ABSTRACT
Interactions between the gp120 and gp41 subunits of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) trimer maintain the metastable unliganded form of the viral spike. Binding of gp120 to the receptor, CD4, changes the Env conformation to promote gp120 interaction with the second receptor, CCR5 or CXCR4. CD4 binding also induces the transformation of Env into the prehairpin intermediate, in which the gp41 heptad repeat 1 (HR1) coiled coil is assembled at the trimer axis. In nature, HIV-1 Envs must balance the requirements to maintain the noncovalent association of gp120 with gp41 and to evade the host antibody response with the need to respond to CD4 binding. Here we show that the gp41 HR1 region contributes to gp120 association with the unliganded Env trimer. Changes in particular amino acid residues in the gp41 HR1 region decreased the efficiency with which Env moved from the unliganded state. Thus, these gp41 changes decreased the sensitivity of HIV-1 to cold inactivation and ligands that require Env conformational changes to bind efficiently. Conversely, these gp41 changes increased HIV-1 sensitivity to small-molecule entry inhibitors that block Env conformational changes induced by CD4. Changes in particular gp41 HR1 amino acid residues can apparently affect the relative stability of the unliganded state and CD4-induced conformations. Thus, the gp41 HR1 region contributes to the association with gp120 and regulates Env transitions from the unliganded state to downstream conformations.
IMPORTANCE The development of an efficient vaccine able to prevent HIV infection is a worldwide priority. Knowledge of the envelope glycoprotein structure and the conformational changes that occur after receptor engagement will help researchers to develop an immunogen able to elicit antibodies that block HIV-1 transmission. Here we identify residues in the HIV-1 transmembrane envelope glycoprotein that stabilize the unliganded state by modulating the transitions from the unliganded state to the CD4-bound state.
KEYWORDS: HIV-1, gp120, intrinsic envelope reactivity, soluble CD4, antibody resistance, HR1, gp41
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) entry into the host cell is mediated by the viral envelope glycoproteins (Envs), which are derived by proteolytic cleavage of a trimeric gp160 Env precursor (1, 2). The resulting mature Envs, gp120 (SU) and gp41 (TM), constitute a metastable trimeric complex that is anchored on the virion surface by the membrane-spanning segments of gp41 (3–6). The gp120 subunit is retained on the trimer via labile, noncovalent interactions with the gp41 ectodomain (7). The HIV-1 Env trimer can sample multiple conformations, either spontaneously or in response to receptor binding. The gp120 glycoprotein binds the initial receptor, CD4 (8, 9); this binding triggers conformational changes in gp120 that promote its interaction with one of the chemokine receptors, CCR5 or CXCR4 (10–17). CD4 binding also induces the prehairpin intermediate, in which the heptad repeat 1 (HR1) coiled coil in the gp41 ectodomain is formed and exposed (18–21). In the process of making the transition from its “ground” unliganded conformation (state 1) to the CD4-bound conformation (state 3), the HIV-1 Env trimer passes through a functional intermediate conformation (state 2) (22, 23). Chemokine receptor binding results in the formation of a stable six-helix bundle composed of the HR1 and HR2 heptad repeat regions, promoting fusion of the viral and target cell membranes (3, 5, 24).
In state 1 prior to receptor engagement, Env gp120 must maintain its noncovalent association with gp41 and prevent gp41 from prematurely undergoing transitions to lower-energy conformations. For primary HIV-1, state 1 is the predominant conformation sampled by Env on the surfaces of viral particles or infected cells in the absence of ligands (i.e., in the absence of CD4, anti-Env antibodies, etc.). Mutagenesis and structural studies suggest that, in the unliganded Env complex, the gp120 inner domain β-sandwich and its N- and C-terminal extensions play a major role in mediating gp120-gp41 association (7, 25, 26). The cognate HIV-1 gp41 elements that potentially interact with gp120 have been studied. The gp41 ectodomain consists of an N-terminal fusion peptide, the HR1 region, a disulfide loop, the HR2 region, and the membrane-proximal external region. Peptides from the gp41 HR2 region have been suggested to bind isolated gp120 monomers (27). However, mutagenesis data implicate other regions of gp41, particularly the HR1 region and disulfide loop, in the noncovalent association of gp120 with the mature Env trimer (28–31). Here, we comprehensively alter the gp41 HR1 region and evaluate the resulting Env phenotypes in light of current information on the conformational states of the HIV-1 Env trimer.
Changes in the gp41 ectodomain can influence HIV-1 sensitivity to gp120 ligands, even when the gp120 epitope for those inhibitory ligands is apparently intact (32–42). The mechanisms underlying these examples of HIV-1 neutralization escape are unknown. HIV-1 neutralization results both from ligand binding to the Env trimer and from the consequences of ligand binding to Env conformation (35, 43). The inhibition of HIV-1 by antibodies and small molecules is influenced by envelope reactivity and the perturbation factor of the Env ligand (43). Envelope reactivity describes the propensity of the HIV-1 Env to move from the unliganded state 1 conformation to downstream conformations, such as that of state 2 (35); Env reactivity is inversely related to the activation barriers that separate state 1 and state 2 (23, 35). The perturbation factor describes the degree of Env conformational change required for the ligand to bind the Env trimer and influences the impact of Env reactivity on HIV-1 neutralization by the ligand (43). In the course of our mutagenesis studies of the HIV-1 gp41 glycoprotein, we identified several Env variants in which gp41 changes influence sensitivity to gp120-directed inhibitors. We investigate the mechanisms underlying the effects of these gp41 changes on HIV-1 neutralization sensitivity.
RESULTS
Env residues important for gp120 association with the trimer.
Upon proteolytic maturation, gp120 is held on the Env trimer by a noncovalent association with gp41 and, through the trimer association domain, by interactions with the gp120 subunits in the other protomers (7, 26, 44–48). To complement the existing data on gp120 and gp41 elements important for subunit association (7, 25, 26, 31, 49–52), we comprehensively altered HIV-1YU2 gp41 ectodomain residues 547 to 590, which are located in the HR1 region. Several of these gp41 mutants exhibited decreases in the association of gp120 with the Env trimer complex (Table 1). The ability of these mutants to mediate the formation of syncytia often exceeded their ability to support virus entry. This phenotype is commonly seen in HIV-1 Env mutants with decreased gp120-gp41 association and results from differences between the two assays' temporal requirements for Env spike stability (26, 53). Thus, several gp41 residues in the HR1 region contribute to the noncovalent association of gp120 with the Env trimer.
TABLE 1.
Characterization of HIV-1YU2 Env variantsa
| Envelope glycoprotein mutationb | Residue position in HR1 | Association indexc | Processing indexd | Relative infectivitye | Cell-to-cell fusion | sCD4 neutralization IC90 (nM) | DMJ-II-121 neutralization IC50 (μM) | TS-II-224 neutralization IC50 (μM) | Cold inactivation IC50 (h) |
|---|---|---|---|---|---|---|---|---|---|
| None (wild-type YU2) | 1.00 | 1.00 | 1.00 | 1.00 | 50 | 10.76 | 5 | >48 | |
| G547A | c | 0.76 | 1.13 | 1.22 | 0.63 | 25 | 5.44 | ND | >48 |
| I548A | d | 0.96 | 0.70 | 3.68 | 4.10 | 40 | 5.23 | 4.5 | >48 |
| V549A | e | 1.14 | 0.82 | 0.94 | 0.95 | 60 | 26.36 | 7.5 | >48 |
| Q550A | f | 0.79 | 0.81 | 2.00 | 0.90 | 50 | 5.39 | ND | >48 |
| Q551A | g | 1.16 | 0.76 | 0.29 | 1.11 | 50 | 6.67 | 1 | >48 |
| Q552A | a | 0.25 | 0.18 | 0.03 | 0.17 | 45 | 8.11 | ND | >48 |
| N553A | b | 0.77 | 0.66 | 1.35 | 0.54 | 50 | 20.23 | ND | >48 |
| N554A | c | 1.28 | 1.52 | 1.81 | 0.37 | 50 | 5.37 | ND | >48 |
| L555A | d | 0.74 | 0.17 | 0.03 | 0.11 | 30 | 16.49 | ND | >48 |
| L556A | e | 0.51 | 0.79 | 0.39 | 0.84 | 40 | 7.12 | 10 | >48 |
| R557A | f | 1.53 | 1.02 | 2.15 | 0.71 | 50 | 39.88 | ND | >48 |
| A558G | g | 0.46 | 0.64 | 0.09 | 0.48 | 20 | ND | ND | >48 |
| I559A | a | 0.39 | 0.22 | 0.02 | 0.04 | ND | ND | ND | ND |
| E560A | b | 0.70 | 0.95 | 0.98 | 1.15 | 50 | 5.45 | 10 | >48 |
| A561G | c | 0.43 | 3.13 | 0.23 | 0.35 | ND | 27.37 | ND | ND |
| Q562A | d | 0.35 | 0.68 | 0.39 | 1.37 | 40 | 6.54 | 0.5 | >48 |
| Q563A | e | 0.92 | 1.29 | 1.31 | 0.79 | 50 | 8.36 | 0.5 | >48 |
| H564A | f | 0.79 | 1.00 | 1.45 | 0.96 | 95 | 24.86 | ND | >48 |
| L565A | g | 0.87 | 0.63 | 0.87 | 0.39 | 80 | 25.73 | ND | >48 |
| L565R | g | 1.02 | 0.90 | ND | 0.09 | ND | ND | ND | ND |
| L566A | a | 0.49 | 0.55 | 0.06 | 0.66 | ND | ND | ND | ND |
| Q567A | b | 0.68 | 0.67 | 2.05 | 1.03 | 40 | 11.20 | 1 | >48 |
| L568A | c | 0.57 | 0.99 | 0.14 | 1.13 | 50 | 8.29 | 3.6 | >48 |
| T569A | d | 0.37 | 0.30 | <0.01 | 0.10 | ND | ND | ND | ND |
| V570A | e | 0.41 | 1.13 | 0.08 | 0.17 | 25 | 6.50 | ND | >48 |
| W571A | f | 0.54 | 0.91 | 0.02 | 0.01 | ND | ND | na | ND |
| W571F | f | 0.32 | 0.68 | 0.09 | 0.02 | ND | ND | ND | ND |
| G572A | g | 0.52 | 0.50 | <0.01 | 0.03 | ND | ND | ND | ND |
| I573G | a | 0.99 | 1.27 | 0.09 | 0.14 | 40 | 26.21 | ND | >48 |
| K574A | b | 0.35 | 0.53 | <0.01 | 0.59 | ND | ND | ND | ND |
| K574D | b | 0.62 | 0.31 | <0.01 | 0.01 | ND | ND | ND | ND |
| K574R | b | 0.84 | 1.00 | 1.28 | 0.85 | 50 | 24.10 | 85 | >48 |
| Q575A | c | 1.22 | 0.60 | 0.28 | 0.98 | 50 | 19.91 | 1.75 | >48 |
| L576A | d | 1.32 | 0.37 | <0.01 | 0.59 | ND | ND | ND | ND |
| Q577A | e | 0.58 | 0.64 | 0.92 | 0.91 | 50 | 6.77 | 0.75 | >48 |
| A578G | f | 0.49 | 0.49 | 0.69 | 1.00 | 25 | 8.43 | ND | >48 |
| R579A | g | 0.37 | 0.84 | 0.01 | 0.44 | ND | ND | ND | ND |
| V580A | a | 0.66 | 1.04 | <0.01 | 1.14 | ND | ND | ND | ND |
| L581A | b | 0.46 | 2.21 | 1.51 | 1.06 | 25 | 5.36 | ND | >48 |
| A582T | c | 1.40 | 0.72 | 1.27 | 2.11 | >200 | 32.45 | 30 | >48 |
| A582S | c | ND | ND | 0.1 | 1.78 | 56.46 | 19.32 | ND | ND |
| A582V | c | ND | ND | 0.23 | 2.12 | >200 | 29.74 | ND | ND |
| A582Q | c | 0.57 | 0.75 | 0.94 | 1.08 | >200 | 25.07 | ND | ND |
| V583A | d | 0.39 | 0.89 | 0.08 | 0.77 | 50 | 12.55 | ND | >48 |
| E584A | e | 0.69 | 0.85 | <0.01 | 0.28 | ND | ND | ND | ND |
| R585A | f | 0.58 | 0.93 | 1.47 | 1.03 | 80 | 18.64 | ND | >48 |
| Y586A | g | 0.61 | 0.79 | 0.58 | 0.80 | 50 | 19.77 | 1 | >48 |
| L587A | a | 1.11 | 0.67 | 0.30 | 0.59 | >200 | 47.96 | >100 | >48 |
| R588A | b | 0.72 | 0.99 | 2.17 | 0.87 | 20 | 7.85 | ND | >48 |
| R588E | b | 0.97 | 0.85 | 1.53 | 1.17 | 50 | 5.57 | 1 | >48 |
| D589A | c | 0.60 | 0.93 | 0.50 | 0.27 | 50 | 24.34 | ND | >48 |
| Q590A | d | 0.53 | 0.61 | 0.28 | 0.87 | 50 | 15.61 | ND | >48 |
Values presented in this table represent the means of data from at least two independent experiments, with experimental variation typically not more than 20% of the value reported. ND, not determined.
Residues are numbered according to those of the prototypic HXBc2 sequence, as per current convention (88). The mutations result in the substitution of the amino acid residue shown on the left for the amino acid residue shown on the right of the number.
The association index is calculated as follows: association index = ([mutant gp120]cell × [wild-type gp120]supernatant)/([mutant gp120]supernatant × [wild-type gp120]cell) and was previously described (7, 26, 46, 52, 53, 79).
The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120, relative to that of the wild-type Env. The processing index was calculated by the following formula: processing index = ([total gp120]mutant × [gp160]wild type)/([gp160]mutant × [total gp120]wild type) and was previously described (7, 26, 46, 52, 53, 79).
The relative infectivity represents the ratio of mutant to wild-type virus infectivity (based on luciferase activity).
Virus infectivity was significantly affected by changes in most of the gp41 residues at the a and d positions of the HR1 coiled coil, consistent with the importance of this structure to membrane fusion (3, 5, 24, 54). In most, but not all, cases, the syncytium-forming ability of these mutants was reduced relative to that of wild-type Env. Two other mutants (the L565R and E584A mutants) also exhibited reductions in membrane-fusing activity that did not result from altered gp160 processing or subunit association. These residues occupy the g and e positions of the HR1 coiled coil, which contribute to the interaction with the HR2 helix in the formation of the gp41 six-helix bundle (3, 5, 24, 54). Thus, changes in the gp41 HR1 residues (a, d, g, and e positions) that contribute to the formation of the six-helix bundle lead to fusion-defective phenotypes.
Sensitivities of viruses with mutant Envs to neutralization by sCD4 and CD4-mimetic compounds.
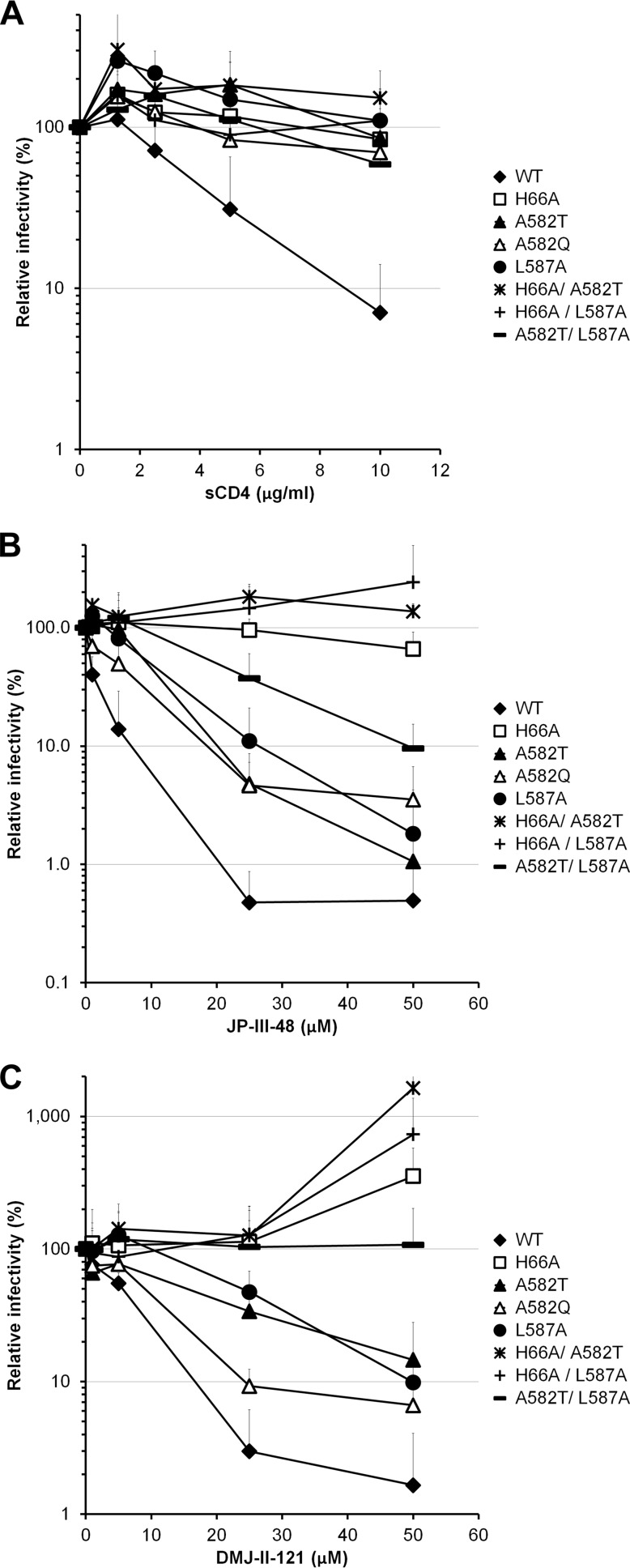
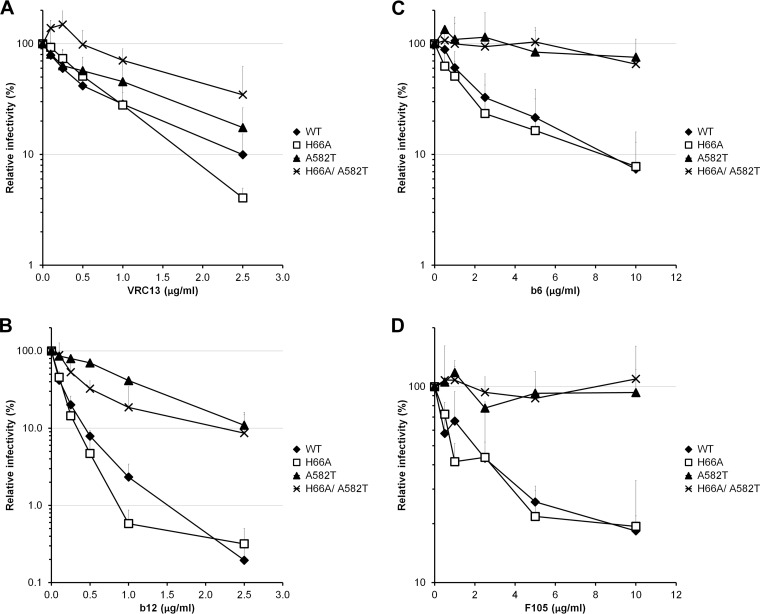
Changes in the gp41 ectodomain have been shown to modulate HIV-1 sensitivity to neutralization by soluble CD4 (sCD4) and anti-gp120 antibodies (32, 34, 36, 38, 39, 55, 56). We evaluated the sensitivities of the gp41 mutants to neutralization by a number of inhibitors directed against gp120 (sCD4, CD4-mimetic compounds, and anti-gp120 antibodies). Some of the gp41 mutants (the I559A, L566A, T569A, W571A, W571F, G572A, K574A, K574D, L576A, R579A, V580A, and E584A mutants) could not be tested in these assays due to their inability to produce infectious viruses. With four exceptions (the A582Q, A582T, A582V, and L587A mutants), no significant differences in sCD4 neutralization profiles were observed between the mutant and wild-type Envs (Fig. 1A and Table 1). Infection mediated by these four mutant Envs was minimally inhibited even by high concentrations of sCD4.
FIG 1.
Soluble CD4 (sCD4) and CD4mc inhibition of HIV-1YU2 Env variants with alterations of the gp41 ectodomain. Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant HIV-1YU2 Envs were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of sCD4 (A), JP-III-48 (B), or DMJ-II-121 (C) at 37°C for 1 h prior to infection of Cf2Th-CD4 CCR5 cells. Infectivity at each dilution of sCD4 or inhibitor tested is shown as the percentage of infection without sCD4 or inhibitor for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results obtained in at least 4 (A) or 3 (B and C) independent experiments. The error bars represent the standard deviations.
Small-molecule CD4-mimetic compounds (CD4mc) induce conformational changes in the HIV-1 Env similar to those induced by sCD4 (57–60). However, because of their small size, the CD4-mimetic compounds are less susceptible to steric constraints that influence the binding of sCD4 and antibodies to the Env trimer. Moreover, because of their relatively small interface with gp120, the inhibitory potency of CD4-mimetic compounds may be more sensitive to changes in Env reactivity than that of sCD4 (35). The replication-competent subset of HIV-1YU2 gp41 ectodomain mutants was tested for sensitivity to the CD4mc analogues JP-III-48, TS-II-224, and DMJ-II-121 (59). Both the A582T and L587A viruses were relatively resistant to TS-II-224, compared with wild-type HIV-1YU2 (Table 1). Another mutant, the K574R mutant, was also relatively resistant to TS-II-224. We also identified three gp41 mutants, the Q562A, Q563A, and Q577A viruses, that were more sensitive to TS-II-224 inhibition than the wild-type virus was. The V549A, R557A, A561G, H564A, L565A, I573G, K574R, A582Q/T, and L587A mutants were also relatively resistant to the CD4-mimetic analogue DMJ-II-121 (Fig. 1C and Table 1). All together, these data suggest that gp41 ectodomain changes can both increase and decrease HIV-1 sensitivity to inhibition by CD4mc. Thus, some changes in the gp41 ectodomain can influence HIV-1 sensitivity to gp120-directed inhibitors like sCD4 and CD4-mimetic compounds. The HIV-1YU2 A582T and L587A variants were selected for further analysis.
Some changes in HIV-1 gp120 inner domain layer 1 have also been shown to decrease virus sensitivity to sCD4, even though layer 1 does not directly contact CD4 (26, 51). In this case, sCD4 resistance results from an increased off rate that decreases the affinity of the sCD4-gp120 interaction (26). We compared the sCD4 sensitivities of the two gp41 ectodomain mutants with that of a gp120 inner domain layer 1 mutant, H66A, which decreases the efficiency of the conformational transition in gp120 that slows the sCD4 off rate (26, 61). The A582T, L587A, and H66A mutants were all less sensitive than wild-type HIV-1YU2 to sCD4 inhibition (Fig. 1A). Similarly, the H66A A582T, H66A L587A, and A582T L587A double mutants were also resistant to sCD4 neutralization (Fig. 1A).
Since the A582T, A582Q, L587A, and H66A mutants and the double mutants were less sensitive to sCD4 neutralization, they were further tested for sensitivity to the CD4mc JP-III-48 (Fig. 1B) and DMJ-II-121 (Fig. 1C). The H66A mutant was extremely resistant to neutralization by JP-III-48 compared with the wild-type virus. The A582T, A582Q, and L587A variants exhibited an intermediate sensitivity to JP-III-48 and DMJ-II-121. Interestingly, the combination of two gp41 changes in the A582T L587A mutant resulted in lower CD4mc susceptibility than those of the single mutants. Moreover, the combination of A582T or L587A with H66A resulted in an even more pronounced resistance to both CD4mc. The infectivity of these double mutants was slightly enhanced at the higher concentrations of the compounds. Of note, the CD4mc resistance resulting from the H66A change appears to be additive with that resulting from either of the gp41 ectodomain changes, suggesting that the gp41 changes mediate their effect by a mechanism that differs from that of the gp120 inner domain layer 1 variant.
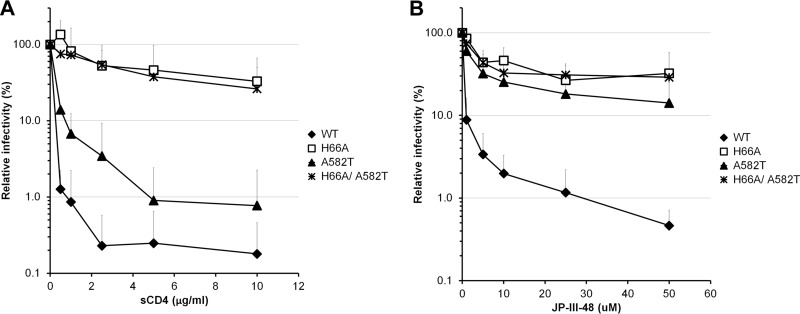
The H66A, A582T, and L587A changes were also introduced in HIV-1HXBc2, a laboratory-adapted, CXCR4-tropic virus. As in the HIV-1YU2 background, the H66A mutant was resistant to sCD4 and JP-III-48 neutralization (Fig. 2A and B). The L587A mutant was poorly infectious in the HIV-1HXBc2 context, and therefore, we were unable to test its sensitivity to the different inhibitors. The HIV-1HXBc2 A582T mutant demonstrated an increased resistance to neutralization by sCD4 compared with that of wild-type HIV-1HXBc2 (Fig. 2A). The combination of the H66A and A582T changes resulted in a virus that was as resistant to neutralization by sCD4 and the CD4mc JP-III-48 as the H66A virus (Fig. 2A and B). This result supports a model in which changes in the gp120 layer 1 residue histidine 66 and in the gp41 HR1 residue alanine 582 limit Env conformational changes induced by CD4 and CD4mc.
FIG 2.
sCD4 and CD4mc inhibition of HIV-1HxBc2 Env variants with alterations of the gp41 ectodomain. Recombinant HIV-1 expressing luciferase and bearing wild-type (WT) or mutant HIV-1HXBc2 Envs were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of sCD4 (A) or JP-III-48 (B) at 37°C for 1 h prior to infection of Cf2Th-CD4 CXCR4 cells. Infectivity at each dilution of inhibitor tested is shown as the percentage of infection without inhibitor for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results from 3 independent experiments. The error bars represent the standard deviations.
Sensitivities of viruses with mutant Envs to neutralization by BMS-806 and 484.
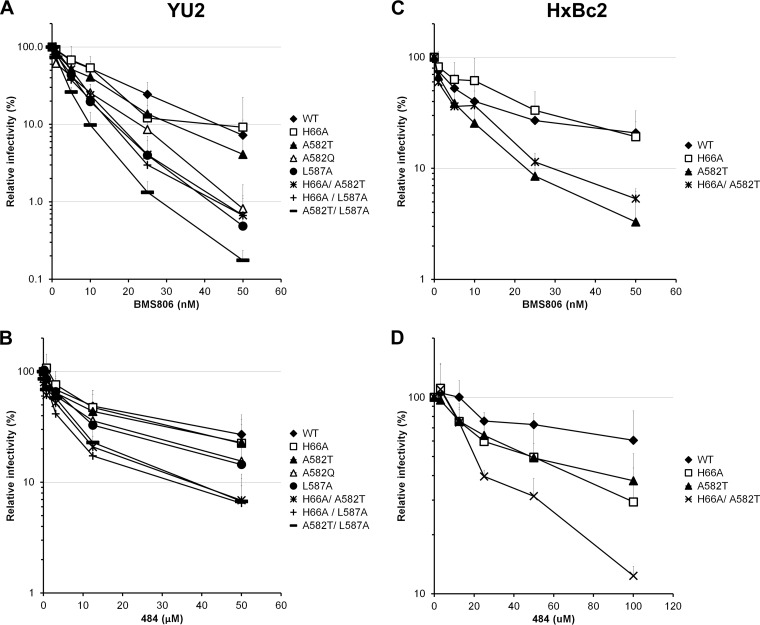
BMS-806 and 484 are two small HIV-1 entry inhibitors that bind unliganded gp120 and block conformational changes induced by CD4 (21, 23, 62). We tested the abilities of these compounds to inhibit the HIV-1YU2 and HIV-1HXBc2 Env mutants. As shown in Fig. 3A and B, the single changes A582Q, A582T, and L587A slightly increased the susceptibility of HIV-1YU2 to BMS-806 and 484 neutralization. Introduction of the H66A change generally enhanced the susceptibility of the A582T and L587A variants to BMS-806 or 484 neutralization (Fig. 3A and B). The combination of A582T with L587A resulted in an HIV-1 Env variant that was more susceptible than the individual mutants to these inhibitors. In HIV-1HXBc2, L587A variants could not be tested due to poor infectivity. However, the A582T change rendered HIV-1HXBc2 more susceptible to BMS-806 and 484 (Fig. 3C and D). The H66A change, individually and in combination with A582T, resulted in an HIV-1HXBc2 strain that exhibited increased sensitivity to 484 but not to BMS-806. These results support a model in which some changes in His 66, Ala 582, and Leu 587 result in a decreased tendency for Env to undergo conformational changes from state 1; this renders these Envs more susceptible to inhibition by blockers of conformational change.
FIG 3.
Effect of gp41 ectodomain changes on inactivation mediated by ligands that prefer the unbound conformation of HIV-1 Env. Recombinant HIV-1 expressing luciferase and bearing wild-type or mutant Envs from HIV-1YU2 (A and B) or HIV-1HXBc2 (C and D) were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of BMS806 (A and C) or 484 (C and D) at 37°C for 1 h prior to infection of Cf2Th-CD4 CCR5 (A and B) or Cf2Th-CD4 CXCR4 cells (C and D). Infectivity at each dilution of inhibitor tested is shown as the percentage of infection without inhibitor for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results from 4 (A), 3 (B), or 2 (C and D) independent experiments. The error bars represent the standard deviations.
Infection of CD4-negative cells by viruses with mutant Envs.
Soluble CD4 induces a transient activated state in HIV-1 Env (63). During this transient period of activation, viruses can infect CD4-negative, CCR5-expressing cells (63). The short half-life (5 to 7 min at 37°C) of the activated wild-type HIV-1 Env and the slow diffusion of the virus to the target cell can make the detection of this transient activated state difficult (63). The H66A change in the gp120 inner domain allows the sCD4 enhancement of HIV-1 infectivity for CD4-negative, CCR5-expressing target cells to be detected readily, compared with the situation for the wild-type virus (26). Therefore, to examine the effect of the gp41 ectodomain changes on sCD4-mediated activation of infection, we introduced the A582T and L587A changes into the wild-type and H66A HIV-1YU2 Envs. Recombinant viruses bearing wild-type or mutant Envs were incubated with CD4-negative, CCR5-expressing Cf2Th cells in the presence of different concentrations of sCD4. In the absence of sCD4, none of the viruses detectably infected the Cf2Th-CCR5 cells (Fig. 4). As reported previously (26), the H66A mutant virus exhibited an enhancement of infection even at high sCD4 concentrations. Remarkably, introduction of the A582T or L587A gp41 change into the H66A virus completely abolished sCD4 activation of infection in this assay (Fig. 4). These results support a model in which the A582T and L587A changes impede the HIV-1 Env conformational transition from the unliganded state to the CD4-bound state. In contrast to the H66A virus, the viruses with the individual A582T and L587A changes or with the combined A582T L587A changes were minimally activated by sCD4. This observation is consistent with the A582T and L587A changes exerting their effects on CD4-mediated activation of HIV-1 infection by a mechanism distinct from that of the H66A change, which increases the off rate of CD4 binding (26).
FIG 4.
sCD4 activation of HIV-1YU2 Env variants with alterations of the gp41 ectodomain. Effect of soluble CD4 on HIV-1 infection of CD4-negative, CCR5-expressing cells. Recombinant luciferase-expressing HIV-1 with the indicated HIV-1 Env variants were normalized by reverse transcriptase activity and incubated with serial dilutions of sCD4 at 37°C for 1 h prior to infection of Cf2Th-CCR5 cells. The level of infection is reported in relative light units (RLU). For each data point, the infection was performed in quadruplicate; data shown are representative of those obtained in at least two independent experiments.
Effect of gp41 ectodomain changes on HIV-1 sensitivity to neutralization by antibodies.
The A582T alteration has previously been reported to decrease HIV-1 sensitivity to neutralization by sera from HIV-1-infected individuals and by CD4-binding site (CD4BS) and CD4-induced (CD4i) antibodies (32, 34, 36, 38–40). Interestingly, only minor differences in the levels of binding of these antibodies to the wild type and the A582T mutant were reported (36); these phenotypes are reminiscent of those observed for HIV-1 Env variants that differ in envelope reactivity (35, 43). The impact of envelope reactivity on HIV-1 neutralization is highly dependent on the amount of conformational change in Env required for antibody binding (i.e., the perturbation factor of the antibody) (43). Therefore, we examined the sensitivity of wild-type HIV-1 and the A582T and L587A mutants to neutralization by antibodies that differ in their perturbation factors (43). These studies were carried out using Env variants derived from HIV-1HXBc2, which has a high envelope reactivity (35, 43, 61).
Compared with wild-type HIV-1HXBc2, the A582T virus was only slightly less sensitive to neutralization by the VRC13 antibody (Fig. 5A), an antibody with a low perturbation factor. The A582T mutant was relatively resistant to antibodies with higher perturbation factors, such as b12, b6, and F105 (Fig. 5B to D) (43). Interestingly, introduction of the H66A change in the A582T context further enhanced resistance to VRC13 but not to b12, b6, and F105. These results are consistent with a model in which the A582T change decreases Env reactivity.
FIG 5.
Sensitivities of HIV-1 variants with alterations of the gp41 ectodomain to neutralization by antibodies. Recombinant HIV-1 strains expressing luciferase and bearing wild-type or mutant HIV-1HXBc2 Envs were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of VRC13 (A), b12 (B), b6 (C), or F105 (D) at 37°C for 1 h prior to infection of Cf2Th-CD4 CXCR4 cells. Infectivity at each concentration of antibody tested is shown as the percentage of infection without inhibitor for each particular mutant. Quadruplicate samples were analyzed in each experiment. Data shown are the means of results from 3 (A, B, and D) or 2 (C) independent experiments. The error bars represent the standard deviations.
Cold sensitivity of the A582T mutant.
HIV-1 Env variants with increased reactivity often demonstrate greater sensitivity to incubation at 4°C (35, 61, 64). To test the cold sensitivity of the wild-type and A582T mutant HIV-1HXBc2 Envs, viruses with these Envs were incubated for various lengths of time on ice, and virus infectivity on Cf2Th-CD4 CCR5 cells was assessed. Relative to wild-type HIV-1HXBc2, which was inactivated in the cold, the A582T and H66A mutant viruses were cold resistant (Fig. 6). This phenotype is consistent with a lower reactivity of the A582T Env.
FIG 6.
Effect of gp41 ectodomain changes on cold inactivation of HIV-1. Recombinant HIV-1 expressing luciferase and bearing wild-type or mutant Envs from HIV-1HXBc2 were normalized by reverse transcriptase activity. Equal amounts of viruses were kept at −80°C (initially) or incubated on ice for the indicated times, as indicated in Materials and Methods, and used to infect Cf2Th-CD4 CXCR4 cells. The infectivity relative to that observed in the absence of incubation on ice is shown. Quadruplicate samples were analyzed; data shown are representative of those obtained in at least two independent experiments.
Conformational changes in Env induced by CD4 binding.
CD4 binding induces the formation/exposure of the gp41 HR1 coiled coil in HIV-1 Env (18–21). To evaluate whether this conformational change occurs in the HIV-1 Env variants studied herein, cells expressing the cytoplasmic-tail-deleted HIV-1JR-FL H66A, A582T, L587A, or S375W Env (EnvΔCT) or wild-type Env were either mock treated or incubated with sCD4 and then assessed for the ability to bind fluorescent C34-immunoglobulin (Ig). C34-Ig corresponds to the gp41 HR2 peptide fused to an immunoglobulin heavy chain (21). The cell surface expression of the Env variants was evaluated using the 2G12 monoclonal antibody, allowing normalization of the results. The wild-type HIV-1JR-FL Env demonstrated significant increases in C34-Ig binding following incubation with sCD4 (Fig. 7). C34-Ig binding also increased for the S375W mutant, which undergoes CD4-induced conformational changes more readily than the wild-type Env due to the substituted tryptophan residue filling the Phe 43 cavity of gp120 (65). Relative to the level of C34-Ig induction by sCD4 for the wild-type Env, the induction of C34-Ig binding for the H66A, A582T, and L587A Env mutants was decreased (Fig. 7). Measurement of CD4-Ig binding indicated that, relative to the wild-type Env, the H66A Env bound less well, as reported previously (26, 53), whereas the levels of binding of the A582T and L587A Envs were similar to that of the wild-type Env (Fig. 7D). These observations suggest that the A582T and L587A mutants bind CD4 but do not undergo CD4-induced conformational changes as efficiently as the wild-type Env. Of note, a previous study showed that in the context of the development of resistance to a C-peptide fusion inhibitor, the A582T change in HIV-1 Env affects C-peptide binding (42). This observation underscores the importance of this gp41 residue in modulating Env conformational transitions.
FIG 7.
Effect of Env changes on gp41 HR1 exposure. 293T cells were transfected with an empty pcDNA3.1 plasmid or a plasmid expressing the indicated cytoplasmic-tail-deleted HIV-1JR-FL Env variants. (A) Two days posttransfection, Env expression was evaluated with the 2G12 antibody, which recognizes the gp120 outer domain, by flow cytometry as described in Materials and Methods. (B) HR1 exposure was assessed with an Alexa Fluor 647 (AF647)-conjugated C34-Ig recombinant protein in the absence (gray) or presence (green) of 10 μg/ml of sCD4 by flow cytometry, as described in Materials and Methods. (C) Means and standard deviations of AF647-C34-Ig recognition of Env variants in the presence (green) or absence (gray) of sCD4, presented as the mean fluorescence intensity (MFI) detected with AF647-C34-Ig divided by the MFI detected with the 2G12 antibody. Data are the averages from at least two independent experiments. Statistical significance was evaluated using a two-way analysis of variance (ANOVA), with multiple comparisons. ****, P < 0.0001; ns, not significant. (D) Means of CD4-Ig recognition of Env variants are presented as the MFI detected with CD4-Ig divided by the MFI detected with the 2G12 antibody. Data are the averages from four independent experiments. Statistical significance was evaluated using Student's t test. *, P < 0.05; **, P < 0.01; ns, not significant.
DISCUSSION
The metastable HIV-1 Env precursor is maintained in the unliganded conformation by intrasubunit, intersubunit, and interprotomer interactions that create activation barriers for transitions into lower-energy states. Env reactivity, which is inversely related to these activation barriers (23, 35), describes the propensity of Env to change conformation from the unliganded state (state 1). Here we demonstrate how alteration of specific gp41 ectodomain residues can modulate Env reactivity, with consequences for HIV-1 sensitivity to a number of inhibitory ligands that interact with gp120. Several observations support the conclusion that the viral phenotypes result from changes in Env reactivity. First, the A582T and L587A mutants exhibit decreased sensitivity to ligands (sCD4, JP-III-48, TS-II-224, DMJ-II-121, some antibodies) that have a high perturbation factor (i.e., they require significant conformational change in Env for efficient binding). This pattern is consistent with that expected for the impact of Env reactivity on HIV-1 neutralization (43). Second, sCD4 activation of HIV-1 infection of CD4-negative cells expressing CCR5 was abrogated by the A582T and L587A changes. The ability of HIV-1 to mediate CD4-independent infection is typically associated with high Env reactivity (35, 66–69). Third, the sensitivity of HIV-1HXBc2, a virus with high Env reactivity (35, 61, 64), to cold inactivation was decreased by the A582T change. Fourth, the phenotypes of combination mutants indicate that the A582T and L587A changes act through mechanisms that are distinct from that of the gp120 inner domain change, H66A, which indirectly increases the off rate of the CD4-gp120 interaction (26, 64). A model explaining these observations is that the A582T and L587A changes increase the activation barriers, contributing to the maintenance of unliganded state 1 in the absence of receptor binding (Fig. 8A).
FIG 8.
(A) Model for modulation of HIV-1 sensitivity to gp120-directed inhibitors by changes in the gp41 ectodomain. (A) In the unliganded conformation, the envelope glycoproteins of primary HIV-1 mostly sample state 1. The proposed effects of the indicated Env changes on Env conformation are shown. The A582T change in gp41 is proposed to stabilize state 1 (indicated by a green arrow), whereas the L587A change in gp41 is suggested to destabilize state 2 (indicated by a red arrow). The H66A change in gp120 decreases CD4 binding by increasing the off rate of the gp120-CD4 interaction (26, 51, 61) (red arrow). All three changes (A582T, L587A, and H66A) act in different ways to predispose Env to assume a state 1 conformation. (B) Sequences of the gp41 ectodomain in primate immunodeficiency virus Envs. Primary sequence alignment of gp41 HR1 region residues from representative HIV-1 B (GenBank accession number K03455), HIV-1 C (accession number U46016), simian immunodeficiency virus strain cpz (SIVcpz) (accession number DQ373064), HIV-2 (accession number AF082339), SIVmac/smm (accession number M33262), SIVtan (accession number U58991), SIVagm (accession number M30931), and SIVsyk (accession number L06042) isolates (7, 25). The shading highlights residues that are conserved in all primate immunodeficiency virus lineages (85). Heptad repeat positions are shown in lowercase letters on top of the amino acid sequence alignment.
The phenotypes of gp120 shedding and decreased Env reactivity associated with our gp41 HR1 mutants are manifest in the context of the mature, unliganded Env trimer. No detailed structural model of the unliganded, mature Env is currently available. A 6-Å cryoelectron microscopy structure of an uncleaved, unliganded Env solubilized from membranes suggests that the gp41 ectodomain consists of a bundle of small helices constrained by the gp120 inner domain and N/C termini (44, 70). Higher-resolution structures will be required to determine how the gp41 changes in this study might influence the gp120-gp41 interaction in this Env conformation. Crystal structures of a soluble gp140 SOSIP.664 trimer from HIV-1BG505 complexed with neutralizing antibody Fab fragments have been solved (45, 47). Cryoelectron microscopy studies of soluble gp140 SOSIP trimers have also been conducted (71, 72). To allow the production of stable, homogeneous soluble gp140 SOSIP.664 trimers, the gp41 ectodomain was modified by an I559P change and a C-terminal truncation of 19 residues. An artificial disulfide bond between gp120 and gp41 was introduced (73–77). The impact of these modifications on the structure of the soluble gp41 SOSIP.664 trimer is unknown, but there are indications that these changes are not without consequence. For example, the I559P change has been shown to alter significantly the conformation of HIV-1 Env trimers (78, 79). This is consistent with the substantial negative effects on Env processing and subunit association of the more conservative I559A change studied herein (Table 1). In the soluble gp140 SOSIP.664 crystal structure (47), the gp41 region surrounding the altered isoleucine 559 (now proline) is disordered and falls outside the density of tomographic maps of the native, mature Env spike on HIV-1 virions (79, 80). Because of the Env modifications required for the stabilization and crystallization of the HIV-1BG505 SOSIP.664 glycoprotein trimer, caution should be exercised when extrapolating from this structure to that of the native unliganded Env trimer on viruses. Consistently with the proposed effect of the I559P change on the premature formation of the gp41 HR1 coiled coil, it has recently been shown that sCD4 binding to the sgp140 SOSIP.664 trimer does not change gp41 conformation (81); this result contrasts with the marked effects of sCD4 binding on the formation/exposure of the gp41 HR1 coiled coil in the native membrane Env (18, 19, 21).
The structure of a membrane-anchored, cytoplasmic-tail-deleted HIV-1JR-FL Env (EnvΔCT) glycoprotein bound to two Fab fragments of the PGT151-neutralizing antibody has been analyzed by cryoelectron microscopy (48). The EnvΔCT trimer in this complex is asymmetric, with PGT151 Fab fragments bound to two of the three protomeric subunits. The unliganded EnvΔCT protomer closely resembles the soluble gp140 SOSIP.664 structure (45, 47, 71, 72), indicating that this represents a preferred conformation of the HIV-1 Env ectodomain. Both the gp41 membrane-proximal external region and the transmembrane segment are disordered in the PGT151-EnvΔCT complex structure (48). As both of these gp41 regions have been implicated in maintaining the conformation of the native HIV-1 Env in state 1 (35, 41, 55, 82–84), the observed disorder makes it unlikely that the state 1 conformation is represented by the EnvΔCT structure (or, on the basis of its close resemblance, by the soluble gp140 SOSIP.664 structure).
One aspect of the EnvΔCT-PGT151 structure differs from that of the soluble gp140 SOSIP.664 trimer (45, 47, 48, 71, 72). The disordered gp41 ectodomain segment from residues 548 to 568 in the soluble gp140 SOSIP.664 structure is a helical structure in the EnvΔCT glycoprotein. This is consistent with the observation that helix-breaking substitutions (Pro and Gly) at Ile 559 stabilize soluble gp140 trimers better than other amino acid substitutions (74, 77). However, the current soluble gp140 SOSIP.664 and EnvΔCT structures fail to explain how disruption of the gp41 helix near Ile 559 and the creation of the disordered segment from residues 548 to 568 increase the stability of the soluble Env trimers. One solution to this quandary is that the gp41 subunits of HIV-1 Envs in state 1 exhibit a conformation distinct from that of either the soluble gp140 SOSIP.664 or the EnvΔCT structure, as proposed previously (79). According to this proposed model, the major conformational alterations in the gp41 ectodomain resulting from the I559P change or from the binding of PGT151 disrupted state 1 to yield the soluble gp140 SOSIP.664 and EnvΔCT states, stabilized by the newly created gp41 α7 helical coiled coil (79). Given the similarity of these structures, the gp41 subunit in these structures potentially represents a preferred intermediate, possibly state 2 (22, 23).
Cognizant of the cautionary evidence discussed above, we also recognize that the only detailed HIV-1 Env trimer structures currently available are those of the soluble gp140 SOSIP.664 and PGT151-bound EnvΔCT glycoproteins (45, 47, 48). Therefore, we asked whether the gp41 subunit in these structures sufficiently resembled the native state 1 gp41 to explain the observed phenotypes of our gp41 HR1 mutants. With one exception (Arg 557), the gp41 residues implicated in subunit association or Env reactivity are conserved in group M HIV-1 strains, and many are conserved, at least in character, among all primate immunodeficiency viruses (Fig. 8B) (85). Although some changes in these residues resulted in isolate-specific phenotypes, we expect that the subunit interactions that govern these properties will be similar among most HIV-1 strains. Using the soluble gp140 SOSIP.664 and PGT151-EnvΔCT structures (47, 48) as models for the unliganded Env trimer, we evaluated whether the observed phenotypes of our gp41 mutants were consistent with these structures. The results are summarized in Table 2. Many of the gp41 residues that modulate Env reactivity or gp120 association with the Env trimer are located in the disordered gp41 segment (residues 548 to 568) that surrounds the I559P change in the soluble gp140 SOSIP.664 structure (47). In agreement with previous studies (31, 49, 50), our results suggest that this conserved gp41 element plays an important role in modulating the physical and functional interaction with gp120. Given this, the disordered state of this gp41 segment observed in the soluble gp140 SOSIP.664 crystal structure (47) is unexpected and unlikely to exist in the native Env trimer. Although some α-helical structure is evident in the gp41 segment between residues 548 and 568 in the EnvΔCT trimer (48), few of the identified residues influencing gp120 association or Env reactivity in this region demonstrate contacts with gp120. Thus, the soluble gp140 SOSIP.664 and PGT151-EnvΔCT structures fail to provide an explanation for the phenotypes associated with most of the changes in gp41 residues in this region.
TABLE 2.
Interpretation of gp41 phenotypes based on the HIV-1BG505 soluble gp140 SOSIP.664 or the HIV-1JR-FL EnvΔCT structures
| gp41 amino acid residue | Conservation among PIVsa | Phenotype(s) | Location on soluble gp140 SOSIP.664 structure (4TVP) | Structural explanation of observed phenotypeb: |
|
|---|---|---|---|---|---|
| 4TVP | 5FUUc | ||||
| Val 549 | Ch PIV | Partial decrease in Env reactivity | Disordered gp41 segment | No explanation | No explanation |
| Gln 552 | PIV | gp120 shedding | Disordered gp41 segment | No explanation | No explanation |
| Arg 557 | Mod Var | Partial decrease in Env reactivity | Disordered gp41 segment | No explanation | No explanation |
| Ala 558 | Ch PIV | gp120 shedding | Disordered gp41 segment | No explanation | No explanation |
| Ile 559 | Ch PIV | gp120 shedding | Disordered gp41 segment | No explanation | Contacts (3.9Å) Pro 76 of gp120 |
| Ala 561 | HIV-1 | gp120 shedding and partial decrease in Env reactivity | Disordered gp41 segment | No explanation | No explanation |
| Gln 562 | HIV-1 | gp120 shedding | Disordered gp41 segment | No explanation | No explanation |
| His 564 | HIV-1 | Partial decrease in Env reactivity | Disordered gp41 segment | No explanation | No explanation |
| Leu 565 | Ch PIV | Partial decrease in Env reactivity | Disordered gp41 segment | No explanation | Met 565 contacts (3.8Å) Glu 106 of gp120 |
| Leu 566 | PIV | gp120 shedding | Disordered gp41 segment | No explanation | No explanation |
| Thr 569 | Ch PIV | gp120 shedding | Loop | No contacts with gp120 | No explanation |
| Val 570 | Ch PIV | gp120 shedding | Loop | Contacts (3.5 Å) Gln 114 of gp120 | No explanation |
| Trp 571 | PIV | gp120 shedding | Loop | Trp 571 makes backbone contacts near gp120 residue 71, between layer 1 and layer 2 of the gp120 inner domain | Trp 571 makes backbone contacts with gp120 between layer 1 and layer 2 of the gp120 inner domain |
| Gly 572 | PIV | gp120 shedding | Loop | No contacts with gp120 | No contacts with gp120 |
| Lys 574 | Ch PIV | gp120 shedding, decreased infectivity or cell-to-cell fusion | α7 helix | Forms a salt bridge with Asp 107 of gp120 α1 helix; an Arg substitution at 574 is less disruptive of the phenotypes than Ala or Asp substitutions, consistent with the prediction | No contacts with gp120 (Asp 107 to Arg 574 distance, ∼4.8 Å) |
| Ala 578 | Ch PIV | gp120 shedding | α7 helix | Potentially interacts with Thr 51 of gp120 | No contacts with gp120 |
| Arg 579 | Ch PIV | gp120 shedding | α7 helix | Forms a salt bridge with Glu 584 in gp41 but has no contacts with gp120 | Interacts with Asn 553 in gp41 but has no contacts with gp120 |
| Leu 581 | HIV-1 | gp120 shedding | α7 helix | Contacts Phe 223 on the outer surface of the gp120 inner domain β-sandwich | Contacts Phe 223 on the outer surface of the gp120 inner domain β-sandwich |
| Ala 582 | Ch PIV | Decreased Env reactivity | α7 helix | Substitutions of Thr and Gln may increase gp41 contacts with gp120 segment from residues 220–223, stabilizing the unliganded state | Substitutions of Thr and Gln could increase gp41 contacts with gp120 segment from residues 220–223, stabilizing the unliganded state |
| Val 583 | Ch PIV | gp120 shedding | α7 helix | No contacts with gp120 | No contacts with gp120 |
| Arg 585 | Mod Var | Wild type | α7 helix | Predicted to interact with Gly 222 and Ile 491 of gp120; Arg 585 is not conserved among HIV-1 strains | Predicted to interact (3.9 Å) with Glu 492 of gp120 |
| Leu 587 | Ch PIV | Decreased Env reactivity | α7 helix | A gp41 at position a; substitution of an Ala residue should destabilize the unliganded state more than it would destabilize the complete HR1 coiled coil | A gp41 at position a; substitution of an Ala residue should destabilize the unliganded state more than it would destabilize the complete HR1 coiled coil |
| Asp 589 | Ch PIV | Partial decrease in Env reactivity | α7 helix | Contacts (3.8 Å) Pro 493 of gp120; an Ala substitution is predicted to increase Env reactivity | Contacts (3.6 Å) Pro 493 of gp120; an Ala substitution is predicted to increase Env reactivity |
| Gln 590 | PIV | gp120 shedding | α7 helix | Contacts (3.2 Å) Tyr 40 of gp120 | No contacts with gp120 |
The conservation of the amino acid residue is indicated as follows: PIV, conserved in all primate immunodeficiency viruses; Ch PIV, the character of the amino acid residue is conserved among primate immunodeficiency viruses; HIV-1, conserved in all group M HIV-1 strains; and Mod Var, moderately varied in group M HIV-1 strains.
Potential explanations of the observed phenotypes of gp120 shedding or decreased Env reactivity were sought by evaluating direct (<4-Å) contacts between the gp41 residue and gp120 in the HIV-1BG505 soluble gp140 SOSIP.664 trimer (4TVP) or the HIV-1JR-FL EnvΔCT trimer (5FUU) (47, 48).
Because the EnvΔCT trimer in the HIV-1JR-FL EnvΔCT trimer is asymmetric (48), the analysis was performed using the Env protomer that was unliganded (i.e., not bound to the PGT151 Fab fragments).
Between the disordered gp41 region and the α7 helix in the soluble gp140 SOSIP.664 structure (47), there is a loop (residues 569 to 572), changes in which result in gp120 shedding (Table 1). In the soluble gp140 SOSIP.664 structure and in the PGT151-bound EnvΔCT trimer (47, 48), highly conserved Trp 571 contacts gp120 between layer 1 and layer 2 of the inner gp120 domain. In the soluble gp140 SOSIP.664 structure but not in the PGT151-bound EnvΔCT trimer structure, the neighboring residue, Val 570, also contacts gp120 at Gln 114 in the inner domain. The other gp41 residues in the loop implicated in gp120 association, Thr 569 and Gly 572, do not contact gp120 in either trimer structure. The gp120-gp41 contacts in the loop may potentially transmit conformational changes induced by CD4 binding from the gp120 inner domain to the gp41 HR1 region (26, 51).
Several of the functionally important gp41 residues identified in this study are located in the α7 helix in the soluble gp140 SOSIP.664 and PGT151-bound EnvΔCT structures (47, 48). In these structures, the α7 helices of the three gp41 protomers form a coiled coil surrounding the Env trimer axis. This α7 coiled coil represents the C-terminal portion of the complete HR1 coiled coil that forms after CD4 binding. Four of the gp41 residues (Lys 574, Ala 578, Leu 581, and Gln 590) implicated in gp120 association with the Env trimer contact gp120 in the soluble gp140 SOSIP.664 structure (47), although of these, only Leu 581 contacts gp120 in the EnvΔCT structure (48). In the soluble gp140 SOSIP.664 glycoprotein (47), Lys 574 forms a salt bridge with Asp 107 in the gp120 α1 helix; consistently with this, an arginine substitution is less disruptive of the phenotype than alanine or aspartic acid substitutions. In both Env trimer structures, Leu 581 contacts Phe 223 on the outer surface of the gp120 inner domain β-sandwich, which has been implicated in the association of gp120 with the Env trimer (25). On the other hand, some of the gp41 α7 residues (Arg 579 and Val 583) in which changes resulted in gp120 shedding do not contact gp120 in either Env trimer structure (47, 48). Conversely, alteration of Arg 585, which is predicted by both Env trimer structures to interact with gp120, resulted in a wild-type phenotype; this phenotype is consistent with the modest level of variation tolerated in this gp41 residue among HIV-1 strains (85).
Ala 582 and Leu 587, changes in which resulted in decreased Env reactivity, and Asp 589, associated with a partial decrease in Env reactivity, are located in the α7 helix in the soluble gp140 SOSIP.664 and PGT151-bound EnvΔCT structures (47, 48). Substitutions of larger amino acids for Ala 582 may increase the contacts of gp41 with the gp120 β4-β5 loop (residues 220 to 223), stabilizing the unliganded state. In contrast, if gp41 in the soluble gp140 SOSIP.664 and EnvΔCT structures represent the conformation of gp41 in the unliganded state (state 1) of Env, the decreases in Env reactivity associated with the L587A and D589A substitutions are more difficult to explain. Leu 587 occupies position a of the coiled coil; the alanine substitution is expected to destabilize the short α7 coiled coil more than the complete HR1 coiled coil, thereby increasing movement of the unliganded Env into downstream conformations. Likewise, an alanine substitution should diminish the contacts of Asp 589 with gp120 Pro 493 and increase Env reactivity. Additional detailed structures of HIV-1 Env trimers and assignment of structures to the various Env conformations on (or off) the virus entry pathway should allow a more complete understanding of the phenotypes observed in this study.
Despite the relatively well conserved nature of the HIV-1 gp120-gp41 interface, the phenotypes of some gp41 mutants depended on the HIV-1 strain from which the Env was derived. For example, the A582T change in the HIV-1YU2 Env conferred a marked decrease in sCD4 sensitivity, whereas the same change in the Env of the laboratory-adapted HIV-1HXBc2 strain had only a minor effect (36). The A582T change rendered the HIV-1HXBc2 Env resistant to cold but did not detectably affect the cold sensitivity of the cold-resistant HIV-1YU2 Env. Likewise, the L587A change did not affect the cold sensitivity of the HIV-1YU2 Env; the L587A mutant of HIV-1HXBc2 Env was not infectious and thus could not be tested. As Env reactivity can be regulated by changes in a different part of Env, the phenotypes resulting from a particular change can likely be influenced by the basal strength of the interprotomer-intersubunit interactions. Improved understanding of the variation in Env trimer structure among HIV-1 strains should shed light on the observed phenotypic differences.
MATERIALS AND METHODS
Cell lines.
293T human embryonic kidney cells, Cf2Th canine thymocytes (American Type Culture Collection), and TZM-bl cells (NIH AIDS Research and Reference Reagent Program) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin (Mediatech, Inc.). The TZM-bl cell line is a HeLa cell line stably expressing high levels of CD4 and CCR5 and possessing an integrated copy of the luciferase gene under the control of the HIV-1 long terminal repeat (86). Cf2Th cells stably expressing human CD4 and CCR5 or CD4 and CXCR4 (87) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.2 mg/ml of hygromycin B (Roche Diagnostics). Cf2Th-CCR5 cells were grown in medium supplemented with 0.4 mg/ml G418.
Site-directed mutagenesis.
Mutations were introduced into the pSVIIIenv plasmid expressing the full-length HIV-1YU2 or HIV-1HXBc2 Envs using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was confirmed by DNA sequencing. All residues are numbered according to those of the prototypic HXBc2 sequence, as per current convention (88). The mutants are designated with the amino acid residue to the right of the number replacing the amino acid residue to the left of the number.
Immunoprecipitation of envelope glycoproteins.
For pulse-labeling experiments, 3 × 105 293T cells were cotransfected by the calcium phosphate method with pLTR-Tat, a plasmid expressing the HIV-1 Tat protein, and the pSVIIIenv plasmid, expressing the HIV-1YU2 or HIV-1HXBc2 Envs. Beginning 1 day after transfection, the cells were metabolically labeled for 16 h with 100 μCi/ml [35S]methionine/cysteine (35S protein labeling mix; Perkin-Elmer) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Cells were subsequently lysed in RIPA buffer (140 nM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, 0.05% sodium dodecyl sulfate [SDS]). Precipitation of radiolabeled HIV-1 Envs from cell lysates or medium was performed with a mixture of sera from HIV-1-infected individuals in the presence of 50 μl of 10% protein A-Sepharose (American BioSciences) for 1 h at 4°C. All precipitated proteins were boiled for 5 min before being analyzed by SDS-PAGE.
Processing was performed and association indices were determined by precipitation of radiolabeled cell lysates and supernatants with mixtures of sera from HIV-1-infected individuals, as previously described (7, 26, 46, 52, 53, 79). The association index is a measure of the ability of the gp120 molecule to remain associated with the mutant Env trimer complex on the expressing cell, relative to that of the wild-type Env. The association index is calculated as follows: association index = ([mutant gp120]cell × [wild-type gp120]supernatant)/([mutant gp120]supernatant × [wild-type gp120]cell). The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120, relative to that of the wild-type Env. The processing index was calculated by the following formula: processing index = ([total gp120]mutant × [gp160]wild type)/([gp160]mutant × [total gp120]wild type).
Recombinant luciferase viruses.
Recombinant HIV-1 expressing the firefly luciferase gene was produced by calcium phosphate transfection of 293T cells with the molecular clone pNL4.3 (Env−) Luc and the pSVIIIenv plasmid expressing the wild-type or mutant HIV-1YU2 or HIV-1HXBc2 Envs at a weight ratio of 4:1. Two days after transfection, the cell supernatants were harvested. The reverse transcriptase activities of all virus preparations were measured as described previously (89). Each virus preparation was frozen and stored in aliquots at −80°C until use.
Infection by single-round luciferase-expressing HIV-1 viruses.
Twenty-four hours before infection, Cf2Th-CD4 CCR5 or Cf2Th-CD4 CXCR4 target cells were seeded at a density of 6 × 103 cells/well in white 96-well luminometer-compatible tissue culture plates (Dynex). Recombinant viruses (10,000 reverse transcriptase units) in a final volume of 100 μl were then added in quadruplicate to the target cells. After a 48-hour incubation at 37°C, the medium was removed from each well and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. After the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (BD Pharmingen), the luciferase activity in each well was measured with an EG&G Berthold microplate luminometer, LB 96V. The relative infectivity represents the ratio of mutant to wild-type virus infectivity (based on luciferase activity).
Neutralization and sCD4 activation assays.
Luciferase-expressing viruses bearing either wild-type or mutant envelope glycoproteins were incubated for 1 h at 37°C with serial dilutions of inhibitor in a total volume of 200 μl. The recombinant viruses were then incubated in quadruplicate with Cf2Th-CD4 CCR5 (YU2 viruses) or Cf2Th-CD4 CXCR4 (HXBc2 viruses) cells (neutralization assay), as described above. Luciferase activity in the cells was measured 2 days later. The soluble CD4 (sCD4) 50% or 90% inhibitory concentration (IC50 or IC90) represents the amount of sCD4 needed to inhibit 50% or 90% of the infection of Cf2Th-CD4 CCR5 cells by recombinant luciferase-expressing HIV-1 bearing the indicated Env.
Cold inactivation assay.
To assess the effect of cold on virus infectivity, virus preparations equalized for reverse transcriptase activity were incubated on ice for 0, 8, 24, or 48 h. At the end of the incubation, aliquots were removed and transferred to a −80°C freezer until infection. To measure the infectivity of the virus, aliquots were thawed at 37°C just before infection and added to target cells in quadruplicate.
Cell-to-cell fusion.
To assess cell-to-cell fusion, 3 × 105 293T cells were cotransfected by the calcium phosphate method with an HIV-1 Tat-expressing plasmid, pLTR-Tat, and the pSVIIIenv plasmid expressing the HIV-1YU2 or HIV-1HXBc2 envelope glycoproteins. Two days after transfection, 3 × 104 293T cells were added to TZM-bl target cells that were seeded at a density of 3 × 104 cells/well in 96-well luminometer-compatible tissue culture plates (Dynex) 24 h before the assay. Cells were coincubated for 6 h at 37°C, after which they were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. Luciferase activity in each well was measured as described above.
Evaluation of gp41 HR1 exposure by flow cytometry.
To express the wild-type and mutant HIV-1JR-FL Envs for flow cytometric analysis, a stop codon was introduced into the expressor plasmid for the codon-optimized env, replacing the codon for Gly 711. This modification results in the expression of Envs with a deletion of the cytoplasmic tail, which increases the level of Env on the cell surface. Using the standard calcium phosphate method, the Env expressor plasmids were transfected into 3 × 105 293T cells along with a pIRES-GFP vector at a ratio of 2 μg Env expressor to 0.5 μg green fluorescent protein (GFP) expressor. At 16 h after transfection, cells were washed with fresh medium, and HR1 exposure was evaluated 24 h later. The recombinant C34-Ig protein (21, 62) was used to detect HR1 exposure and was conjugated to an Alexa-Fluor 647 (AF647) probe (Invitrogen) according to the manufacturer's instructions. The transfected 293T cells were incubated with either the 2G12 antibody (1 μg/ml), which recognizes the gp120 outer domain, or AF647-C34-Ig (10 μg/ml) in the absence or presence of 10 μg/ml sCD4 at room temperature for 1 h. The 2G12 antibody was detected using a goat anti-human antibody coupled to Alexa Fluor 647 (Invitrogen). CD4 binding was independently assessed by measuring the binding of CD4-Ig to cells expressing the cytoplasmic-tail-deleted HIV-1JR-FL Env variants. 2G12, AF647-C34-Ig, and CD4-Ig binding was detected by gating on GFP-positive cells with an LSRII cytometer (BD Biosciences, Mississauga, ON, Canada). Data analysis was performed using FlowJo vX.0.7 (Tree Star, Ashland, OR, USA).
ACKNOWLEDGMENTS
We thank Yvette McLaughlin, Elizabeth Carpelan, and Julia Barnes for manuscript preparation.
This work was supported by grants from the National Institutes of Health (AI24755, AI24982, GM56550, and AI67854), by the International AIDS Vaccine Initiative, by a grant from the late William F. McCarty-Cooper to J.S., by Canada Foundation for Innovation Program Leader grant 29866, and by CIHR Foundation grant 352417 to A.F. A.F. was supported by phases I and II of the amfAR Mathilde Krim Fellowship in Basic Biomedical Research (107963-49-RKVA). A.F. is the recipient of a Canada Research Chair on Retroviral Entry (grant 950-229930). N.A. is the recipient of a King Abdullah scholarship for higher education from the Saudi Government. N.M. was supported by amfAR grant 107431-45-RFNT, NIH grant AI090682, and a Ragon Institute Innovation Award. A.H. is the recipient of an amfAR Mathilde Krim Fellowship in Basic Biomedical Research (108501-53-RKNT) and was also supported by a phase II amfAR research grant (109285-58-RKVA) for independent investigators.
We have no conflicts of interest to report.
REFERENCES
- 1.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 2.Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 3.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 4.Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol 72:7620–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhu P, Chertova E, Bess J Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 65:2119–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 9.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 10.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. doi: 10.1016/S0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 17.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 18.Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 19.He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J Virol 77:1666–1671. doi: 10.1128/JVI.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshiba T, Chan DC. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J Biol Chem 278:7573–7579. doi: 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- 21.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB III, Sodroski JG. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A 101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herschhorn A, Ma X, Gu C, Ventura JD, Castillo-Menendez L, Melillo B, Terry DS, Smith AB III, Blanchard SC, Munro JB, Mothes W, Finzi A, Sodroski J. 2016. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. mBio 7:e01598-16. doi: 10.1128/mBio.01598-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117–125. doi: 10.1016/S0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 26.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Pang HB, Kay MS. 2008. Peptide mimic of the HIV envelope gp120-gp41 interface. J Mol Biol 376:786–797. doi: 10.1016/j.jmb.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol 67:2747–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maerz AL, Drummer HE, Wilson KA, Poumbourios P. 2001. Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J Virol 75:6635–6644. doi: 10.1128/JVI.75.14.6635-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poumbourios P, Maerz AL, Drummer HE. 2003. Functional evolution of the HIV-1 envelope glycoprotein 120 association site of glycoprotein 41. J Biol Chem 278:42149–42160. doi: 10.1074/jbc.M305223200. [DOI] [PubMed] [Google Scholar]
- 31.Sen J, Yan T, Wang J, Rong L, Tao L, Caffrey M. 2010. Alanine scanning mutagenesis of HIV-1 gp41 heptad repeat 1: insight into the gp120-gp41 interaction. Biochemistry 49:5057–5065. doi: 10.1021/bi1005267. [DOI] [PubMed] [Google Scholar]
- 32.Reitz MS Jr, Wilson C, Naugle C, Gallo RC, Robert-Guroff M. 1988. Generation of a neutralization-resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell 54:57–63. doi: 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- 33.Robert-Guroff M, Reitz MS Jr, Robey WG, Gallo RC. 1986. In vitro generation of an HTLV-III variant by neutralizing antibody. J Immunol 137:3306–3309. [PubMed] [Google Scholar]
- 34.Wilson C, Reitz MS Jr, Aldrich K, Klasse PJ, Blomberg J, Gallo RC, Robert-Guroff M. 1990. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J Virol 64:3240–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog 7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol 68:674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klasse PJ, McKeating JA, Schutten M, Reitz MS Jr, Robert-Guroff M. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(→ Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332–337. doi: 10.1006/viro.1993.1484. [DOI] [PubMed] [Google Scholar]
- 39.Stern TL, Reitz MS Jr, Robert-Guroff M. 1995. Spontaneous reversion of human immunodeficiency virus type 1 neutralization-resistant variant HXB2thr582: in vitro selection against cytopathicity highlights gp120-gp41 interactive regions. J Virol 69:1860–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watkins BA, Buge S, Aldrich K, Davis AE, Robinson J, Reitz MS Jr, Robert-Guroff M. 1996. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J Virol 70:8431–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley T, Trama A, Tumba N, Gray E, Lu X, Madani N, Jahanbakhsh F, Eaton A, Xia SM, Parks R, Lloyd KE, Sutherland LL, Scearce RM, Bowman CM, Barnett S, Abdool-Karim SS, Boyd SD, Melillo B, Smith AB III, Sodroski J, Kepler TB, Alam SM, Gao F, Bonsignori M, Liao HX, Moody MA, Montefiori D, Santra S, Morris L, Haynes BF. 31 August 2016. Amino acid changes in the HIV-1 gp41 membrane proximal region control virus neutralization sensitivity. EBioMedicine 12:196–207. doi: 10.1016/j.ebiom.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hermann FG, Egerer L, Brauer F, Gerum C, Schwalbe H, Dietrich U, von Laer D. 2009. Mutations in gp120 contribute to the resistance of human immunodeficiency virus type 1 to membrane-anchored C-peptide maC46. J Virol 83:4844–4853. doi: 10.1128/JVI.00666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. doi: 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol 84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poumbourios P, Wilson KA, Center RJ, El Ahmar W, Kemp BE. 1997. Human immunodeficiency virus type 1 envelope glycoprotein oligomerization requires the gp41 amphipathic alpha-helical/leucine zipper-like sequence. J Virol 71:2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.York J, Nunberg JH. 2004. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J Virol 78:4921–4926. doi: 10.1128/JVI.78.9.4921-4926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding S, Tolbert WD, Prevost J, Pacheco B, Coutu M, Debbeche O, Xiang SH, Pazgier M, Finzi A. 2016. A highly conserved gp120 inner domain residue modulates Env conformation and trimer stability. J Virol 90:8395–8409. doi: 10.1128/JVI.01068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang SH, Mao Y, Sodroski J, Finzi A. 2013. The highly conserved layer-3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J Virol 87:2549–2562. doi: 10.1128/JVI.03104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan K, Liu J, Wang J, Shen S, Lu M. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci U S A 94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blish CA, Nguyen MA, Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med 5:e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Back NK, Smit L, Schutten M, Nara PL, Tersmette M, Goudsmit J. 1993. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J Virol 67:6897–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E. 2006. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, Do Kwon Y, Huang CC, Wyatt R, Kwong PD, Freire E, Smith AB III, Sodroski J. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lalonde JM, Le-Khac M, Jones DM, Courter JR, Park J, Schon A, Princiotto AM, Wu X, Mascola JR, Freire E, Sodroski J, Madani N, Hendrickson WA, Smith AB III. 2013. Structure-based design and synthesis of an HIV-1 entry inhibitor exploiting X-ray and thermodynamic characterization. ACS Med Chem Lett 4:338–343. doi: 10.1021/ml300407y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madani N, Princiotto AM, Schon A, LaLonde J, Feng Y, Freire E, Park J, Courter JR, Jones DM, Robinson J, Liao HX, Moody MA, Permar S, Haynes B, Smith AB III, Wyatt R, Sodroski J. 2014. CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J Virol 88:6542–6555. doi: 10.1128/JVI.00540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herschhorn A, Gu C, Espy N, Richard J, Finzi A, Sodroski JG. 2014. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat Chem Biol 10:845–852. doi: 10.1038/nchembio.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB III, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog 5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB III, Sodroski J. 2009. Identification of a human immunodeficiency virus (HIV-1) envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. 2002. Mutagenic stabilization/disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus (HIV-1) gp120 envelope glycoprotein. J Virol 76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol 75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol 76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas ER, Shotton C, Weiss RA, Clapham PR, McKnight A. 2003. CD4-dependent and CD4-independent HIV-2: consequences for neutralization. AIDS 17:291–300. doi: 10.1097/00002030-200302140-00002. [DOI] [PubMed] [Google Scholar]
- 69.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol 76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol 19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–14909. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders RW, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore JP, Binley JM. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol 74:5091–5100. doi: 10.1128/JVI.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol 87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kesavardhana S, Varadarajan R. 2014. Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J Virol 88:9590–9604. doi: 10.1128/JVI.00494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alsahafi N, Debbeche O, Sodroski J, Finzi A. 2015. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One 10:e0122111. doi: 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci U S A 113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lovelace E, Xu H, Blish CA, Strong R, Overbaugh J. 2011. The role of amino acid changes in the human immunodeficiency virus type 1 transmembrane domain in antibody binding and neutralization. Virology 421:235–244. doi: 10.1016/j.virol.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ringe R, Bhattacharya J. 2012. Association of enhanced HIV-1 neutralization by a single Y681H substitution in gp41 with increased gp120-CD4 interaction and macrophage infectivity. PLoS One 7:e37157. doi: 10.1371/journal.pone.0037157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dev J, Park D, Fu Q, Chen J, Ha HJ, Ghantous F, Herrmann T, Chang W, Liu Z, Frey G, Seaman MS, Chen B, Chou JJ. 2016. Structural basis for membrane anchoring of HIV-1 envelope spike. Science 353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B (ed). 2013. HIV sequence compendium. LA-UR-13-26007 Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 86.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.LaBonte JA, Patel T, Hofmann W, Sodroski J. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J Virol 74:10690–10698. doi: 10.1128/JVI.74.22.10690-10698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG. The Human Retroviruses and AIDS 1998 Compendium, part III, Analyses. Los Alamos National Laboratory, Los Alamos, NM: https://hfv.lanl.gov/content/sequence/HIV/COMPENDIUM/98compendium.html. [Google Scholar]
- 89.Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360. doi: 10.1016/0042-6822(81)90642-5. [DOI] [PubMed] [Google Scholar]