ABSTRACT
The Middle East respiratory syndrome coronavirus (MERS-CoV) nonstructural protein 16 (nsp16) is an S-adenosyl-l-methionine (SAM)-dependent 2′-O-methyltransferase (2′-O-MTase) that is thought to methylate the ribose 2′-OH of the first transcribed nucleotide (N1) of viral RNA cap structures. This 2′-O-MTase activity is regulated by nsp10. The 2′-O methylation prevents virus detection by cell innate immunity mechanisms and viral translation inhibition by the interferon-stimulated IFIT-1 protein. To unravel the regulation of nsp10/nsp16 2′-O-MTase activity, we used purified MERS-CoV nsp16 and nsp10. First, we showed that nsp16 recruited N7-methylated capped RNA and SAM. The SAM binding promotes the assembly of the enzymatically active nsp10/nsp16 complex that converted 7mGpppG (cap-0) into 7mGpppG2′Om (cap-1) RNA by 2′-OH methylation of N1 in a SAM-dependent manner. The subsequent release of SAH speeds up nsp10/nsp16 dissociation that stimulates the reaction turnover. Alanine mutagenesis and RNA binding assays allowed the identification of the nsp16 residues involved in RNA recognition forming the RNA binding groove (K46, K170, E203, D133, R38, Y47, and Y181) and the cap-0 binding site (Y30, Y132, and H174). Finally, we found that nsp10/nsp16 2′-O-MTase activity is sensitive to known MTase inhibitors, such as sinefungin and cap analogues. This characterization of the MERS-CoV 2′-O-MTase is a preliminary step toward the development of molecules to inhibit cap 2′-O methylation and to restore the host antiviral response.
IMPORTANCE MERS-CoV codes for a cap 2′-O-methyltransferase that converts cap-0 into cap-1 structure in order to prevent virus detection by cell innate immunity mechanisms. We report the biochemical properties of MERS-CoV 2′O-methyltransferase, which is stimulated by nsp10 acting as an allosteric activator of the nsp16 2′-O-methyltransferase possibly through enhanced RNA binding affinity. In addition, we show that SAM promotes the formation of the active nsp10/nsp16 complex. Conversely, after cap methylation, the reaction turnover is speeded up by cap-1 RNA release and nsp10/nsp16 complex dissociation, at the low intracellular SAH concentration. These results suggest that SAM/SAH balance is a regulator of the 2′-O-methyltransferase activity and raises the possibility that SAH hydrolase inhibitors might interfere with CoV replication cycle. The enzymatic and RNA binding assays developed in this work were also used to identify nsp16 residues involved in cap-0 RNA recognition and to understand the action mode of known methyltransferase inhibitors.
KEYWORDS: RNA processing, RNA virus, biochemistry
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV) is an emerging zoonotic Betacoronavirus that was initially isolated from a 60-year-old Saudi Arabian man in September 2012 (1). Since then, MERS-CoV is steadily spreading in the Arabian Peninsula and contained secondary outbreaks have occurred in Europe, Africa, Asia, and North America (2). As of April 2016, 1,728 cases of infection have been confirmed, with a fatality rate of 36%.
MERS-CoV infection may be asymptomatic or result in clinical symptoms ranging from mild respiratory illness to severe acute pneumonia, renal failure, systemic disorder (1), and severe neurological syndrome (3). MERS-CoV belongs to the lineage C of the genus Betacoronavirus and is phylogenetically related to the bat coronaviruses HKU4 and HKU5 (4–7). Bats are the host of a variety of coronaviruses, including Betacoronavirus genetically very closely related to the MERS-CoV which crossed the species barrier into dromedary camels (2). The virus was detected in Camelus dromedarius milk and nasal swabs, suggesting that camels are the probable source for zoonotic transmission of the virus to humans (6, 8). Recent works found at least five lineages of MERS-CoV in camels and identified six recombination events in MERS-CoV which may raise the virus' pathogenicity (9). Human-to-human transmission requires close contact and occurred principally in health care settings (8, 10). At present, there are no approved antiviral treatments or vaccines available against MERS-CoV infection.
After binding of MERS-CoV spike envelope proteins to the dipeptidyl peptidase-4 receptor (DPP4; also known as CD26) (11), the virus genome is released in the host cytoplasm. MERS-CoV genome is a polycistronic positive-stranded RNA 30,119 nucleotides (nt) in length, organized in 11 open reading frames (ORFs). The 3′ part of the genome contains at least nine ORFs that encode structural and virus accessory proteins, which are translated from a set of subgenomic RNAs. The two large ORFs (1a and b) that encode nonstructural proteins (nsp's) are in the 5′ proximal two-thirds of the genome (12). After viral infection, the 5′ region of capped genomic RNA is directly translated into the polyprotein pp1a and pp1ab, which are the precursors of the CoV nsp's. The pp1ab protein synthesis involves a ribosomal frameshift during RNA translation. These precursors proteins are cleaved by viral proteinases into 11 and 16 nsp's, respectively, that form the replication-transcription complex (RTC) (12, 13). The RTC harbors the catalytic activities required for the synthesis of genomic and subgenomic RNAs and contains most of the enzymes involved in the formation of the cap structure that decorates the 5′ end of viral mRNA.
Capping is a common modification of the 5′ end of eukaryotic mRNA. The cap structure consists of a methylated guanosine at position 7 linked to the first transcribed nucleotide by a 5′-5′ triphosphate bridge (7mGpppN). The cap is cotranscriptionally added to the 5′ end of nascent mRNA after synthesis of the first 20 to 30 nt (14, 53). Cap structure synthesis involves four sequential reactions catalyzed by an RNA 5′ triphosphatase (RTPase), a guanylyltransferase (GTase), a guanine N7 methyltransferase (N7-MTase), and a 2′-O-MTase (14). The cap structure plays several key roles in mRNA functionality. First, capping is essential for mRNA stability and limits its degradation by cellular 5′-3′ exoribonucleases, such as XRN1 (15). In the cytoplasm, the cap-0 structure (7mGpppN) ensures efficient mRNA recognition by the eukaryotic translation initiation factor 4E (eIF4E) (14). RNA capping also provides a molecular signature for the discrimination between self and non-self RNA. Indeed, viral double-stranded RNA, 5′-triphosphate RNA and incorrectly capped RNA are detected by intracellular pathogen recognition receptors (PRRs). Among these PRRs, retinoic acid-inducible gene (RIG)-like receptors, such as RIG-I and melanoma differentiation-associated protein 5 (Mda5), can detect uncapped 5′-triphosphate RNA and also cap-0 RNA (16–19). Detection of erroneously capped RNA in the cytoplasm induces a signal transduction cascade that initiates an antiviral response through interferon production (20). Among the interferon-stimulated genes (ISGs), interferon-induced protein with tetratricopeptide repeats 1 (IFIT 1) can recognize mis-capped RNA and inhibit its translation (21, 22).
Like other CoVs, MERS-CoV replicates in the cytoplasm of infected cells, where it should produce its own capping machinery. CoVs seem to adopt the eukaryotic canonical capping pathway with four sequential events that involve several viral nsp's: (i) first, the 5′-3′helicase/nucleoside triphosphatase (NTPase) nsp13 hydrolyzes the γ-phosphate from nascent 5′-triphosphate RNA (23); (ii) then, a GMP molecule is transferred to 5′-diphosphate RNA by an as-yet-unknown GTase, thus forming a primitive cap structure (GpppN); (iii) then, the guanosine is methylated, at the N7 position, by nsp14 in the presence of the methyl donor S-adenosyl-l-methionine (SAM) to produce the cap-0 structure (7mGpppN) and S-adenosyl-homocysteine (SAH) as by-product (24); and (iv) finally, the nsp10/nsp16 complex methylates the 2′-OH group (ribose) of the first transcribed nucleotide of the viral RNA, leading to the conversion of the cap-0 into a cap-1 structure (7mGpppN2′om) (25, 26). Thus, in CoVs, RNA cap methylation might involve at least three proteins (nsp10, nsp14, and nsp16), and it is known that both nsp14 and nsp16 can interact with nsp10 (27).
CoV 2′-O-MTase activity is mediated by nsp16 that contains both a conserved K-D-K-E catalytic tetrad, which is characteristic of SAM-dependent 2′-O-MTases, and a conserved SAM-binding site (28). CoV 2′-O-MTase activity was first demonstrated using in vitro biochemical assays with purified feline CoV (FCoV) nsp16 (29). However, recombinant severe acute respiratory syndrome coronavirus (SARS-CoV) nsp16 was inactive using similar experimental conditions. It was then demonstrated that SARS-CoV nsp16 needs to interact with nsp10 to become catalytically active (25). The nsp10/nsp16 complex MTase activity was demonstrated using synthetic capped N7-methylated RNA and longer RNAs that mimic the 5′ end of the SARS-CoV genome (25). In contrast, RNA with an unmethylated cap structure (Gppp-RNA) was not recognized by the nsp10/nsp16 complex, and no enzymatic activity was detected. Several mutagenesis studies of SARS-CoV nsp10 and nsp16 confirmed the key role of the K-D-K-E catalytic tetrad for the 2′-O-MTase activity (30) and demonstrated that the interaction between nsp10 and nsp16 is absolutely required for this activity (30, 31). The molecular basis governing nsp16 stimulation by nsp10 was partially elucidated by the crystal structure determination of the SARS-CoV nsp10/nsp16 complex (26, 30). The nsp16 adopts the catechol O-MTase fold containing alternating β strands (β1 to β7) and α helices (αZ and αA to αE) that form a seven-stranded β sheet with three α helices on each side (30, 32). In addition, nsp10 binds to nsp16 through a 930-Å2 activation area on nsp10 and stabilizes nsp16 (25, 26, 30, 33). Moreover, structural and biochemical analyses also suggest that nsp10 binding extends and narrows the RNA binding groove to accommodate the RNA substrate and enhances nsp16 RNA- and SAM-binding properties.
Although cap 2′-O-MTases are present in different virus families (15), the exact role of this RNA-cap modification was only recently unraveled through reverse genetic studies performed, among others, on CoV (16–19). Single point mutations in the conserved K-D-K-E tetrad of nsp16 have revealed a barely attenuated phenotype in infected cells (16, 34). In contrast, infection of small animal models by viruses expressing nsp16 with active-site substitution showed a robust reduction of viral titers and less severe infection symptoms (weight loss, lung histology, and breathing function) concomitantly with a strong antiviral response, possibly linked to stimulation of the innate immunity. Indeed, it was shown that incompletely capped RNA can be detected by RIG-I or Mda5 (16, 18, 19), leading to the initiation of a signaling cascade that stimulates the secretion of type I interferon (IFN) and induces an antiviral response in neighboring cells. Altogether, these observations indicate that the inhibition of the viral 2′-O-MTase activity might stimulate the detection of viral RNAs by RIG-I or Mda5 and restore the host antiviral response mediated by ISGs such as IFIT proteins (35).
In this study, we assessed the biochemical activity of the MERS-CoV nsp10/nsp16 complex MTase. We show that MERS-CoV nsp16 possesses 2′-O-MTase activity. As previously shown, nsp16 is stimulated by nsp10 and leads to 2′-O-methylation of cap-0 RNA (33). Biochemical assays were used to decipher the fine-tuning of the reaction turnover. Mutagenesis combined with RNA binding assays identified the residues essential for cap-0 recognition and those forming the RNA binding groove. Finally, using our radioactive enzymatic assay, we screened a small subset of MTase inhibitors blocking nsp10/nsp16 MTase activity, and the compounds blocking RNA recognition were identified using a fluorescence polarization assay.
RESULTS
MERS-CoV nsp16 assembles with nsp10 to constitute an active SAM-dependent cap-0 2′-O-methyltransferase.
The CoV nsp16 2′-O-MTase contains both a conserved K-D-K-E catalytic tetrad, which is characteristic of SAM-dependent 2′-O-MTase, and a conserved SAM-binding site (28). Although purified feline CoV (FCoV) nsp16 shows a weak 2′-O-MTase activity in the absence of nsp10, SARS-CoV nsp16 2′-O-MTase activity is strictly dependent on its interaction with nsp10 (29, 33). MERS-CoV nsp16 shows ∼65 and ∼53% amino acid residue similarity with SARS-CoV and FCoV nsp16, respectively, and requires nsp10 for nsp16 2′-O-MTase activity (33). To assess the mechanism of fine-tuning of MERS-CoV nsp16 2′-O-MTase activity by nsp10, recombinant nsp10 and nsp16 with an N-terminal His6-tag were produced and purified, and SDS-PAGE analysis showed that nsp10 and nsp16 migrate at the expected molecular masses (Fig. 1A, 16 and 37 kDa, respectively), and their identity was confirmed by mass spectrometry after trypsin digestion (not shown). The nsp10/nsp16 complex was obtained by colysis of an equal volume of His6-nsp10 and His6-nsp16 expressing bacteria and purification by immobilized metal affinity chromatography (IMAC) and size exclusion chromatography. The chromatogram (Fig. 1B) indicated that the nsp10/nsp16 complex was detected as one major peak eluting at a volume corresponding to an nsp10/nsp16 heterodimer (∼50 kDa).
FIG 1.
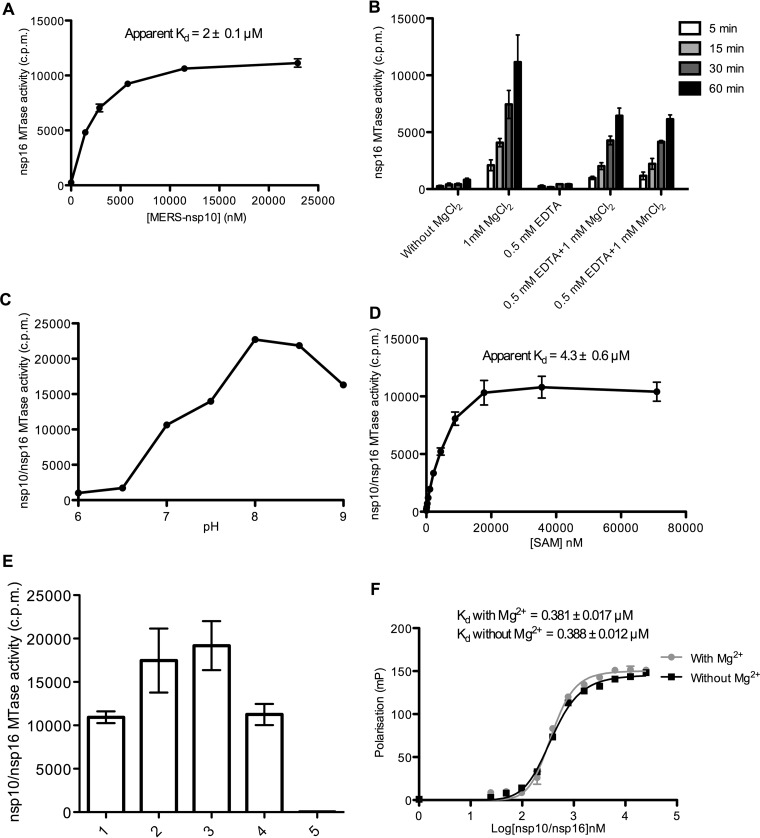
MERS-CoV nsp16 2′-O-MTase activity is promoted by nsp10. Recombinant MERS-CoV nsp10, nsp16, and nsp10/nsp16 complex were expressed in E. coli and purified by affinity chromatography on IMAC columns. (A) After SDS-PAGE separation of purified proteins (0.2 μg), gels were stained with Coomassie blue. MW, molecular mass markers. (B) The nsp10/nsp16 complex was purified by gel filtration using a S200-16/60 column. The elution chromatogram (optical density, 280 nm) shows one main peak that eluted at 90 ml and corresponded to the nsp10/nsp16 complex. (C) The methyltransferase activities (MTase) of nsp16 (1.2 μM) and nsp10 (2 μM) alone or together and of the nsp10/nsp16 complex (1 μM) were determined by monitoring the transfer of 3H-CH3 from SAM to RNA oligonucleotides with sequences corresponding to the 5′ end of the MERS-CoV genome with various cap modifications (pppGAUUUAA, GpppGAUUUAA, 7mGpppGAUUUAA, GpppG2′omAUUUAA, and 7mGpppG2′omAUUUAA). Assays were stopped after 5, 30, 60, and 180 min, and the radioactivity associated with the RNA was determined by a DEAE-filter binding assay. The bar graph presents the means and standard deviations of three independent experiments.
Then, in vitro MTase assays were performed using RNA oligonucleotides with sequences corresponding to the 5′ end of the MERS-CoV genome with various 5′-end modifications (pppGAUUUAA, GpppGAUUUAA, 7mGpppGAUUUAA, GpppG2′omAUUUAA, and 7mGpppG2′omAUUUAA) in the presence of nsp10 or nsp16 alone or as a complex and of 3H-SAM as methyl donor. Quantification of the amount of [3H]CH3 transferred to RNA over time using a DEAE filter-binding assay indicated the absence of MTase activity when nsp10 or nsp16 alone was used (Fig. 1C). Conversely, RNA carrying a cap-0 structure (7mGpppGAUUUAA) was methylated in a time-dependent manner when incubated with both nsp16 and nsp10 or with the purified nsp10/nsp16 complex. Since MTase activity was detected only using 7mGpppGAUUUAA, but not GpppG2′omAUUUAA or 7mGpppG2′omAUUUAA, we hypothesized that the MERS-CoV MTase converts cap-0 into cap-1 RNA. The addition of a single methyl group on 7mGpppGAUUUAA was confirmed by comparing the molecular mass of 7mGpppGAUUUAA before and after incubation with nsp10/nsp16 and SAM. We detected a mass increase of about ∼14.2 Da by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry analysis indicating that one methylation event has occurred. We conclude that MERS-CoV nsp16 carries the cap 2′-O-MTase activity and is stimulated by nsp10. The nsp10 stimulation effect on nsp16 was confirmed by assessing nsp16 2′-O-MTase activity in the presence of increasing concentrations of nsp10. An apparent dissociation constant for the nsp10/nsp16 complex could be derived from the stimulation curves (Kd = 2 ± 0.1 μM, Fig. 2A). Moreover, the nsp10/nsp16 MTase activity required the addition of divalent ions (Mg++ or Mn++) and was inhibited by EDTA (Fig. 2B). The optimum of activity peaked between pH 8 and 8.5 (Fig. 2C). The optimal concentration of methyl-donor (SAM) (Kd = 4.3 ± 0.6 μM) was also determined by measuring the MTase activity of the complex in the presence of increasing SAM concentrations (Fig. 2D). Finally, MTase assays performed using the optimal conditions deduced from these biochemical assays and cap-0 RNAs with sequences corresponding to the 5′ ends of various viruses showed that the nsp10/nsp16 complex methylated cap-0 RNAs that started with either an A or a G (Fig. 2E). Taken together, these results suggest that MERS-CoV nsp16 acts as an nsp10-dependent 2′-O-MTase that converts cap-0 into cap-1 RNA structures with no obvious specific recognition mechanism of the MERS-CoV sequence. Moreover, the absence of 2′-O-MTase activity using GpppRNA suggests that nsp16 can discriminate the cap-0 structure from an unmethylated cap structure.
FIG 2.
Biochemical characterization of the MERS-CoV nsp10/nsp16 2′-O-MTase. The 2′-O-MTase activity of MERS-CoV nsp16 was characterized by filter binding assay as described in Fig. 1C. (A) nsp16 2′-O-MTase activity is promoted by nsp10. nsp16 (2 μM) was incubated with 7mGpppGAUUUAA (0.7 μM) in the presence of increasing concentrations of nsp10. The apparent Kd of the nsp10-nsp16 interaction (2 ± 0.1 μM; mean ± the standard error of the mean [SEM] of three independent experiments) was deduced by curve fitting using Hill slope equations. (B) Effect of divalent ions on nsp16 2′-O-MTase activity. MTase assays were performed in the presence or not of divalent ions (MgCl2). Inhibition of the MTase reaction by EDTA was assessed, and the activity was recovered by addition of MgCl2 or MnCl2. The methyl transfer was measured after 5, 15, 30, and 60 min at 30°C (mean ± the SD). (C) Optimum pH of the 2′-O-MTase activity mediated by nsp10/nsp16. (D) nsp10/nsp16 MTase activity in the presence of increasing concentrations of SAM. The apparent Kd of SAM (4.3 ± 0.6 μM) was deduced by curve fitting using a Hill equation and the GraphPad Prism program. (E) Measurement of nsp10/nsp16 MTase activity on cap-0 RNA sequences that correspond to the 5′-end of various viruses: (1) MERS-CoV (7mGpppGAUUUAAGUGAAUA), (2) West Nile virus (7mGpppAGUAGUUCGCCUG), (3) dengue virus (7mGpppAGUUGUUAGUCUA), (4) Ebola virus (7mGpppGAUGAAGAUUAAG) after 30 min of incubation at 30°C (n = 3, mean ± SEM), and (5) without RNA. (F) Magnesium ions do not influence nsp10-nsp16 interaction with cap-0 RNA. 7mGpppGAUUUAA-cy5 was incubated with increasing concentrations of the nsp10/nsp16 complex with or without 1 mM MgCl2. The interaction with the cy5-labeled RNA was followed by fluorescent polarization, and the apparent calculated Kd values are indicated at the top of the graph.
The MERS-CoV nsp10/nsp16 complex recognizes specifically a cap-0 structure, and downstream RNA nucleotides promote loading of cap-0 substrate and their 2′O methylation.
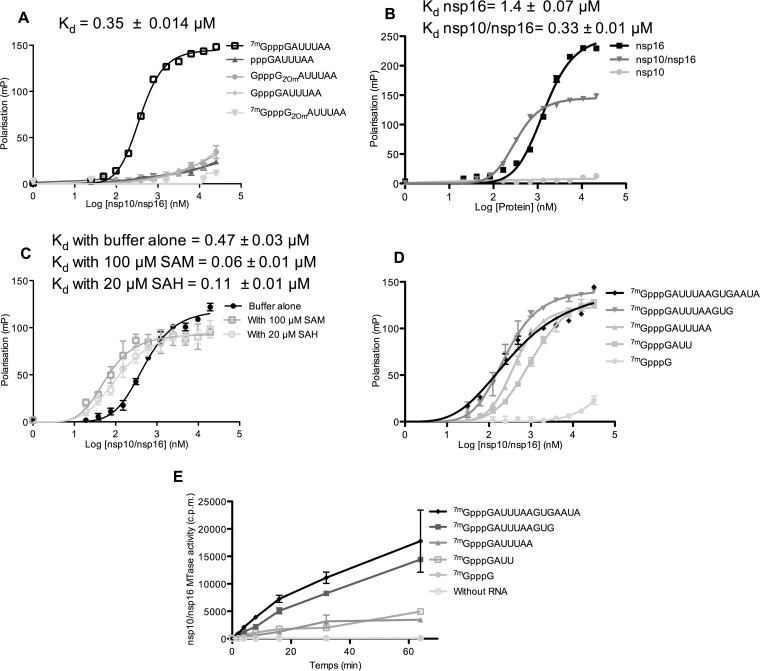
RNA binding assays were then developed to determine why the nsp10/nsp16 complex was active only on cap-0 RNA. For this purpose, the 3′ ends of various 5′-end-modified RNAs were labeled with pCp-cy5 (where cy5 represents cyanin 5), and then their binding to freshly purified MERS-CoV nsp10/nsp16 complex was measured in fluorescence polarization assays. The nsp10/nsp16 complex recognized only cap-0 RNA (7mGpppRNA) with a Kd of ∼0.35 μM (Fig. 3A). No significant binding was detected using the other RNAs (pppRNA or GpppRNA, GpppG2′omRNA, and 7mGpppG2′omRNA). This observation is consistent with the MTase assay results (Fig. 1C) and indicates that the absence of nsp10/nsp16 2′-O-MTase activity using GpppRNA is due to the lack of recognition of such capped RNA. It also suggests that the nsp10/nsp16 complex has an RNA binding site that recognizes specifically cap-0 RNA. Moreover, the ability of this binding site to recognize cap-0 RNA is strongly regulated by the 2′-O-methylation status of the substrate RNA. Indeed, 7mGpppG2′omRNA, which corresponds to the nsp16 2′-O-MTase reaction product, did not bind to the nsp10/nsp16 complex (Fig. 3A). This indicates that cap-1 RNA generated by the nsp10/nsp16 2′-O-MTase activity is actively released, promoting the reaction turnover.
FIG 3.
RNA binding properties of MERS-CoV nsp10, nsp16, and the nsp10/nsp16 complex. (A) nsp10/nsp16 recognizes cap-0 RNA. Different short RNA oligonucleotides (pppGAUUUAA, GpppGAUUUAA, 7mGpppGAUUUAA, GpppG2′omAUUUAA, or 7mGpppG2′omAUUUAA) corresponding to the 5′ end of the MERS-CoV genome were 3′ end labeled with pCp-cy5 and incubated with increasing concentrations of nsp10/nsp16 complex. The interaction between the nsp10/nsp16 complex and each RNA was monitored by measuring the fluorescence polarization signal at 675 nm. The apparent affinity constant (Kd) was calculated by nonlinear regression analysis with a Hill slope equation and is indicated at the top of the graph. (B) nsp16 and the nsp10/nsp16 complex recognize cap-0 RNA. 7mGpppGAUUUAA-cy5 was incubated with increasing concentrations of nsp10, nsp16, or nsp10/nsp16 complex. The interaction with cy5-labeled RNA was monitored by measuring the fluorescence polarization, as described in panel A, and the calculated Kd is indicated at the top of the graph. (C) SAM and SAH enhance the interaction of the nsp10/nsp16 complex with cap-0 RNA. 7mGpppGAUUUAA-cy5 was incubated with increasing concentrations of nsp10/nsp16 complex in the presence or not (buffer alone) of 100 μM SAM or 20 μM SAH. The interaction with cy5-labeled RNAs was monitored by measuring the fluorescence polarization, and the calculated Kd is indicated at the top of the graph. (D) RNA length effect on nsp10/nsp16 binding was analyzed using viral native 7mGppp RNAs of different lengths (7mGpppG, 7mGpppGAUU, 7mGpppGAUUUAA, 7mGpppGAUUUAAGUG, and 7mGpppGAUUUAAGUGAAUA) in the presence of increasing concentrations of nsp10/nsp16. The interaction with cy5-labeled RNAs was monitored by fluorescence polarization, and the calculated Kd values are indicated in Table 1. (E) Kinetics of MTase activity of the nsp10/nsp16 complex (0.25 μM) on 7mGpppG-RNAs (0.7 μM) of increasing length was measured by filter binding assay after 2, 4, 8, 16, 32, and 64 min of incubation at 30°C in the presence of 3H-SAM. The curve presents the means and standard errors of the means for three independent experiments. All the fluorescent polarization experiments were performed twice independently, and the Kd values of each condition were calculated using GraphPad Prism (n = 2, mean ± the SEM) using one site-specific binding equation with the Hill slope.
As nsp16 2′-O-MTase activity strictly depends on its interaction with nsp10 (Fig. 1C and 2A), we then sought to determine whether nsp10 regulates nsp16 RNA-binding properties. Comparison of nsp10, nsp16, and nsp10/nsp16 complex binding to cap-0 RNA (7mGpppRNA) using a fluorescence polarization assay showed that nsp10 barely interacted with cap-0 RNA (Fig. 3B). Conversely, both nsp16 and nsp10/nsp16 complex bound to cap-0 RNA with apparent Kd values of ∼1.4 and ∼0.33 μM, respectively. The polarization signal detected after nsp16 binding is higher than that obtained in the presence of the nsp10/nsp16 complex. This suggests that the RNA might bind to nsp16 dimers. Our result indicates that only nsp16 harbors the cap-0 binding site and that RNA binding is enhanced by nsp16 interaction with nsp10. Since nsp16 alone binds cap-0 RNA (Fig. 3B) but is not methylated (Fig. 1C), we suggest that nsp10 is an allosteric regulator of nsp16 2′-O-MTase activity. In addition, the methyl donor SAM and, to a lesser extent, the reaction by-product SAH increased the affinity for the RNA substrate (Fig. 3C). Conversely, magnesium ions (Mg2+), which stimulate nsp10/nsp16 MTase activity (Fig. 2B), did not seem to affect RNA binding (Fig. 2F) in these experimental conditions, indicating that magnesium ions are not required for RNA recognition.
The MTase assay showed that the nsp10/nsp16 complex is able to methylate cap-0 RNA of variable RNA sequences (Fig. 2E), starting with either mGpppA or mGpppG. However, the MTase activity is weaker using short RNA substrates (Fig. 3E). Therefore, we tested whether nucleotides downstream from the cap structure contributed to nsp10/nsp16 complex binding to RNA. To do this, fluorescence polarization assays were performed using 3′-end-labeled RNA sequences of increasing length that mimicked the 5′ end of the MERS-CoV genome sequence. Quantification of their interaction with the nsp10/nsp16 complex (Fig. 3D and Table 1) showed that the cap analogue 7mGpppG-cy5, which was not methylated by nsp10/nsp16 (Fig. 3E), barely interacted with the nsp10/nsp16 complex. In contrast, a significant interaction was observed using 7mGpppGAUU-cy5. The affinity then increased with the RNA length, and the optimal affinity was observed with 7mGpppGAUUUAAGUG-cy5 (Fig. 3D). The MTase activity measured using a filter-binding assay followed the same trend (Fig. 3E).
TABLE 1.
Kd values of the interaction of the nsp10/nsp16 complex with pCp-Cy5-labeled RNA sequences of increasing length measured by fluorescence polarizationa
| RNA | Mean Kd (μM) ± SEM |
|---|---|
| 7mGpppG | >20 |
| 7mGpppGAUU | 1.07 ± 0.09 |
| 7mGpppGAUUUAA | 0.42 ± 0.01 |
| 7mGpppGAUUUAAGUG | 0.251 ± 0.02 |
| 7mGpppGAUUUAAGUGAAUA | 0.278 ± 0.05 |
See also Fig. 3D.
Identification of nsp16 residues playing a key role in RNA recognition and MTase activity.
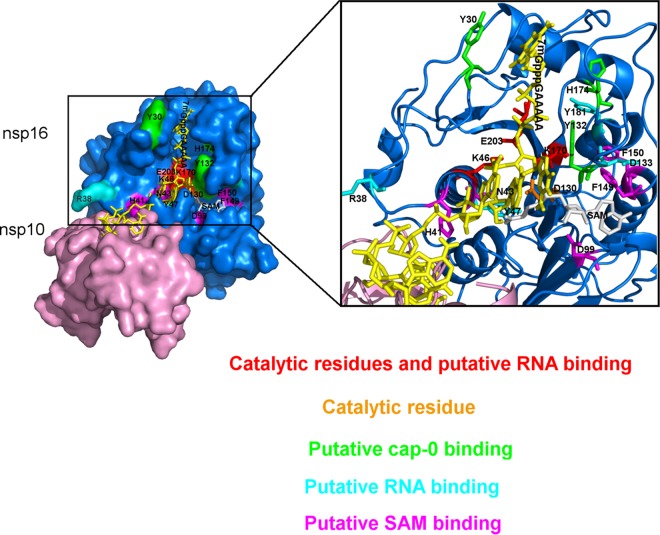
The crystal structure of the SARS-CoV nsp10/nsp16 complex with SAH has been reported at 2.0-Å resolution (30). Since no structure with a cap analogue or substrate RNA is available yet, the nsp16 residues involved in RNA binding were tentatively inferred by modeling the RNA in the catalytic site (Fig. 4: structural model based on the SARS-CoV and vaccinia MTase VP39 structures). Based on the structural model, alanine mutagenesis was performed, and the effects of nsp16 mutations on MTase activity (Fig. 5B) and on RNA binding properties (Fig. 5A) were determined by filter binding assay and fluorescent polarization assay, respectively. Alanine substitution of the conserved residues in the catalytic tetrad (K-D-K-E) almost completely abolished the 2′-O-MTase activity, as expected. With the exception of D130A, these mutations also strongly reduced RNA binding, indicating that these catalytic residues also participated in RNA binding process. Mutation of the residues adjacent to the putative SAM binding site (H41, N43, D99, F149, and F150) drastically reduced MTase activity but had no significant effect on RNA binding. Mutation of K46, K170, E203, D133, R38, Y47, or Y181 concomitantly reduced MTase activity (80 to 100%) and RNA binding. These residues are localized in the putative RNA binding groove of nsp16 (Fig. 4). In addition, mutation into alanine of Y30 and Y132, which are within two mobile α helices (26-38 and 130-148) in close proximity to the cap in the nsp10/nsp16 model (Fig. 4), and of H174, which is close to the 130-148 loop, also decreased nsp16 RNA binding properties and MTase activity. These observations confirm that the cap-0 RNA binding site involves these three aromatic residues.
FIG 4.
3D structural model of the nsp10/nsp16 complex interaction with 7mGppp-RNA. The nsp10/nsp16 model was built by alignment of SARS-CoV (PDB entry 3R24) and MERS-CoV nsp10/nsp16 sequences using the Swiss model and the PyMOL software. The cap-0 RNA/nsp16 model was built by aligning the MERS-CoV nsp16 sequence with that of the vaccinia virus MTase VP39 (PDB entry 1AV6) using the PyMOL software. The surface of nsp16 residues is indicated in blue, and that of nsp10 is indicated in pink. The nsp16 catalytic residue D130 is depicted in orange, while the catalytic and putative RNA binding residues K46, K170, and E203 are depicted in red. The putative cap-0 RNA binding site (light green) is delimited by Y30 and Y132, which are localized in the two mobile α helices (26-38 and 130-148, respectively), and by H174 close to the last helix. The residues of the putative RNA binding groove are indicated in cyan (Y181, D133, Y47, and R38). The SAM binding pocket involves the residues N43, D99, F149, F150, and H41 (magenta); SAM is indicated in gray, and cap-0 RNA is indicated in yellow. The zoom enlargement focuses on the RNA binding groove and the SAM binding domain (ribbon structure).
FIG 5.
MERS-CoV nsp16 mutations that affect RNA recognition and/or MTase activity. (A) Alanine scanning mutagenesis. Mutations were introduced in the nsp16 clone and, after colysis, the nsp10/nsp16 complex was purified by affinity chromatography on IMAC. The affinity (Ka = 1/Kd) of each mutant protein for 7mGpppGAUUUAA-cy5 was measured by fluorescence polarization as described in Fig. 3. The affinity (Ka = 1/Kd) of each mutant protein for 7mGpppGAUUUAA-cy5 was measured by fluorescence polarization as described for Ka in Fig. 3. The Ka values were estimated from the Hill equation using GraphPad Prism (n = 2, mean ± SEM). The Ka values were only estimated for the mutants indicated by an asterisk because the RNA binding plateau was not reached in the fluorescence polarization experiment. (B) The MTase activity of each mutant (1 μM) was determined after incubation with 7mGpppGAUUUAA at 30°C in the presence of 3H-SAM for 60 min. The bar graph presents the mean and the standard deviation for three independent experiments.
SAM and SAH levels regulate nsp10-nsp16 interaction.
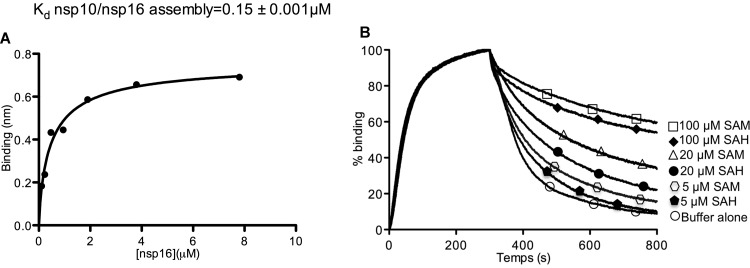
nsp16 2′-O-MTase activity requires interaction with nsp10. To determine how cosubstrate (SAM) availability might regulate the RNA-capping reaction pathway, the nsp16 interaction with nsp10 was analyzed using biolayer interferometry. nsp10 was biotinylated before immobilization on a streptavidin-coated biosensor. Binding of nsp16 was then monitored in the presence of various SAM/SAH concentrations, and the apparent affinity constants were determined (Fig. 6A and Table 2). The nsp16 interaction with biotinylated nsp10 increased in the presence of 100 μM SAM or SAH. The interaction kinetics was characterized by the same association rate (same kon) in the presence or in the absence of SAM or SAH. Conversely, the addition of SAM or SAH increased nsp10/nsp16 complex stability by decreasing the dissociation kinetics (koff). This suggests that both SAM and SAH induce nsp10/nsp16 complex assembly. To further analyze how SAM or SAH regulated nsp10-nsp16 interaction, the dissociation of the nsp10/nsp16 complex, formed in the presence of 100 μM SAM, was assessed using dissociation buffers containing various concentrations of SAM or SAH (Fig. 6B). The dissociation rate of the nsp10/nsp16 complex depended on SAM or SAH concentration. In the presence of 100 μM SAM, a concentration that mimics its intracellular level (36), the nsp10/nsp16 complex stability was highest, as indicated by the slow nsp16 release. In contrast, after MTase reaction, which converts SAM into SAH, the latter is released from the enzyme at the lower intracellular concentration of SAH (∼20 μM [37]). In turn, the release of SAH will speed up the dissociation of the nsp10/nsp16 complex. Altogether, these data indicate that nsp10 and nsp16 form a dynamic complex, the association of which is promoted by SAM binding. The complex will dissociate when SAH, the reaction by-product, is released, thus favoring the reaction turnover.
FIG 6.
SAM and SAH modulate nsp10-nsp16 interaction. (A) nsp10/nsp16 steady-state assembly: real-time nsp16 binding to biotinylated nsp10 was measured by Octet biolayer interferometry. Streptavidin biosensors coated with 100 nM biotinylated nsp10 were used to measure nsp16 (0 to 7.8 μM) association and dissociation. The reference sensor (0 μM nsp16) values were subtracted from the sample traces. All curves were fitted with a 1:1 model by using the Octet biolayer program, and then the steady-state curve was traced using the site-specific binding equation on GraphPad Prism. The different Kd values of the nsp10-nsp16 interaction in the presence or not of 100 μM SAM or 20 or 100 μM SAH are presented in Table 2. (B) Effects of SAM and SAH on the dissociation kinetics of nsp16 from immobilized nsp10. A 7.8 μM concentration of nsp16 was bound to sensor-immobilized biotinylated nsp10 in the presence of 100 μM SAM. The sensor was then moved to dissociation buffer alone or containing 5, 20, or 100 μM SAM or SAH. nsp16 binding to immobilized nsp10 was normalized for each trace.
TABLE 2.
Kd, Kon, and Koff values of nsp10-nsp16 interaction measured by Octet biolayer interferometry in the presence or not of SAM or SAHa
| Condition(s) | Mean ± SD |
||
|---|---|---|---|
| Kd, nsp10-nsp16 interaction (μM) | Kon (M−1 s−1) | Koff (s−1) | |
| Without SAM, SAH | 1.24 ± 0.015 | 3.77 × 103 ± 4.35 | 4.68 × 10−3± 0.016 × 10−3 |
| With 100 μM SAM | 0.15 ± 0.001 | 3.98 × 103 ± 2 | 6.23 × 10−4 ± 0.031 × 10−4 |
| With 20 μM SAH | 0.48 ± 0.003 | 3.10 × 103±1.97 | 1.51 × 10−3 ± 0.004 × 10−3 |
| With 100 μM SAH | 0.19 ± 0.0013 | 3.3 ×103± 2.2 | 6.51 × 10−4 ± 0.041 × 10−4 |
See also Fig. 6A.
MERS-CoV nsp16 MTase activity inhibition.
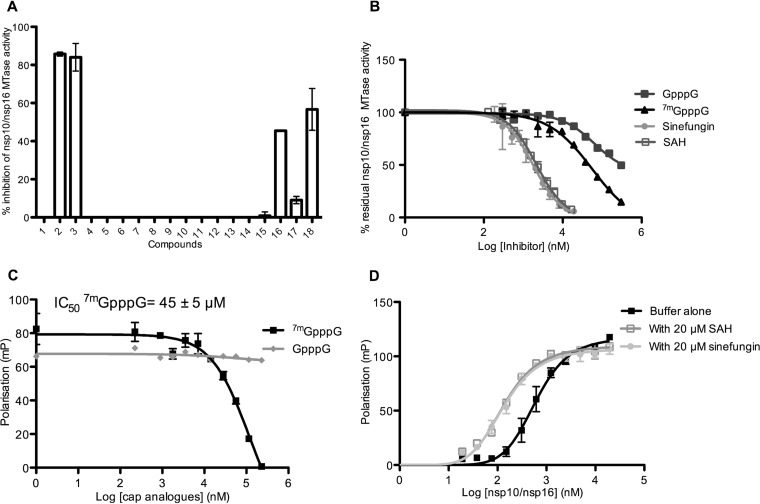
It was previously shown that 2′-O-methylation of viral RNA cap structures limited the infected host's antiviral response (38), suggesting that molecules blocking nsp16 2′-O-MTase activity might favor viral clearance in infected animals (35). In MTase inhibition assays, we tested 18 molecules known to block MTase activity (25, 39). Four of these compounds (Fig. 7A) inhibited nsp10/nsp16 2′-O-MTase activity (between 45 and 100%) at a concentration of 50 μM. Among them, two SAM analogues (sinefungin and SAH, compounds 2 and 3 in Fig. 7A) and showed the most potent inhibition. Cap-0 analogues (7mGpppG and 7mGpppA, compounds 18 and 16) also inhibited nsp16 2′-O-MTase activity (40 to 45%), whereas GpppG and GpppA (compounds 17 and 15) did not. This suggests that cap analogues block the binding of the RNA substrate in nsp16 RNA or cap binding site, in agreement with the weak interaction (Kd > 20 μM) of these compounds with the nsp10/nsp16 complex (Fig. 3D).
FIG 7.
Inhibition of nsp10/nsp16 complex MTase activity and RNA binding. (A) Bar graph showing the MTase inhibition activity (percentage) of each candidate inhibitor (final concentration, 50 μM). An MTase assay was performed as in Fig. 1C by incubating 0.5 μM nsp10/nsp16 complex with 0.7 μM 7mGpppGAUUUAA and 3H-SAM at 30°C for 30 min. The methyl transfer to RNA was measured by a filter binding assay in the absence (5% DMSO) or presence of each inhibitor (n = 2, mean ± the SD): 1, DMSO; 2, sinefungin; 3, SAH; 4, SIBA; 5, ribavirin; 6, ribavirin-TP; 7, N-(5-chloro-2-methoxyphenyl)-3,4-dimethoxy-N-[2-(4-methyl-1-piperazinyl)-2-oxoethyl]benzenesulfonamide; 8, 2-[[4-benzyl-5-(3-pyridinyl)-4H-1,2,4-triazol-3-yl]thiol]-N-(5-methyl-1,3,4-thiadiazol-2-yl) acetamide; 9, N-1-cyclopropyl-N-2-(2,5-dimethoxyphenyl)-N-2-(methylsulfonyl) glycinamide; 10, 4-bromophenyl 3-(3,5-dioxo-4-azatetracyclo [5.3.2.0-2,6-0.0-8,10-] dodec-11-en-4-yl) propanoate; 11, 1′-[(4-tert-butylphenyl)sulfonyl]-1,4′-bipiperidine-4′-carboamide; 12, N′-(3,4-dichlorobenzylidene)-2-(2-pyridinylthio) acetohydrazide; 13, GTP; 14, 7mGTP; 15, GpppA; 16, 7mGpppA; 17, GpppG; and 18, 7mGpppG. (B) Dose response of the inhibitory effect of sinefungin, SAH, and cap analogues on nsp10/nsp16 MTase activity (MTase assays were performed as described for panel A [n = 2, mean ± the SEM]). The IC50s deduced by using GraphPad Prism and the log (inhibitor) versus response variable slope equation are shown in Table 3. (C) The competition effect of cap analogues on 7mGpppGAUUUAA-cy5 binding to the nsp10/nsp16 complex was analyzed by fluorescence polarization, as described for Fig. 3. Measurements were performed in the presence of increasing concentrations of cap analogues. (D) The effect of 20 μM SAH, 20 μM sinefungin, or buffer alone on the nsp10/nsp16 complex interaction with 7mGpppGAUUUAA-cy5 was assessed by fluorescence polarization, as described in Fig. 3C. The Kd values deduced from the curve fitting are given in Results (n = 2, mean ± the SEM).
To better define the 2′-O-MTase activity inhibition of sinefungin, SAH, 7mGpppG and 7mGpppA, the nsp10/nsp16 complex was preincubated with increasing concentrations of each inhibitor, and then the MTase reaction was started by the addition of 3H-SAM and the RNA substrates. The dose-response curves (Fig. 7B) indicated that sinefungin and SAH inhibited nsp10/nsp16-mediated 2′-O-MTase activity, with 50% inhibitory concentrations (IC50s) in the micromolar range (7.4 and 7.0 μM, respectively, Table 3). The IC50s of the cap analogues 7mGpppG and GpppG (45 and 274 μM, respectively) (Table 3) suggest that 7mGpppG is a competitive inhibitor that blocks specifically the RNA cap-0 binding site. This was confirmed by measuring the effect of these compounds on nsp10/nsp16 RNA binding activity. 7mGpppG blocked RNA binding with an IC50 of 45 μM, which is in the same range of that of the inhibition of the MTase activity (Fig. 7C). Conversely, the SAM analogues (SAH and sinefungin), which are supposed to enter in the nsp16 SAM binding site, barely affected RNA binding properties, as expected. The Kd values for the nsp10/nsp16 complex and 7mGpppRNA interactions in the presence or not of SAH or sinefungin were measured by fluorescence polarization (Fig. 7D) under the following conditions: buffer alone, 0.47 ± 0.03 μM; with 20 μM SAH, 0.11 ± 0.01 μM; and with 20 μM sinefungin, 0.09 ± 0.009 μM. Thus, the radioactive assay provides an efficient method to screen molecules to identify inhibitors of SAM-dependent MTase activity of the MERS-CoV nsp10/nsp16 complex. The fluorescence polarization assay developed here represents a tool to analyze their mode of action.
TABLE 3.
nsp10/nsp16 2′-O-MTase inhibition by sinefungin, SAH, and cap analogsa
| Inhibitor | Mean IC50 (μM) ± SEM |
|---|---|
| Sinefungin | 7.4 ± 0.9 |
| SAH | 7 ± 0.4 |
| GpppG | 274.4 ± 140 |
| 7mGpppG | 45 ± 3.5 |
Calculated from the dose-response curve of Fig. 7B.
DISCUSSION
MERS-CoV is the most recently discovered zoonotic virus that causes pathology in humans. Upon cell infection, MERS-CoV starts its replication by translation of ORFs 1a and 1b to produce 16 nsp's of the replication/transcription complex (RTC) (12). In this study, we characterized the MERS-CoV 2′-O-MTase activity of nsp16 and show that nsp10 is a cofactor required for nsp16 2′-O-MTase enzymatic activity, as previously reported for SARS and MERS-CoV (25, 33). The substrate of the reaction is RNA with a cap-0 structure. This observation is consistent with RNA binding assays showing that N7-methylated cap RNA is recognized by the nsp10/nsp16 complex, whereas unmethylated RNA is not. Thus, MERS-CoV cap methylation follows an obligatory sequence in which 2′-O-methylation might occur after N7 methylation mediated by nsp14. Consequently, it is likely that RNA cap synthesis by MERS-CoV follows the canonical capping pathway observed in eukaryotic cells and in some viruses, such as SARS-CoV, dengue virus, and West Nile virus (20).
Although the cap-0 structure gives RNA binding specificity, cap analogues barely bind to the nsp10/nsp16 complex at the micromolar concentration of 7mGpppG-cy5 used in our assay and are not a good substrate for the 2′-O-MTase. The presence of additional nucleotides downstream of the cap structure increases the substrate recognition and 2′-O-MTase activity. These nucleotides are thus required for substrate stabilization in the RNA binding groove. The optimal recognition is reached with 10 nt of capped RNA. This is consistent with the model proposed for the SARS-CoV nsp10/nsp16 complex with 7mGpppA-RNA in which the first nucleotides are held by nsp16 and the following nucleotides might be positioned in a RNA binding groove stabilized by nsp10 (26, 30). In addition, it is likely that the MERS-CoV 2′-O-MTase recognizes and methylates cap-0 RNA with no obvious sequence specificity. We also observed that the nsp10/nsp16 complex methylated similarly cap-0 RNA with A or G as the first nucleotide. This observation is in contrast to SARS-CoV 2′-O-MTase that preferentially binds and methylates RNA with A at N1 position ((7mGpppA-capped RNA, [26]). Using synthetic RNAs that mimic the 5′ end of the MERS-CoV genome, we did not observe any additional methylation (N2) or internal methylation, as indicated by the absence of enzymatic activity on cap-1 RNA or pppRNA sequences. The absence of internal methylation was also confirmed using longer (∼500 nt) pppRNA substrates (not shown). This observation contrasts with findings on other viral MTases (flavivirus NS5-MTase) that can perform internal methylation of adenosines (40). Nevertheless, the absence of internal methylation with the nsp10/nsp16 complex is consistent with the fact that neither cap-1 RNAs nor pppRNAs interact with the nsp10/nsp16 complex.
To further understand the mechanism whereby nsp10 promotes the stimulation of nsp16 2′-O-MTase activity by nsp10, we determined whether nsp10 is required for substrate recognition process. Our findings indicate that nsp16 can recognize cap-0 RNA even in the absence of nsp10 as already reported for FCoV nsp16 (29). Nevertheless, nsp10 increases nsp16 RNA binding properties of nsp16 (3-fold) and, in turn, the 2′-O-MTase activity can be detected only in the presence of both proteins under this condition. These observations suggest that nsp10 acts as an allosteric regulator of nsp16 2′-O-MTase activity rather than as an RNA binding module. A similar allosteric effect was reported for the vaccinia virus and human RNA N7-MTases. Indeed, in the vaccinia virus family, the D1 catalytic subunit alone is unstable and inactive, and its stability and MTase activity are enhanced by the presence of D12 through the increase in GTP, SAM, and GpppA binding affinity (41, 42). Similarly, the human RNA N7 MTase (RNMT) is allosterically activated by RNMT activating miniprotein that stabilizes its structure and favors the recruitment of methyl donors (43).
Since nsp10 is not the main player involved in RNA binding, we mapped the nsp16 residues involved in cap-0 RNA recognition. We could not determine the structure of nsp10/nsp16 with RNAs or cap analogues, although we performed many attempts (not shown). Therefore, we used alanine scanning mutagenesis to identify the residues involved in cap-0 RNA specific recognition and nsp16 MTase activity. Our results suggest that the cap-0 RNA structure is stacked between two flexible loops, 26-38 and 130-148, by the aromatic residues Y30 and Y132, respectively, and the H174 residue in close proximity. The alanine scanning results also indicate that the RNA chain of the substrate might be held by K46, K170, E203, D133, R38, Y47, and Y181 because both RNA binding and MTase activity decreased upon mutation of these residues. Interestingly, three residues belonging to the catalytic tetrad (K46, K170, and E203) might directly participate in RNA recognition. These results corroborate the model of MERS-CoV nsp10/nsp16 (Fig. 4) in which the RNA was positioned using vaccinia MTase VP39 structures (PDB entry 1AV6) (26, 30, 44). The mutagenesis study also allowed identifying residues that strongly reduce MTase activity with a weak effect on RNA recognition (D99, F149, F150, H41, and N43). On the basis of their position in the structural model, we suggest that these residues might participate in SAM binding.
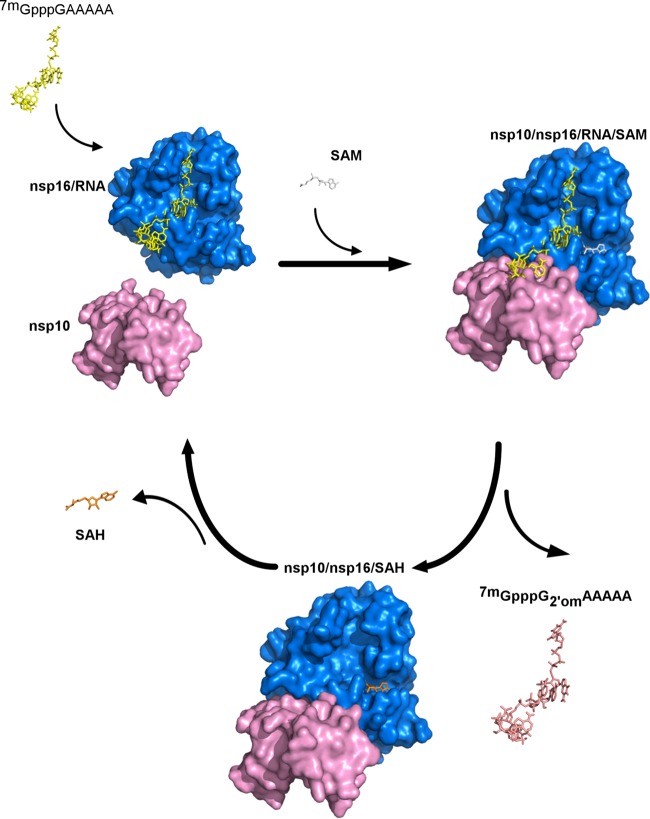
We then demonstrated that SAM/SAH balance plays an important role in nsp10-nsp16 interaction. The methyl donor SAM, strengthens the interaction between nsp10 and nsp16 at the physiological intracellular concentration (100 μM [36]) and, in turn, the RNA binding properties of nsp16 are enhanced. This suggests that SAM stabilizes or induces small conformational changes of the enzyme leading to an increase in both RNA affinity and methylation by nsp10/nsp16. This observation is also consistent with thermal shift assay indicating that the nsp10/nsp16 complex is stabilized in the presence of SAM (delta melting temperature [ΔTm] ≈ 1°C). During RNA methylation, SAM is converted into SAH. The SAH will be next released by nsp16 stimulating the dissociation of the nsp10/nsp16 complex. In addition, the reaction product (cap-1 RNA) barely interacts with the nsp10/nsp16 complex and is released. Overall, these observations suggest that cap-0 RNA methylation is linked to the association and dissociation of the nsp10/nsp16 complex (Fig. 8). These data also suggest that the SAM/SAH balance in infected cells is a key factor for cap RNA methylation. Since SAH hydrolase is a key enzyme for maintaining low levels of SAH in cells, inhibitors of cellular SAH hydrolases might have an antiviral effect on CoV replication. Interestingly, the SAH hydrolase inhibitor 3-deazaneplanocin A has previously been shown to inhibit several viruses (45). It was recently demonstrated to limit Ebola virus replication and to lead to IFN-α production in virus-infected mice (46, 47). The SAH hydrolase inhibitors may thus confer a selective antiviral activity that differ from one virus to another depending on their RNA methylation levels (48). The inhibition may have the direct effect of an SAH concentration increase on the viral MTase or have indirect effects on cellular mRNA cap methylation, which may then be recognized as “non-self” and thus trigger an IFN response.
FIG 8.
Model of the nsp10/nsp16-7mGpppRNA reaction turnover regulated by the SAM/SAH balance. The nsp10/nsp16 model was built as described previously in Fig. 4. The three-dimensional representation of nsp16 structure is indicated in blue, 7mGpppGAAAAA is indicated in yellow, and nsp10 is indicated in pink. According to our model, nsp16 first binds to cap-0 RNA and then loads a SAM molecule (gray, 100 μM in infected cells), thus allowing the recruitment of the nsp10 allosteric activator. The nsp16 then catalyzes cap RNA methylation and converts cap-0 into cap-1 RNA (indicated in salmon). Methylated RNA is released from the nsp16 cap-binding site. Since SAH (indicated in orange) is supposed to be at lower concentration than SAM in infected cells (20 μM versus 100 μM), SAH would be released from the nsp10/nsp16 complex, and this should favor its dissociation and reaction turnover.
The viral 2′-O-MTase was highlighted as a potential antiviral target (38). Indeed, such RNA methylation could be a marker of “self,” thus avoiding detection of viral RNA by Mda5/RIG-I sensors (16, 18, 19). In addition, one IFN-stimulated gene product, IFIT1, sequesters 2′O-unmethylated viral RNA and thereby blocks its translation, which results in inhibition of viral replication (49). Thus, compounds blocking the viral 2′-O-MTase could limit viral replication in infected animals and elicit a strong antiviral response, favoring viral clearance. Here, we show that SAM analogues, such as SAH and sinefungin, limit 2′-O-MTase activity, with IC50s in the micromolar range (7.0 and 7.4 μM, respectively) with no inhibition of RNA binding, as expected. Conversely, a N7-methylated cap analogue (7mGpppG) inhibits both 2′-O-MTase activity and RNA recognition, with an IC50 of 45 μM. This indicates that 7mGpppG acts as a competitive inhibitor that blocks cap-0 RNA recognition by the nsp10/nsp16 complex. This was confirmed by the absence of inhibition in the presence of unmethylated cap analogues. Altogether, these results might help in developing an efficient assay to screen compounds that inhibit the SAM-dependent MERS-CoV nsp10/nsp16 complex. The biochemical tools and the RNA binding assay allow the determination of whether inhibitors are competing directly with the RNA substrate. It remains to be tested whether targeting the 2′-O-MTase activity can limit viral replication in cells or animal models that harbor the Mda5/RIG-I antiviral pathway (33).
MATERIALS AND METHODS
Plasmid constructs.
The expression vectors pDEST-14/6 His-nsp10 and pDEST-14/6 His-nsp16 containing the coding sequence of the human betacoronavirus 2c EMC/2012 (GenBank accession no. JX869059.2), a MERS-CoV strain, were provided by Eric Snijder's team. For alanine scanning, nsp16 mutants were generated by single-site amino acid substitution to alanine using the pDEST-14/6His-nsp16 plasmid and a QuikChange site-directed mutagenesis kit (Agilent) according to the manufacturer's instructions (unpublished data). The mutant numbering starts at the beginning of the nsp16 sequence: Y30A, K31A, R38A, H41A, N43A, K46A, Y47A, K76A, D99A, N101A, D130A, Y132A, D133A, T136A, K137A, F149A, F150A, K170A, H174A, Y181A, and E203A. All constructs were verified by DNA sequencing (Eurofins MWG Operon).
Expression and purification of the MERS-CoV nsp10, nsp16 and nsp10/nsp16 proteins.
MERS-CoV nsp10 and nsp16 fusion proteins (N-terminal hexahistidine tag) were expressed in Escherichia coli C2566 cells that contain the pRARE-2 plasmid. Transformed bacterial cells were grown at 37°C in Luria-Bertani and 2YT media containing 100 μg/ml ampicillin and 17 μg/ml chloramphenicol. Protein expression was induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After overnight incubation at 17°C, cells expressing nsp10, nsp16, or a mixture of nsp10 and nsp16 (in equal volumes) were pelleted by centrifugation (13,000 × g, 10 min) and frozen before resuspension in lysis buffer (50 mM HEPES [pH 7.5], 300 mM NaCl, 30 mM imidazole, 10% glycerol supplemented with 1 mM phenylmethylsulfonyl fluoride, 0.25 mg/ml lysozyme, 10 μg/ml DNase I). After sonication and clarification (80,000 × g, 4°C, 30 min), the supernatants were incubated with HisPur cobalt resin (Thermo Scientific) at 4°C with gentle shaking for 30 min. After washing in buffer W [50 mM HEPES (pH 7.5), 40 mM imidazole, 10% glycerol, and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl)] containing 300 or 500 mM NaCl, bound proteins were eluted with buffer W supplemented with 250 mM imidazole. Recombinant nsp10 and nsp16 proteins were then concentrated by ultrafiltration devices with molecular mass cutoffs of 5 (Millipore) and 10 (Sartorius) kDa, respectively.
The nsp10/nsp16 complex was purified on a Superdex 200 column 16/60 (GE) equilibrated with buffer W containing 500 mM NaCl (GE ÄKTA purifier). After peak collection, fractions containing the nsp10/nsp16 complex were concentrated by ultrafiltration using 10-kDa Vivaspin 15 (Sartorius). All purified proteins were analyzed by SDS-PAGE, followed by Coomassie blue staining. Purified enzymes were stored at −20°C in 50% glycerol for enzymatic assays.
Methyltransferase assays.
Assay to test the MTase activity were carried out in a reaction mixture (40 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol [DTT], 1 mM MgCl2, 2 μM SAM, and 0.33 μM 3H-SAM [Perkin-Elmer]) in the presence of 0.7 μM synthetic RNA corresponding to the 5′ extremity of the MERS-CoV genome with various 5′ end modifications (triphosphorylated, pppGAUUUAA; cap, GpppGAUUUAA; cap-0, 7mGpppGAUUUAA; or 2′-O-methylated cap, GpppG2′OmAUUUAA). Purified nsp10 was added at a final concentration of 2 μM in the presence of 1.2 μM nsp16 since the apparent interaction Kd is ∼2 μM (Fig. 2A). The purified nsp10/nsp16 complex was used at 1 μM. Reaction mixtures were incubated at 30°C and stopped at the indicated time points by diluting the reaction mixture with a 10-fold excess of 20 microM ice-cold SAH (AdoHcy, New England BioLabs). Samples were then transferred to DEAE filters (Perkin-Elmer) by using a Filtermat Harvester apparatus (Packard Instruments). Unincorporated 3H-SAM was removed from the DEAE filters by several washes with 0.01 M ammonium formate (pH 8.0), H2O, and absolute ethanol. After drying, filters were incubated with BetaplateScint (Wallac) scintillation fluid before quantification of the 3H methylation of the RNA substrates using a Wallac 1450 MicroBetaTriLux liquid scintillation counter (results were expressed as counts per minute [cpm]).
For the MTase inhibition assays, 0.5 μM nsp10/nsp16 complex was mixed with 50 μM each candidate inhibitor before addition of SAM (AdoMet; New England BioLabs) and 7mGpppGAUUUAA to start the reaction. The final dimethyl sulfoxide (DMSO) concentration in the reaction mixtures was lower than 5%, and control reaction mixtures contained a similar DMSO concentration. Reaction mixtures were incubated at 30°C for 30 min and then analyzed by filter binding assay, as described above.
The IC50s of SAH (AdoHcy), sinefungin, and cap analogues (7mGpppA and 7mGpppG) were determined with GraphPad Prism by using a log (inhibitor) versus a response variable slope equation.
Fluorescence polarization assays. (i) RNA labeling.
For RNA labeling, 10 μM RNA was labeled by ligation of 12.5 μM pCp-cyanine 5 (pCp-cy5; Jena Bioscience) at the 3′ extremity using 1 mM T4 RNA ligase-1 (New England BioLabs) in 50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM DTT, and 1 mM ATP at 16°C overnight, according to the manufacturer's instruction. T4 RNA ligase-1 was removed using Strata-Clean Resin (Agilent) and labeled RNA was collected after centrifugation in a Micro-Spin G-25 column (GE health care) to eliminate the free pCp-Cy5 dye.
(ii) Polarization assay.
For the polarization assay, pCp-Cy5-labeled RNA was mixed with increasing concentrations of freshly purified nsp10/nsp16 complex, nsp10, or nsp16 in binding buffer (50 mM Tris-HCl [pH 8], 1 mM DTT) in a 384-well opaque microplate (Greiner Bio-One). Fluorescence polarization was measured using a microplate reader (PHERAstar) with a 590/675 fluorescence polarization optic module (i.e., 590-nm excitation and 675-nm emission wavelengths). The dissociation constants (Kd) were determined using nonlinear regression analysis: site-specific binding with Hill slope equation (GraphPad Prism). The competitive effect of cap analogues on 7mGpppGAUUUAA-cy5 binding by 20 μM nsp10/nsp16 complex were determined by using increasing concentrations of GpppG or 7mGpppG (New England BioLabs) in binding buffer. Data were plotted in GraphPad Prism 5.0 and a nonlinear sigmoidal dose-response curve fitted in order to determine the IC50 values for RNA binding inhibition.
Interferometry for nsp10-nsp16 interaction. (i) nsp10 biotinylation.
nsp10 was biotinylated (at room temperature for 30 min) using EZ-Link NHS-PEG4-biotin (Thermo Fisher) at a molar ratio of 1:1 in biotinylation buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM TCEP). Biotinylated nsp10 was separated from free biotin using NAP-5 columns equilibrated with biotinylation buffer.
(ii) Octet analysis.
Assays were performed using streptavidin sensors preincubated in 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1 mM TCEP, and 0.5 mg/ml bovine serum albumin (BSA). Biotinylated nsp10 (100 nM) was loaded on streptavidin-coated biosensors for 5,000 s to achieve 0.8-nm binding. Various nsp16 concentrations (0 to 7.8 μM) were used for the association step (300 s) performed in 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1 mM TCEP, and 0.5 mg/ml BSA. The dissociation step was followed for 500 s. Real-time nsp10-nsp16 interaction kinetics were recorded by using biolayer interferometry (Octet RED96). The Kd, Kon, and Koff values were calculated using forteBio software after subtraction of the reference sensor from the sample traces. All curves were fitted using a 1:1 interaction model. The steady-state curve of nsp10-nsp16 interaction was traced using one site-specific binding with the Hill slope equation (GraphPad Prism).
Synthesis of RNA substrates.
RNAs were chemically synthesized on a solid support using an ABI 394 synthesizer. After RNA elongation with 2′-O-pivaloyloxymethyl phosphoramidite monomers (Chemgenes, USA) (50), the 5′-hydroxyl group was phosphorylated, and the resulting H-phosphonate derivative was oxidized and activated into a phosphoroimidazolidate derivative to react with pyrophosphate (pppRNA) (51) or GDP (GpppRNA) (52). After deprotection and release from the solid support, pppRNAs and GpppRNAs were purified by ion exchange high-pressure liquid chromatography (IEX-HPLC), and their purity (>95%) confirmed by MALDI-TOF spectrometry. N7 methylation of purified GpppRNAs was then performed enzymatically using an N7-hMTase (52).
Mass spectrometry and IEX-HPLC analysis.
Crude pppRNAs and GpppRNAs were analyzed and purified by IEX-HPLC on a Dionex Ultimate 3000 apparatus equipped with an UV detector at the wavelength of 260 nm and an anion-exchange DionexDNAPac PA200 column (4 by 250 mm for analysis or 9 by 250 mm for semipreparative purpose) with a flow rate of 1.5 ml/min for analysis or 5 ml/min for semipreparative purposes. The following eluents were used: buffer A (20% CH3CN in 25 mM Tris-HCl [pH 8]) and buffer B (20% CH3CN containing 200 mM NaClO4 in 25 mM Tris-HCl [pH 8]). After purification, pure fractions containing the desired RNA were pooled in a 100-ml round-bottomed flask and concentrated to dryness under reduced pressure. The residues were desalted on a C18 cartridge (Sep-Pak Classic).
MALDI-TOF mass spectra were recorded on a Voyager-DE (PerSeptive Biosystems, USA) or on an AXIMA Assurance spectrometer (Shimadzu Corp., Japan) equipped with an N2 laser (337 nm) using 2,4,6-trihydroxyacetophenone as a saturated solution in a mixture of acetonitrile and 0.1 M ammonium citrate solution (1:1, vol/vol) for the matrix. The ionization mode was a linear negative ion mode. Analytical samples were mixed with the matrix in a 1:5 (vol/vol) ratio, crystallized on a 100-well stainless steel plate, and analyzed.
ACKNOWLEDGMENTS
We thank Clara Posthuma and Eric Snijder for the cloning of MERS-CoV nsp10 and nsp16; Isabelle Imbert, Axelle Collet, Maria Mathe, and Marion Sevajol for technical support; and Bruno Coutard and Barbara Selisko for scientific contributions and for critical reviews of the manuscript.
This work was supported by the European Union Seventh Framework Program (Project Silver; grant 200644), the French National Research agency (grants ANR-12-BSV3-007-01 and ANR-DGA MTase IN), the Fondation pour la Recherche Médicale (FRM; grant FDT20160434967) and the Fondation Infectiopôle Sud (student fellowship).
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arabi YM, Harthi A, Hussein J, Bouchama A, Johani S, Hajeer AH, Saeed BT, Wahbi A, Saedy A, Al Dabbagh T, Okaili R, Sadat M, Balkhy H. 2015. Severe neurologic syndrome associated with Middle East respiratory syndrome coronavirus (MERS-CoV). Infection 43:495–501. doi: 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durai P, Batool M, Shah M, Choi S. 2015. Middle East respiratory syndrome coronavirus: transmission, virology, and therapeutic targeting to aid in outbreak control. Exp Mol Med 47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusken CB, Farag EA, Jonges M, Godeke GJ, El-Sayed AM, Pas SD, Raj VS, Mohran KA, Moussa HA, Ghobashy H, Alhajri F, Ibrahim AK, Bosch BJ, Pasha SK, Al-Romaihi HE, Al-Thani M, Al-Marri SA, AlHajri MM, Haagmans BL, Koopmans MP. 2014. Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro Surveill 19:1–5. [DOI] [PubMed] [Google Scholar]
- 7.Raj VS, Osterhaus AD, Fouchier RA, Haagmans BL. 2014. MERS: emergence of a novel human coronavirus. Curr Opin Virol 5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr AR, Channappanavar R, Perlman S. 2016. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu Rev Med 68:annurev-med-051215–031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, Qureshi MI, Abu-Zeid M, Zhang Y, Khiyami MA, Alharbi NS, Hajrah NH, Sabir MJ, Mutwakil MH, Kabli SA, Alsulaimany FA, Obaid AY, Zhou B, Smith DK, Holmes EC, Zhu H, Guan Y. 2016. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 10.van Doremalen N, Bushmaker T, Munster VJ. 2013. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill 18:1–4. [DOI] [PubMed] [Google Scholar]
- 11.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJM, Osterhaus ADME, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Boheemen S, De Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus A, Haagmans BL, Gorbalenya A, Snijder E, Fouchier R. 2012. Genomic characterization of newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 14.Cougot N, Van Dijk E, Babajko S, Séraphin B. 2004. “Cap-tabolism.” Trends Biochem Sci 29:436–444. [DOI] [PubMed] [Google Scholar]
- 15.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J 19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor MDA5. Nat Immunol 12:137–143. doi: 10.1038/nrm3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, Goldeck M, Schmidt T, Schmid-Burgk JL, Kerber R, Wolter S, Stümpel JP, Roth A, Bartok E, Drosten C, Coch C, Hornung V, Barchet W, Kümmerer BM, Hartmann G, Schlee M. 2015. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity 43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, Patel SS, Marcotrigiano J. 2016. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A 113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyde JL, Diamond MS. 2015. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 479-480:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decroly E, Ferron F, Lescar J, Canard B. 2012. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond MS. 2014. IFIT1: a dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 25:543–550. doi: 10.1016/j.cytogfr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daugherty MD, Schaller AM, Geballe AP, Malik HS. 2016. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 5:1–22. doi: 10.7554/eLife.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov KA, Ziebuhr J. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J Virol 78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. 2010. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog 6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, Betzi S, Imbert I, Canard B, Guillemot JC, Lécine P, Pfefferle S, Drosten C, Snijder EJ, Decroly E, Morelli X. 2014. Coronavirus nsp10, a critical cofactor for activation of multiple replicative enzymes. J Biol Chem 289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Grotthuss M, Wyrwicz LS, Rychlewski L. 2003. mRNA Cap-1 methyltransferase in the SARS genome. Cell 113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B. 2008. Coronavirus nonstructural protein 16 is a Cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol 82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lugari A, Betzi S, Decroly E, Bonnaud E, Hermant A, Guillemot JC, Debarnot C, Borg JP, Bouvet M, Canard B, Morelli X, Lécine P. 2010. Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J Biol Chem 285:33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin JL, McMillan FM. 2002. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol 12:783–793. doi: 10.1016/S0959-440X(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Sun Y, Wu A, Xu S, Pan R, Zeng C, Jin X, Ge X, Shi Z, Ahola T, Chen Y, Guo D. 2015. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J Virol 89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menachery VD Jr, Y BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Züst R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K. 2013. Rational design of a live attenuated dengue vaccine: 2′-O-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finkelstein JD. 1990. Methionine metabolism in mammals. J Nutr Biochem 23:228–237. [DOI] [PubMed] [Google Scholar]
- 37.Svardal M, Uelands M. 1987. Compartmentalization of S-adenosylhomocysteine in rat liver. J Biol Chem 262:15413–15417. [PubMed] [Google Scholar]
- 38.Ferron F, Decroly E, Selisko B, Canard B. 2012. The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res 96:21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Aq̊vist J. 2007. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2′O)-methyltransferase. Bioorg Med Chem 15:7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 40.Dong H, Chang DC, Hua MHC, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao X, Shuman S. 1994. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. J Biol Chem 269:24472–24479. [PubMed] [Google Scholar]
- 42.De la Peña M, Kyrieleis OJ, Cusack S. 2007. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J 26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varshney D, Petit AP, Bueren-Calabuig JA, Jansen C, Fletcher DA, Peggie M, Weidlich S, Scullion P, Pisliakov AV, Cowling VH. 2016. Molecular basis of RNA guanine-7 methyltransferase (RNMT) activation by RAM. Nucleic Acids Res 44:10423–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodel AE, Gershon PD, Quiocho FA. 1998. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell 1:443–447. doi: 10.1016/S1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 45.De Clercq E. 1987. S-Adenosylhomocysteine hydrolase inhibitors as broad spectrum antiviral agents. Biochem Pharmacol 36:2567–2575. [DOI] [PubMed] [Google Scholar]
- 46.De Clercq E. 2001. Molecular targets for antiviral agents. J Pharmacol Exp Ther 297:1–10. [PubMed] [Google Scholar]
- 47.Bray M, Raymond JL, Geisbert T, Baker RO. 2002. 3-Deazaneplanocin A induces massively increased interferon-α production in Ebola virus-infected mice. Antiviral Res 55:151–159. doi: 10.1016/S0166-3542(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 48.De Clercq E. 2002. Strategies in the design of antiviral drugs. Nat Rev Drug Discov 1:13–25. [DOI] [PubMed] [Google Scholar]
- 49.Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, Thiel V, Pichlmair A. 2013. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog 9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavergne T, Bertrand JR, Vasseur JJ, Debart F. 2008. A base-labile group for 2′-OH protection of ribonucleosides: a major challenge for RNA synthesis. Chem Eur J 14:9135–9138. doi: 10.1002/chem.200801392. [DOI] [PubMed] [Google Scholar]
- 51.Zlatev I, Lavergne T, Debart F, Vasseur J-J, Manoharan M, Morvan F. 2010. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org Lett 12:2190–2193. doi: 10.1021/ol1004214. [DOI] [PubMed] [Google Scholar]
- 52.Thillier Y, Decroly E, Morvan F, Canard B, Vasseur J-J, Debart F. 2012. Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N7)-methyltransferase. RNA 18:856–868. doi: 10.1261/rna.030932.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shatkin AJ. 1976. Capping of eucaryotic mRNAs. Cell 9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]