΔfleQ mutant strain compared with that in the wild-type strain. This increase is consistent with a negative role for FleQ on cheOp2 expression. Unexpectedly, we detected a low-abundance transcript in both strains that initiates approximately 155 nt upstream of the translational start of RSWS8N_13355. The amounts of transcript are similar for the two samples.
DISCUSSION
Our work shows that the expression of chemotaxis proteins in R. sphaeroides is coordinated with the expression of the cognate flagellar system. In many bacteria, chemotaxis genes are expressed under the control of the flagellar regulator proteins, and in E. coli and other related bacteria, their expression is dependent on σ28 (FliA), indicating that these genes belong to the lower class of the flagellar hierarchy. In contrast, chemotaxis genes in other bacteria are expressed under the control of the master regulator of the flagellar hierarchy; therefore, the chemotaxis proteins are synthesized during the early steps of flagellar formation. These opposing patterns suggest that the timing of chemotaxis protein expression may be irrelevant for the function of the system or that this regulation may reflect different regulatory needs. Nonetheless, the coordinated expression of flagellar proteins and chemotaxis proteins seems to be a ubiquitous scenario, and this scheme prevails in the case of R. sphaeroides, even though one flagellar system was horizontally acquired, indicating a strong pressure to coordinate the presence of the flagellar and chemotaxis proteins. In agreement with this view, it has been proposed that the gene encoding σ54-2 (rpoN2) is a duplication from the vertically inherited rpoN1 gene (37).
In R. sphaeroides, the expression of the fla2 genes is dependent on the transcriptional activator CtrA (21). We demonstrated that cheOp1 expression is also dependent on CtrA. Our data identified a 652-bp fragment that contains the cheOp1 promoter and revealed the possible site through which CtrA mediates transcriptional activation. Our results also showed that RSWS8N_02465 is part of cheOp1; the product of this gene has yet to be implicated in chemotaxis, but its presence in cheOp1 suggests its potential involvement in this response. Interestingly, a gene containing a STAS domain is also located upstream of cheY3 in cheOp3, strengthening the idea that the products of these genes might be involved in chemotaxis. In addition, our results show that the cheOp1, CheY5, CheY2, and McpA gene products are not expressed when the CckA/ChpT/CtrA two-component system is inactive (in the WS8N strain or its derivative, SP13 [ΔfleQ::kan]), further supporting the notion that cheOp1 is expressed only when the fla2 genes are expressed.
In contrast to the Fla2 flagella, where the expression of all the CheY proteins involved in its control are dependent on the master flagellar regulator CtrA, the expression of the chemotaxis proteins that control Fla1 appears to be more complex. The expression of cheOp3, in which CheY6 is encoded, was previously suggested to depend on the sigma factor σ54-3 (27); however, our results show that cheOp3 expression is dependent on σ54-2 and the master regulator FleQ. We speculate that a mistake in the nomenclature of the σ54 factors might be behind this disparity. The relevant point that should be stressed is that the expression of cheOp3, which encodes CheY6, the only CheY able to bind the Fla1 flagellar motor, is controlled by the master flagellar regulator FleQ and the σ54-2 factor.
We also observed a reduction in the amount of CheY6 in cell extracts from the ΔfleT::aadA (SP12) and ΔfliA::kan (SP15) strains (Fig. 4A), suggesting that cheOp3 should have an additional σ28-dependent promoter and that cheOp3 might be expressed from both a FleQ-dependent promoter and from a σ28 (FliA)-dependent promoter. If this hypothesis is correct, the observed reduction of CheY6 in the absence of FleT and FliA can be explained by the absence of σ28 in these two strains, leaving only the FleQ promoter active. The fact that CheY6 expression depends on the master flagellar regulator FleQ and probably also on σ28 (FliA) highlights the importance of keeping a high and sustained level of this protein as long as the Fla1 flagellum is present, consistent with the idea that CheY6 is the main chemotaxis regulator of Fla1 (34).
The response regulators CheY3 and CheY4, which act along with CheY6 to control Fla1, are also expressed through the action of the transcriptional factors that regulate the Fla1 gene hierarchy. We observed that CheY4 expression is dependent on σ28 (FliA), and consequently we did not detect this protein in the absence of either σ28 or the FleQ, FleT, and σ54-2 proteins, which are required to transcribe fliA (19). Accordingly, no RT-PCR product of cheY4 was obtained from RNA of the cheY4::aadA strain, and a sequence similar to the σ28 consensus promoter was identified upstream of cheY4.
The expression of CheY3, which is encoded in cheOp2, depends on σ70 and σ28, and a part of its expression is dependent on the σ28 promoter (27). Our results also showed a reduction in the level of CheY3 in the ΔfliA mutant strain. However, the lack of a reduction in the ΔfleQ and ΔfleT strains suggests that these proteins have a negative effect on CheY3 expression. A primer extension experiment designed to detect expression by the two cheOp2 promoters showed a 26% increase in the amount of the σ70-dependent transcript in the absence of FleQ, which could explain the Western blot results. The mechanism of this negative regulation remains to be determined, but the sustained expression of cheOp2 mediated by σ70, as well as its possible increase in the absence of FleQ, makes this operon unique among the four chemotaxis operons, suggesting that the products of cheOp2 might be required to accomplish another physiological role.
Figure 7 presents a model summarizing the conclusions of this work. This model illustrates the transcriptional relationship between the expression of flagellar genes and their cognate chemosensory proteins. Notably, only one transcriptional regulator directly controls the expression of the proteins involved in the chemotaxis response of Fla2, whereas the chemosensory proteins that control Fla1 are directly or indirectly regulated by two different transcription factors, FleQ and σ28. Whether there is a physiological reason for this multiple regulation remains to be elucidated, but obviously, a pathway controlled by several regulators offers multiple checkpoints for finely tuned responses. Nevertheless, it should be kept in mind that this global regulation may simply reflect the evolutionary history of these genes.
FIG 7.
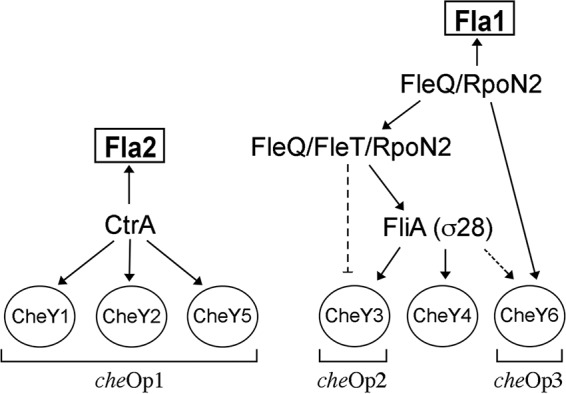
Model for the transcriptional control of genes encoding the CheY proteins of R. sphaeroides through the master regulators of fla1 and fla2. Solid arrows represent the confirmed regulatory pathways established here and in reference 27. Broken lines represent the regulation inferred from our results. The brackets at the bottom indicate the chemotaxis operon to which each cheY gene belongs.
MATERIALS AND METHODS
Genome sequences, plasmids, bacterial strains, and growth conditions.
All plasmids and bacterial strains used in this work are listed in Table 1. The genome sequence analyzed in this work corresponds to R. sphaeroides WS8N (NCBI reference sequence NZ_CM001161.1, GI:332561612). R. sphaeroides WS8N (38) and its derivatives were grown chemoheterotrophically at 30°C in Sistrom's minimal medium (39). AM1 and its derivatives were grown photoheterotrophically in Sistrom's minimal medium with 80 μM succinic acid as the carbon source. Photoheterotrophic liquid cultures were grown under continuous illumination and in completely filled screw-cap tubes. Heterotrophic liquid cultures were incubated in the dark with orbital shaking at 200 rpm. Escherichia coli was grown in Luria-Bertani (LB) medium (40) at 37°C. When required, antibiotics were added at the indicated concentrations: for R. sphaeroides, kanamycin (25 μg/ml), tetracycline (1 μg/ml), spectinomycin (50 μg/ml), and hygromycin (20 μg/ml for liquid cultures and 150 μg/ml for plates); for E. coli, kanamycin (50 μg/ml), spectinomycin (50 μg/ml), ampicillin (100 μg/ml), and hygromycin (20 μg/ml for liquid cultures and 200 μg/ml for plates).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Rhodobacter sphaeroides strains | ||
| AE1 | AM1 derivative; ΔfleQ::kan cckAL391F ΔctrA::aadA | 21 |
| AM1 | WS8N derivative; ΔfleQ::kan cckAL391F | 25 |
| JV1 | AM1 derivative; ΔfleQ::kan cckAL391F mcpA-Flag | This work |
| JV2 | JV1 derivative but ΔctrA::hyg | This work |
| JV3 | AM1 derivative; mcpB::uidA-aadA | This work |
| JV4 | AM1 derivative; mcpB::uidA-aadA ΔctrA::hyg | This work |
| LC7 | AM1 derivative; ΔctrA::hyg | This work |
| RS1Y3 | WS8N derivative; cheY3::aadA | 25 |
| RS1Y4 | WS8N derivative; ΔcheY4::aadA | 25 |
| RS1Y6 | WS8N derivative; ΔcheY6::aadA | 25 |
| RS2Y1 | AM1 derivative; cheY1::aadA | 25 |
| RS2Y2 | AM1 derivative; cheY2::aadA | 25 |
| RS2Y5 | AM1 derivative; cheY5::aadA | 25 |
| SP7 | WS8N derivative; ΔrpoN2::kan rpoN2 (RSWS8N_05555) | 17 |
| SP8 | WS8N derivative; ΔrpoN1::aadA rpoN1 (RSWS8N_07950) | 17 |
| SP12 | WS8N derivative; ΔfleT::aadA | 19 |
| SP13 | WS8N derivative; ΔfleQ::kan | 19 |
| SP15 | WS8N derivative; ΔfliA::kan | 19 |
| SP16 | WS8N derivative; ΔnifA::kan | 30 |
| SP21 | WS8N derivative; ΔrpoN3::kan rpoN3 (RSWS8N_19584) | 17 |
| SP22 | WS8N derivative; ΔrpoN4::kan rpoN4 (RSWS8N_04590) | 17 |
| SP23 | WS8N derivative; ΔRSWS8N_RS13335::hyg | This work |
| SP24 | WS8N derivative; ΔRSWS8N_RS04480::kan | This work |
| WS8N | Wild-type strain; spontaneous Nalr | 38 |
| Escherichia coli strains | ||
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7 | 53 |
| TOP10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pB45 | Plasmid source of the hygromycin cassette | 42 |
| pBBMCS53 | Transcriptional uidA fusion vector; Gmr | 35 |
| pBBMCS53_cheA4 | pBBMCS53 carrying the cheOp3 promoter | This work |
| pBBMCS53_cheOp1L | pBBMCS53 carrying 652 bp upstream of cheOp1 | This work |
| pBBMCS53_cheOp1S | pBBMCS53 carrying 591 bp upstream of cheOp1 | This work |
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pFlgC1 | Integration vector used to create a C-terminal fusion with the FLAG tag epitope; Spcr | 45 |
| pJQ200_ΔctrA::hyg | pJQ200mp18 carrying ΔctrA::hyg | This work |
| pJQ200_mcpB::uidA-aadA | pJQ200mp18 carrying mcpB::uidA-aadA | This work |
| pJQ_ΔRSWS8N_RS04480::kan | pJQ200mp18 carrying ΔRSWS8N_RS04480::kan | This work |
| pJQ_ΔRSWS8N_13335::hyg | pJQ200 mp18 carrying ΔRSWS8N_13335::hyg | This work |
| pJQ200mp18 | Mobilizable suicide vector for R. sphaeroides; Gmr | 41 |
| pRK_ctrA | pRK415 derivative expressing ctrA | 21 |
| pTZ_ctrAupdown | pTZ19R Bam− carrying ctrAupdown | 21 |
| pTZ EcoRI− SacI− | pTZ19R without EcoRI and SacI sites | Laboratory collection |
| pTZ_Eco− SstI− ΔRSWS8N_RS04480::kan | pTZ19R EcoRI− SacI− carrying ΔRSWS8N_RS04480::kan | This work |
| pTZ_Eco− SacI− RSWS8N_RS04480 | pTZ19R EcoRI− SacI− carrying RSWS8N_RS04480 | This work |
| pTZ19R | Cloning vector; Apr | Thermo Scientific |
| pTZ19R Bam− | pTZ19R without BamHI site | Laboratory collection |
| pTZ19R BamHI−_ΔRSWS8N_RS13335::hyg | pTZ19R Bam− carrying ΔRSWS8N_RS13335::hyg | This work |
| pTZ19R BamHI−_RSWS8N_RS13335 | pTZ19R Bam− carrying RSWS8N_RS13335 | This work |
| pTZ18R_mcpB | pTZ18R carrying mcpB | 26 |
| pTZ18R_mcpB::uidA-aadA | ptZ18R carrying mcpB::uidA-aadA | This work |
| pUC4K | Plasmid source of the kanamycin cassette | Pharmacia |
| pWM5 | Vector source of the uidA-aadA cassette | 46 |
Oligonucleotides.
The oligonucleotides used in this work are listed in Table S1 in the supplemental material.
Isolation of mutant strains.
In general terms, to inactivate RSWS8N_RS13335 and RSWS8N_RS04480, which encode proteins with characteristic bEBP signatures, a suicide vector (41) carrying the target gene disrupted with an antibiotic resistance gene was used to perform allelic exchange. Specifically, to inactivate RSWS8N_RS13335, which encodes a 446-residue protein, the oligonucleotides 159-1 and 159-2 (both carrying an XbaI recognition site at the 5′ end), were used to PCR amplify a 2,541-bp product containing the complete coding region of RSWS8N_RS13335, as well as regions upstream and downstream of this gene. This product was cloned into pTZ19RBam−, and the resulting plasmid was digested with BamHI and partially digested with BglII to remove a 573-bp portion of RSWS8N_RS13335 (residues 229 to 420). The resulting fragment was ligated to a BamHI-digested, 1.4-kb hygromycin resistance cassette (hyg), obtained by PCR from the pB45 plasmid (42). The plasmid carrying the ΔRSWS8N_RS13335::hyg allele was digested with XbaI, and the fragment containing the mutant allele was subcloned into pJQ200mp18 (41). This plasmid was then introduced into E. coli S17-1 (43) and subsequently transferred to R. sphaeroides by conjugation (44). Since pJQ200mp18 cannot replicate in R. sphaeroides, the double-recombination event was selected directly on LB agar plates in the presence of hygromycin and 5% sucrose. The mutant was verified by PCR.
A similar strategy was used to inactivate RSWS8N_RS04480, which encodes a 645-residue protein. The oligonucleotides 187-1 and 187-2 (both carrying an XbaI recognition site at the 5′ end) were used to PCR amplify a 2,968-bp product containing the complete coding region of RSWS8N_RS04480, as well as regions upstream and downstream of the target gene. This product was cloned into pTZ18REcoRI− SacI−, and the resulting plasmid was digested with EcoRI and SacI to remove a 1,098-bp segment of RSWS8N_RS04480 (residues 4 to 370). The resulting fragment was end repaired with T4 DNA polymerase and ligated with a kanamycin resistance cassette (kan) obtained from the pUC4K plasmid. The plasmid carrying the ΔRSWS8N_RS04480::kan allele was digested with XbaI, and the fragment containing the mutant allele was subcloned into pJQ200mp18. This plasmid was then introduced into E. coli S17-1 and subsequently transferred to R. sphaeroides by conjugation. The mutant strains were selected, and the gene replacement was verified by PCR.
To obtain strain JV1 (mcpA-Flag), a 764-bp PCR product corresponding to the 3′ end of mcpA without its stop codon but including an extra guanine was obtained using the oligonucleotides mcpAtagFw and mcpAtagRv; the McpA-Flag fusion was obtained by cloning the PCR product into the pFlgC1 plasmid (45). The resulting plasmid was introduced into R. sphaeroides by conjugation, and integration into the chromosome by single recombination was selected. The Spcr transconjugants were tested by PCR and Western blotting by using monoclonal antibodies against the Flag epitope. Subsequently, the ctrA+ allele in the JV1 strain was replaced with the ΔctrA::hyg allele by using the plasmid pJQ200_ΔctrA::hyg. This plasmid was obtained by inserting a BglII fragment carrying the hyg cassette into the BamHI site of the pTZ_ctrAupdown plasmid (21).
To obtain the mutant strains JV3 (mcpB::uidA-aadA) and JV4 (mcpB::uidA-aadA ΔctrA::hyg), we used the plasmid pTZ18R_mcpB (26), which carries a 1,830-bp fragment of the coding region of mcpB plus 107 bp upstream of the start codon and 40 bp downstream of the stop codon. This plasmid was digested with NcoI, end repaired with T4 DNA polymerase, and ligated to a 4-kb SmaI fragment obtained from pWM5, which carries the uidA-aadA cassette (46). The fragment containing the mcpB::uidA-aadA allele was subcloned into pJQ200mp18. This plasmid was introduced into AM1 and LC7 strains to replace the chromosomal mcpB gene by following the procedure described above.
Plasmids used in this work.
A transcriptional fusion between the regulatory region of cheOp1 and the reporter gene uidA was obtained by cloning a 652-bp fragment of the upstream region of cheOp1 into the pBBMCS53 plasmid (35). This fragment was obtained by PCR using the oligonucleotides cheOpromFw and cheOpromRv. The resulting plasmid was named pBBMCS53_cheOp1L. The pBBMCS53 plasmid was designed to generate transcriptional fusions with the uidA gene, which encodes the enzyme β-glucuronidase (35). The plasmid pBBMCS53_cheOp1S was obtained by cloning a 591-bp PCR product carrying the regulatory region of cheOp1 (without the putative CtrA binding site identified in this work) into the pBBMCS53 plasmid. The PCR product in this plasmid was generated using the oligonucleotides cheOprommut and cheOpromRv. Both plasmids were introduced into AM1 and EA1 strains by conjugation.
Molecular biology techniques.
Standard methods were used to obtain chromosomal or plasmid DNA (40). Restriction and other DNA-modifying enzymes were acquired from New England BioLabs (NEB), Roche, or Invitrogen. For sequencing, plasmids were purified using the Illustra plasmidPrep mini spin kit (GE Healthcare Life Sciences). Chromosomal or plasmid DNA was amplified with the appropriate oligonucleotides using PrimeSTAR HS DNA polymerase (TaKaRa Bio Inc.) according to the manufacturer's recommendations.
RNA isolation and RT-PCR assays.
Total RNA was isolated from cells grown to an optical density at 600 nm (OD600) of 0.5 in Sistrom's minimal medium using hot phenol (47). After purification, the samples were incubated with RNase-free DNase I using the DNA-free DNA removal kit (Thermo Fisher Scientific). Reverse transcription and PCR amplification were performed with the Access RT-PCR system (Promega) according to the manufacturer's instructions. A control reaction mixture lacking reverse transcriptase was run in parallel. The PCR products were analyzed by agarose or acrylamide gel electrophoresis. The oligonucleotides cheY5XbaFw and cheY5PstRev were used to test the region between RSWS8N_02465 and cheY5. CheY6UP and tlpTdown were used to test the region between cheY6 and tlpT. The oligonucleotides cheY4Fw and cheY4Rev were used to amplify an internal region of cheY4. FlgHFw and flgHRv were used as a control to amplify an internal region of flgH.
Primer extension.
Total RNA was purified as previously described (47). The oligonucleotide cheOp2B was 5′ end labeled with [γ-32P]ATP. Then, 50 μg of total RNA was hybridized with the labeled oligonucleotide at 42°C in the presence of 50% formamide. The hybrid RNA-DNA was ethanol precipitated and resuspended in a 30-μl reaction volume. The cDNA was synthetized using avian myeloblastosis virus (AMV) reverse transcriptase. The products were subjected to electrophoresis on a 6% polyacrylamide–8 M urea gel. As a molecular marker, we used DNA from PhiX174 digested with HinfI (Promega) and 5′ end labeled with [γ-32P]ATP. The images were visualized using a Typhoon scanner (GE Healthcare Life Science) and quantified using ImageJ software (48).
Western blot analysis.
Samples were obtained from exponentially growing cultures at an OD600 of 0.6 from heterotrophically grown cultures and at an OD600 of 0.3 for photoheterotrophic cultures grown in 80 μM succinic acid. Whole bacterial cells were lysed by boiling in a solution containing 2% SDS, 1% β-mercaptoethanol, and 50 mM Tris (pH 7.5). These samples were separated on 12% polyacrylamide gels. The proteins were electrophoretically transferred to nitrocellulose membranes (49). The membranes were then probed with antibodies raised in rabbits against CheY2, CheY3, CheY4, CheY5, and CheY6 (1:3,000) or probed with a commercial mouse antibody against the Flag epitope tag (1:10,000) (Sigma-Aldrich). Anti-rabbit or anti-mouse IgG antibodies (1:30,000) (Sigma-Aldrich) coupled to alkaline phosphatase were used for chemiluminescent detection with CDP-star as the substrate (Applied Biosystems). His-tagged CheY proteins were purified by following previously reported protocols (50). Rabbit immunizations were performed as reported elsewhere (49). Representative results of three independent experiments are shown in Fig. 2A, S1, 4A, 5A, and 6A.
β-Glucuronidase activity assay.
β-Glucuronidase assays employed 4-methylumbelliferyl-β-d-glucuronide (MUG) as the substrate along with sonicated cell extracts, as previously described (51). Samples of 100 μl were taken at three time points between 5 and 20 min and then mixed with 0.9 ml of stop buffer (0.2 M Na2CO3). Fluorimetric determinations were made on a Perkin-Elmer LS-5 apparatus (excitation, 360 nm; emission, 446 nm). The fluorimeter was calibrated using 4-methylumbelliferone (Sigma-Aldrich) standards. Specific enzyme activity in cell extracts was expressed as picomoles of methylumbelliferone per minute per milligram of protein. Protein content was determined with a Bio-Rad protein assay kit, using bovine serum albumin as the standard.
Identification of the CtrA and σ28 binding sites.
The sequence upstream of cheY4 was analyzed using a position weight matrix (PWM) built using the σ28 promoters from E. coli (36) with the tool matrix-scan (52) included in the Regulatory Sequence Analysis Tools (RSAT) (http://embnet.ccg.unam.mx/rsa-tools/). The sequence upstream of the first gene in the cheOp1 operon, RSWS8N_02465, was analyzed using the same tool, with a PWM built from the regulatory region of 54 genes identified to be controlled by CtrA in C. crescentus (31).
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Ana Martinez del Campo for the RT-PCR experiments depicted in Fig. 3B and to Aurora Osorio and Teresa Ballado for their helpful technical assistance. We thank the Molecular Biology Unit IFC-UNAM for sequencing facilities. Language editing of the manuscript was done by Springer Nature Services.
This work was partially supported by DGAPA-UNAM (PAPIIT-IN204614) and CONACyT (CB2014-235996).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00670-16.
REFERENCES
- 1.Baker MD, Wolanin PM, Stock JB. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 2.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 3.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He K, Bauer CE. 2014. Chemosensory signaling systems that control bacterial survival. Trends Microbiol 22:389–398. doi: 10.1016/j.tim.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnosti DN, Chamberlin MJ. 1989. Secondary sigma factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci U S A 86:830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang TC, Leu YW, Chang-Chien HC, Hu RM. 2009. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J Bacteriol 191:2266–2275. doi: 10.1128/JB.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A 99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. 2014. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci U S A 111:3217–3224. doi: 10.1073/pnas.1400421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 16.Poggio S, Abreu-Goodger C, Fabela S, Osorio A, Dreyfus G, Vinuesa P, Camarena L. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J Bacteriol 189:3208–3216. doi: 10.1128/JB.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poggio S, Osorio A, Dreyfus G, Camarena L. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol Microbiol 46:75–85. doi: 10.1046/j.1365-2958.2002.03158.x. [DOI] [PubMed] [Google Scholar]
- 18.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol 169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poggio S, Osorio A, Dreyfus G, Camarena L. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol Microbiol 58:969–983. doi: 10.1111/j.1365-2958.2005.04900.x. [DOI] [PubMed] [Google Scholar]
- 20.de la Mora J, Uchida K, del Campo AM, Camarena L, Aizawa S, Dreyfus G. 2015. Structural characterization of the Fla2 flagellum of Rhodobacter sphaeroides. J Bacteriol 197:2859–2866. doi: 10.1128/JB.00170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vega-Baray B, Domenzain C, Rivera A, Alfaro-Lopez R, Gomez-Cesar E, Poggio S, Dreyfus G, Camarena L. 2015. The flagellar set Fla2 in Rhodobacter sphaeroides is controlled by the CckA pathway and is repressed by organic acids and the expression of Fla1. J Bacteriol 197:833–847. doi: 10.1128/JB.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah DS, Porter SL, Martin AC, Hamblin PA, Armitage JP. 2000. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J 19:4601–4613. doi: 10.1093/emboj/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter SL, Armitage JP. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol 324:35–45. doi: 10.1016/S0022-2836(02)01031-8. [DOI] [PubMed] [Google Scholar]
- 24.Porter SL, Wadhams GH, Martin AC, Byles ED, Lancaster DE, Armitage JP. 2006. The CheYs of Rhodobacter sphaeroides. J Biol Chem 281:32694–32704. doi: 10.1074/jbc.M606016200. [DOI] [PubMed] [Google Scholar]
- 25.del Campo AM, Ballado T, de la Mora J, Poggio S, Camarena L, Dreyfus G. 2007. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J Bacteriol 189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-del Campo A, Ballado T, Camarena L, Dreyfus G. 2011. In Rhodobacter sphaeroides, chemotactic operon 1 regulates rotation of the flagellar system 2. J Bacteriol 193:6781–6786. doi: 10.1128/JB.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin AC, Gould M, Byles E, Roberts MA, Armitage JP. 2006. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54. J Bacteriol 188:7932–7940. doi: 10.1128/JB.00964-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Darbari VC, Zhang N, Lu D, Glyde R, Wang YP, Winkelman JT, Gourse RL, Murakami KS, Buck M, Zhang X. 2015. Structures of the RNA polymerase-σ54 reveal new and conserved regulatory strategies. Science 349:882–885. doi: 10.1126/science.aab1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poggio S, Osorio A, Dreyfus G, Camarena L. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J Biol Chem 281:27205–27215. doi: 10.1074/jbc.M601735200. [DOI] [PubMed] [Google Scholar]
- 31.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouimet MC, Marczynski GT. 2000. Analysis of a cell-cycle promoter bound by a response regulator. J Mol Biol 302:761–775. doi: 10.1006/jmbi.2000.4500. [DOI] [PubMed] [Google Scholar]
- 34.Porter SL, Wadhams GH, Armitage JP. 2008. Rhodobacter sphaeroides: complexity in chemotactic signalling. Trends Microbiol 16:251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Girard L, Brom S, Davalos A, Lopez O, Soberon M, Romero D. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol Plant Microbe Interact 13:1283–1292. doi: 10.1094/MPMI.2000.13.12.1283. [DOI] [PubMed] [Google Scholar]
- 36.Grote A, Klein J, Retter I, Haddad I, Behling S, Bunk B, Biegler I, Yarmolinetz S, Jahn D, Munch R. 2009. PRODORIC (release 2009): a database and tool platform for the analysis of gene regulation in prokaryotes. Nucleic Acids Res 37:D61–D65. doi: 10.1093/nar/gkn837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domenzain C, Camarena L, Osorio A, Dreyfus G, Poggio S. 2012. Evolutionary origin of the Rhodobacter sphaeroides specialized RpoN sigma factors. FEMS Microbiol Lett 327:93–102. doi: 10.1111/j.1574-6968.2011.02459.x. [DOI] [PubMed] [Google Scholar]
- 38.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp 53:473–479. [Google Scholar]
- 39.Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J Gen Microbiol 28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 40.Ausubel FM, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1987. Current protocols in molecular biology John Wiley and Sons, New York, NY. [Google Scholar]
- 41.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 42.Servín-Gonzalez L, Sampieri AI, Cabello J, Galván L, Juárez V, Castro C. 1995. Sequence and functional analysis of the Streptomyces phaeochromogenes plasmid pJV1 reveals a modular organization of Streptomyces plasmids that replicate by rolling circle. Microbiology 141(Part 10):2499–2510. doi: 10.1099/13500872-141-10-2499. [DOI] [PubMed] [Google Scholar]
- 43.Priefer UB, Simon R, Pühler A. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol 163:324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis J, Donohue TJ, Kaplan S. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol 170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metcalf WW, Wanner BL. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17–25. doi: 10.1016/0378-1119(93)90691-U. [DOI] [PubMed] [Google Scholar]
- 47.Aiba H, Adhya S, de Crombrugghe B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 256:11905–11910. [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harlow E, Lane D. 1988. Antibodies. A Laboratory Manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 50.Ferre A, De La Mora J, Ballado T, Camarena L, Dreyfus G. 2004. Biochemical study of multiple CheY response regulators of the chemotactic pathway of Rhodobacter sphaeroides. J Bacteriol 186:5172–5177. doi: 10.1128/JB.186.15.5172-5177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jefferson RA, Burgess SM, Hirsh D. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A 83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. 2008. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc 3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.