ABSTRACT
The latency-related (LR) RNA encoded by bovine herpesvirus 1 (BoHV-1) is abundantly expressed in latently infected sensory neurons. Although the LR gene encodes several products, ORF2 appears to mediate important steps during the latency-reactivation cycle because a mutant virus containing stop codons at the amino terminus of ORF2 does not reactivate from latency in calves. We recently found that the Wnt/β-catenin signaling pathway is regulated during the BoHV-1 latency-reactivation cycle (Y. Liu, M. Hancock, A. Workman, A. Doster, and C. Jones, J Virol 90:3148–3159, 2016). In the present study, a β-catenin coactivator, high-mobility group AT–hook 1 protein (HMGA1), was detected in significantly more neurons in the trigeminal ganglia of latently infected calves than in those of uninfected calves. Consequently, we hypothesized that HMGA1 cooperates with ORF2 and β-catenin to maintain latency. In support of this hypothesis, coimmunoprecipitation studies demonstrated that ORF2 stably interacts with a complex containing β-catenin and/or HMGA1 in transfected mouse neuroblastoma (Neuro-2A) cells. Confocal microscopy provided evidence that ORF2 was relocalized by HMGA1 and β-catenin in Neuro-2A cells. ORF2 consistently enhanced the ability of HMGA1 to stimulate β-catenin-dependent transcription, suggesting that interactions between ORF2 and a complex containing β-catenin and HMGA1 have functional significance. An ORF2 stop codon mutant, an ORF2 nuclear localization mutant, or a mutant lacking the 5 protein kinase A or C phosphorylation sites interfered with its ability to stimulate β-catenin-dependent transcription. Since the canonical Wnt/β-catenin signaling pathway promotes neurogenesis (synapse formation and remodeling) and inhibits neurodegeneration, interactions between ORF2, HMGA1, and β-catenin may be important for certain aspects of the latency-reactivation cycle.
IMPORTANCE The lifelong latency of bovine herpesvirus 1 (BoHV-1) requires that significant numbers of infected sensory neurons survive infection and maintain normal functions. Consequently, we hypothesize that viral products expressed during latency cooperate with neuronal factors to maintain latency. Our studies revealed that a β-catenin coactivator, high-mobility group AT–hook 1 protein (HMGA1), was readily detected in a subset of trigeminal ganglion neurons in latently infected calves but not in uninfected calves. A viral protein (ORF2) expressed in latently infected neurons interacted with β-catenin and HMGA1 in transfected cells, which resulted in the nuclear localization of β-catenin. This interaction correlated with the ability of ORF2 to stimulate the coactivator functions of HMGA1. These findings are significant because the canonical Wnt/β-catenin signaling pathway promotes neurogenesis and inhibits neurodegeneration.
KEYWORDS: bovine herpesvirus 1, HMGA1, β-catenin, latency
INTRODUCTION
Bovine herpesvirus 1 (BoHV-1) infections frequently lead to upper respiratory tract disease (1, 2) and erosion of mucosal surfaces. Epithelial cell damage and immune suppression occur after viral infection, thus allowing commensal bacteria in the upper respiratory tract to colonize the lung and cause disease. A BoHV-1 entry receptor, poliovirus receptor related 1, was identified to be a bovine respiratory disease complex susceptibility gene for Holstein calves (3), confirming the role of BoHV-1 as a cofactor in this important polymicrobial disease.
Following acute infection of mucosal linings in the oral, nasal, or ocular cavity, BoHV-1 establishes latency in sensory neurons within trigeminal ganglia (TG). The ability to establish, maintain, and reactivate from latency is crucial for virus transmission. The latency-related (LR) gene, which encodes two microRNAs and at least two proteins (ORF2 and ORF1), is the only viral gene abundantly expressed during latency. Both microRNAs can reduce expression of the viral regulatory protein bovine infected cell protein 0 (bICP0) in transfected cells (4), which is likely important during the transition from acute infection to the establishment and maintenance of latency. ORF2 inhibits apoptosis, interacts with the signaling molecule Notch, and interferes with Notch-mediated signaling (5–7). A virus with a mutant LR gene containing three stop codons near the initiating methionine of ORF2 does not reactivate from latency or replicate efficiently in certain tissues (8, 9), suggesting that ORF2 regulates crucial steps during the latency-reactivation cycle.
We recently discovered that a cellular transcription factor, β-catenin, is readily detected in TG neurons from latently infected calves but not in TG neurons from uninfected calves or during stress-induced reactivation from latency (10). Most β-catenin-positive (β-catenin+) neurons express ORF2 but not the lytic cycle regulatory protein (bICP0). Additional studies revealed that ORF2 enhances β-catenin-dependent transcription and cooperates with β-catenin to promote the survival of neuroblastoma cells. Consequently, β-catenin may be important during the establishment and maintenance of latency because the canonical Wnt/β-catenin pathway regulates axonal growth and the navigation of axons to their synaptic targets (reviewed in references 11, to ,15). Expression of a soluble Wnt antagonist (Dickkopf-1) was induced more than 10-fold during dexamethasone (DEX)-induced reactivation from latency (10). Interestingly, Dickkopf-1 mediates stress-induced neuronal death (16, 17). Collectively, the results of these studies suggest that the Wnt/β-catenin signaling pathway is regulated during the BoHV-1 latency-reactivation cycle.
In this study, we provide evidence that the RNA encoding the high-mobility group AT–hook 1 protein (HMGA1) was differentially expressed in TG latently infected with BoHV-1 compared to its expression in the uninfected control TG. Furthermore, HMGA1 protein expression was readily detected in a subset of TG neurons from latently infected calves but not TG neurons from uninfected calves. Based on these observations, we hypothesized that HMGA1 plays a role in maintaining a latent infection because HMGA1 is a known coactivator of β-catenin (18–20), a cellular transcription factor that promotes neurogenesis and interferes with neurodegeneration (reviewed in references 11 to 15). Additional studies demonstrated that ORF2 and HMGA1 were associated with β-catenin in transfected mouse neuroblastoma cells. ORF2, HMGA1, and β-catenin also partially colocalized in the nucleus and enhanced β-catenin-dependent transcriptional activity. In summary, these studies suggest that HMGA1 likely regulates certain aspects of the BoHV-1 latency-reactivation cycle.
RESULTS
Identification of differentially expressed genes in TG of latently infected calves relative to those of uninfected calves.
Microarray studies were performed to identify cellular genes differentially expressed in uninfected calves versus latently infected calves (3 calves in each group). The top 6 genes that were either stimulated or repressed in the TG of latently infected calves relative to their expression in the TG of uninfected calves are shown in Table 1. In general, the fold change in expression seen in the TG of uninfected calved versus those of latently infected calves was not as dramatic as that seen when expression during latency was compared to that after dexamethasone-induced reactivation from latency (21). The top gene induced in latently infected TG is the high-mobility group AT–hook 1 protein (HMGA1) (the level of expression of which was 2.8-fold times higher in latently infected TG than uninfected TG; P < 0.05). The HMGA1 gene encodes a nuclear protein that binds AT-rich DNA sequences, interacts with β-catenin, is induced by the Wnt/β-catenin signaling pathway (18, 19), and stimulates β-catenin-dependent transcription in cancer cells (20). Expression of another β-catenin regulator, frizzled homolog 8 (FZD8), was repressed 2.4-fold in the TG of latently infected calves compared to its expression in the TG of uninfected calves. FZD8 encodes a soluble cytoplasmic protein that has been reported to block Wnt/β-catenin signaling and can increase apoptosis in dopaminergic neurons (22). Conversely, FZD8 has also been reported to positively affect lung tumor cell growth and is upregulated in non-small cell lung cancer (23). Together with the findings of past studies (10), these results suggest that the canonical Wnt/β-catenin signaling pathway is regulated during BoHV-1 latency and during DEX-induced reactivation from latency (21).
TABLE 1.
Summary of cellular genes differentially expressed in TG of latently infected calves and uninfected calves
| Gene name | Relative gene expression during latency | Gene description |
|---|---|---|
| HMGA1 | +2.8 | High-mobility group AT–hook 1 |
| MAN2B1 | +2.2 | Mannosidase, alpha, class 2B, member 1 |
| DSG2 | +2.1 | Desmoglein-2 |
| SLC12A3 | +2 | Solute carrier family 12 (sodium/chloride transporters), member 3 |
| IGF2BP3 | +2 | Insulin-like growth factor 2 mRNA binding protein 3 |
| Cytochrome b-561 | +1.9 | An ascorbate-dependent oxidoreductase family member involved with electron transport |
| C1QB | −6.7 | Complement component 1, q subcomponent, B chain |
| BRB | −4.8 | Brain RNase |
| LOC788911 | −4.2 | Similar to autotaxin isoform preproprotein |
| GMFG | −3.6 | Glia maturation factor, gamma |
| APOA1 | −3.6 | Apolipoprotein A-I |
| FZD8 | −2.4 | Frizzled homolog 8 (Drosophila) |
aThe top 6 genes that were induced and the top 6 genes that were repressed in TG during latency are listed. The changes in the levels of expression along with a brief description of the gene are included. To determine the genes differentially expressed over time, we compared the values for latently infected TG from 3 calves to those for uninfected TG from 3 calves using a random-variance F test and then selected genes with a false discovery rate of <10%. Fold changes in expression for each gene differentially expressed in latently infected calves relative to the value for the group of uninfected calves were calculated.
Examination of HMGA1 in TG neurons of calves latently infected with BoHV-1.
Since the primary focus of this study was to provide additional insight into how the Wnt/β-catenin signaling pathway is regulated during the BoHV-1 latency-reactivation cycle, immunohistochemistry (IHC) studies were performed to confirm that genes known to be directly involved with the Wnt pathway were differentially expressed in the TG of latently infected calves versus those of uninfected calves. An FZD8 antibody that was useful for IHC studies was not identified. Consequently, we compared HMGA1 protein expression in the TG neurons of latently infected calves and those of uninfected calves. HMGA1 was readily detected in the nucleus of a subset of TG neurons from three latently infected calves (Fig. 1A; arrows denote HMGA1-positive [HMGA1+] neurons). In contrast, HMGA1+ neurons were not readily detected in TG prepared from three uninfected calves. TG neurons from uninfected calves that contained visible nuclei also had nucleoli that were clearly counterstained: conversely, the HMGA1 antibody did not stain the nuclei (Fig. 1A, closed circles). There were significantly higher numbers of HMGA1+ neurons in the TG of latently infected calves than in those of uninfected calves (Fig. 1B). In contrast to the results obtained with the HMGA1 antibody, bICP0 was not readily detected in TG neurons from uninfected or latently infected calves but was readily detected during reactivation from latency (data not shown) (6, 10, 24, 25). In summary, HMGA1, a β-catenin coactivator, was readily detected in a subset of TG neurons from latently infected calves but not in TG neurons from uninfected calves.
FIG 1.
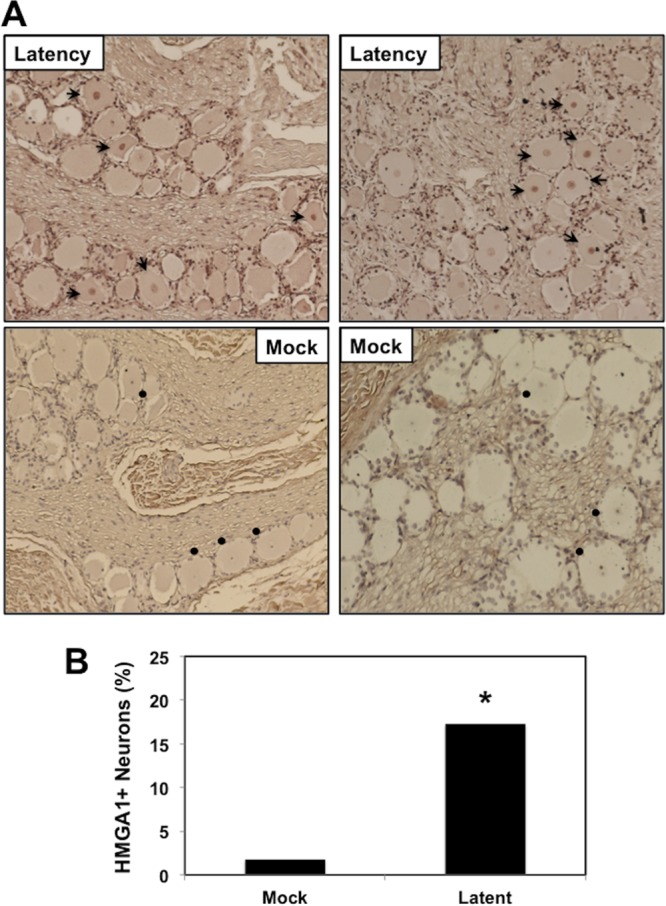
Detection of HMGA1 in TG neurons during latency. (A) TG were collected from 3 uninfected calves (denoted as Mock) or latently infected calves (at least 60 days postinfection). Thin sections were cut from formalin-fixed paraffin-embedded TG sections. The HMGA1 antibody used for this study was purchased from Abcam (catalog number ab129153) and was diluted 1:400. Biotinylated goat anti-rabbit IgG (Vector Laboratories) was used as a secondary antibody. Arrows, HMGA1-positive neurons; circles, TG neurons in uninfected calves that contain a visible nucleus in which the nucleolus was counterstained (however, the HMGA1 antibody did readily stain the nucleus). (B) The percentage of HMGA1-positive neurons among 490 total neurons was estimated from sections derived from three latently infected calves or three uninfected calves. *, significant differences (P < 0.05) in the numbers of HMGA1-positive neurons, as determined by a Student t test.
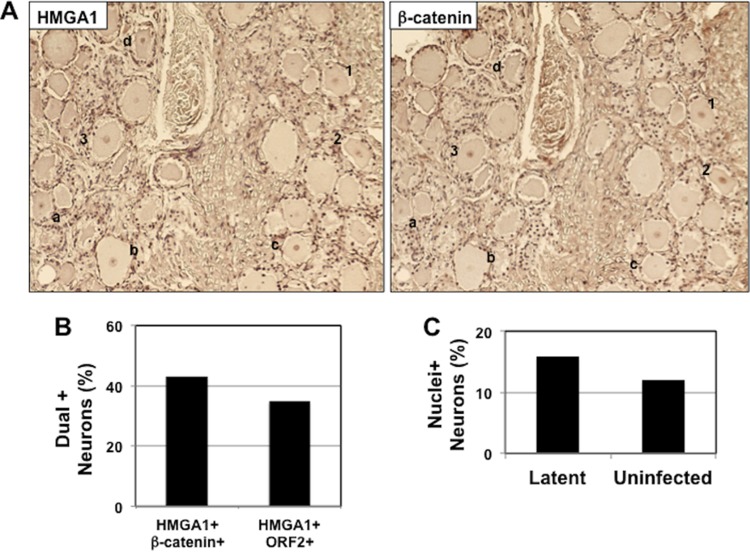
Additional studies tested whether HMGA1+ neurons also expressed β-catenin and ORF2 because a previous study demonstrated that nearly all β-catenin+ neurons contain ORF2 (10). For these studies, consecutive sections were cut, and one section was stained with the HMGA1 antibody and the other was stained with an antibody that recognized β-catenin or ORF2. A subset of HMGA1+ neurons also expressed β-catenin (Fig. 2A, neurons numbered 1 to 3) and ORF2 (data not shown). It was also clear that certain neurons were stained by the HMGA1 antibody but not the β-catenin antibody (neurons denoted a to d in Fig. 2A). The proportion of HMGA1+ neurons stained by antibodies detecting β-catenin or ORF2 was less than 50% (Fig. 2B). In TG sections from latently infected calves, 63 neurons out of 400 total neurons (15.8%) contained visible nuclei, and in a TG section from an uninfected calf, 48 neurons out of 400 neurons (12%) contained visible nuclei (Fig. 2C). Since HMGA1 was detected only in the nuclei of latently infected neurons, the results in Fig. 2C suggest that the number of dual-positive neurons may be underestimated because TG thin sections contain a low percentage of neurons with visible nuclei.
FIG 2.
Analysis of HMGA1+ neurons that express β-catenin or ORF2 in consecutive sections. (A) Consecutive sections from TG of calves latently infected with BoHV-1 were prepared, and one section was stained with an antibody that recognizes HMGA1. The adjacent section was stained with an antibody that recognizes β-catenin. The neurons numbered 1 to 3 were stained with both antibodies in adjacent sections. The letters a to d denote neurons that were stained by the HMGA1 antibody but not the β-catenin antibody. These results are representative of those for TG from 2 different calves latently infected with BoHV-1. (B) Dually stained TG neurons in adjacent sections. The number of β-catenin+ or ORF2+ neurons among 150 HMGA1+ neurons was calculated, and the results are expressed as the percentage of neurons stained by both antibodies. (C) TG sections were counterstained with methyl green. The percentage of neurons in which the nucleus was clearly detected was counted. For this study, 400 TG neurons from a calf latently infected with BoHV-1 and TG from an uninfected calf were counted.
ORF2 is associated with β-catenin and HMGA1 in transfected neuroblastoma cells.
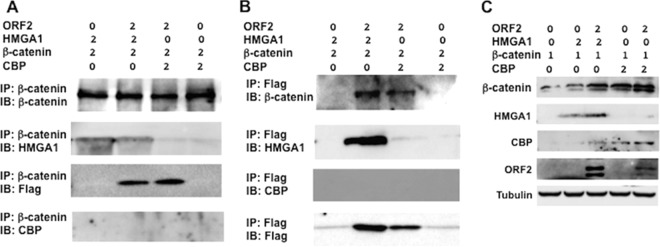
To test whether ORF2 interacts with β-catenin or HMGA1, coimmunoprecipitation (co-IP) studies were performed with cells of a mouse neuroblastoma cell line (Neuro-2A) transfected with plasmids expressing ORF2, β-catenin lacking the Flag tag, and HMGA1. Neuro-2A cells were used for these studies because they are readily transfected, ORF2 is consistently detected in these cells following transfection with an expression plasmid, ORF2 stimulates neurite formation in cells (6, 26), and β-catenin is not readily detected in nontransfected Neuro-2A cells (10). For these studies, the β-catenin-specific antibody was initially used to immunoprecipitate proteins associated with β-catenin. Following separation of the proteins in an SDS-polyacrylamide gel, Western blotting assays were performed to identify proteins associated with β-catenin. These studies, which were performed using an antibody that specifically recognizes HMGA1 and the Flag antibody, which detects the ORF2-Flag fusion protein, revealed that HMGA1 and ORF2 are associated with β-catenin (Fig. 3A). We also tested whether CREB-binding protein (CBP) was associated with ORF2 and β-catenin because it was reported to be a β-catenin transcriptional coactivator (27). In contrast to HMGA1, CBP was not readily detected in the immunoprecipitate (Fig. 3A), suggesting that it was not stably associated with ORF2 and β-catenin. When ORF2 was not included in the transfection mixture, the Flag antibody did not detect a protein with the same molecular weight as ORF2 (Fig. 3A).
FIG 3.
ORF2 is stably associated with HMGA1 and β-catenin. Neuro-2A cells were transfected with the designated plasmids using the indicated DNA concentrations (in micrograms). (A) At 48 h after transfection, IP was performed using total cell lysate (400 μg protein) and a β-catenin-specific antibody (catalog number ab32572; Abcam). Immunoprecipitated proteins bound to magnetic protein A beads (catalog number S1425S; Invitrogen) were washed extensively, suspended in SDS-PAGE buffer, and separated in a 10% SDS-polyacrylamide gel. After the proteins were transferred to a polyvinylidene difluoride membrane, the immunoblot (IB) was probed with the designated antibody: anti-β-catenin antibody (catalog number ab32572; Abcam), anti-HMGA1 antibody (catalog number ab129153; Abcam), anti-Flag monoclonal antibody (catalog number F1804; Sigma), or anti-CBP antibody (catalog number sc-25748; Santa Cruz Biotech). (B) IP was performed using an anti-Flag M2 affinity gel (catalog number A2220; Sigma) to precipitate proteins associated with ORF2. Immunoblotting of the precipitated proteins was performed using the designated antibodies after the beads were washed as described in the Materials and Methods. (C) The levels of the denoted proteins in the total cell lysate (30 μg protein) of transfected Neuro-2A cells were examined at 48 h after transfection using the antibodies described above. The results are representative of those from three independent experiments.
The Flag-tagged monoclonal antibody was also used to immunoprecipitate ORF2 and the proteins associated with ORF2 identified by Western blotting (Fig. 3B). These studies confirmed that ORF2 is associated with β-catenin and HMGA1. When the cell lysate was immunoprecipitated with the Flag-tagged antibody and then blotted with the CBP antibody, we were unable to detect CBP in the immunoprecipitate regardless of whether ORF2 was used to transfect Neuro-2A cells (Fig. 3B). Western blot studies of the total cell lysate demonstrated that the respective proteins were expressed in transfected cells (Fig. 3C). HMGA1, CBP, and ORF2 stabilized β-catenin steady-state levels. In several experiments, we found higher steady-state CBP levels in the total cell lysate following transfection with ORF2, β-catenin, and HMGA1. These studies also revealed that ORF2 slightly increased the steady-state levels of HMGA1 in transfected Neuro-2A cells (Fig. 3C). In summary, these studies indicated that ORF2 is stably associated with β-catenin and HMGA1.
Localization of ORF2, HMGA1, or β-catenin in transfected Neuro-2A cells.
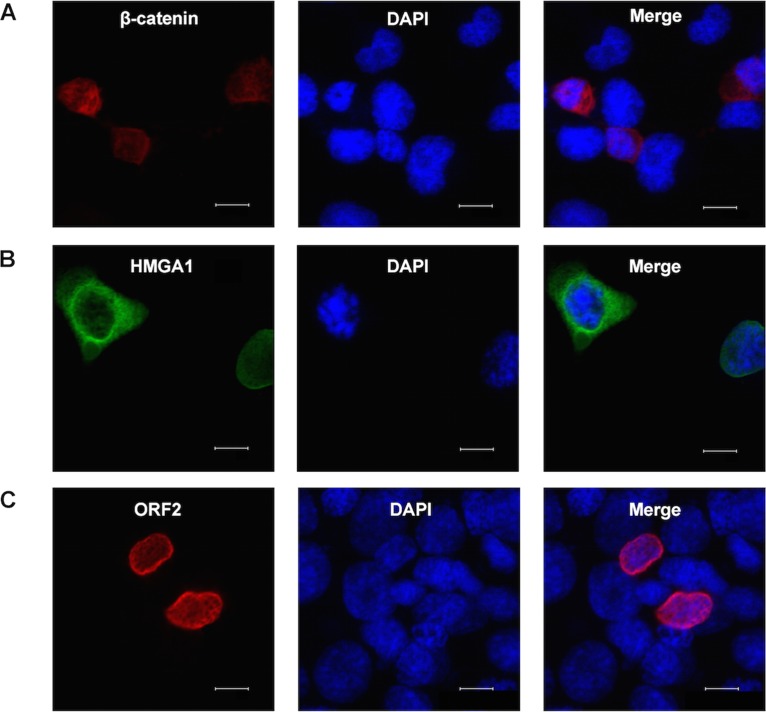
To confirm that ORF2 was associated with β-catenin and HMGA1 in transfected Neuro-2A cells (Fig. 3), confocal microscopy was performed. Initial studies examined the localization of the individual proteins in transfected cells. Most of the Flag-tagged β-catenin protein (S33Y) was localized in the cytoplasm of transfected Neuro-2A cells (Fig. 4A), suggesting that β-catenin was not active in these cells. Similar results were obtained with the non-Flag-tagged β-catenin construct (data not shown). HMGA1 was localized to the perinuclear region and the cytoplasm of most transfected cells (Fig. 4B). As previously reported (6, 7, 28), ORF2 was primarily localized at the rim of the nucleus (Fig. 4C).
FIG 4.
Localization of ORF2, β-catenin, and HMGA1 in transiently transfected Neuro-2A cells. Neuro-2A cells were transfected with 1 μg of a plasmid that expresses Flag-tagged β-catenin (S33Y) (A), HMGA1 (B), or Flag-tagged ORF2 (C) by use of the Lipofectamine 2000 transfection reagent (Invitrogen). At 48 h posttransfection, cells were stained with anti-Flag antibody (red), anti-HMGA1 antibody (green), and DAPI (4′,6-diamidino-2-phenylindole) to visualize β-catenin, ORF2, HMGA1, and the nucleus. The images were observed using confocal microscopy. These images are representative of those from four independent experiments. Bars, 10 μm.
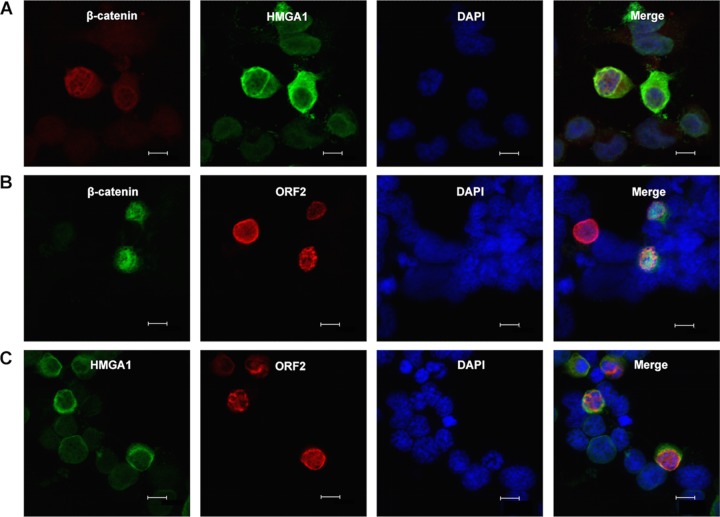
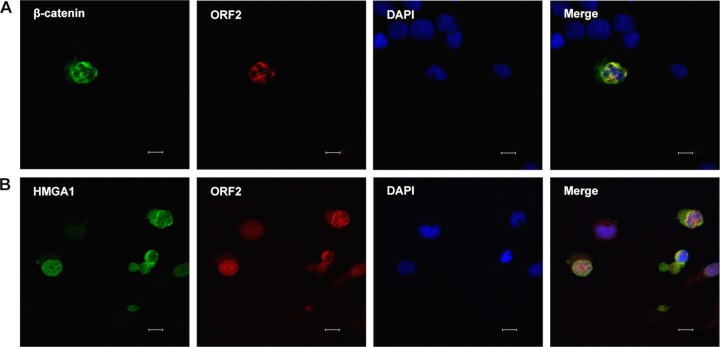
We next examined the localization of two of the three proteins in transfected Neuro-2A cells. For these studies, a non-Flag-tagged β-catenin construct was used when cells were cotransfected with ORF2 to allow visualization of both proteins in transfected cells. Thus, β-catenin was detected using an antibody that specifically recognizes β-catenin. When β-catenin was cotransfected with HMGA1, β-catenin was primarily detected in the nucleus, whereas HMGA1 was still localized to the perinuclear region (Fig. 5A). A subset of HMGA1 and β-catenin also appeared to colocalize near the rim of the nucleus. Cotransfection of ORF2 with β-catenin led to the relocalization of ORF2 and the partial colocalization of β-catenin and ORF2 in the nucleus (Fig. 5B). The partial colocalization of ORF2 and HMGA1 was also detected in cells expressing both proteins (Fig. 5C).
FIG 5.
Examination of ORF2 with β-catenin or HMGA1 in transiently cotransfected Neuro-2A cells. Neuro-2A cells were cotransfected with 0.5 μg of two of the following plasmids that express Flag-tagged β-catenin (S33Y) by use of the Lipofectamine 2000 transfection reagent (Invitrogen): a β-catenin construct that was not Flag tagged, HMGA1, or Flag-tagged ORF2. At 48 h posttransfection, cells were fixed and then stained with the designated primary antibodies. (A) The S33Y construct that expresses Flag-tagged β-catenin was stained with the Flag monoclonal antibody (red) and HMGA1 (green). (B) A β-catenin-specific antibody (catalog number ab32572; Abcam) was used to detect non-Flag-tagged β-catenin (green) and Flag to detect ORF2 (red). (C) HMGA1 (green) and Flag to detect ORF2 (red). Nuclear DNA was stained with DAPI. The images were observed by confocal microscopy. The selected images are representative of those from four experiments (at least 100 stained cells were examined). Bars, 10 μm.
Plasmids expressing all three proteins were cotransfected into Neuuro-2A cells. For these studies, we could examine only two proteins at a time because the antibodies used were made in rabbits or mice. These studies revealed that ORF2, β-catenin, and HMGA1 were primarily localized to the nucleus (Fig. 6A and B). Furthermore, ORF2 was partially colocalized with β-catenin and HMGA1, as judged by the appearance of yellow staining in the nucleus. In summary, these studies indicated that a subset of ORF2 colocalized with HMGA1 and β-catenin in transfected Neuro-2A cells.
FIG 6.
Examination of ORF2, β-catenin, and HMGA1 in transiently transfected Neuro-2A cells. Neuro-2A cells were cotransfected with 0.4 μg of each plasmid expressing β-catenin (non-Flag-tagged protein), HMGA1, and Flag-tagged ORF2 by use of the Lipofectamine 2000 transfection reagent (Invitrogen). At 48 h posttransfection, cells were prepared for confocal microscopy. (A) Neuro-2A cells were stained with primary antibodies against β-catenin (green) and Flag to detect ORF2 (red). (B) Neuro-2A cells were stained with a primary antibody directed against HMGA1 (green) or Flag to detect ORF2 (red). Nuclear DNA was stained with DAPI. Images were observed by confocal microscopy, and these images are representative of those from four experiments. Bars, 10 μm.
ORF2 stimulates β-catenin-dependent transcription in the presence of HMGA1 and CBP.
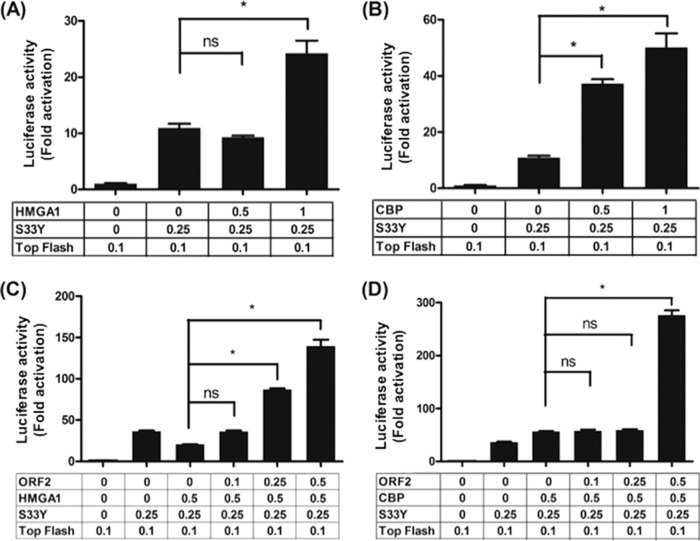
Since ORF2 stably interacted with a complex containing HMGA1 and β-catenin, we tested whether ORF2 influenced the ability of HMGA1 to function as a coactivator of β-catenin-dependent transcription. Initial studies tested whether HMGA1 and CBP functioned as coactivators of β-catenin-dependent transcription in Neuro-2A cells. For these studies, we utilized a promoter construct containing 8 T-cell factor (TCF) binding sites upstream of a minimal promoter (Super 8× Top-Flash) and the Flag-tagged β-catenin expression plasmid construct (S33Y). The Super 8× Top-Flash reporter accurately measures β-catenin-dependent transcription because when β-catenin is localized to the nucleus, it interacts with a TCF family member bound to the consensus site AGATCAAGG (reviewed in references 29 and 30). β-Catenin binding to TCF displaces bound corepressors and recruits transcriptional coactivators, thus stimulating the transcription of promoters containing TCF binding sites (18). HMGA1 stimulated β-catenin-dependent transcription more than 2-fold when 1 μg of HMGA1 was used in the transfection but had little effect when a lower concentration (0.5 μg) was used (Fig. 7A). CBP significantly stimulated β-catenin-dependent transcription in a dose-dependent manner in Neuro-2A cells (Fig. 7B).
FIG 7.
ORF2 stimulates coactivator enhancement of β-catenin-dependent transcription. (A) Neuro-2A cells were cotransfected with 0.1 μg of the Top-Flash luciferase reporter and 0.25 μg of β-catenin (S33Y). Where indicated, increasing concentrations of a plasmid that expresses HMGA1 (0.25 or 0.5 μg plasmid) were used to examine the effect that these coactivators have on TCF promoter activity. (B) Neuro-2A cells were cotransfected with 0.1 μg of the Top-Flash luciferase reporter and 0.25 μg of β-catenin (S33Y). Where indicated, increasing concentrations of a plasmid expressing CBP (0.25 or 0.5 μg plasmid) were used to examine the effect that these coactivators have on TCF promoter activity. (C) The effect of ORF2 on the ability of HMGA1 to stimulate β-catenin-dependent transcription was examined. Neuro-2A cells were transfected with the designated plasmids. (D) The effect of ORF2 on CBP to stimulate β-catenin-dependent transcription was examined in Neuro-2A cells. Neuro-2A cells were transfected with the designated plasmids. Dual-luciferase assays were performed at 48 h after transfection. The results for cells transfected with 0.1 μg of the Top-Flash luciferase reporter and the Renilla luciferase under the control of a minimal herpesvirus TK promoter (0.05 μg DNA) were normalized to a value of 1, and the fold activation was calculated for the other samples. For the studies whose results are presented in panels A to D, plasmid DNA concentrations for each transfection contained the same amount of DNA by adding empty expression plasmid (pCDNA3.1). Furthermore, Neuro-2A cells were transfected with the TransIT Neural transfection reagent (catalog number MIR2145; Mirus) according to the manufacturer's instructions. The results are averages from three independent experiments, and error bars denote standard errors. *, significant differences (P < 0.05) relative to the promoter activity of samples containing Top-Flash and β-catenin using a one-way analysis of variance and Fisher's least-significant-difference multiple-means comparison tests; ns, the differences between samples were not significant.
We then tested whether ORF2 mediated the ability of HMGA1 to stimulate β-catenin-dependent transcription by cotransfecting increasing levels of ORF2 with 0.5 μg HMGA1, S33Y, and the Super 8× Top-Flash promoter. This concentration of HMGA1 did not significantly stimulate β-catenin-dependent transcription (Fig. 7A and data not shown). When 0.25 or 0.5 μg of the ORF2 expression plasmid was included in the transfection, significantly higher levels of β-catenin-dependent transcription were observed in Neuro-2A cells (Fig. 7C). Although ORF2 cooperated with CBP to stimulate β-catenin-dependent transcription approximately 4-fold (Fig. 7D), the significant difference was observed only at the highest concentration of ORF2. In the absence of HMGA1 or CBP, ORF2 has little effect on β-catenin-dependent transcription (10) (data not shown).
ORF2 protein expression is important for enhancing β-catenin-dependent transcription.
To determine whether ORF2 expression was important for enhancing β-catenin-dependent transcription in the presence of HMGA1, we initially examined an ORF2 mutant in which the first in-frame methionine residue was deleted and three stop codons were added (the ORF2-stop mutant) (see Fig. 8A for a schematic of ORF2). In contrast to wild-type (wt) ORF2, the ORF2-stop mutant did not significantly increase the level of β-catenin-dependent transcription compared to that in cells transfected with the empty vector (Fig. 8C). ORF2 contains a nuclear localization signal (NLS) (Fig. 8A) that, when deleted (the ORF2ΔNLS mutant), expresses a protein that localizes to the plasma membrane (28). The ORF2ΔNLS mutant was not able to enhance β-catenin-dependent transcription more effectively than the empty vector. Finally, ORF2 contains 5 consensus protein kinase A (PKA) or protein kinase C (PKC) phosphorylation sites (Fig. 8A). Mutation of these 5 serine or threonine residues to alanine residues generated a protein that is more stable and is localized to the nucleus (the ORF2-P mutant) (28). Interestingly, the ORF2-P mutant was unable to enhance β-catenin-dependent transcription better than the other mutants examined or the empty vector (Fig. 8C). In summary, these studies provide evidence that wt ORF2 protein expression is required for enhancing β-catenin-dependent transcription in the presence of HMGA1.
FIG 8.
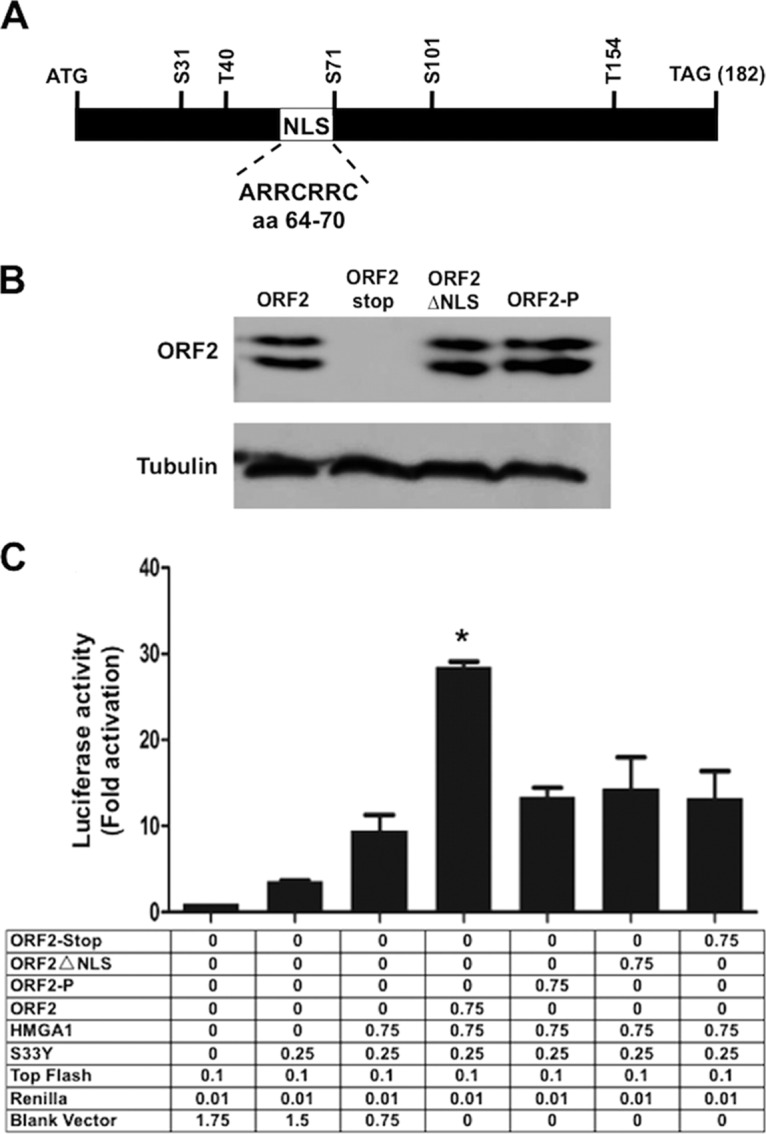
wt ORF2 but not the ORF2 mutants cooperate with HMGA1 to stimulate β-catenin-dependent transcription. (A) Schematic of ORF2 and positions of NLS and PKA/PKC phosphorylation sites. The ORF2-stop construct lacks the first in-frame methionine codon and contains three stop codons at the beginning of ORF2. The ORF2-ΔNLS construct lacks the nuclear localization signal and localizes to the plasma membrane in transfected Neuro-2A cells. The ORF2-P construct contains alanine substitutions in the 5 consensus PKA or PKC phosphorylation sites. (B) Neuro-2A cells were transfected with the designated plasmids (1.0 μg DNA) using the Lipofectamine 2000 transfection reagent. At 48 h after transfection, the cell lysate was prepared and the levels of ORF2 expressed by the respective plasmids were determined by Western blotting using the Flag antibody. (C) Neuro-2A cells were transfected with the designated plasmids using Lipofectamine 2000. The efficiency of transfection of Neuro-2A cells with Lipofectamine 2000 was less than that with the TransIT Neural reagent used in the assay whose results are presented in Fig. 7; consequently, the activation levels were also lower. However, the overall trends were the same. The amount of plasmid DNA in each transfection was the same as that in the empty expression plasmid (pCMV2-Tag-2B) used to construct the ORF2-expressing plasmid. Dual-luciferase assays were performed at 48 h after transfection. The results for cells transfected with 0.1 μg of the Top-Flash luciferase reporter and the Renilla luciferase under the control of a minimal herpesvirus TK promoter (0.05 μg DNA) were normalized to a value of 1, and the fold activation was calculated for the other samples. These results are the averages from three independent experiments. *, a significant difference (P < 0.05) compared to the result for the indicated control by one-way analysis of variance and Fisher's least-significant-difference multiple-means comparison tests.
DISCUSSION
In this study, we provide evidence that HMGA1 RNA is differentially expressed in the TG of latently infected calves. IHC studies confirmed that the HMGA1 protein is differentially expressed and further revealed that it is expressed in the nucleus of a subset of neurons from latently infected calves but not those from uninfected calves. HMGA1 is not typically expressed in adult, differentiated tissues but is abundantly expressed in hematopoietic stem cells, in poorly differentiated cancers, and during various stages of embryogenesis (31–33). Thus, it was unexpected to detect HMGA1 in terminally differentiated sensory neurons of calves latently infected with BoHV-1. Since HMGA1 is induced by the Wnt/β-catenin signaling pathway and activates this pathway (18–20), HMGA1 may cooperate with ORF2 and β-catenin to maintain neuronal survival and normal neuronal functions during latency. The relevance of the other cellular genes differentially regulated during latency will require additional studies, and evaluation of their relevance was not in the scope of the present study.
Co-IP studies provided evidence that ORF2 is stably associated with HMGA1 and/or β-catenin but not CBP. The conformation of the co-IP studies came from confocal microscopy studies demonstrating that ORF2 partially colocalized with β-catenin and HMGA1 in the nucleus of Neuro-2A cells. These studies indicated that interactions between ORF2 and β-catenin as well as HMGA1 correlated with increased levels of β-catenin-dependent transcription. Although we believe that ORF2 stabilizes the Wnt/β-catenin signaling pathway and HMGA1 expression during latency, other LR gene products may be involved because this locus encodes at least two microRNAs and a protein downstream of ORF2 (reviewed in reference 34).
Canonical β-catenin-dependent transcription is tightly regulated by at least 19 Wnt family members, 11 human Wnt receptors, numerous Wnt antagonists, and a β-catenin destruction complex that degrades β-catenin in the cytoplasm (reviewed in references 30, 35, and 36). In the absence of Wnt, β-catenin-dependent transcription is extinguished because transcriptional repressors are bound to TCF family members (denoted by X in Fig. 9A). Following Wnt-mediated activation of β-catenin, β-catenin enters the nucleus, displaces transcriptional repressors, and recruits transcriptional coactivators (denoted by Y), including HMGA1 (Fig. 9B). ORF2 is not a direct transcriptional coactivator of β-catenin-dependent transcription because in the absence of a coactivator it has little to no effect on transcription in transfected Neuro-2A cells (10). However, ORF2 can enhance the ability of a coactivator (HMGA1 and CBP) to stimulate β-catenin-dependent transcription, even though ORF2 is not stably associated with CBP. The finding that the ORF2-P mutant was unable to enhance β-catenin-dependent transcription may provide insight into how ORF2 enhances the stimulation of β-catenin-dependent transcription because this mutant preferentially binds double-stranded DNA, whereas wt ORF2 preferentially binds single-stranded DNA (37). We suggest that the DNA binding properties of wt ORF2 (i) stabilize or initiate interactions between β-catenin and TCF family members or (ii) stabilize interactions between HMGA1 and AT-rich DNA and/or β-catenin (Fig. 9C). It is not clear whether HMGA1 and β-catenin cooperate with ORF2 to stimulate the expression of all or a subset of cellular genes in TG neurons that are regulated by β-catenin.
FIG 9.
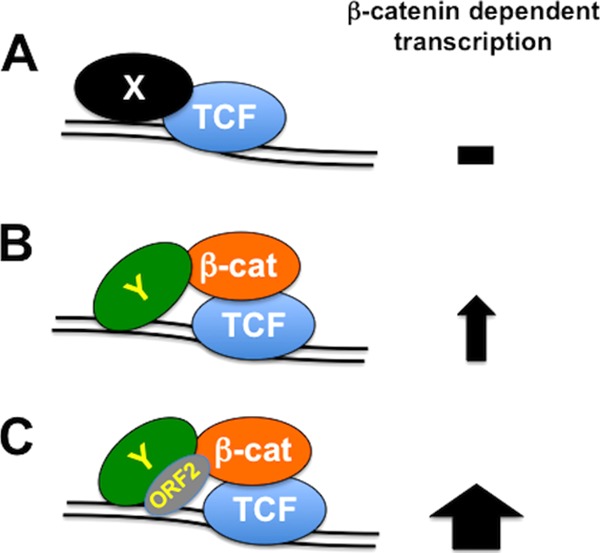
Schematic of β-catenin-dependent transcription. (A) In the absence of Wnt, β-catenin is degraded in the cytoplasm. Consequently, TCF family members are bound to transcriptional repressors (denoted X) and β-catenin-dependent transcription is silenced. (B) Following Wnt binding to its receptor(s), β-catenin (β-cat) enters the nucleus, binds to TCF family members, displaces transcriptional repressors, and recruits transcriptional coactivators, for example, HMGA1 (denoted Y), which culminates in the stimulation of β-catenin-dependent transcription. (C) ORF2, by virtue of its ability to interact with HMGA1 and β-catenin, stabilizes β-catenin in the nucleus. These interactions lead to enhancement of the coactivator (denoted Y); HMGA1 and CBP, for example, mediated stimulation of β-catenin-dependent transcription. The ability of ORF2 to preferentially interact with single-stranded DNA may be important for these activities.
HMGA1 has additional functions that may be important for a latent infection and are independent of β-catenin-dependent transcription. For example, HMGA1 has been reported to relieve p53-dependent repression of Bcl-2 promoter activity, thus interfering with p53-dependent apoptosis (38). Furthermore, HMGA1 promotes the activation of Akt, a protein kinase known to enhance cell survival (39), in pancreatic adenocarcinoma cells (40). In a neuronal culture model of herpes simplex virus 1 latency, reactivation from latency occurs when Akt is inhibited (41), suggesting that Akt may also mediate certain aspects of the BoHV-1 latency-reactivation cycle. Finally, HMGA1 localizes to AT-rich G/Q and C bands as well as sites of chromosome attachment to the nuclear matrix, indicating that HMGA1 is involved with chromosome condensation during the cell cycle (reviewed in reference 42). Whether the association between ORF2 and HMGA1 regulates the conformation of the viral chromosome in latently infected neurons or enhances AKT activation is not known.
MATERIALS AND METHODS
Infection of calves.
All TG samples from the calves used for this study were previously described (21). In brief, BHV-1-free crossbred calves (weight, ∼200 kg) were inoculated with 107 PFU of BHV-1 in the ocular and nasal cavities as described previously (8, 9, 43–46). At 60 days postinfection, calves were not shedding virus and considered to be latently infected. Calves were housed under strict isolation and given antibiotics to prevent secondary bacterial infections. Calves (latently infected or uninfected) were transported to the University of Nebraska Veterinary Diagnostic Lab. The calves were then anesthetized with xylazine (Rompun), followed by electrocution. Experiments were performed in accordance with the American Association of Laboratory Animal Care guidelines and the University of Nebraska IACUC (approval A3459). Following euthanasia, TG were collected and minced into small pieces, and then a portion of the TG was formalin fixed and paraffin embedded. The remainder of both TG from each calf was minced into small pieces and placed into a single 50-ml conical tube, and the tube was placed in a dry ice-ethanol bath and then stored at −80°C. After decapitation, it took 5 to 10 min to collect TG, mince TG, place TG pieces in formalin or in a 50-ml conical tube, and submerge TG pieces in 50-ml conical tubes in a dry ice-ethanol bath.
Microarray analysis.
Details of the microarray study, which was performed using a bovine gene chip (Affymetrix, Santa Clara, CA) which contains more than 23,000 genes, were previously described (21). To determine differentially expressed genes, we compared latently infected versus uninfected TG using a random-variance F test and selected genes with a false discovery rate of less than 10%. The fold change in gene expression in latently infected TG relative to that in uninfected TG was calculated for each differentially expressed gene.
IHC analysis.
Immunohistochemistry (IHC) studies were performed using an ABC kit (Vector Laboratories) according to the specifications of the manufacturer. Thin sections (4 to 5 μm) of TG were cut, mounted on glass slides, and processed as described previously (6, 10, 47). The slides were subsequently incubated with β-catenin (1:250; catalog number ab32572; Abcam), HMGA1 (1:400; catalog number ab129153; Abcam), or ORF2 rabbit polyclonal antibody at a 1:1,000 dilution overnight in a humidified chamber at 4°C. On the next day, the slides were washed in 1× Tris-buffered saline (TBS) and incubated in biotinylated goat anti-rabbit IgG (catalog number PK-6101; Vector Laboratories) for 30 min at room temperature in a humidified chamber. Avidin-biotinylated enzyme complex was added to the slides for 30 min of incubation at room temperature. After three washes in 1× TBS, the slides were incubated with freshly prepared substrate (catalog number SK-4800; Vector Laboratories), rinsed with distilled water, and counterstained with methyl green. The number of HMGA1-positive neurons was estimated from 490 neurons by counting the HMGA1-positive neurons in a blind fashion.
Cells, plasmids, and antibodies.
Murine neuroblastoma (Neuro-2A) cells (CCL-131) were obtained from ATCC (Manassas, VA, USA) and were grown in Earle's modified Eagle's medium (EMEM) supplemented with 10% fetal calf serum (FCS), penicillin (10 U/ml), and streptomycin (100 μg/ml).
The ORF2 expression construct was generated in the pCMV-Tag-2B vector (Stratagene) and was described previously (5, 6, 26). A Flag epitope is present at the N terminus of ORF2, and the human immediate early (IE) cytomegalovirus (CMV) promoter drives its expression. Sequences derived from ORF2 with a nuclear localization signal (NLS) deletion (amino acids [aa] 64 to 70; the ORF2ΔNLS mutant) or the PKA/PKC phosphorylation sites converted to alanine (the ORF2-P mutant) were synthesized by Integrated DNA Technologies (IDT; Coralville, IA) and cloned into the pCMV-Tag-2B plasmid using BamHI-HindIII restriction enzymes. For the location of the NLS and PKA/PKC sites in ORF2, see Fig. 8A.
The human β-catenin expression construct (S33Y), which expresses a Flag-tagged β-catenin protein, was a gift from Bert Vogelstein (Addgene plasmid number 16519) (48). The mouse non-Flag-tagged β-catenin expression construct (pcDNA3.1/nV5-DEST-beta catenin) was a gift from Valeri Vasioukhin and was obtained from Addgene (plasmid number 20140). The HMGA1 expression plasmid (pIRES-HMGA1) was a gift from Edward Whang and was obtained from Addgene (plasmid number 13466). M50 Super 8× Top-Flash contains a simple promoter that is stimulated by β-catenin and was a gift from Randall Moon (Addgene plasmid number 12456) (49). All plasmids were transfected into Neuro-2A cells in 6-well plates using the Lipofectamine 2000 transfection reagent (catalog number 11668019; Invitrogen) or the TransIT Neural transfection reagent (catalog number MIR2145; Mirus) according to the manufacturers' instructions. The anti-β-catenin antibody (catalog number ab32572; Abcam), anti-HMGA1 antibody (catalog number ab129153; Abcam), anti-Flag monoclonal antibody (catalog number, F1804; Sigma), or anti-CBP antibody (catalog number sc-25748; Santa Cruz Biotech) was used for the Western blot, confocal microscopy, and immunoprecipitation (IP) studies.
Co-IP studies and Western blot analysis.
For coimmunoprecipitation (co-IP) studies, Neuro-2A cells grown in 10-cm plates were transfected with the designated plasmids using the Lipofectamine 2000 transfection reagent (catalog number 11668019; Invitrogen). At 48 h after transfection, cells were lysed with 1 ml of radioimmunoprecipitation assay (RIPA) buffer (1× phosphate-buffered saline [PBS], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitor cocktail (Roche). The cell lysate was clarified by centrifugation at 13,000 rpm for 10 min. The clarified supernatant (approximately 400 μg protein) was incubated with the anti-Flag M2 affinity gel with gentle rotation by a roller shaker for 2 h at 4°C. The anti-Flag M2 affinity gel (catalog number A2220; Sigma) was prepared according to the manufacturer's specification. After the beads were washed three times with 0.5 ml of PBS, they were boiled in SDS loading buffer and subsequent Western blotting was performed to detect the designated proteins. When the IP was performed with β-catenin, immunoprecipitated proteins were collected using magnetic protein A beads (catalog number S1425S; Invitrogen). The beads were washed extensively, suspended in SDS-PAGE buffer, and separated in a 10% SDS-polyacrylamide gel.
Neuro-2A cells in 60-mm dishes were transfected with the designated plasmids. At 48 h after transfection, the cells were collected, washed once with PBS, and then lysed in RIPA buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) with protease and phosphatase inhibitors (Thermo Scientific). The respective samples were boiled in Laemmli sample buffer for 5 min, and all samples were separated on an 8% or 10% SDS-polyacrylamide gel. Immunodetection of the respective proteins was performed using the antibodies described above.
Dual-luciferase reporter assay.
Neuro-2A cells (8 × 105) were seeded into 60-mm dishes containing EMEM with 10% FCS at 24 h prior to transfection. At 2 h before transfection, the medium was replaced with fresh EMEM containing 0.5% FCS to lower the basal levels of promoter activity. Cells were cotransfected with the designated plasmids and a plasmid carrying Renilla luciferase under the control of a minimal herpesvirus thymidine kinase (TK) promoter (10 ng). To maintain equal plasmid amounts in the transfection mixtures, an empty expression vector was added as needed. At 40 h after transfection, cells were harvested and protein extracts were subjected to a dual-luciferase assay by using a commercially available kit (catalog number E1910; Promega) according to the manufacturer's instructions. Luminescence was measured by using a GloMax 20/20 luminometer (catalog number E5331; Promega).
ACKNOWLEDGMENTS
This research was supported by a grant from the USDA-NIFA Competitive Grants Program (13-01041), funds derived from the Sitlington Endowment, and support from the Oklahoma Center for Respiratory and Infectious Diseases (National Institutes of Health Centers for Biomedical Research Excellence grant number P20GM103648). L.Z. was partially supported by the China Scholarship Council, a Chinese National Science Foundation grant (no. 31472172), and the National Key Research Program (no. 2016YFD0500704).
REFERENCES
- 1.Hodgson PD, Aich P, Manuja A, Hokamp K, Roche FM, Brinkman FSL, Potter A, Babiuk LA, Griebel PJ. 2005. Effect of stress on viral-bacterial synergy in bovine respiratory disease: novel mechanisms to regulate inflammation. Comp Funct Genomics 6:244–250. doi: 10.1002/cfg.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones C, Chowdhury S. 2010. Bovine herpesvirus type 1 (BHV-1) is an important cofactor in the bovine respiratory disease complex. Vet Clin North Am Food Anim Pract 26:303–321. doi: 10.1016/j.cvfa.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser JN, Neupane M, Womack JE, Van Eenennaam A, Hagevoort GR, Lehenbauer TW, Aly S, Davis J, Taylor JF, Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team. 2014. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics 15:1164. doi: 10.1186/1471-2164-15-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaber T, Workman A, Jones C. 2010. Small noncoding RNAs encoded within the bovine herpesvirus 1 latency-related gene can reduce steady-state levels of infected cell protein 0 (bICP0). J Virol 84:6297–6307. doi: 10.1128/JVI.02639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen W, Jones C. 2008. Open reading frame 2, encoded by the latency-related gene of bovine herpesvirus 1, has antiapoptotic activity in transiently transfected neuroblastoma cells. J Virol 82:10940–10945. doi: 10.1128/JVI.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinani D, Frizzo da Silva L, Jones C. 2013. A bovine herpesvirus 1 protein expressed in latently infected neurons (ORF2) promotes neurite sprouting in the presence of activated Notch1 or Notch3. J Virol 87:1183–1192. doi: 10.1128/JVI.02783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Workman A, Sinani D, Pittayakhajonwut D, Jones C. 2011. A protein (ORF2) encoded by the latency related gene of bovine herpesvirus 1 interacts with Notch1 and Notch3. J Virol 85:2536–2546. doi: 10.1128/JVI.01937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inman M, Lovato L, Doster A, Jones C. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J Virol 75:8507–8515. doi: 10.1128/JVI.75.18.8507-8515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inman M, Lovato L, Doster A, Jones C. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 disrupts the latency reactivation cycle in calves. J Virol 76:6771–6779. doi: 10.1128/JVI.76.13.6771-6779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Hancock M, Workman A, Doster A, Jones C. 2016. Beta-catenin, a transcription factor activated by canonical Wnt signaling, is expressed in sensory neurons of calves latently infected with bovine herpesvirus 1. J Virol 90:3148–3159. doi: 10.1128/JVI.02971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas PC. 2012. Wnt signaling in the vertebrate central nervous system: from axon guidance to synaptic function. Cold Spring Harb Perspect Biol 4:a008003. doi: 10.1101/cshperspect.a008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purro SA, Galli S, Salinas PC. 2014. Dysfunction of Wnt signaling and synaptic disassembly in neurodegenerative diseases. J Mol Cell Biol 6:75–80. doi: 10.1093/jmcb/mjt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhardwaji D, Nager M, Camats J, David M, Benguira A, Dopazo A, Canti C, Herreros J. 2013. Chemokines induce axon outgrowth downstream of hepatocyte growth factor and TCF/beta-catenin signaling. Front Cell Neurosci 7:52. doi: 10.3389/fncel.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murase S, Mosser E, Schuman EM. 2002. Depolarization drives beta-catenin into neuronal spines promoting changes in synaptic structure and function. Neuron 35:91–105. doi: 10.1016/S0896-6273(02)00764-X. [DOI] [PubMed] [Google Scholar]
- 15.Bamji SX, Rico B, Kimes N, Reichardt LF. 2006. BDNF mobilizes synaptic vesicles and enhances synaptic vesicles and enhances synapse formation by disrupting cadherin-beta-catenin interactions. J Cell Biol 174:289–299. doi: 10.1083/jcb.200601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matrisciano F, Buscetti CL, Bucci D, Orlando R, Caruso A, Molinaro G, Cappuccion I, Riozzi B, Gradini R, Motolese M, Caraci F, Copani A, Scaccianoce S, Melchiorri D, Bruno V, Battaglia G, Nicoletti F. 2011. Induction of the Wnt antagonist Dickkopf-1 is involved in stress-induced hippocampal damage. PLoS One 6:e16447. doi: 10.1371/journal.pone.0016447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moors M, Bose R, Johansson-Haque K, Edoff K, Okret S, Ceccatelli S. 2012. Dickkopf mediates glucocorticoid-induced changes in human neural progenitor cell proliferation and differentiation. Toxicol Sci 125:488–495. doi: 10.1093/toxsci/kfr304. [DOI] [PubMed] [Google Scholar]
- 18.Bush BM, Brock AT, Deng JA, Nelson RA Jr, Sumter TF. 2013. The Wnt/β-catenin/TCF pathway upregulates HMGA1 expression in colon cancer. Cell Biochem Funct 3:228–236. doi: 10.1002/cbf.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akaboshi S-I, Watanabe S, Hino Y, Sekita Y, Xi Y, Araki K, Yamamura K-I, Oshima M, Ito T, Baba H, Nako M. 2009. HMGA1 is induced by Wnt/β-catenin pathway and maintains cell proliferation in gastric cancer. Am J Pathol 175:1675–1685. doi: 10.2353/ajpath.2009.090069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing J, Cao G, Fu C. 2014. HMGA1 interacts with beta-catenin to positively regulate Wnt/beta-catenin signaling in colorectal cancer cells. Pathol Oncol Res 20:847–851. doi: 10.1007/s12253-014-9763-0. [DOI] [PubMed] [Google Scholar]
- 21.Workman A, Eudy J, Smith L, Frizzo da Silva L, Sinani D, Bricker H, Cook E, Doster A, Jones C. 2012. Cellular transcription factors induced in trigeminal ganglia during dexamethasone-induced reactivation from latency stimulate bovine herpesvirus 1 productive infection and certain viral promoters. J Virol 86:2459–2473. doi: 10.1128/JVI.06143-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. 2003. Differential regulation of midbrain dompaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci U S A 100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bravo DT, Yang Y-L, Kuchenbecker K, Hung M-S, Xu Z, Jablons DM, You L. 2013. Frizzled-8 receptor is activated by the Wnt-2 ligand in non-small cell lung cancer. BMC Cancer 13:316. doi: 10.1186/1471-2407-13-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kook I, Doster A, Jones C. 2015. Bovine herpesvirus 1 regulatory proteins are detected in trigeminal ganglionic neurons during the early stages of stress-induced escape from latency. J Neurovirol 21:585–591. doi: 10.1007/s13365-015-0339-x. [DOI] [PubMed] [Google Scholar]
- 25.Kook ICH, Meyer F, Hoffmann F, Jones C. 2015. Bovine herpesvirus 1 productive infection and the immediate early transcription unit 1 are stimulated by the synthetic corticosteroid dexamethasone. Virology 484:377–385. doi: 10.1016/j.virol.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Sinani D, Liu Y, Jones C. 2014. Analysis of a bovine herpesvirus 1 protein encoded by an alternatively spliced latency related (LR) RNA that is abundantly expressed in latently infected neurons. Virology 464–465:244–252. doi: 10.1016/j.virol.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 27.Takemaru K-T, Moon RT. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol 149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinani D, Jones C. 2011. Localization of sequences in a protein encoded by the latency related gene of bovine herpesvirus 1 (ORF2) that inhibits apoptosis and interferes with Notch1 mediated trans-activation of the bICP0 promoter. J Virol 85:12124–12133. doi: 10.1128/JVI.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biechele TL, Kulikauskas RM, Toroni RA, Lucero OM, Swift RD, James RG, Robin NC, Dawson DW, Moon RT, Chien AJ. 2012. Wnt/beta-catenin signaling and AXIN1 regulate apoptosis mediated by inhibition of BRAFV600E kinase in human melanoma. Sci Signal 5:ra3. doi: 10.1126/scisignal.2002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell 149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Porath I, Thompson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou G, Chen J, Lee S, Clark T, Rowley JD, Wang SM. 2001. The pattern of gene expression in human CD34+ stem/progenitor cells. Proc Natl Acad Sci U S A 98:13966–13971. doi: 10.1073/pnas.241526198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, Belton A, Huso DL, Resar LMS. 2012. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One 7:e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones C, da Silva LF, Sinani D. 2011. Regulation of the latency-reactivation cycle by products encoded by the bovine herpesvirus 1 (BHV-1) latency-related gene. J Neurovirol 17:535–545. doi: 10.1007/s13365-011-0060-3. [DOI] [PubMed] [Google Scholar]
- 35.Hayward P, Kalmar T, Arlas AM. 2008. Wnt/Notch signalling and information processing during development. Development 135:411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 36.Zorn AM. 2001. Wnt signaling: antagonistic Dickkopf. Curr Biol 11:R592–R595. doi: 10.1016/S0960-9822(01)00360-8. [DOI] [PubMed] [Google Scholar]
- 37.Pittayakhajonwut D, Sinani D, Jones C. 2013. A protein (ORF2) encoded by the latency related gene of bovine herpesvirus 1 interacts with DNA. J Virol 87:5493–5501. doi: 10.1128/JVI.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esposito F, Tornincasa M, Chieffi P, De Martino I, Pierantoni GM, Fusco A. 2010. High-mobility group A1 proteins regulate p53-mediated transcription of Bcl-2 gene. Cancer Res 70:5379–5388. doi: 10.1158/0008-5472.CAN-09-4199. [DOI] [PubMed] [Google Scholar]
- 39.Franke TF, Kaplan DR, Cantley LC. 1997. PI3K: downstream AKTion blocks apoptosis. Cell 88:435–437. doi: 10.1016/S0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 40.Liau S-S, Jazag A, Whang EE. 2007. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer 96:993–1000. doi: 10.1038/sj.bjc.6603654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camarena V, Kobayashi M, Kim JK, Roehm P, Perez R, Gardner J, Wilson AC, Mohr I, Chao MV. 2010. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe 8:320–330. doi: 10.1016/j.chom.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleyenen I, van de Ven WJM. 2008. The HMGA proteins: myriad of functions (review). Int J Oncol 32:289–305. [PubMed] [Google Scholar]
- 43.Lovato L, Inman M, Henderson G, Doster A, Jones C. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J Virol 77:4848–4857. doi: 10.1128/JVI.77.8.4848-4857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez S, Inman M, Doster A, Jones C. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J Clin Microbiol 43:393–401. doi: 10.1128/JCM.43.1.393-401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez S, Lovato L, Zhou J, Doster A, Jones C. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild type BHV-1 versus a virus strain containing a mutation in the LR (latency-related) gene. J Neurovirol 12:392–397. doi: 10.1080/13550280600936459. [DOI] [PubMed] [Google Scholar]
- 46.Perez S, Meyer F, Saira K, Doster A, Jones C. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J Gen Virol 89:1338–1345. doi: 10.1099/vir.0.83481-0. [DOI] [PubMed] [Google Scholar]
- 47.Sinani D, Cordes E, Workman A, Thunuguntia P, Jones C. 2013. Stress-induced cellular transcription factors expressed in trigeminal ganglionic neurons stimulate the herpes simplex virus 1 ICP0 promoter. J Virol 87:13042–13047. doi: 10.1128/JVI.02476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin PJ, Sparks AB, Korinek V, Barker N, Clever H, Vogelstein B, Kinzler KW. 1997. Activation of B-cetenin-TCF signaling in colon cancer by mutations in B-cetenin or APC. Science 275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 49.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13:680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]