ABSTRACT
Whole-genome sequences of representative highly pathogenic avian influenza A(H5) viruses from Vietnam were generated, comprising samples from poultry outbreaks and active market surveillance collected from January 2012 to August 2015. Six hemagglutinin gene clades were characterized. Clade 1.1.2 was predominant in southern Mekong provinces throughout 2012 and 2013 but gradually disappeared and was not detected after April 2014. Clade 2.3.2.1c viruses spread rapidly during 2012 and were detected in the south and center of the country. A number of clade 1.1.2 and 2.3.2.1c interclade reassortant viruses were detected with different combinations of internal genes derived from 2.3.2.1a and 2.3.2.1b viruses, indicating extensive cocirculation. Although reassortment generated genetic diversity at the genotype level, there was relatively little genetic drift within the individual gene segments, suggesting genetic stasis over recent years. Antigenically, clade 1.1.2, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c viruses remained related to earlier viruses and WHO-recommended prepandemic vaccine strains representing these clades. Clade 7.2 viruses, although detected in only low numbers, were the exception, as indicated by introduction of a genetically and antigenically diverse strain in 2013. Clade 2.3.4.4 viruses (H5N1 and H5N6) were likely introduced in April 2014 and appeared to gain dominance across northern and central regions. Antigenic analyses of clade 2.3.4.4 viruses compared to existing clade 2.3.4 candidate vaccine viruses (CVV) indicated the need for an updated vaccine virus. A/Sichuan/26221/2014 (H5N6) virus was developed, and ferret antisera generated against this virus were demonstrated to inhibit some but not all clade 2.3.4.4 viruses, suggesting consideration of alternative clade 2.3.4.4 CVVs.
IMPORTANCE Highly pathogenic avian influenza (HPAI) A(H5) viruses have circulated continuously in Vietnam since 2003, resulting in hundreds of poultry outbreaks and sporadic human infections. Despite a significant reduction in the number of human infections in recent years, poultry outbreaks continue to occur and the virus continues to diversify. Vaccination of poultry has been used as a means to control the spread and impact of the virus, but due to the diversity and changing distribution of antigenically distinct viruses, the utility of vaccines in the face of mismatched circulating strains remains questionable. This study assessed the putative amino acid changes in viruses leading to antigenic variability, underscoring the complexity of vaccine selection for both veterinary and public health purposes. Given the overlapping geographic distributions of multiple, antigenically distinct clades of HPAI A(H5) viruses in Vietnam, the vaccine efficacy of bivalent poultry vaccine formulations should be tested in the future.
KEYWORDS: highly pathogenic avian influenza, Vietnam, antigenicity, evolution, reassortment, vaccine selection
INTRODUCTION
Measures to control the 2003 epizootics of highly pathogenic avian influenza (HPAI) A(H5N1) virus in Vietnam included the stamping out of infected and neighboring flocks and restriction of poultry movements. Vaccination of poultry has also been a major component of the disease control strategy since September 2005. Despite these efforts to control HPAI A(H5N1) in Vietnam, eradication has not been achieved, likely due in part to the multiple introductions and spread of viruses with distinct genetic and antigenic properties from neighboring countries. Phylogenetic analyses of whole-genome sequences (WGSs) of H5N1 viruses isolated from poultry outbreaks in Vietnam have identified a total of 48 genotypes and circulation of at least 15 distinct hemagglutinin (HA) clades (1–4). Until recently, the geographic distribution of H5 virus clades across Vietnam suggested nonoverlapping transmission zones; virus descendants from clade 2.3.4 lineages were detected exclusively in the north, with frequent lineage replacement via introduction of new viruses from bordering countries, whereas clade 1 lineages circulated exclusively in the Mekong River Delta and neighboring Cambodia (5). In addition, clade 7 virus was occasionally detected in poultry that had been illegally imported from China and then transported to live-bird markets (LBMs) located in northern Vietnam (6–8). Emergence of clade 2.3.2.1 and three clusters within this group (i.e., A/Hubei/1/2010-like [Hubei-like, 2.3.2.1a], A/barn swallow/Hong Kong/1161/2010-like [BS-like, 2.3.2.1b], and A/Hong Kong/6841/2010-like [HK-like, 2.3.2.1c]) was reported in Vietnam in 2009 and continued to cause outbreaks through 2012 (3). Significant antigenic diversity between these viruses suggested possible vaccine “breakthrough.” Evidence of suboptimal protection of poultry vaccinated with Re-1, which has HA and neuraminidase (NA) genes derived from A/goose/Guangdong/1/1996 (clade 0), Navet-Vifluvac, which has HA and NA genes derived from A/Vietnam/1194/2004 (clade 1), and Re-5, which has HA and NA genes derived from A/duck/Anhui/1/2006 (clade 2.3.4 H5N1 virus), led to a temporary suspension of the national avian influenza vaccination program in mid-2012 (9, 10). In addition, during 2012 and 2013, clade 2.3.2.1 viruses were detected in an expanding range, which accompanied the disappearance of the previously dominant clade 2.3.4 viruses in northern and central Vietnam (1). Subsequently, the Re-6 vaccine, which has HA and NA genes derived from A/duck/Guangdong/S1322/2010 (clade 2.3.2.1b), was produced in China and approved for use in Vietnam (10). Currently, both Re-5 and Re-6 vaccines are used in poultry (11).
Outbreaks of HPAI A(H5) virus associated with high mortality are sporadically reported in poultry throughout Vietnam. Despite ongoing detection in birds, the number of human case reports has declined over the last several years, with no cases detected in 2015 (12). Importantly, the lack of recent human cases of H5 viruses suggests that control measures to prevent zoonotic infections are effective. However, both passive surveillance in response to H5 poultry outbreaks and active surveillance through monthly sampling in live-bird markets (LBM) continue to detect circulation of the virus throughout the country (13). These surveillance platforms provide essential components for monitoring viral diversity and guiding effective application of disease control interventions. Here we report on the epidemiological situation in Vietnamese poultry and molecular evolution of HPAI A(H5) virus in Vietnam from 2012 through 2015. Full-genome sequence analysis of samples collected across the country, together with antigenic characterization, demonstrated a shifting distribution and introduction of virus clades and antigenic variants with an increasing number of reassortant viruses resulting from cocirculation of multiple clades and genotypes. Molecular clock analysis was performed to estimate the timing of these events and to assess variability among the rates of evolution of the viruses detected. Hemagglutination inhibition (HI) assays using panels of ferret antisera raised against previously circulating strains and prepandemic candidate vaccine viruses (CVVs) were performed to address the issue of antigenic matches between newly detected viruses and available vaccines.
RESULTS
Sample collection, identification, and sequencing.
Between September 2012 and August 2015, over 19,000 samples from 55 provinces were analyzed at the National Center for Veterinary Diagnostics (NCVD), Hanoi, Vietnam, and regional Department of Animal Health (DAH) offices; 666 samples were from poultry outbreaks, whereas 18,405 were collected through market-based surveillance (Table 1). A total of 1,448 A(H5)-positive specimens were detected by real-time reverse transcription-PCR (RT-PCR) (259 H5 outbreak samples, 17.89%; 1,189 H5 market samples, 82.11%) (Table 1). Virus isolation in embryonated chicken eggs yielded 214 isolates from 45 provinces (see Tables S1 and S2 in the supplemental material). Among the species, 59.8% (128 isolates) were from ducks (unknown species), Muscovy ducks, and geese, 35.5% (76 isolates) were from chickens, and 4.7% were from other species (6 quail, 2 pheasant, 1 swiftlet, and 1 tiger from a zoological collection). Of these, 90.6% (194 isolates) were H5N1 and 9.4% (20 isolates) were H5N6. A total of 119 full-length HA and NA sequences and 95 partial HA1 sequences were generated from the 214 isolates. A total of 106 whole-genome sequences (WGSs) were generated from isolates selected for sequencing based on representative collection locations and dates (see Table S2 in the supplemental material). Additional partial genomes were obtained from 11 viruses. The complete data set used for analysis here comprised the 106 novel Vietnamese H5 WGSs from 2012 to 2015, plus 169 WGSs from 2010 to 2012 that were available and had been described in previous studies (1), for a total of 275 WGSs.
TABLE 1.
Passive and active sample collection and A (H5) detection from September 2012 to August 2015
| Period of collection | Sourcea | Total no. of samples | Total no. of H5-positive samples | % of positive samples | No. of provinces |
|---|---|---|---|---|---|
| September to December 2012 | Market-based surveillance | 2,546 | 125 | 4.91 | 21 |
| Outbreak | 109 | 56 | 51.38 | 15 | |
| 2013 | Market-based surveillance | 4,904 | 336 | 6.85 | 44 |
| Outbreak | 123 | 83 | 67.48 | 20 | |
| 2014 | Market-based surveillance | 8,926 | 655 | 7.34 | 30 |
| Outbreak | 311 | 84 | 27.01 | 33 | |
| January to August 2015 | Market-based surveillance | 2,029 | 73 | 3.60 | 14 |
| Outbreak | 123 | 36 | 29.27 | 13 | |
| Total | Market-based surveillance | 18,405 | 1,189 | 6.46 | 50 |
| Outbreak | 666 | 259 | 38.89 | 46 |
Market-based surveillance, active surveillance through monthly sampling in live-bird markets; outbreak, passive surveillance in response to poultry outbreak.
Highly pathogenic avian influenza A(H5) virus identification.
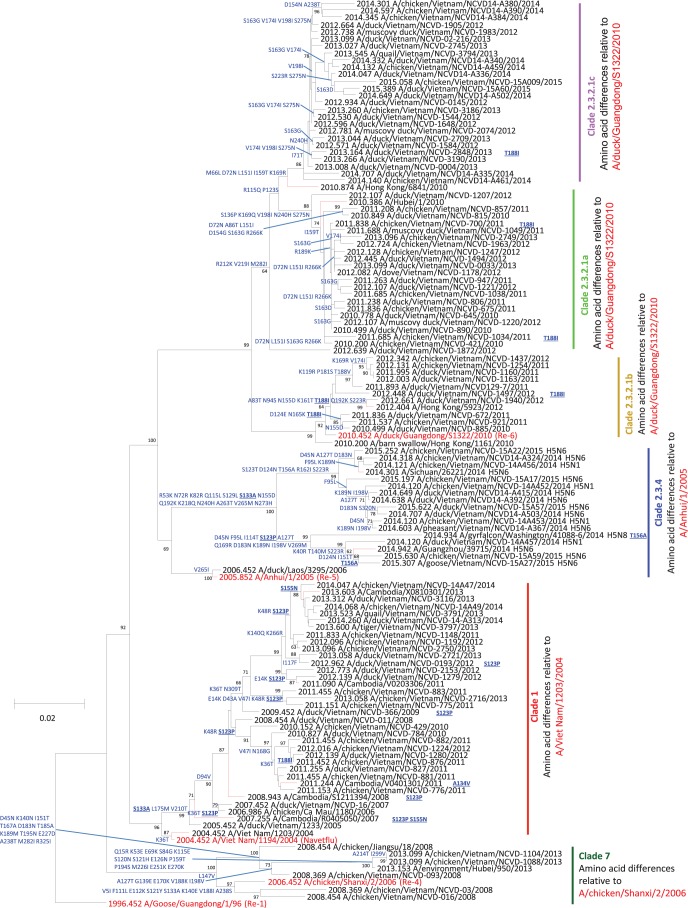
The viruses identified belonged to six different clades, including 1.1.2, 2.3.2.1a, 2.3.2.1b, 2.3.2.1c, 2.3.4.4, and 7.2 (Fig. 1; see also Fig. S2a in the supplemental material). With the exception of a few rare detections of clade 2.3.2.1a, 2.3.2.1b, and clade 7.2 viruses, the predominant H5N1 clades detected throughout this period were clades 1.1.2 and 2.3.2.1c. Twenty-three clade 1.1.2 viruses were identified from October 2012 to April 2014; the majority of those (n = 21) were detected in the southern provinces of the Mekong Delta. The two remaining clade 1.1.2 viruses were found in the north region (Bac Ninh) and central region (Khanh Hoa). The average level of pairwise nucleotide divergence within clade 1.1.2 viruses was 2.1%. The last detection of 1.1.2 viruses was in April 2014. Clade 2.3.2.1 HA viruses were classified into three subclades (2.3.2.1a, 2.3.2.1b, and 2.3.2.1c). Clade 2.3.2.1a viruses previously circulated widely throughout Vietnam and Southeast Asia between 2010 and 2012. However, they were gradually replaced by clade 2.3.2.1c viruses, starting in July 2012 (1). Here we report detection of four clade 2.3.2.1a viruses in early 2013 but none after August 2013. Clade 2.3.2.1b never achieved a wider circulation beyond Hong Kong SAR and China, and the last detection was in August 2013. In contrast, clade 2.3.2.1c viruses were found in 16 provinces in the north of Vietnam, 12 provinces in the center, and 9 provinces in the south. Although clade 2.3.2.1c viruses were identified in 37 provinces, they were highly related to each other, forming a distinct monophyletic group within the larger multicountry 2.3.2.1c lineage. To date, clade 7.2 viruses (n = 3) have been found only in North Vietnam (Ha Noi province) and were detected in poultry that had likely been smuggled across northern Vietnam borders. In February 2014, an emerging subclade of 2.3.4 A(H5N1), termed 2.3.4.4, was detected in central Vietnam (Quang Ngai and Kon Tum), followed shortly thereafter by detection of the same clade 2.3.4.4 HA gene but with an N6 NA gene in April 2014. H5N6 viruses were then reported in 11 provinces in northern (n = 6) and central (n = 5) regions and were detected throughout the rest of the study period. Phylogenic analysis indicated that the H5N6 viruses originated through reassortment with Vietnamese H5N1 viruses (Fig. 1; see also Fig. S2a). To date, two diverse clusters of Vietnamese H5N6 have been identified. The first cluster included recent (2015) viruses (n = 6) that were closely related to the Jiangxi-like virus in China (A/duck/Jiangxi/NCDZT1126/2014-like), and the second group (n = 14) included viruses more closely related to Sichuan-like viruses (A/Sichuan/26221/2014-like).
FIG 1.
Maximum likelihood tree of the HA1 genes of H5 highly pathogenic avian influenza A(H5) viruses. The nearest reassortant WHO candidate vaccine viruses (CVV) for each clade are colored red. Amino acid differences at branch nodes indicate shared HA1 substitutions relative to the reference strains. Mutations listed to the right of each strain name indicate amino acid changes found only in that individual virus. Mutations highlighted with underlining indicate previously recognized molecular markers. Branches on the tree with HA sequences from human cases are colored red. Bootstraps with values greater than 50 generated from 1,000 replicates are shown at branch nodes. The scale bar represents the number of nucleotide substitutions per site.
Reassortment of HPAI H5 internal genes.
Phylogenetic analyses of the neuraminidase (NA) gene and internal genes (PB2, PB1, PA, NP, M, and NS) revealed numerous instances of reassortment among the 106 WGSs (Fig. 2; see also Fig. S2a to h). Similarly to the H5 HA gene phylogeny, multiple lineages of N1 NA and internal genes cocirculated in Vietnam. Eleven (of 19) clade 1.1.2 viruses were of the typical VN3 genotype, 3 of 4 clade 2.3.2.1a viruses were genotype VN12, 1 clade 2.3.2.1b virus was genotype VN45, 42 (of 66) clade 2.3.2.1c viruses were genotype VN48, and 3 clade 7.2 viruses were genotype VN43.
FIG 2.
Schematic diagram of genome constellations of H5 viruses in Vietnam, 2012 to 2015. Surface and/or internal protein gene derivatives of clade 1.1.2 viruses are shown in red, clade 2.3.2.1a viruses in light green, clade 2.3.2.1b viruses in brown, clade 2.3.2.1c viruses in purple, clade 2.3.4.4 H5N6 viruses in blue, clade 7.2 viruses in dark green, H6N6 LPAI PB2 viruses in black, H7N7 LPAI PB2 viruses in light blue, and N6 with a stalk deletion in orange.
Each gene segment for the H5 subtype viruses in Vietnam formed gene constellations related to the HA clades: clade 1.1.2, clade 2.3.2.1a to 2.3.2.1c, clade 7.2, and clade 2.3.4.4 H5N6. Five previously described genotypes were detected (VN3, VN12, VN45, VN48, and VN43) (1, 3, 4), and eight newly identified genotypes were detected (VN49 [n = 8], VN50 [n = 1], VN51 [n = 2], VN52 [n = 15], VN53 [n = 7], VN54 [n = 9], VN55 [n = 3], and VN56 [n = 1]). NA and internal gene sequences of clade 1.1.2, clade 2.3.2.1a, and clade 2.3.2.1b viruses were closely related to the corresponding genes found in viruses isolated before September 2012 in Vietnam, China, and Cambodia (1, 3, 4).
NA and internal genes of the majority of clade 1.1.2 viruses shared a common ancestor defined by the progenitor strain A/duck/Cambodia/202W6M1/2013. However, the six internal gene sequences found in eight 2013 clade 1.1.2 viruses were related to those found in clade 2.3.2.1a viruses, indicating reassortment and generation of genotype VN49. Clade 2.3.2.1c viruses also revealed evidence of reassortment, and whole-genome analysis revealed 3 new genotypes in this clade with internal gene sequences previously found in the clade 2.3.2.1a or clade 2.3.2.1b viruses (Fig. 2).
Analyses indicated the presence of a clade 2.3.2.1a reassortant collected in 2012 with HA, PB2, and PA genes from clade 2.3.2.1a and PB1, NP, NA, MP, and NS genes from clade 2.3.2.1b (VN50). Further genetic reassortment was evident among clade 2.3.4.4 A(H5N6) viruses. The NA gene of all 13 H5N6 viruses reported here most likely originated from related strains of H6N6 viruses circulating in the Fujian and Guangdong provinces of China (Fig. 2; see also Fig. S2f). The internal genes of 9 viruses were related to clade 2.3.2.1b viruses, similarly to strains found in domestic ducks from south-central and eastern China (Fig. S2b to h) (VN54). Three of the H5N6 viruses had 5 internal genes related to clade 2.3.2.1b viruses, but the PB2 was distinct and related to A/duck/Yamagata/061004/2014 (H6N6) (termed VN55). Similarly, one H5N6 virus (A/chicken/Vietnam/NCVD-15A22/2015) had the same five internal genes from clade 2.3.2.1b but had a PB2 gene more closely related to that of A/chicken/Jiangxi/10784/2014 (H7N7) (VN56).
Geotemporal distribution of clades and genotypes in Vietnam.
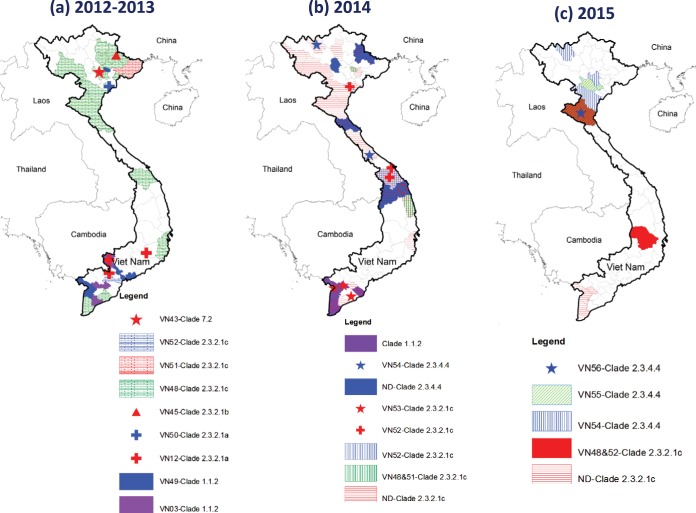
The geographic distribution of H5 HA clades and genotypes detected between September 2012 and August 2015 was analyzed, and the results reveal the expansion of clade 2.3.2.1c (genotypes VN48 and VN52) throughout the country (Fig. 3). Clade 2.3.2.1c VN53 viruses were detected only in the south, while VN51 viruses were found only in the north. Only VN50 virus was detected in a north province (Nam Dinh) in 2012; however, the older original clade 2.3.2.1a VN12 strain circulated in all three regions during 2013. Clade 2.3.2.1b (VN45) and clade 7.2 (VN43) were identified from the north early in early 2013 but did not circulate persistently. Clade 1.1.2 (VN3 and VN49) were found in southern provinces, with the last reported detection in April 2014 (Fig. 3). Clade 2.3.4.4 A(H5N6) viruses (VN54, VN55, and VN56) were detected in the central and northern provinces from 2014 to August 2015 and appear to have been gaining dominance toward the end of 2015 (data not shown).
FIG 3.
Spatial distribution of H5 virus clades and genotypes in Vietnam. (a) 2012 to 2013. (b) 2014. (c) 2015. Maps drawn using ArcGIS software (Esri, Redlands, CA).
Substitution rates and divergence times.
To further distinguish the evolutionary features of the major clades detected and discern potential differences in selective pressures acting on each genomic segment, we estimated nucleotide substitution rates by Bayesian analysis of each gene segment (Table 2; see also Fig. S1). Clade 2.3.2.1c HA genes exhibited a higher mean substitution rate (6.87E−3) than clade 1.1.2 or clade 2.3.4.4 genes; however, the overlapping ranges of the 95% highest posterior density (HPD) intervals indicated that these differences were likely not significant and could also have been due to a larger population having been sampled. Analysis of substitution rates by segment showed values that ranged from 2.63E−3 to 5.61E−3 substitutions/site/year; the mean rate for HA was highest among the gene segments, followed by those for NA and NS. However, these overlapping HPD intervals suggested that the differences were not statistically significant. Substitution rates of viral polymerase genes (PB2, PB1, and PA) fell in the range of 3.83E−3 to 4.55E−3, and the MP gene had the lowest rate (mean, 2.63E−3), which would be expected due to the highly conserved nature of this gene.
TABLE 2.
Estimation of rate of evolution of Vietnamese H5 viruses, 2012 to 2015
| Stratification | Uncorrelated relaxed clock model | Mean substitution rate (×10−3) | 95% HPDa interval (×10−3) |
|---|---|---|---|
| Clade | |||
| 1.1.2 | Lognormal | 5.69 | 4.72 to 6.69 |
| 2.3.2.1c | Lognormal | 6.87 | 5.39 to 8.40 |
| 2.3.4.4 | Lognormal | 5.98 | 4.90 to 7.16 |
| Gene | |||
| PB2 | Exponentialb | 4.55 | 3.54 to 5.78 |
| PB1 | Lognormal | 4.18 | 3.61 to 4.76 |
| PA | Lognormal | 3.83 | 3.27 to 4.37 |
| HA | Lognormal | 5.61 | 4.98 to 6.25 |
| NP | Lognormal | 3.39 | 2.70 to 4.12 |
| NA | Lognormal | 4.83 | 4.25 to 5.42 |
| MP | Lognormal | 2.63 | 2.10 to 3.13 |
| NS | Lognormal | 4.80 | 3.085 to 6.66 |
HPD, highest posterior density.
An exponential model was used due to the small effective sample size with the lognormal model.
Bayesian estimates of the divergence times of clades 1.1.2, 2.3.2.1c, and 2.3.4.4 through calculation of times to the most recent common ancestor (tMRCA) indicated that the HA gene for clade 1.1.2 viruses diverged in October 2011 from that of closely related viruses from Cambodia (Fig. S1a to c). The estimated tMRCA for the clade 2.3.2.1c HA gene indicated that most viruses diverged in June 2013 from closely related viruses from Khanh Hoa province (central region; see Fig. S1b). From mid-2012 to early 2013, the population size of this clade expanded as the virus spread and caused outbreaks in new regions across the country (Fig. S1b). All Vietnam viruses of subtype A(H5N6) appear to have diverged from Chinese ancestors in July 2012. The viruses in the first cluster, including the most recent viruses collected in 2015, were estimated to have emerged during September 2014 in Vietnam from a closely related Jiangxi-like virus (A/duck/Jiangxi/NCDZT1126/2014). The second and larger group of A(H5N6) viruses was estimated to have diverged from ancestral strains around July 2012 and emerged in Vietnam in February 2014. This cluster was related to 2013/2014 Chinese viruses (e.g., A/Sichuan/26221/2014) (Fig. S1c). Although the larger genetic lineage of A(H5N6) viruses had a stable population size from 2011 through 2013, the emergence of these clusters in Vietnam and the coinciding expansion and spread of the clade 2.3.4.4 HA genes in Europe and North America during 2014 resulted in an increased population size (Fig. S1c).
Molecular markers and putative antigenic sites of the HA1 protein.
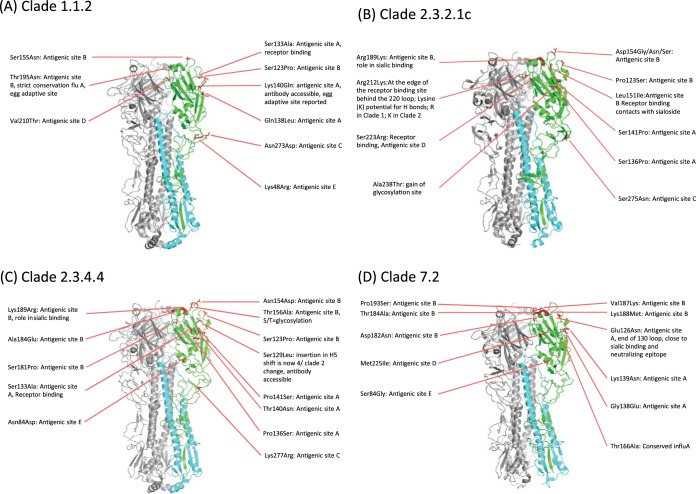
The multibasic cleavage motif was present in all H5 HA protein sequences; however, there were consistently conserved differences across clades. The motif PQREERRKKR↓G was identified in 19/21 sequences from clade 1.1.2; PQRERRRKR↓G was found in clade 2.3.2.1c (n = 73/77) and clade 2.3.2.1a (n = 4/4); clade 2.3.2.1b had PQIERRRRKR↓G; clade 2.3.4.4 had PLREKRRKR↓G; and clade 7.2 had PQIEGRRRKRG↓G. Three viruses had additional mutations in comparison to their respective clades: two clade 1.1.2 viruses had PQREGRRKKR↓G and PQRKERRKKR↓G, respectively, and one clade 2.3.2.1c virus had PQKERRRKR↓G. All viruses showed replication in MDCK cells in the absence of trypsin. Conserved amino acid mutations in the HA1 protein of each clade with respect to the closest H5 candidate vaccine virus are shown in Fig. 1 and 4 (see also Tables S3 to S8 in the supplemental material). Clade 1.1.2 viruses averaged 13 amino acid substitutions relative to A/Viet Nam/1203/2004, 9 changes relative to A/Cambodia/R0405050/2007, and 12 changes relative to A/Vietnam/1194/2004 (Navet-Vifluvac; clade 1 Vietnamese poultry vaccine) (Fig. 4A; see also Table S3). Clade 2.3.2.1a viruses had an average of 6 amino acid substitutions relative to A/Hubei/1/2010, 5 of which occurred in predicted antigenic sites, and 19 amino acid substitutions relative to the poultry vaccine Re-6 (A/duck/Guangdong/S1322/2010) (Table S4). Clade 2.3.2.1b viruses, which belong to the same clade as the vaccine virus, A/duck/Guangdong/S1322/2010 (Re-6), had 10 conserved amino acid substitutions in HA1 relative to the WHO CVVs A/barn swallow/Hong Kong/1151/2010 and A/duck/Guangdong/S1322/2010 (Re-6), with 5 occurring in epitope regions (Table S5). Similarly, clade 2.3.2.1c viruses possessed an average of 9 coding changes relative to A/Hubei/1/2010 and 17 amino acid mutations relative to Re-6, 17 of which were identified in putative epitope regions (Fig. 4B; see also Table S6). The greatest number of changes in relation to the nearest WHO CVV or available poultry vaccine was identified in clade 2.3.4.4 A(H5N6) viruses, with an average 27 and 40 mutations relative to Re-5 and Re-6, respectively (Fig. 4C; see also Table S7). Finally, clade 7.2 HA1 sequences had an average of 34 amino acid substitutions compared to the sequence of the A/chicken/Shanxi/2/2006 (Re-4) vaccine strain (Fig. 4D; see also Table S8).
FIG 4.
HA1 substitutions identified at putative antigenic sites on the HA trimeric structure. Amino acid differences between the consensus sequence of each clade and the sequence of the nearest poultry vaccine virus are displayed for clade 1.1.2 versus A/Vietnam/1203/2004 (A), clade 2.3.2.1c versus A/duck/Guangdong/S1322/2010 (Re-6) (B), clade 2.3.4.4 versus A/Anhui/1/2005 (Re-5) (C), and clade 7.2 versus A/chicken/Shanxi/2/2006 (Re-4) (D).
All 119 HA sequences had glycosylation sites at position 286. The majority had additional putative glycosylation sites at positions 10 (n = 99), 11 (n = 100), 23 (n = 101), 165 (n = 101), 484 (n = 108), and 543 (n = 108). Analysis of the glycosylation motif at position 154 showed that all clade 1.1.2 and 7.2 sequences retained the motif, whereas the glycosylation was ablated in clade 2.3.4.4 sequences and clade 2.3.2.1a to -c sequences, except for one recently analyzed clade 2.3.2.1c sequence (A/chicken/Vietnam/NCVD14-A380/2014).
Molecular markers within NA and internal genes.
Substitutions in NA proteins associated with altered susceptibility to neuraminidase inhibitors (NAIs) were identified in numerous clade 1.1.2 viruses but not in viruses from other clades (Table 3). A V129A substitution (or residue 149 in N2 numbering) associated with reduced susceptibility to zanamivir (1, 14) was found in 17 clade 1.1.2 viruses collected in 2012 (n = 2) and 2013 (n = 15). Another marker of reduced susceptibility to oseltamivir associated with an I203V mutation (or residue 222 in N2 numbering) was found in a single clade 1.1.2 virus. Mutation of R430Q, associated with reduced susceptibility to NAIs, was found in a single clade 1.1.2 virus (A/chicken/Vietnam/NCVD-2750/2013). The PB2 K627 residue has been implicated in significant phenotypic changes in polymerase activity, which imparts replicative fitness advantages and virulence in mammals (15, 16). PB2-E627 was identified in 111 viruses, suggesting a lack of mammalian adaptation in these viruses. Similarly, substitutions of R368Q have been associated with reduced replication efficiency in cell culture and virulence in mice and ferrets. This residue was identified in 91 viruses. A T339K substitution which enhanced polymerase activity and increased virulence in mice was found in 91 viruses (17). An N328K or N375S substitution, which decreased replication efficiency and virulence in ferrets, was found in the PB1 protein encoded by a single clade 2.3.2.1c virus and two clade 1.1.2 viruses. The M2 protein of one clade 2.3.2.4 virus had a V27A mutation, and one clade 2.3.2.1c virus encoded an A30S mutation, both of which were associated with reduced susceptibility to rimantadine. The substitution N31K in the M2 protein, which may reduce susceptibility to rimantadine, was encoded by 8, 68, and 13 clade 1.1.2, 2.3.2.1c, and 2.3.4.4 viruses, respectively (Table 3). All viruses encoded NS1 proteins with the deletion of amino acids 80 to 84 associated with altered virulence in mice. An D92E substitution in the NS1 protein, also associated with altered virulence in mice, was encoded by all viruses, except for the clade 1.1.2 viruses. A T47A substitution in the NS2 protein, which was associated with decreased antiviral response in the host, was encoded by all clade viruses, except for two clade 2.3.2.1c viruses. Finally, clade 7.2 viruses encoded a protein with a mutation of M51I associated with decreased antiviral response.
TABLE 3.
Molecular markers identified in H5 viruses in Vietnam
| Gene | Mutation location/phenotypic consequence (s) | Mutation | Molecular marker (% of viruses with substitution) or amino acids |
|||
|---|---|---|---|---|---|---|
| Clade 1 | Clade 2.3.2.1c | Clade 2.3.4.4 (H5N6) | Clade 7.2 | |||
| HA | Antigenic site C | D43A | A (4.8) | |||
| Antigenic site E | K48R | R (95.2) | ||||
| Antigenic site E | S84G | G (100) | ||||
| Antigenic site B; increased α2-6 binding | S123P | P (90.5) | S (85.7) | P (30.7) | ||
| Antigenic site A | E126N | N (100) | ||||
| Antigenic site A; increased α2-6 binding | S133A | A (100) | (100) | |||
| Antigenic site A | Q138L | L (19.0) | ||||
| Antigenic site A | G138E | E (100) | ||||
| Antigenic site A | K140N/Q | Q (90.5); N (4.8) | ||||
| Antigenic site A | S141P | P (3.9) | P (7.7) | |||
| Antigenic site B; receptor binding | L151I | I (87.0) | T (31.0) | |||
| Antigenic site B | D154N/G | N (10.4); G (2.6) | ||||
| Antigenic site B | S155N | N (47.6) | ||||
| Antigenic site B; predicted loss of glycosylation; increased α2-6 binding; increased transmission in mammalian models | T156A | A (100) | ||||
| Antigenic site B | S181P | P (23.08) | ||||
| Antigenic site B | T182A | A (100) | ||||
| Antigenic site B | D183N | N (61.53) | ||||
| Antigenic site B | A184E | E (23.08) | ||||
| Antigenic site B | V184K | K (100) | ||||
| Antigenic site B | K187M | M (100) | ||||
| Antigenic site B | P188S | S (100) | ||||
| Antigenic site B; increased virus binding to α2-6 | R189K | K (3.9) | ||||
| Antigenic site B | Q192K | K (100) | ||||
| Antigenic site B | P193S | S (100) | ||||
| Antigenic site B | T195N | N (9.5) | ||||
| Antigenic site D | V210T | T (100) | ||||
| Edge of the receptor binding site | R212K | K (87.0) | ||||
| Increased virus binding to α2-6; antigenic site D | S223R | R (6.5) | R (100) | |||
| Antigenic site D | M225I | I (100) | ||||
| Antigenic site C | N273D | D (4.8) | ||||
| Antigenic site C | S275N | N (79.2) | ||||
| Highly pathogenic cleavage peptides | PQRE/KE/GRRKKR↓G | PQR/KERRRKR↓G | PLREKRRKR↓G | PQIEGRRRKRG↓G | ||
| NA | Stalk deletion | 59–69del | del (30.8) | |||
| Reduced susceptibility to zanamivir | V129A | A (80.9) | ||||
| Reduced susceptibility to oseltamivir | I203V | A (4.8) | ||||
| Associated with reduced inhibition by NAIs | R430Q | Q (4.8) | ||||
| PB2 | Enhanced polymerase activity; increased virulence in mice | T339K | K (42.1) | K (94.4) | K (61.5) | K (100) |
| Reduction in replication efficacy in cell culture and virulence in mice and ferrets | R368Q | Q (42.1) | Q (97.2) | Q (68.2) | ||
| Reduction in replication efficiency in cell culture and virulence in mice and ferrets | Q391E | E (100) | E (100) | E (100) | E (100) | |
| Increased replication efficiency in cell culture and enhanced virulence in mice; mammalian host adaptation | K627E | E (100) | E (100) | E (100) | E (100) | |
| PB1 | Decreased replication efficiency and virulence in ferrets | N328K | K (1.41) | |||
| Decreased replication efficiency and virulence in ferrets | N375S | S (10.5) | ||||
| PB1-F2a | Increased pathogenicity in mice | |||||
| PA | K356R | R (1.0) | ||||
| M2 | Reduced susceptibility to amantadine/rimantadine | V27A | A (7.69) | |||
| Reduced susceptibility to amantadine/rimantadine | A30S | S (1.41) | ||||
| Reduced susceptibility to amantadine/rimantadine | N31S | S (42.1) | S (100) | S (100) | ||
| NS1b | Altered virulence in mice | D92E | E (100) | E (100) | E (100) | |
| Altered virulence in mice, PDZ motif | 227–230 | |||||
| NS2 | Decreased antiviral response in host | T47A | A (100) | A (100) | A (100) | |
| Decreased antiviral response in host | M51I | I (100) | ||||
Protein lengths for clade 1, 90 amino acids (aa) (80% density) and 57 aa (20% density); protein length for clade 2.3.2.1c, 57 aa (100% density); protein length for clade 2.3.4.4 (H5N6), 57 aa (100% density); protein length for clade 7.2, 90 aa (100% density).
NS1 also showed a deletion of amino acids 80 to 84 that was associated with altered virulence in mice for all four clades.
Antigenic characterization of H5 viruses circulating in Vietnam.
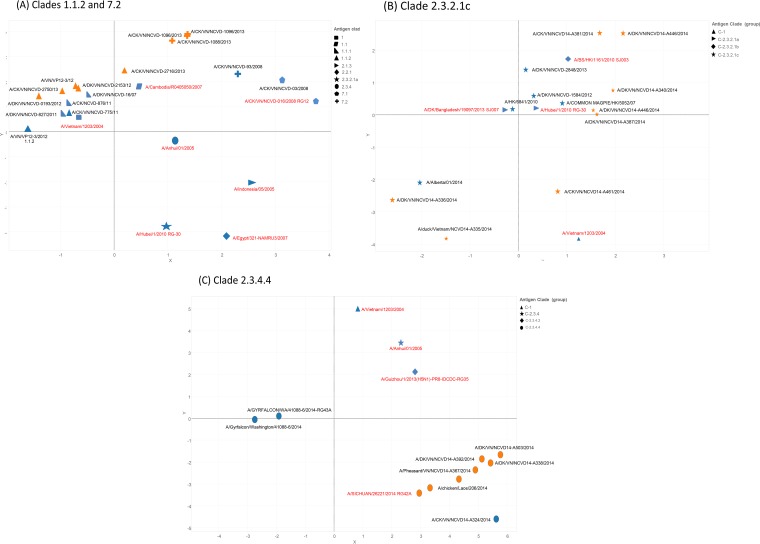
The HI assay was used to assess the antigenic relatedness of poultry viruses in Vietnam (Fig. 5). Most clade 1 viruses tested with antiserum raised to the CVV A/Vietnam/1203/04 virus showed HI titers 4-fold lower than the homologous titer, but approximately 20% of viruses tested had ≥8-fold reductions in heterologous titers. Antiserum raised against A/Cambodia/X0810301/2013, the most recently developed clade 1.1.2 CVV, reacted with 8 of 11 viruses collected from 2013 to 2014 in Vietnam with HI titers that were within 4-fold or less of the homologous virus titer. Three viruses collected in 2013 to 2014 from Vietnam reacted with antiserum at titers that were 8-fold to 32-fold lower than the homologous virus titer (Fig. 5A). The proteins encoded by the tested viruses collected in 2012 to 2014 differed by 8 to 12 amino acids from those of the A/Vietnam/1203/2004 and A/Vietnam/1194/2004 candidate vaccine viruses but by only 5 to 6 amino acids from those of the candidate vaccine virus, A/Cambodia/X0810301/2013 (Table 2 and Fig. 4A).
FIG 5.
Antigenic mapping for clades 1.1.2 and 7.2 (A), clade 2.3.2.1c (B), and clade 2.3.4.4 (C). Each grid indicates 1 unit of antigenic distance, a 2-fold dilution in HI titer.
Antigenic analyses of 34 clade 2.3.2.1a avian isolates from Vietnam revealed that 23 of 34 viruses showed titers that were within 8-fold of the homologous HI titer with antiserum to A/cm/HK/5052/2007. Twelve of 17 clade 2.3.2.1a viruses showed ≥8-fold differences in HI titers in comparison to the titers seen with antiserum against the A/Hubei/1/10 vaccine candidate (relative to homologous antigen). These viruses also showed reduced titers with antiserum to the A/VN/NCVD700/11 strain, albeit the differences were in the 4-fold range. Antiserum produced to the A/Hubei/1/2010 CVV reacted with three viruses from Vietnam at titers that were at least 16-fold higher than the homologous titers. New antiserum produced to a more closely related Vietnamese clade 2.3.2.1a virus (A/chicken/Vietnam/NCVD-1255/2012) reacted within 2-fold or less with the same Vietnam viruses (Fig. 5B). Six clade 2.3.2.1b viruses were analyzed here. The HI titers of antiserum to A/barn swallow/HK/D10-1161/2010 tested against these viruses showed ≤4-fold differences from those seen with the homologous antigen (Fig. 5B).
Antiserum to A/HK/6841/2010 (clade 2.3.2.1c) showed a majority of viruses (33 of 34) with titers that differed from that of the homologous virus by <4-fold. Seventeen clade 2.3.2.1c avian isolates collected in 2013 from Vietnam were characterized. Ferret antiserum raised against the CVV, A/duck/Vietnam/NCVD-1584/2012, reacted with all viruses with HI titers that were within 2-fold or less of the titer with homologous virus. Ferret antisera against A/duck/Vietnam/NCVD-1584/2012 inhibited well a majority of 2014 poultry viruses from Vietnam also, with HI titers within 4-fold of the homologous virus titers. One exception was a poultry virus which possessed the R189K HA1 mutation and exhibited an 8-fold-reduced HI titer compared to the HI titer to the homologous virus (Fig. 4B and 5B). Three 2013 clade 7.2 viruses collected in Vietnam revealed no detectable HI titers with sera produced against clade 7.1 CVVs (Fig. 5A). Extensive amino acid sequence divergence continues to be detected in clade 7 viruses from 2013 relative to previous CVVs, A/chicken/Vietnam/NCVD-016/2008 and A/chicken/Vietnam/NCVD-03/2008 (Table S8), suggesting the reason for the antigenic distance between these viruses and the CVVs (Fig. 5A).
Ferret antiserum raised against clade 2.3.4.4 A/gyrfalcon/Washington/41088-6/2014 (H5N8) reacted with titers that were within 2-fold of or were equivalent to the homologous and heterologous virus titers of related H5N8 viruses, indicating antigenic similarity. In contrast, this antiserum did not cross-react well with H5N6 viruses from Vietnam. Similarly, antiserum raised to A/chicken/Vietnam/NCVD-14-A324/2014 (H5N6) reacted poorly with H5N8 and H5N2 viruses detected in Europe and North America; heterologous virus HI titers were greater than 8-fold less than the homologous virus titer (Fig. 5C). The HA1 proteins of H5N8 and H5N2 viruses detected in the United States differed from those of the H5N6 viruses by 11 to 12 substitutions identified at 4 potential antigenic sites (data not shown). The HA1 proteins of poultry H5N6 viruses recently circulating in Vietnam, Laos, and China differed from that of the H5N6 CVV by four or fewer amino acids but had on average 13 substitutions compared to those of the RE-5 poultry vaccine (Fig. 4C). Notably, the Vietnamese H5N6 antiserum reacted at a level that was within 4-fold of the level seen with A/chicken/Laos/206/2014 (H5N6). HI titers of antisera raised to the A/Sichuan/26211/2014 (H5N6) vaccine strain were reduced (>8-fold) against H5N2 and H5N8 viruses detected in the United States. However, this H5N6 antiserum reacted at a level that was within 2-fold of the levels seen with A/chicken/Laos/206/2014 (H5N6) and well-inhibited H5N6 viruses detected in Vietnamese poultry (Fig. 5C).
DISCUSSION
Given the cocirculation of several clades and genotypes of A(H5N1) viruses in Vietnam during recent years and the difficulties in achieving optimum vaccination coverage under field conditions, there is a need to continuously monitor and characterize circulating H5N1 viruses to promptly detect changes in genetic and antigenic characteristics. This study focused on the molecular epidemiology of HPAI H5 viruses collected from poultry outbreaks and active surveillance at live-bird markets (LBMs) in Vietnam during the period from 2012 to 2015. The antigenic relatedness of newly circulating viruses to older strains as well as to prepandemic candidate vaccine viruses recommended by the WHO was determined. Full-genome sequencing and dated phylogenic analysis were performed to identify further expansion and reassortment of H5 viruses in Vietnam and to estimate the time periods during which these events occurred. Molecular changes associated with the diversification of each major clade and the emergence of novel genotypes identified specific HA mutations associated with antigenic drift and substitutions previously correlated with increased pandemic risk. Throughout the study period, several key events, such as the emergence of clade 2.3.2.1c, the likely disappearance of clade 1.1.2, and the emergence and reassortment of clade 2.3.4.4 viruses, were evaluated to assess the need for development of public health and poultry vaccines with improved antigenic matches.
The comprehensive phylogenetic analysis of Vietnamese H5 viruses presented here revealed significant diversification, reassortment, and lineage replacement over the last 3 years. Sequence analyses of Vietnamese viruses compared to those from other countries revealed that clade 1.1.2 viruses were closely related only to those found in Cambodia, confirming previous reports of sustained enzootic circulation of these viruses within the Mekong Delta region. Despite this apparent geographic restriction, clade 1.1.2 viruses continued to diversify genetically through 2013 and reassorted most likely with clade 2.3.2.1a viruses, acquiring their internal genes and giving rise to a novel genotype (VN49) also identified in Cambodia (18). Clade 1 viruses have not been detected by either active or passive surveillance since April 2014 in Vietnam. It was unclear on the basis of available sequence data whether or not this clade continued to circulate on the Cambodian portion of the delta beyond 2014. Interestingly, the most recent clade 1.1.2 viruses detected in Vietnam possessed all internal genes from clade 2.3.2.1 viruses, suggesting a possible loss of fitness of clade 1.1.2 viruses with this genome arrangement. Clade 2.3.2.1c viruses, most likely introduced into Vietnam in July 2012, became predominant throughout Vietnam during 2013 and gradually displaced clade 1.1.2 viruses from the Mekong Delta region, where clade 1 (VN3) had been predominant since 2003.
The broad geographic distribution and genetic diversity of the three groups of clade 2.3.2.1 in Vietnam suggested multiple separate introductions rather than endemic genetic drift. The tMRCA estimates also support the idea of a series of independent introductions. Clade 2.3.2.1a viruses appeared as early as 2009 and continued to be detected from 2012 to 2013 throughout the northern, central, and southern regions, during which time this group was also identified in Bangladesh, Bhutan, Laos, India, Nepal, and Myanmar. Both clades 2.3.2.1a and 2.3.2.1b appear to have been displaced by clade 2.3.2.1c viruses by 2014. During this surveillance period, clade 2.3.2.1c viruses became dominant in both the south and center of Vietnam, causing sporadic outbreaks, and were identified in LBMs through August 2015. 2.3.2.1c viruses disseminated widely throughout Southeast Asia, China, Japan, South Korea, Mongolia, Russia, and Europe, indicating widespread circulation likely mediated by large outbreaks in wild-bird populations (19–24). Despite their detection in 37 of 63 provinces in Vietnam, the clade 2.3.2.1c viruses showed limited diversity, suggesting that they originated from a single progenitor source. The long phylogenetic branch lengths separating the Vietnamese clade 2.3.2.1c viruses from those found in other countries make it difficult to speculate about potential origin.
The cocirculation of the three subgroups of clade 2.3.2.1 led to reassortment between these viruses and emergence of several novel genotypes with the HA of clade 2.3.2.1c and different combinations of internal genes derived from 2.3.2.1a and 2.3.2.1b (genotypes VN48, VN51, VN52, and VN53). Although these reassortments generated genetic diversity at the genotype level, to date there has been relatively little drift within the HA and NA and not much evidence for diversification of the internal genes of VN51 to VN53, suggesting that these reassortants originated from a not-so-distant ancestor. Clade 2.3.4.4 viruses were detected in the northern and central regions of the country in April 2014 and have since gained dominance in many provinces throughout the country. These viruses, which also circulated in China, Laos, and South Korea during 2014 and 2015, generated several new reassortant 2.3.2.1 genotypes in Vietnam. The PB2 genes of several clade 2.3.4.4 viruses were derived from low-pathogenicity avian influenza (LPAI) viruses, indicating their cocirculation as well. The lack of reassortment between clade 1.1.2 and 2.3.4.4 viruses in Vietnam could be explained by the displacement of clade 1.1.2 from the Mekong region prior to or close to the incursion of clade 2.3.4.4, with no substantial period of cocirculation among the two viruses.
Although clade 1.1.2 viruses appear to have been displaced during this study period, antigenic testing of viruses revealed that existing prepandemic vaccine candidates belonging to this clade covered circulating viruses fairly well. The same was true for clade 2.3.2.1c viruses, which cross-reacted with the most closely related vaccine strain with little to no reduction in serum HI titers. The few viruses that were not inhibited well by antisera raised against this clade had an R189K substitution in antigenic site B of the mature HA1 protein. This change has previously been associated with a loss of antigenic cross-reactivity. Interestingly, characterization of the dominant immunogenic portion of the 2.3.2.1c surface protein (mature HA1) indicated a significant number of amino acid substitutions in other positions that did not appear to affect antigenicity in relation to the WHO CVV. Clade 7.2 viruses were not inhibited by ferret antisera developed against earlier clade 7.1 viruses. As a consequence of this and in recognition of evidence of continued circulation of the clade 7.2 in China in 2014 and 2015, a recently proposed WHO CVV against clade 7.2 is under development (25). Finally, clade 2.3.4.4 presents a unique challenge for optimizing antigenic matching due to the expanding range and diversity of these viruses. HI testing demonstrated that recent clade 2.3.4.4 viruses from Vietnam reacted less well with postinfection ferret antiserum raised to the North American clade 2.3.4.4 A/gyrfalcon/Washington/41088-6/2014 (H5N8), the virus from which a clade 2.3.4.4 CVV was developed. This finding indicated significant antigenic distances between North American and East Asian clade 2.3.4.4 viruses and suggested low vaccine coverage by the North American candidate vaccine virus. In contrast, the A/Sichuan/26221/2014 strain, also developed as a CVV, cross-reacted well with the 2014 H5N6 viruses tested, demonstrating the need for more than one clade 2.3.4.4 vaccine candidate despite their recent global emergence and spread. In fact, increasing genetic and antigenic diversity among clade 2.3.4.4 viruses detected in China and Vietnam has also led to the WHO recommendation to develop two additional A(H5N6) clade 2.3.4.4 CVVs (25). Because of the antigenic diversity among even genetically related WHO prepandemic vaccine candidates, these findings demonstrate that systematic antigenic testing of H5 clade variants in the context of current and proposed poultry vaccine strains is also urgently needed to assess potential impacts on poultry vaccine efficacy. Antigenic characterization of circulating viruses with antisera raised specifically to vaccine strains in avian hosts (e.g., chickens and/or ducks) would allow for timely assessment of antigenic matching. Routine identification of A(H5) antigenic variants may also help to guide selection of challenge viruses used in experimental vaccine efficacy trials in poultry. Together, these data have the potential to inform poultry vaccine strain selection in a manner akin to the WHO pandemic vaccine strain selection process.
In summary, contemporary H5 viruses circulating in Vietnamese poultry demonstrate the ever-changing genetic and antigenic characteristics of these viruses and their propensity to continue evolving both at the individual gene level and over the genome as a whole. Given the overlapping geographic distributions of H5 clade variants and their antigenic divergence, the vaccine efficacy of multivalent vaccine formulations containing representatives of both clade 2.3.2.1c and 2.3.4.4 viruses should be tested to determine their ability to protect poultry in Vietnam.
MATERIALS AND METHODS
Viruses used in this study.
Viruses used in this study included HPAI A(H5) virus isolates obtained from poultry outbreaks and active surveillance in LBMs derived from national programs implemented by the National Centre for Veterinary Diagnostics (NCVD) and Department of Animal Health (DAH) between September 2012 and August 2015. Original samples from poultry outbreaks comprised tissue homogenates from poultry carcasses (typically brain or lung specimens from deceased birds). Market samples were pooled oropharyngeal duck swabs (typically five swabs per pool). Spatial analyses of sample collection locations were conducted in ArcGIS 9.3 (26) using the Vietnamese national spatial database (Fig. 3; see also Table S1 in the supplemental material) (27). Screening for detection of influenza A(H5) by real-time RT-PCR and virus isolation in embryonated eggs was performed as previously described (7). Representative samples were shipped to the Centers for Disease Control and Prevention (CDC), Influenza Division (Atlanta, GA, USA), for virus isolation and/or passaging and further characterization. The majority of whole-genome sequences (WGS) were generated at the CDC by Sanger sequencing as previously described (1). A subset of 2015 outbreak samples were deep sequenced using Illumina MiSeq in Vietnam with protocols adopted from St. Jude Children's Research Hospital (28). Sequences were submitted to the GenBank sequence database (see accession numbers in Table S2). Handling of infectious materials at CDC was performed in compliance with biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agents program (http://www.cdc.gov/od/ohs/biosfty/bmbl5/bmbl5toc.htm).
Molecular analyses.
Analysis of H5 clade distribution was based on all HA sequences generated during the study period (n = 214). HA sequences were aligned via MUSCLE (29) and trimmed to the mature H5 HA coding region using BioEdit v7.0 (30). A maximum likelihood tree was constructed using MEGA 5.05. The tree was rooted to A/goose/Guangdong/1/1996 (Fig. 1). Amino acid differences on each branch reflect differences from the nearest Vietnamese poultry vaccine, and underlined mutations indicate previously recognized residues associated with antigenic epitopes, changes in receptor affinity, mammalian adaptation, and increased virulence or transmissibility as identified from automated search tools within the Rainbow software package (5, 17). Branches with viruses sequenced from human cases are colored red. Bootstraps with numbers greater than 50 generated from 1,000 replicates are shown at branch nodes. The scale bar represents nucleotide substitutions per site. For genotyping, we compiled a comprehensive data set of all previously available WGS from Vietnam and combined these with the novel WGS generated here. Alignments were created for each gene segment. Additional non-Vietnamese A(H5) sequences were included as outgroups (Cambodian clade 1.1.2; Bangladesh clade 2.3.2.1a; China clade 2.3.4.4 and 7.2; H5N8 clade 2.3.4.4 from the United States). Neighbor-joining trees for each gene were implemented in MEGA5 with the Kimura 2-parameter substitution model and with 1,000 bootstrap replications (data not shown). Trees were either rooted to A/goose/Guangdong/1/1996 or were mid-point rooted. HA gene lineages were defined using criteria from the WHO/OIE/FAO H5 Evolution Working Group (WHO/OIE/FAO H5N1, 2008, 2012, 2014, and 2015), and genotype assignments followed conventions previously established for Vietnamese H5 virus characterization (Fig. 2) (1, 3, 4). Complete amino acid codon alignments of the NA and internal proteins were also used to identify amino acid residues conferring previously described viral phenotypes (Table 3) (17).
Evolutionary rates and time of most recent common ancestor.
For analysis of the time to the most recent common ancestor (tMRCA), we prepared a reduced data set of unique WGSs (n = 140) that remained representative of geographic regions, dates of collection, and phylogenetic clusters. Dates of collection (day/month/year) were transformed into decimal values. For viruses without precise collection dates, data representing the middle of the collection year were used. Markov chain Monte Carlo (MCMC) runs were performed in BEAST v1.8.1 (31, 32), with convergence evaluated in Tracer v1.5 after removal of 10% burn-in (33). Each analysis was repeated with the lognormal relaxed and exponential relaxed clock models and mean substitution rates with a normal prior distribution until the effective sample size (ESS) of each prior was at least 200. The model with the best fit was the SRD06 codon partition model using the general time-reversible (GTR) substitution model for each partition with unlinked frequencies. We used the lognormal relaxed clock (uncorrelated) model for all genes except for the PB2 gene (exponential model). The tree priors used were coalescent Bayesian Skyline with 10 groups and a piecewise constant skyline model with a randomly generated tree as a start. Each MCMC run had a chain length of 50 million and was sampled every 5,000 generations. For each tMRCA, a probability interval (Bayesian confidence interval) was estimated as the highest posterior density (HPD of 95%) that represented an interval in the domain of a posterior probability distribution. Maximum clade credibility (MCC) trees were generated in TreeAnnotator 1.7.1 (31) with 10% burn-in removed, posterior probabilities on 0.5, and median heights. Trees were visualized in FigTree version 1.3.1 (33), and posterior probabilities are shown on or above nodes.
Hemagglutination inhibition assay.
Antigenic characterization of HPAI H5N1 and H5N6 viruses isolated from poultry in Vietnam was performed using the hemagglutination inhibition (HI) assay with polyclonal ferret immune sera produced against representative circulating strains and vaccine candidates. A panel of antiserum raised against representative Vietnamese H5N1 viruses and WHO prepandemic H5N1 vaccine candidates was used to test the antigenic relatedness of circulating viruses collected during this study (34). HI assays were performed separately for clade 1.1.2, 2.3.2.1c, 7.2, and 2.3.4.4 viruses as previously described using turkey erythrocytes (35).
Antigenic cartography.
The HI results of each data set (by clades) were formatted for processing and were imported to ACMACS to generate x/y coordinate output files (https://acmacs-web.antigenic-cartography.org/). Metadata associated with all reference and test antigens were prepared in Excel spreadsheets with an antigen table (test, antigen identifier [ID], strain name, clade, and abbreviation) and an antisera table (test, serum ID, serum strain, serum clade, abbreviation, and serum type). Antigenic maps were generated from ACMACS output files using Tableau version 9.1 by importing x/y coordinate output files and linking each coordinate with reference or test antigen metadata (Fig. 5). HA1 substitutions identified at putative antigenic sites between the consensus sequence of each clade and nearest poultry vaccine virus sequence were displayed on models of the HA trimeric structure using Pymol version 7.2 (Fig. 4).
Accession number(s).
Sequences determined in this work have been submitted to the GenBank sequence database; the GenBank accession numbers are listed in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
The findings and conclusions in this report are ours and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
This work was funded, in part, by the United States Department of Health and Human Services, Centers for Disease Control and Prevention. Contributions of D.T.N. were supported by the Wellcome Trust and Oxford University, UK.
We are grateful to the Vietnamese Department of Animal Health and the Food and Agriculture Organization of the United Nations for support and implementation of national poultry surveillance systems. We thank staff of DAH, NCVD, RAHOs, and SDAHs of Vietnam and field veterinary staff for their support in collecting and testing samples. We thank Rogier van Doorn for critical reviews of early drafts of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01708-16.
REFERENCES
- 1.Creanga A, Diep Thi N, Gerloff N, Hoa Thi D, Balish A, Hoang Dang N, Jang Y, Vui Thi D, Thor S, Jones J, Simpson N, Shu B, Emery S, Berman L, Nguyen HT, Bryant JE, Lindstrom S, Klimov A, Donis RO, Davis CT, Tung N. 2013. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology 444:12–20. doi: 10.1016/j.virol.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Tung DH, Van Quyen D, Nguyen T, Xuan HT, Nam TN, Duy KD. 2013. Molecular characterization of a H5N1 highly pathogenic avian influenza virus clade 2.3.2.1b circulating in Vietnam in 2011. Vet Microbiol 165:341–348. doi: 10.1016/j.vetmic.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen T, Rivailler P, Davis CT, Hoa do T, Balish A, Dang NH, Jones J, Vui DT, Simpson N, Huong NT, Shu B, Loughlin R, Ferdinand K, Lindstrom SE, York IA, Klimov A, Donis RO. 2012. Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology 432:405–416. doi: 10.1016/j.virol.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Wan XF, Nguyen T, Davis CT, Smith CB, Zhao ZM, Carrel M, Inui K, Do HT, Mai DT, Jadhao S, Balish A, Shu B, Luo F, Emch M, Matsuoka Y, Lindstrom SE, Cox NJ, Nguyen CV, Klimov A, Donis RO. 21 October 2008. Evolution of highly pathogenic H5N1 avian influenza viruses in Viet Nam between 2001 and 2007. PLoS One doi: 10.1371/journal.pone.0003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rith S, Davis CT, Duong V, Sar B, Horm SV, Chin S, Ly S, Laurent D, Richner B, Oboho I, Jang Y, Davis W, Thor S, Balish A, Iuliano AD, Sorn S, Holl D, Sok T, Seng H, Tarantola A, Tsuyuoka R, Parry A, Chea N, Allal L, Kitsutani P, Warren D, Prouty M, Horwood P, Widdowson MA, Lindstrom S, Villanueva J, Donis R, Cox N, Buchy P. 10 September 2014. Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A (H5N1) viruses detected in Cambodia in 2013. J Virol doi: 10.1128/JVI.01887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis CT, Balish AL, O'Neill E, Nguyen CV, Cox NJ, Xu XY, Klimov A, Nguyen T, Donis RO. 2010. Detection and characterization of clade 7 high pathogenicity avian influenza H5N1 viruses in chickens seized at ports of entry and live poultry markets in Vietnam. Avian Dis 54:307–312. doi: 10.1637/8801-040109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen DT, Bryant JE, Davis CT, Nguyen LV, Pham LT, Loth L, Inui K, Nguyen T, Jang Y, To TL. 2014. Prevalence and distribution of avian influenza A (H5N1) virus clade variants in live bird markets of Vietnam, 2011–2013. Avian Dis 58:599–608. doi: 10.1637/10814-030814-Reg. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen T, Davis CT, Stembridge W, Shu B, Balish A, Inui K, Do HT, Ngo HT, Wan XF, McCarron M, Lindstrom SE, Cox NJ, Nguyen CV, Klimov AI, Donis RO. 2009. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology 387:250–256. doi: 10.1016/j.virol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Cha RM, Smith D, Shepherd E, Davis CT, Donis R, Nguyen T, Nguyen HD, Do HT, Inui K, Suarez DL. 2013. Suboptimal protection against H5N1 highly pathogenic avian influenza viruses from Vietnam in ducks vaccinated with commercial poultry vaccines. Vaccine 31:4953–4960. doi: 10.1016/j.vaccine.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 10.DAH. 2013. Annual report for animal diseases and control strategies. Department of Animal Health (DAH), Ha Noi, Viet Nam. [Google Scholar]
- 11.DAH. 22 February 2016. Circulation of avian influenza viruses and vaccination campaign for first round 2016 in Viet Nam (translated). Technical guideline number 262/TY-DT. Department of Animal Health (DAH), Ha Noi, Viet Nam. [Google Scholar]
- 12.WHO. 2016. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. World Health Organization (WHO), Geneva, Switzerland. [Google Scholar]
- 13.DAH. 2015. Annual report for animal diseases and control strategies. Department of Animal Health (DAH), Ha Noi, Viet Nam. [Google Scholar]
- 14.Naughtin M, Dyason JC, Mardy S, Sorn S, von Itzstein M, Buchy P. 2011. Neuraminidase inhibitor sensitivity and receptor-binding specificity of Cambodian clade 1 highly pathogenic H5N1 influenza virus. Antimicrob Agents Chemother 55:2004–2010. doi: 10.1128/AAC.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 17.CDC. 2012. H5N1 genetic changes inventory: a tool for influenza surveillance and preparedness. CDC, Atlanta, GA. [Google Scholar]
- 18.Sorn S, Sok T, Ly S, Rith S, Tung N, Viari A, Gavotte L, Holl D, Seng H, Asgari N, Richner B, Laurent D, Chea N, Duong V, Toyoda T, Yasuda CY, Kitsutani P, Zhou P, Bing S, Deubel V, Donis R, Frutos R, Buchy P. 2013. Dynamic of H5N1 virus in Cambodia and emergence of a novel endemic sub-clade. Infect Genet Evol 15:87–94. doi: 10.1016/j.meegid.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert M, Jambal L, Karesh WB, Fine A, Shiilegdamba E, Dulam P, Sodnomdarjaa R, Ganzorig K, Batchuluun D, Tseveenmyadag N. 2012. Highly pathogenic avian influenza virus among wild birds in Mongolia. PLoS One 7:e44097. doi: 10.1371/journal.pone.0044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Hou G, Jiang W, Han C, Liu S, Chen J, Li J, Zhang P, Huang B, Liu Y. 2012. A survey of avian influenza in tree sparrows in China in 2011. PLoS One 7:e33092. doi: 10.1371/journal.pone.0033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam M, Haque M, Giasuddin M, Chowdhury E, Samad M, Parvin R, Nooruzzaman M, Rahman M, Monoura P. 2012. New introduction of clade 2.3. 2.1 avian influenza virus (H5N1) into Bangladesh. Transbound Emerg Dis 59:460–463. doi: 10.1111/j.1865-1682.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 22.Nagarajan S, Tosh C, Smith DK, Peiris JSM, Murugkar HV, Sridevi R, Kumar M, Katare M, Jain R, Syed Z. 2012. Avian influenza (H5N1) virus of clade 2.3. 2 in domestic poultry in India. PLoS One 7:e31844. doi: 10.1371/journal.pone.0031844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakoda Y, Ito H, Uchida Y, Okamatsu M, Yamamoto N, Soda K, Nomura N, Kuribayashi S, Shichinohe S, Sunden Y. 2012. Reintroduction of H5N1 highly pathogenic avian influenza virus by migratory water birds, causing poultry outbreaks in the 2010–2011 winter season in Japan. J Gen Virol 93:541–550. doi: 10.1099/vir.0.037572-0. [DOI] [PubMed] [Google Scholar]
- 24.Uchida Y, Suzuki Y, Shirakura M, Kawaguchi A, Nobusawa E, Tanikawa T, Hikono H, Takemae N, Mase M, Kanehira K. 2012. Genetics and infectivity of H5N1 highly pathogenic avian influenza viruses isolated from chickens and wild birds in Japan during 2010–11. Virus Res 170:109–117. doi: 10.1016/j.virusres.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. September 2016. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. http://www.who.int/influenza/vaccines/virus/201609_zoonotic_vaccinevirusupdate.pdf?ua=1. [PubMed]
- 26.ESRI Inc. 2009. ArcGIS version 9.3. ESRI Inc, Redlands, CA. [Google Scholar]
- 27.NARENC. 2012. Spatial database of Viet Nam. Viet Nam Publishing House of Natural Resources, Environment and Cartography (NARENC), Ha Noi, Viet Nam. [Google Scholar]
- 28.Zaraket H, Baranovich T, Kaplan BS, Carter R, Song M-S, Paulson JC, Rehg JE, Bahl J, Crumpton JC, Seiler J, Edmonson M, Wu G, Karlsson E, Fabrizio T, Zhu H, Guan Y, Husain M, Schultz-Cherry S, Krauss S, McBride R, Webster RG, Govorkova EA, Zhang J, Russell CJ, Webby RJ. 8 April 2015. Mammalian adaptation of influenza A (H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun doi: 10.1038/ncomms7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 31.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23:7–9. [DOI] [PubMed] [Google Scholar]
- 33.Rambaut A. 2009. FigTree v1. 3.1 2006-2009. Retrieved 12 August 2012 http://tree.bio.ed.ac.uk/software/figtree/.
- 34.World Health Organization/World Organization for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. 2014. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses 8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balish AL, Davis CT, Saad MD, El-Sayed N, Esmat H, Tjaden JA, Earhart KC, Ahmed LaE, Abd El-Halem M, Ali AHM, Nassif SA, El-Ebiary EA, Taha M, Aly MM, Arafa A, O'Neill E, Xu X, Cox NJ, Donis RO, Klimov AI. 2010. Antigenic and genetic diversity of highly pathogenic avian influenza A (H5N1) viruses isolated in Egypt. Avian Dis 54:329–334. doi: 10.1637/8903-042909-Reg.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.