ABSTRACT
Human APOBEC3H (A3H) is a cytidine deaminase that inhibits HIV-1 replication. To evade this restriction, the HIV-1 Vif protein binds A3H and mediates its proteasomal degradation. To date, little information on the Vif-A3H interface has been available. To decipher how both proteins interact, we first mapped the Vif-binding site on A3H by functionally testing a large set of A3H mutants in single-cycle infectivity and replication assays. Our data show that the two A3H α-helixes α3 and α4 represent the Vif-binding site of A3H. We next used viral adaptation and a set of Vif mutants to identify novel, reciprocal Vif variants that rescued viral infectivity in the presence of two Vif-resistant A3H mutants. These A3H-Vif interaction points were used to generate the first A3H-Vif structure model, which revealed that the A3H helixes α3 and α4 interact with the Vif β-sheet (β2-β5). This model is in good agreement with previously reported Vif and A3H amino acids important for interaction. Based on the predicted A3H-Vif interface, we tested additional points of contact, which validated our model. Moreover, these experiments showed that the A3H and A3G binding sites on HIV-1 Vif are largely distinct, with both host proteins interacting with Vif β-strand 2. Taken together, this virus-host interface model explains previously reported data and will help to identify novel drug targets to combat HIV-1 infection.
IMPORTANCE HIV-1 needs to overcome several intracellular restriction factors in order to replicate efficiently. The human APOBEC3 locus encodes seven proteins, of which A3D, A3F, A3G, and A3H restrict HIV-1. HIV encodes the Vif protein, which binds to the APOBEC3 proteins and leads to their proteasomal degradation. No HIV-1 Vif-APOBEC3 costructure exists to date despite extensive research. We and others previously generated HIV-1 Vif costructure models with A3G and A3F by mapping specific contact points between both proteins. Here, we applied a similar approach to HIV-1 Vif and A3H and successfully generated a Vif-A3H interaction model. Importantly, we find that the HIV-1 Vif-A3H interface is distinct from the Vif-A3G and Vif-A3F interfaces, with a small Vif region being important for recognition of both A3G and A3H. Our Vif-A3H structure model informs on how both proteins interact and could guide toward approaches to block the Vif-A3H interface to target HIV replication.
KEYWORDS: HIV-1, APOBEC3H, Vif, restriction factor, APOBEC3, A3H, HIV
INTRODUCTION
The human APOBEC3 family consists of seven distinct proteins (A3A to A3H), several of which have potent activity against HIV-1 (1, 2). During infection of the new cell, virion-incorporated APOBEC3 proteins target the process of reverse transcription through directly deaminating the viral single-stranded DNA as well as by deaminase-independent mechanisms (3–6). In turn, HIV-1 counteracts APOBEC3 restriction by expressing an accessory Vif protein that leads to the proteasomal degradation of APOBEC3 in the producer cell (5, 7, 8).
Human A3H encodes at least seven different haplotypes differing considerably in their protein stability and activity to restrict HIV-1 (9–11). A3H haplotypes II, V, and VII potently restrict HIV-1, whereas haplotypes I, III, IV, and VI express unstable proteins and lack anti-HIV activity. We have previously shown that endogenously expressed stable A3H haplotype II restricts HIV-1 variants that are unable to counteract it (12, 13). Moreover, HIV-1 Vif alleles derived from HIV-infected patients encoding unstable A3H haplotypes are more likely to lack anti-A3H activity, indicating that HIV-1 Vif adapts to the stable A3H (12). Further, recently HIV-infected patients with one or two stable A3H alleles displayed reduced plasma viral loads and higher CD4+ T-cell counts in the absence of antiretroviral treatment than patients with virus encoding unstable A3H variants (12). Combined, these data indicate that stable A3H is a bona fide HIV-1 restriction factor.
Despite numerous studies on A3H, it remains unclear how A3H and HIV-1 Vif interact. To date, only a single amino acid at position 121 in A3H has been implicated in the interaction with HIV Vif: mutating aspartic acid (D) at position 121 to a lysine (K) renders A3H resistant to Vif (14, 15). On the Vif side, amino acid positions 39, 48, and 60 to 63 are important for the counteraction of A3H, indicating they are part of the A3H binding site (12, 13, 15, 16).
We used a stepwise selection and counterselection approach, which we successfully used to map the Vif-A3G interaction (17) to decipher the interface between A3H and HIV-1 Vif. We first identified A3H residues critical for resistance to HIV-1 Vif followed by defining the reciprocal Vif mutants that restore the Vif-A3H interaction (Fig. 1A). These specific Vif-A3H pairs were subsequently used as anchor points to dock both proteins and create a Vif-A3H interaction model. This experimental approach revealed that A3H helixes α3 and α4 interact with the Vif β-sheet comprised of β-strands β2 to β5. Of note, A3H and A3G both interact with the Vif β-strand β2. Thus, our Vif-A3H model not only explains previous data (12–16) but also predicts novel interaction points, which will help determine novel targets for therapeutics aimed to disrupt this crucial virus-host interface.
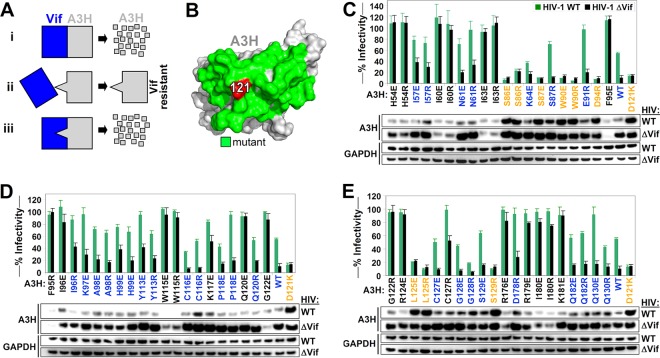
FIG 1.
Identification of the Vif binding site of A3H. (A) Overview of the approach used to map the Vif-A3H interaction. (Scenario i) Vif binds A3H, resulting in its degradation. (Scenario ii) An A3H with a mutation in the Vif-binding site is resistant to Vif and is not degraded. (Scenario iii) A Vif variant with a specific mutation to accommodate the A3H mutation would regain its ability to bind and degrade A3H. The mutation that makes A3H Vif resistant and the accommodating Vif mutation are likely directly interacting. (B) A3H residue 121 is indicated in red, and neighboring surface-exposed residues that were individually mutated for functional studies are colored in green. (C, D, and E) Single-cycle infectivity assay with 20 ng A3H R/E mutants, A3H WT, and A3H-D121K were cotransfected with 500 ng WT HIV or HIV ΔVif, the infectivity was analyzed in TZM-bl reporter cells, and A3H expression was assessed by Western blotting. A3H mutants that were resistant to Vif are colored in orange (HIV-WT/HIV-ΔVif infectivity ratio of <1.5 and restriction of HIV-ΔVif of >50%). Active A3H mutants that are sensitive to Vif are colored in blue (HIV-WT/HIV-ΔVif infectivity ratio of >1.5 and restriction of HIV-ΔVif of >50%). Inactive A3H mutants are indicated in black (restriction of HIV-ΔVif of <50%). The dashed line represents the 50% restriction cutoff. The infectivity of WT HIV and HIV-ΔVif in the absence of A3H was set at 100%, and the average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations.
RESULTS
Identification of the Vif binding site of A3H.
Previous studies demonstrated that mutating the aspartic acid to lysine at A3H position 121 (D121K) renders human A3H resistant to HIV-1 Vif-mediated degradation (14, 15). To further map the HIV-1 Vif-binding site, we introduced positively charged arginine residues and negatively charged glutamate residues at surface-exposed positions around A3H residue 121 (Fig. 1B). These A3H mutants were cotransfected with full-length wild-type HIV (HIV-WT; LAI) and a mutant HIV encoding a nonfunctional mutant Vif (SLQ>AAA; here named ΔVif), and infectivity was assessed by infecting TZM-bl HIV reporter cells. A3H expression was analyzed by Western blotting as described previously (18). HIV-ΔVif served as a control to ensure that the A3H mutants were expressed and retained their antiviral activity. The infectivity of WT HIV and HIV-ΔVif in the absence of A3H was set at 100%. Only A3H mutants that restricted HIV-ΔVif more than 50% (infectivity of <50%) were considered functionally active (50% infectivity is indicated by a dashed line in the figure). We defined functionally active A3H mutants to be HIV-1 Vif resistant if the infectivity ratio of HIV-WT and HIV-ΔVif was lower than 1.5-fold (HIV-WT/HIV-ΔVif infectivity ratio of <1.5). We found that A3H mutants S86E, S86R, S87E, W90R, W90E, D94R, L125R, L125E, and S129R restricted both HIV-WT and HIV-ΔVif to similar levels, indicating they were resistant to HIV-1 Vif in a manner similar to the positive-control mutant D121K (Fig. 1C and D). In addition, the expression of these A3H mutants was not affected by the presence of Vif (Fig. 1C and D). Certain A3H mutants, such as H54E, H54R, I60E, I60R, and others, failed to restrict HIV-ΔVif (HIV-ΔVif infectivity of >50%), suggesting that these mutations disrupted the antiviral function of A3H, and therefore were excluded from all of the follow-up studies. Indeed, most inactive A3H mutants were not efficiently expressed (Fig. 1C and D).
The Vif binding site of A3H consists of α-helixes α3 and α4.
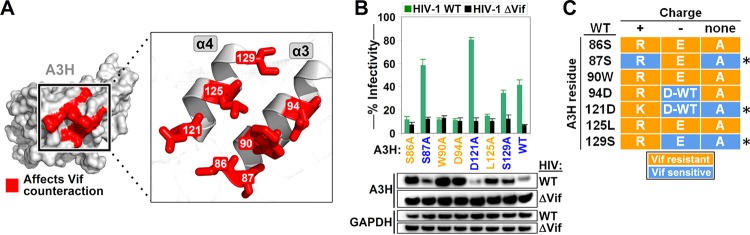
Mapping the HIV-1 Vif-resistant mutations on a predicted A3H structure model showed that they were all located on the α-helixes α3 and α4 (Fig. 2A). To determine whether the specific charge was important for the resistance phenotype, we next introduced alanine substitutions at the seven positions leading to Vif resistance and tested these A3H mutants in a single-cycle infectivity assay and by Western blotting (Fig. 2B). Alanine substitutions at positions 86, 90, 94, and 125 remained resistant to HIV-1 Vif, indicating that the original amino acid identity at these key positions is critical for interacting with Vif. However, A3H variants with alanine substitutions at positions 87, 121, and 129 were efficiently counteracted by HIV-1 Vif, indicating that variation is tolerated at this position (Fig. 2B and C).
FIG 2.
Vif binding site of A3H consists of α-helixes α3 and α4. (A) A3H residues that conferred resistance to HIV Vif are highlighted in red on the A3H protein surface and on α-helixes α3 and α4. (B) Single-cycle infectivity assays with selected A3H alanine mutants. The average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations. (C) Summary of the A3H mutations that confer resistance to Vif. An asterisk indicates the A3H residue positions that do not disrupt its interaction with Vif when mutated to an alanine.
Vif-resistant A3H mutants restrict HIV replication.
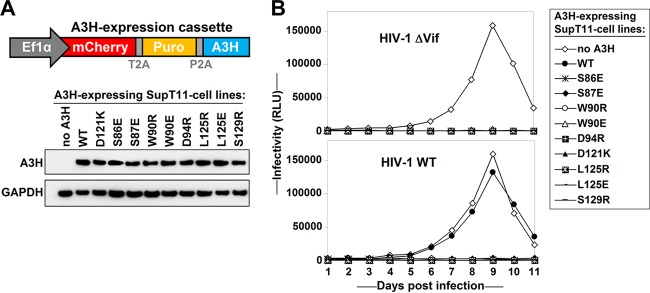
To test whether the Vif-resistant A3H mutants could restrict HIV replication, we generated SupT11 cell lines to stably express untagged A3H-WT or one of the nine identified Vif-resistant A3H mutants (Fig. 1). SupT11 T cells naturally do not express any APOBEC3 proteins (19). To stably express A3H, we used lentiviral constructs expressing A3H as part of a cassette encoding mCherry and the puromycin resistance gene separated by T2A and P2A cleavage sites (Fig. 3A). Of note, because C-terminal tagging of A3H may interfere with Vif binding, we decided to leave A3H untagged (2, 20) and detected A3H expression using an A3H-specific antibody (Fig. 3A) (13). Infection of these T cell lines with HIV-WT and HIV-ΔVif showed that HIV-ΔVif only replicated in the absence of A3H, indicating that all of the A3H variants selected restricted HIV efficiently (Fig. 3B) in this spreading infection model system. HIV-WT replicated to comparable levels in the presence and absence of A3H-WT, indicating that only A3H-WT is counteracted by Vif, whereas all other A3H mutants are resistant to Vif. This observation is in excellent agreement with the results obtained from the single-cycle infectivity experiments (Fig. 1).
FIG 3.
Vif-resistant A3H mutants restrict HIV replication. (A) Schematic representation of the lentiviral A3H expression cassette used to express A3H in SupT11 T cells. The Ef1α promoter drives the expression of mCherry, puromycin resistance, and A3H separated by T2A and P2A cleavage sites. The stable T cell lines were lysed and analyzed by Western blotting. (B) The indicated SupT11 T-cell lines were infected with WT HIV or HIV ΔVif at an MOI of 0.1. RLU, relative light units. Culture supernatants were collected every day and used to infect TZM-bl HIV reporter cells.
HIV adapts to Vif-resistant A3H-S86E mutant by acquiring a Vif-E45Q substitution.
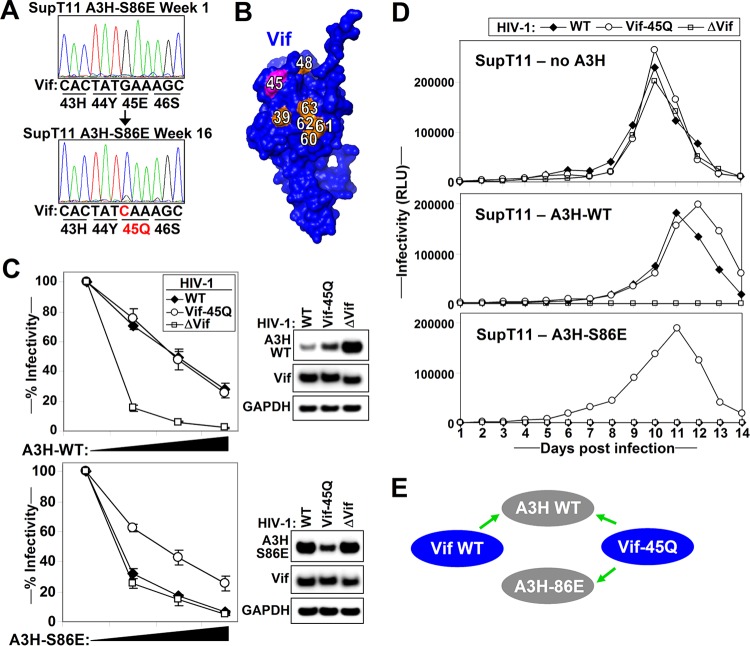
To find specific Vif changes that can rescue infectivity in the presence of the Vif-resistant A3H mutants identified in the previous section, we performed long-term culture experiments to adapt HIV to replicate in the presence of these mutant proteins. We kept the infected cells in culture for 6 months with biweekly microscopic inspections for evidence of viral replication. At week 16, the SupT11 cells expressing A3H-S86E showed syncytia, and sequencing of proviral DNA from these cells revealed a Vif-E45Q change (Fig. 4A). Interestingly, this position is close to other Vif positions important for counteracting A3H (Fig. 3B). In addition, Vif residue 45 has been implicated in A3G neutralization (21), indicating that A3H and A3G binding regions to Vif overlap. Of note, no other Vif adaptations were observed in any of the other cell cultures.
FIG 4.
HIV adapts to Vif-resistant A3H-S86E mutant by a Vif-E45Q mutation. (A) Cellular DNA was extracted from the infected SupT11 T-cell line expressing A3H-S86E at week 1 and week 16 postinfection. DNA was used to PCR amplify the full-length proviral vif open reading frame. The two chromatograms show a G-to-C mutation resulting in a Vif-E45Q change. (B) Vif position 45 is indicated together with Vif residues reported to be important for counteracting A3H. (C) Single-cycle infectivity assays with WT HIV, HIV Vif-45Q, and HIV ΔVif in the presence of increasing amounts (0, 10, 20, and 40 ng A3H) of WT A3H (top) or A3H-S86E (bottom). The infectivity was analyzed by infection of TZM-bl reporter cells. The average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations. Cell lysates corresponding to 10 ng A3H were analyzed by Western blotting. (D) The indicated T-cell lines were infected with WT HIV, HIV Vif-45Q, or HIV ΔVif at an MOI of 0.1, and supernatants were collected every day and used to infect TZM-bl HIV reporter cells. (E) Summary of the results indicating that A3H-86 interacts with Vif-45.
To test whether the Vif-E45Q change allowed HIV-1 Vif to counteract the A3H-S86E mutant, we introduced Vif-E45Q into HIV-WT and compared its infectivity in the presence of A3H-WT and A3H-S86E using single-cycle infectivity assays (Fig. 4C). Both HIV Vif-WT and HIV Vif-E45Q degraded and counteracted WT A3H (Fig. 4C, top), but only HIV Vif-E45Q could counteract A3H-S86E (bottom). In multicycle replication experiments, HIV Vif-WT, HIV Vif-45Q, and HIV-ΔVif replicated similarly in the absence of A3H (Fig. 4D, top), while HIV-WT and HIV-45Q (but not HIV-ΔVif) replicated in the presence of A3H-WT (middle). SupT11 expressing A3H-S86E supported replication of HIV-45Q exclusively (bottom). Combined, our data indicate that Vif-45Q is critical for overcoming A3H-86 restriction (Fig. 4E).
Vif-R63E and Vif-R90E specifically counteract A3H-S129R.
Because one point of contact between HIV-1 Vif and A3H is insufficient to properly orient HIV-1 Vif on A3H, we used a more directed alternative approach to finding additional Vif mutants. We generated a panel of 13 Vif mutants consisting of positively charged arginine and negatively charged glutamate at and around positions deemed important for A3H recognition based on previous work (Fig. 5A) (12–16) and tested their activity against selected Vif-resistant A3H mutants in single-cycle infectivity assays. We found that Vif-WT, Vif-G37R, and Vif-D61R efficiently counteracted A3H-WT, Vif mutants G37E, G60E, A62E, A62R, R63E, and R90E showed reduced anti-A3H activity, and Vif mutants F39E, F39R, E54R, H56E, and R93E completely lost their activity against A3H-WT (Fig. 5B). Of note, all of the Vif mutants efficiently counteracted A3G, indicating they were folded properly (Fig. 5B). A3H-S129R, which is resistant to most Vif variants, was efficiently counteracted by Vif mutants R63E and R90E, while both A3H-S87E and A3H-D121K were resistant to all Vif mutants tested. Interestingly, Vif positions 63 and 90 are located in close proximity to each other on the Vif crystal structure (Fig. 5A).
FIG 5.
Vif-R63E and Vif-R90E specifically counteract A3H-S129R. (A) The Vif residues that were mutated to glutamic acid or arginine are indicated on the Vif structure in green. (B) Single-cycle infectivity assay of the indicated HIV Vif mutants (500 ng) with 20 ng WT A3H, A3H-S87E, A3H-D121K, and A3H-S129R and 20 ng A3G. Infectivity was analyzed by TZM-bl reporter cells. The average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations. (C) Single-cycle infectivity assays with HIV-WT, HIV-Vif-63E, HIV-Vif-90E, and HIV ΔVif in the presence of increasing amounts (0, 10, 20, and 40 ng A3H) of WT A3H (top) or A3H-S129R (bottom). The infectivity was analyzed by TZM-bl reporter cells. The average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations. Cell lysates corresponding to 10 ng A3H were analyzed by Western blotting. (D) The indicated T-cell lines were infected with HIV-WT, HIV-Vif-63E, HIV-Vif-90E, or HIV ΔVif at an MOI of 0.1, and supernatants were collected every day and used to infect TZM-bl HIV reporter cells. (E) Summary of the results indicating that A3H-129 interacts with Vif-63 and Vif-90.
Single-cycle infectivity assays using increasing amounts of A3H-WT and A3H-S129R show that Vif mutants R63E and R90E had diminished activity against WT A3H (Fig. 5C). However, these Vif mutants efficiently counteracted A3H-S129R. Multicycle replication assays in T-cell lines stably expressing A3H-WT or A3H-S129R showed a similar trend, as all viruses replicated efficiently in the absence of A3H. Only WT HIV replicated well on cells expressing WT A3H, while replication of both HIV Vif-R63E and HIV Vif-R90E was delayed in these cells (Fig. 5D). In contrast, HIV Vif-R63E and HIV Vif-R90E replicated efficiently in the presence of A3H-S129R, whereas HIV Vif-WT was fully restricted. Combined, our data indicate that A3H-129 interacts with residues 63 and 90 in Vif (Fig. 5E).
Identification of the Vif-A3H interface by structural docking.
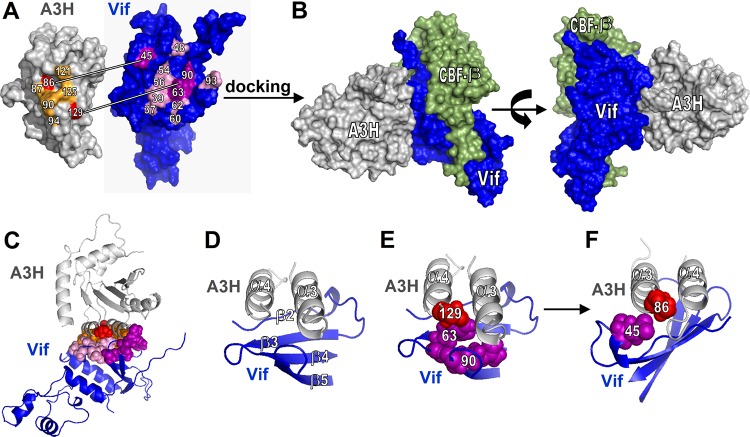
Because no crystal structures are available for A3H and LAI Vif, we first generated structure models using SWISS-MODEL. The A3H structure was predicted using the A3G-CTD structure (PDB entry 2KBO) (22). Using other available APOBEC3 structures, such as A3C, generated similar A3H structures, as all APOBEC3 domains are structurally very similar (data not shown). The LAI Vif structure was predicted using the NL4-3 Vif crystal structure (4N9F) (23).
We next performed protein docking with ClusPro (24) using Vif and A3H structures and constraints that favor interactions between A3H positions 86, 87, 90, 94, 121, 125, and 129 and Vif positions 37, 39, 45, 48, 54, 56, 60, 62, 63, 90, and 93 (Fig. 6A). Sixty-one structures were visually inspected to select those in which A3H-86 and A3H-129 were close to Vif-45, Vif-63, and Vif-90, respectively. We retained seven models that supported the two Vif-A3H pairs but also included all of the other important A3H and Vif residues that have been previously identified (Fig. 6A). Of note, the A3H and CBF-β binding sites on HIV-1 Vif do not overlap (Fig. 6B). A closer look at the Vif-A3H interface reveals that A3H helixes α3 and α4 interact with the Vif β-sheet, which comprises β-strands 2 to 5 (Fig. 6C and D). In addition, A3H-86 and Vif-45 as well as A3H-129 and Vif-63/Vif-90 are in close proximity (Fig. 6E and F).
FIG 6.
Identification of the Vif-A3H interface by structural docking. (A) Overview of A3H and Vif residues important for the Vif-A3H interaction. Vif and A3H were docked using ClusPro. (B) The structure that has A3H positions 86 and 129 in close proximity to Vif positions 45 and 63 to 90 is shown. CBF-β is indicated in green and does not overlap A3H. (C) A cartoon model of the Vif-A3H structure in which the important Vif and A3H are indicated shows that they all localize to the Vif-A3H interface. (D) Close-up of the Vif-A3H interface. A3H α-helixes α3 and α4 interact with the Vif β-sheet consisting of β-strands β2 to β5. The model indicates that Vif-45 and A3H-86 (E) and Vif-63/90 and A3H-129 (F) are in close proximity.
Vif-A3H model validation. (i) A3H position 100 is important for interaction with Vif.
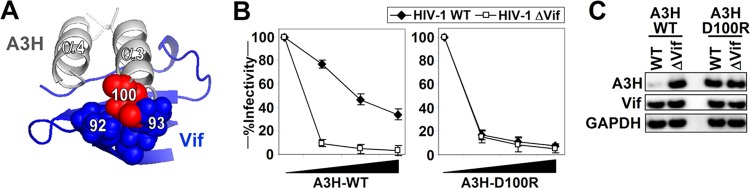
To validate our Vif-A3H interface structure model, we selected A3H and Vif residues that were predicted by the model to be present at the Vif-A3H interface for additional functional testing. A3H-100D, which was not tested before and is relatively distant from the previously identified Vif resistance positions in A3H, is in close proximity to Vif residues K92 and R93 (Fig. 7A). To test our model, we mutated A3H-100D to an arginine and determined its antiviral activity and resistance to Vif. Increasing amounts of A3H-WT potently restricted HIV-ΔVif, and infectivity was largely rescued in the presence of Vif (Fig. 7B, left). A3H-D100R also potently restricted HIV-ΔVif, but infectivity could not be rescued by HIV-1 Vif, indicating that A3H-D100R is no longer recognized by HIV-1 Vif (Fig. 7B, right). This functional Vif resistance of A3H-D100R is further supported by Western blot analysis, which shows that A3H-D100R is not degraded by Vif (Fig. 7C).
FIG 7.
Vif-A3H model validation. A3H position 100 is important for interaction with Vif. (A) Close-up of the Vif-A3H interface shows that A3H-100D is in close proximity to the Vif loop between β4 and β5. (B) Single-cycle infectivity assays with WT HIV and HIV ΔVif in the presence of increasing amounts (0, 10, 20, and 40 ng A3H) of WT A3H (left) or A3H-D100R (right). The infectivity was analyzed with TZM-bl reporter cells. The average relative infectivity values for a triplicate transfection are shown. Error bars represent the standard deviations. (C) Cell lysates corresponding to 10 ng A3H from the single-cycle infectivity assay shown were analyzed by Western blotting.
(ii) Vif β-strand 2 is important for interaction with A3H.
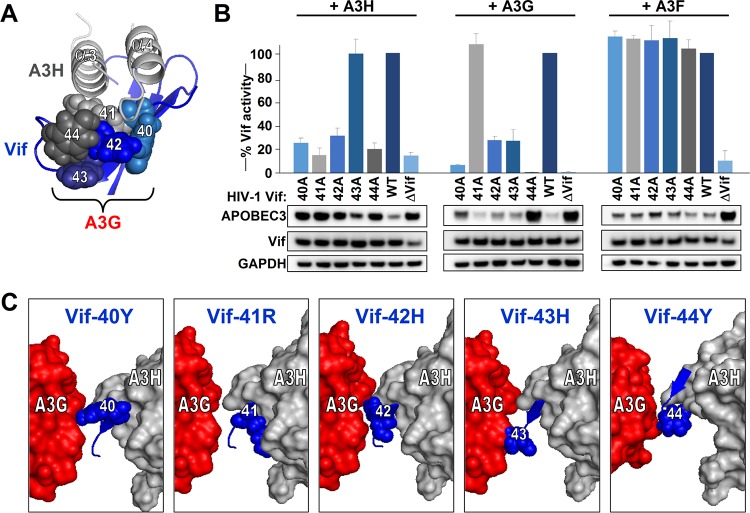
The Vif-A3H structure model also reveals that the Vif β-strand 2, which consists of Vif residues 40 to 44, is part of the Vif-A3H interface (Fig. 8A). Vif positions 40, 41, 42, and 44 are predicted to be close to the N-terminal ends of A3H α-helixes α3 and α4. Interestingly, this Vif stretch is important for counteracting other APOBEC3 proteins and is believed to be A3G specific (17, 25–27). We individually mutated each of the Vif residues at positions 40 to 44 to alanines and tested these mutants against A3H, A3G, and A3F. We included A3F because the A3F-binding site on Vif does not include the Vif 40-44 stretch (17, 28, 29). Vif mutants 40A, 41A, 42A, and 44A lost their ability to counteract A3H (Fig. 8B), while Vif mutants 40A, 42A, 43A, and 44A failed to counteract A3G. Vif-41A was fully functional against A3G. All five Vif mutants tested remained capable of efficiently counteracting A3F, indicating that the Vif 40-44 stretch is not involved in A3F counteraction. Importantly, activity of these Vif mutants against A3F also demonstrates that these five Vif proteins are, per se, functionally active.
FIG 8.
Validation that Vif β-strand 2 is important for interaction with A3H and A3G. (A) Close-up of the A3H α-helixes α3 and α4 interacting with Vif residues 40 to 44. Residue colors correspond with the graph indicated in panel B. (B) Single-cycle infectivity assays of the indicated Vif mutants together with 40 ng A3H, A3G, or A3F. The infectivity was analyzed by TZM-bl reporter cells. Infectivity values are relative to the infectivity of WT HIV in the presence of A3H/A3G/A3F, which was set at 100%. The average relative infectivity values for a triplicate transfection are shown, and error bars represent the standard deviations. Cell lysates were analyzed by Western blotting. (C) The Vif-A3H structure is shown together with the N-terminal domain of A3G as described in Letko et al. (17). The individual Vif amino acids at positions 40 to 44 are indicated in blue together with A3H and A3G.
Modeling both A3H and A3G on Vif shows that their binding sites do not overlap but that they interact with different sides of the Vif stretch containing residues 40 to 44 (Fig. 8C). Indeed, the Vif-A3G-A3H models show that Vif residues 40, 42, and 44 are located between A3H and A3G, while Vif-41 is closer to A3H and Vif-43 is closer to A3G (Fig. 8C).
In summary, the two Vif-A3H pairs identified in this study allowed us to decipher the Vif-A3H interface, which we further validated by testing other residues based on our model. Our data indicate that A3H and A3G both interact with different sides of the Vif 40-44 patch, which previously was thought to be exclusively used by A3G.
DISCUSSION
It is essential for HIV to counteract the different APOBEC3 proteins, such as A3D, A3F, A3G, and A3H, expressed in target cells (2, 20). Despite ample research, no Vif-APOBEC3 crystal or nuclear magnetic resonance (NMR) costructures exist, which hinders our understanding of this important virus-host interface at a molecular level and complicates research into Vif-APOBEC3 inhibitors.
We and others approached this challenge by deciphering the interface between Vif and A3G and between Vif and A3F by finding specific points of contact between the viral protein and the host proteins, followed by subsequent protein structure docking and validation (17, 28, 29). In this report, we investigate both sides of the A3H and HIV-1 Vif interface. We show that the Vif-binding site of A3H consists of α-helixes α3 and α4 (Fig. 4). We also determined the interface of HIV-1 Vif and A3H by identifying specific points of contact between A3H and Vif (A3H-86, Vif-45; A3H-129, Vif-63/90) (Fig. 4 and 5). After careful inspection of different models, we selected a Vif-A3H structure model in which A3H α-helixes α3 and α4 interact with the β-sheet (β2 to β5) of Vif (Fig. 6). This model not only explains existing data but also allowed us to predict and validate new points of contact (Fig. 7 and 8).
Interestingly, our Vif-A3H structure model identified the Vif β-strand 2, which consists of Vif residues 40 to 44, to interact with the N-terminal α-helixes α3 and α4 of A3H (Fig. 8). However, this Vif stretch has also been shown before to be specific for counteracting A3G (17, 25, 26). Until now, our laboratory and others have attributed the effects of mutating this region to A3G (27, 30, 31). Our infectivity assays combined with direct evidence of A3H degradation clearly indicate that this Vif stretch is critical for counteracting both A3G and A3H (Fig. 8). In the future, a more nuanced interpretation is warranted when using these A3G-specific HIV mutants in a cellular environment expressing active A3H variants.
Human A3H is polymorphic, and not every human individual carries the stable A3H haplotypes which exert anti-HIV activity (9, 10, 12, 13). As a result, not every HIV-1 strain is able to counteract A3H. Indeed, we previously showed that HIV-infected patients that express stable A3H are more likely to carry an HIV strain able to counteract A3H, indicating that HIV Vif adapts to the A3H haplotype in vivo (12, 13). We pinpointed the Vif residues responsible for the difference in anti-A3H activity to Vif residues 39 and 48 (12, 15, 16). Indeed, both Vif residues are located at the interface and are predicted to interact in our Vif-A3H model: Vif-39F interacts with A3H amino acids at positions 121, 124, and 125 and Vif-48H is in close proximity to A3H position 90, which we experimentally identified to be important for binding to Vif (Fig. 1C to E).
Although our Vif-A3H interface model helps us understand how HIV-1 Vif and A3H interact, it also has its limitations. First, both the A3H and Vif structures used for docking are NMR or crystal structures but are modeled on existing structures. Second, protein docking provides a rough estimate on how two proteins interact, but structural changes in the interface upon binding remain difficult to predict. We therefore refrain from speculations on the Vif-A3H interface at a molecular level and on specific interactions between Vif and A3H amino acids. Nevertheless, our Vif-A3H structure model currently provides the best insight on how these proteins interact. Further structural analyses, such as cocrystal structures or NMR, are required for a more detailed structure of the Vif-A3H interaction.
In summary, we applied mutagenesis coupled with functional infectivity assays to identify both the Vif-binding site on A3H and the A3H-binding site on HIV-1 Vif. The identification of two specific Vif-A3H amino acid pairs allowed us to generate a detailed Vif-A3H structure model, which we subsequently validated independently. In addition to mapping Vif-APOBEC3 interactions, we believe that our approach of mapping the structural interface between two proteins by finding specific anchor points is suitable to map protein-protein interactions where classical approaches such as crystallography and NMR are technically challenging. Our Vif-A3H model structure will guide us toward improved approaches to generate detailed crystal or NMR structures of the Vif-APOBEC3 interface and also direct pharmacological interventions to obstruct the Vif-A3H interaction in future antiretroviral therapies.
MATERIALS AND METHODS
Plasmids.
The replication-competent full-length HIV-1 molecular clone LAI was obtained from the AIDS Research and Reference Reagent Program (number 2532). Of note, HIV-1 LAI counteracts the stable A3H variants. Mutagenesis of Vif in the full-length LAI clone was performed by standard overlapping PCR using the restriction sites PshAI and NdeI and the In-Fusion cloning kit per the manufacturer's instructions. HIV ΔVif was made by mutating Vif-SLQ to AAA.
The hemagglutinin (HA)-tagged A3H haplotype II, A3F, and A3G expression plasmids were previously described (32). All A3H mutants were C-terminally triple HA tagged and cloned into ptr600 expression vector as previously described (32).
HIV single-cycle infectivity assays.
For titration experiments, increasing amounts of A3H expression plasmids (0, 10, 20, and 40 ng) were cotransfected with HIV-WT or HIV ΔVif (500 ng) in 293T cells as previously described (18). For all other experiments, 20 ng of A3H and A3G was used. Culture medium was replaced 24 h posttransfection, and supernatants were collected 48 h posttransfection. Infectivity was analyzed by infecting TZM-bl reporter cells as previously described (18). Average relative infectivity values and their standard deviations were calculated from representative triplicate transfections. Proteins were analyzed by Western blotting as previously described (18).
Lentiviral vectors and cell transduction.
The mCherry-T2A-Puro-P2A-A3H (untagged) cassettes were constructed by overlap PCR and cloned into the EF1α lentivector backbone (CD527A-1; System Biosciences). Lentiviral vectors were produced in 293T cells according to the manufacturer's recommendations. Restriction of lentiviral constructs by A3H was prevented by cotransfecting a plasmid expressing SIVmac Vif (18). SupT11 cells (kindly provided by Reuben Harris) were transduced and placed under puromycin selection (0.5 μg/ml) 2 days after infection. Cell lysates were analyzed for A3H expression using an A3H-specific antibody (NBP1-91682; Novus Biologicals) (13).
HIV-1 replication assay.
For spreading infection experiments, 0.5 × 106 SupT1 cells were infected (multiplicity of infection [MOI] of 0.01) in a 24-well format (1.5 ml). Supernatants were collected every day, and cultures were replenished with fresh media. Supernatants were used to infect TZM-bl reporter cells as described previously (18).
HIV-1 viral adaptation.
A 25-ml flask with 5.0 × 106 SupT1 cells expressing different A3H mutants was infected with WT LAI (MOI of 1.0). Cells were cultured for 6 months, and new cells were added as needed. Cellular DNA was isolated using the DNeasy DNA isolation kit (Qiagen). Proviral Vif was amplified by PCR and sequenced as previously described (12).
Molecular docking and visualization.
The structure for A3H was predicted using the A3G-CTD solution structure (PDB entry 2KBO) (22). The LAI Vif structure was predicted using the Vif crystal structure (4N9F) (23) using SWISS-MODEL. Vif and A3H structures were submitted to the ClusPro server using constraints that favor A3H positions 86, 87, 90, 94, 121, 125, and 129 and Vif positions 37, 39, 45, 48, 54, 56, 60, 62, 63, 90, and 93 to interact. Protein structures were visualized using the PyMOL molecular graphics system (v.1.3; Schrödinger, LLC.). The Vif-A3H structure in which the two contact points were in close proximity was refined using FiberDock (33).
ACKNOWLEDGMENTS
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV LAI number 2532 and TZM-bl number 8129.
We thank Reuben Harris for the SupT11 T-cell line.
This work was funded by NIH/NIAID grants AI064001 (V.S.) and AI120998 (V.S.) and Public Health Service Institutional Research Training Award T32A107647 (M.L.).
REFERENCES
- 1.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris R, Bishop K, Sheehy A. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 6.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 8.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 9.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol 83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. 2013. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, Brown WL, Reilly C, Emerman M, Harris RS. 2014. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet 10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. 2010. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J Virol 84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooms M, Letko M, Binka M, Simon V. 2013. The resistance of human APOBEC3H to HIV-1 NL4-3 molecular clone is determined by a single amino acid in Vif. PLoS One 8:e57744. doi: 10.1371/journal.pone.0057744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binka M, Ooms M, Steward M, Simon V. 2012. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol 86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M, Booiman T, Kootstra N, Simon V, Ooms M. 2015. Identification of the HIV-1 Vif and human APOBEC3G protein interface. Cell Rep 13:1789–1799. doi: 10.1016/j.celrep.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letko M, Silvestri G, Hahn BH, Bibollet-Ruche F, Gokcumen O, Simon V, Ooms M. 2013. Vif proteins from diverse primate lentiviral lineages use the same binding site in APOBEC3G. J Virol 87:11861–11871. doi: 10.1128/JVI.01944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 38:4274–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog 1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, Kobayashi N, Yokoyama S, Takaku H, Katahira M. 2009. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J 28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. 2014. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 24.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. 2004. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res 32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell R, Smith J, Barr R, Bhattacharyya D, Pathak VK. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol 83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell R, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol 81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krisko JF, Begum N, Baker CE, Foster JL, Garcia JV. 2016. APOBEC3G and APOBEC3F act in concert to extinguish HIV-1 replication. J Virol 90:4681–4695. doi: 10.1128/JVI.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards C, Albin JS, Demir O, Shaban NM, Luengas EM, Land AM, Anderson BD, Holten JR, Anderson JS, Harki DA, Amaro RE, Harris RS. 2015. The binding interface between human APOBEC3F and HIV-1 Vif elucidated by genetic and computational approaches. Cell Rep 13:1781–1788. doi: 10.1016/j.celrep.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, Sugiura W, Yokomaku Y, Watanabe N, Iwatani Y. 2016. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol 90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, Takeuchi JS, Misawa N, Izumi T, Kobayashi T, Kimura Y, Iwami S, Takaori-Kondo A, Hu WS, Aihara K, Ito M, An DS, Pathak VK, Koyanagi Y. 2014. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog 10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaipan C, Smith JL, Hu WS, Pathak VK. 2013. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol 87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooms M, Krikoni A, Kress AK, Simon V, Munk C. 2012. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol 86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashiach E, Nussinov R, Wolfson HJ. 2010. FiberDock: a web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Res 38:W457–W461. doi: 10.1093/nar/gkq373. [DOI] [PMC free article] [PubMed] [Google Scholar]