ABSTRACT
Bats harbor a large diversity of coronaviruses (CoVs), several of which are related to zoonotic pathogens that cause severe disease in humans. Our screening of bat samples collected in Kenya from 2007 to 2010 not only detected RNA from several novel CoVs but, more significantly, identified sequences that were closely related to human CoVs NL63 and 229E, suggesting that these two human viruses originate from bats. We also demonstrated that human CoV NL63 is a recombinant between NL63-like viruses circulating in Triaenops bats and 229E-like viruses circulating in Hipposideros bats, with the breakpoint located near 5′ and 3′ ends of the spike (S) protein gene. In addition, two further interspecies recombination events involving the S gene were identified, suggesting that this region may represent a recombination “hot spot” in CoV genomes. Finally, using a combination of phylogenetic and distance-based approaches, we showed that the genetic diversity of bat CoVs is primarily structured by host species and subsequently by geographic distances.
IMPORTANCE Understanding the driving forces of cross-species virus transmission is central to understanding the nature of disease emergence. Previous studies have demonstrated that bats are the ultimate reservoir hosts for a number of coronaviruses (CoVs), including ancestors of severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and human CoV 229E (HCoV-229E). However, the evolutionary pathways of bat CoVs remain elusive. We provide evidence for natural recombination between distantly related African bat coronaviruses associated with Triaenops afer and Hipposideros sp. bats that resulted in a NL63-like virus, an ancestor of the human pathogen HCoV-NL63. These results suggest that interspecies recombination may play an important role in CoV evolution and the emergence of novel CoVs with zoonotic potential.
KEYWORDS: Africa, bats, coronavirus, HCoV-229E, HCoV-NL63, recombination, zoonoses
INTRODUCTION
Coronaviruses (CoVs) (subfamily Coronavirinae, family Coronaviridae, order Nidovirales) are common infectious agents that infect a wide range of hosts, including humans, causing respiratory, gastrointestinal, liver, and neurologic diseases, and that possess the largest genomes of any RNA viruses described to date (1). The subfamily Coronavirinae is currently classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (2). The alphacoronaviruses (alpha-CoVs) and betacoronaviruses (beta-CoVs) are found exclusively in mammals, while the gammacoronaviruses (gamma-CoVs) and deltacoronaviruses (delta-CoVs) are associated mainly with birds. Presently, the greatest diversity of alpha- and beta-CoVs has been documented in bats, which in part reflects the more intensive surveillance of these animals since Rhinolophus sp. bats were implicated as the reservoir hosts for severe acute respiratory syndrome (SARS)-related CoVs (3, 4). This surveillance resulted in the discovery of a potential reservoir host (bat) species for another two human CoVs: human CoV 229E (HCoV-229E), a relative of which is present in Hipposideros bats (5, 6), and Middle East respiratory syndrome coronavirus (MERS-CoV), for which related viruses are present in Pipistrellus, Tylonycteris, and Neoromicia bats (7–10), although the most likely reservoir host of human MERS-CoV identified to date is the dromedary camel (11). Most recently, HCoV-229E-like CoVs were also identified in camels, although their role in human infection is unknown (12).
Africa is a major hot spot of zoonotic emerging diseases. With its rich biodiversity, Africa is inhabited by many bats of different species, including those that serve as reservoirs of important zoonotic diseases, such as Marburg hemorrhagic fever and rabies (13). Our initial screening demonstrated the presence of diverse CoVs in African bats, including those collected in the southern parts of Kenya during 2006 (14, 15) and in other countries, including South Africa, Nigeria, and Ghana (16). Furthermore, recent studies have provided strong evidence that HCoV-229E originated from bat viruses circulating in Africa (5), underscoring the zoonotic potential of bat-borne CoVs from this continent.
One human coronavirus, HCoV-NL63, was first isolated in 2004 from the aspirate of an 8-month-old boy suffering from pneumonia in the Netherlands (17). While the clinical significance of this virus is debated, it has a worldwide distribution and is known to infect both the upper and lower respiratory tracts (18). Based on a phylogeny of the RNA-dependent RNA polymerase (RdRp), HCoV-NL63 is related to another human virus, HCoV-229E, and had no close relatives identified in bats (16). Although Huynh et al. (19) suggested that a virus (ARCoV.2/2010/USA) isolated from the American tricolored bat (Perimyotis subflavus) may share a common ancestry with HCoV-NL63, the genetic distance between the two viruses is large, and their close relationship has not been corroborated in other phylogenetic analyses (16, 20). Nevertheless, the successful passage of HCoV-NL63 in an immortalized bat cell line suggests its potential association with bats (19).
As is well appreciated, recombination leads to rapid changes in genetic diversity in RNA viruses (21). CoVs represent a classic example of viruses with high frequencies of homologous recombination through discontinuous RNA synthesis (22). Indeed, under experimental conditions, the recombination frequency can be as high as 25% for the entire CoV genome (23). Recombination in CoVs is also frequently reported under natural conditions, including in some emerging human pathogens such as SARS-CoV (24, 25), MERS-CoV (11), HCoV-OC43 (26), and HCoV-NL63 (27), although most reports are between closely related viruses.
The Global Disease Detection Program (GDD) of the Centers for Disease Control and Prevention (CDC, Atlanta, GA) is focused on the detection of emerging infectious agents worldwide. One of the GDD projects was directed toward the detection of such potential zoonotic pathogens in African bats. Since the initial study performed during 2006 in Kenya (14, 15), an expanded surveillance of bat CoVs has been performed in the same and other countries, including Kenya, Nigeria, Democratic Republic of Georgia, Democratic Republic of Congo, Guatemala, and Peru. The project included more bat species and geographic locations, allowing a more thorough investigation of the genetic diversity and ecological dynamics of CoVs circulation in bats. In this study, we performed an ecological and evolutionary characterization of CoVs circulating in Kenya and identified distinct CoVs from Triaenops afer and Hipposideros sp. bats that are phylogenetically related to HCoV-NL63 in different parts of the genome. Based on these data, we propose a scenario for the origin and evolutionary history of HCoV-NL63 and related viruses.
RESULTS
Prevalence of CoV in Kenyan bats.
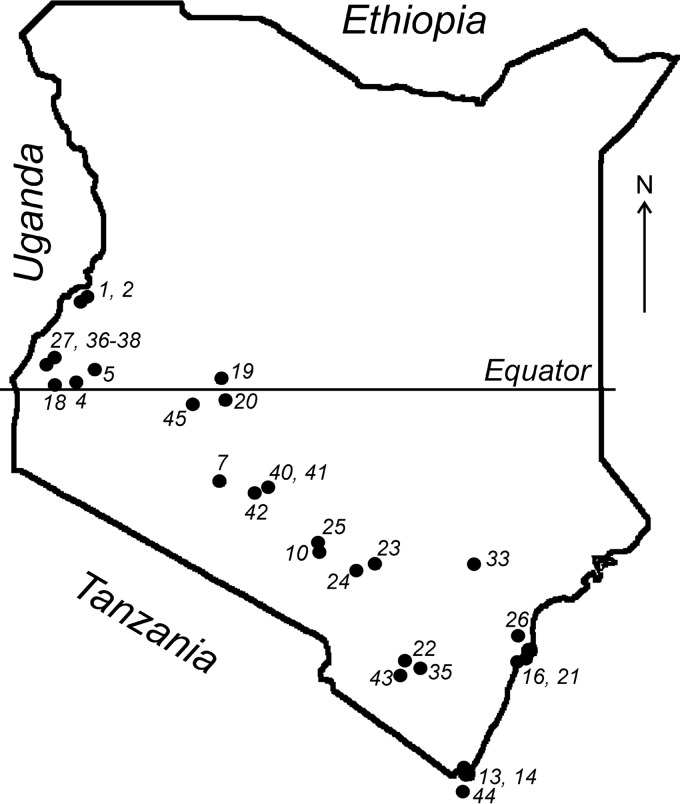
We examined bats from at least 27 species (17 genera) collected over a 4-year period (2007 to 2010) from 30 locations across the southern part of Kenya (Fig. 1). A total of 2,050 bat samples were screened for CoV RNA using a pan-coronavirus reverse transcription (RT)-PCR assay. We found an overall prevalence of 11.7% (240/2,050 bats) (see Table S1 in the supplemental material). This overall prevalence is in line with recent reports of CoVs in bats from numerous locations, including South Africa, Mexico, Philippines, Kenya, United Kingdom, Japan, Italy, and Ghana (6, 14, 15, 28–33).
FIG 1.
Map of Kenya showing the geographic locations of 30 bat collection sites.
Bats of the species tested (Chaerephon pumilus, Coleura afra, Lissonycteris angolensis, Miniopterus africanus, Neoromicia tenuipinnis, Neoromicia sp., Nycteris sp., Pipistrellus sp., and Scotoecus sp.) did not yield CoV-positive samples, although the sample number was limited and might not reflect the real prevalence (Table S1). Conversely, in bats of several other species, the CoV prevalence was high (Cardioderma cor, 25%; Eidolon helvum, 21%; Epomophorus labiatus, 28.6%; Hipposideros sp., 27.6%; Miniopterus minor, 22.6%; Otomops martiensseni, 28.6%; Rhinolophus hildebrandtii, 31.3%; Rhinolophus sp., 28.9%; Triaenops afer, 26.7%). Most species (21/27) were sampled at more than one location. Of note, we detected CoVs in 21% of E. helvum bats tested in Kenya, whereas a previous study in Ghana failed to detect any CoVs in a similar number of bats from this species (6).
Phylogenetic diversity of Kenyan bat CoVs.
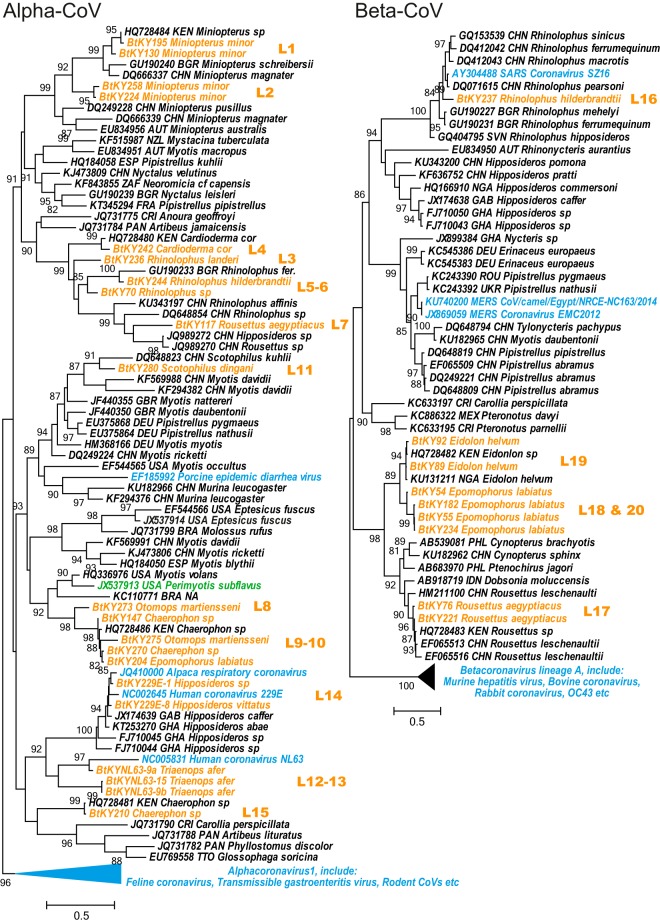
The viral sequences identified in Kenyan bats showed a remarkable diversity within both alpha- and beta-CoVs (Fig. 2). Based on our phylogenetic analysis, the CoVs newly identified here can be grouped into 20 phylogenetic lineages (Fig. 2). Many of the sampled bat genera are associated with more than one viral lineage. Furthermore, in some cases, the divergence of the CoVs within the same host genera may also be associated with possible differences in sample types. For example, we found two lineages of CoV in Rousettus aegyptiacus bats, one of which was present in oral swabs (Fig. 2, L7 Rousettus), while the other one was identified in fecal swabs (L17 Rousettus). The default tissue tropism for bat CoVs is believed to be intestinal, and samples of choice are fecal swabs. In agreement with this, only four viruses were identified from oral swab samples (L7 Rousettus), as indicated in the phylogeny (Fig. 2).
FIG 2.
Phylogeny of RdRps of all CoVs discovered in this study. The host (bat genus), number of sequences, and operational classification (lineage) are shown on the right of the tree. Branches that represent the minority host genera within the lineage defined by a single dominant host genus are indicated in red and labeled with a solid red circle. The tree is midpoint rooted for clarity only, and support values are shown only for internal branches.
Our phylogenetic analyses also revealed a number of cross-species transmission events at the genus level, many of which appeared to be transient spillovers with no evidence of onward transmission. This pattern was observed as CoV sequences recovered from bats of a particular genus located as tree tips within the phylogenetic diversity that is mainly associated with a different bat genus. From our Kenyan data set, there were seven such cross-species transmission events in total, each represented by a single sequence (indicated by red dots in Fig. 2), suggesting that these are most likely viruses with limited transmission within new hosts, although this hypothesis requires confirmation on a larger set of samples.
A more comprehensive and informative phylogeny (Fig. 3) was obtained later, including the representative global CoV sequences from GenBank, which also included the Kenyan viruses previously reported (15). The phylogeny, which included viral sequences recovered from bats of more than 50 species (30 genera), resulted in an accurate phylogenetic assignment of the viruses described in this study (Fig. 3). Importantly, the newly discovered viruses from Kenya have greatly extended our previous work (15) in terms of (i) expanding the diversity of existing lineages, including the Miniopterus-, Rhinolophus-, and Scotophilus-associated CoV clusters in the genus Alphacoronavirus and the Rousettus- and Rhinolophus-associated CoVs clusters in the genus Betacoronavirus, and (ii) the discovery of new viruses from either a novel bat host (i.e., Triaenops) or new divergent CoV clusters in known hosts (i.e., Rhinolophus, Rousettus, Chaerephon, etc.) (Fig. 3).
FIG 3.
Phylogenies of RdRp of alphacoronaviruses and betacoronaviruses. The trees are inferred using representative CoV sequences from this study as well as those obtained from GenBank. The sequences are labeled with accession number/strain name, host (species), and geographic origin (three-letter country code). Different colors are used to distinguish the following groups: orange, Kenyan bat CoVs discovered during this study; blue, CoVs identified from nonbat mammals; green, the Perimyotis subflavus virus previously reported to be related to HCoV-NL63; black, the remaining bat viruses. The lineage information for Kenyan CoVs is shown to the right of the phylogeny and matches that in Fig. 2.
The phylogeny suggests both ancient virus-host codivergence and recent cross-species transmission of CoVs between bats and other mammalian hosts. The phylogeny clearly demonstrates that CoVs from two host groups, one dominated by bats and the other exclusively by nonchiropteran mammals, formed sister clades for both alpha- and beta-CoVs (Fig. 3), suggestive of an ancient divergence between them. Conversely, several nonchiropteran CoVs are nested within the diversity of bat CoVs, suggesting that these viruses are relatively recent introductions from bats. These cross-species transmission events resulted in the emergence of severe (SARS-CoV and MERS-CoV) and mild (HCoV-NL63 and HCoV-229E) human pathogens, as well as animal pathogens (porcine epidemic diarrhea virus [PEDV] and alpaca respiratory CoV). Interestingly, HCoV-NL63, previously thought to be related to the North American tricolored bat (P. subflavus) (19), in our phylogeny is deeply nested within the newly identified CoVs from African Triaenops afer bats (Fig. 3), while the P. subflavus virus (labeled green in Fig. 3) grouped with a North American CoV sampled from a Myotis volans bat (Fig. 3). Therefore, Triaenops afer bats likely represent the most recent chiropteran reservoir host of viruses ancestral to HCoV-NL63. In addition, our results identified 16 additional 229E-like viruses (Fig. 2, L14), providing further evidence that Hipposideros bats in Africa harbor viruses that are ancestral to HCoV-229E (5, 6).
Host and spatial dynamics of bat CoVs in Kenya.
We used Mantel's test to compare the virus and host genetic distance matrices, as well as virus and geographic distance matrices. Notably, the correlation values were positive and highly significant in both comparisons (Table 1), suggesting that both host and geography have shaped the structure of virus genetic diversity. This conclusion remained following partial Mantel analyses and multiple linear regression analyses in which we tested the effect between two matrices while controlling for the third (Tables 1 and 2). Importantly, however, in both simple and partial Mantel analyses, the virus genetic distance matrices had much higher correlation with host genetic distance matrices than with geographic distance matrices (Table 1), indicating that bat CoV diversity is more structured by host genetic distance than by geographic distance.
TABLE 1.
Results of Mantel tests and partial Mantel tests comparing two factors (host genetic distance and geographic distance) that predict the structure of virus genetic diversity
| Modeld | r value for Kenyan bats (P value) |
|---|---|
| Hosta | 0.5265 (<0.0001)c |
| Host | geographyb | 0.5055 (<0.0001)c |
| Geographya | 0.2122 (<0.0001)c |
| Geography | hostb | 0.1285 (0.0005)c |
Mantel test.
Partial Mantel test.
Significant at 0.001.
Vertical lines indicate that the first factor excludes the effect of the second.
TABLE 2.
Multiple regression of virus genetic distance against host genetic distance and geographic distance in Kenyan bat CoVs (2007 to 2010)
| Variable | Correlation coefficient | P value |
|---|---|---|
| Host | 7.58E − 01 | 1.00E − 04 |
| Geography | 1.19E − 06 | 1.00E − 02 |
Next, we used Mantel autocorrelograms to examine the effect of (i) geographic distance (Fig. 4A) and (ii) host genetic distance on virus diversity (Fig. 4B). Host genetic distance decreased from highly significantly positive at short taxonomic distances to highly significantly negative at long distances. Importantly, the crossing-over point was at a host genetic distance of around 0.15 to 0.19, which marks the boundary of intra- and intergenus host diversity (Fig. 4B). However, no obvious clinal patterns in geographic distance were observed within the Kenyan data set.
FIG 4.
Mantel correlograms showing the Kenyan bat CoV RdRp sequences stratified by geographic distances (A) and host genetic distances (B). A Mantel correlation index (r) was calculated for each of the distance classes. Under the null hypothesis of no relationship between the distance matrices, r values would be close to zero. Positive r values suggest smaller genetic distances between case pairs, whereas negative r values suggest larger genetic distances between case pairs. Filled circles, r significantly different from zero; empty circles, r not significantly different from zero. The graph in panel B also shows kernel density plots for intragenus host distance density (solid light gray line) and intergenus host distance density (dotted light gray line). The corresponding y axis for the plot is shown on the right of panel B. The light gray rectangle between the two plots represents the transition area between the intragenus and intergenus host genetic distances.
Full genome characterization and recombination analyses of NL63-like and 229E-like viruses.
To further explore the evolution of the NL63-like and 229E-like viruses, we generated the complete genome sequences of five representative bat-derived CoVs: three (BtKYNL63-9a, BtKYNL63-9b, and BtKYNL63-15) were from the NL63-like group, and two (BtKY229E-1 and BtKY229E-8) were from the 229E-like group (Fig. 2, L12 to L14). For all the viruses newly described here, the genome structures follow an identical open reading frame (ORF) arrangement: ORF1ab-S-ORF4-E-M-N-ORF8 in 229E-related viruses and ORF1ab-S-ORF3-E-M-N-ORFx in NL63-related viruses (Fig. 5; Tables 3 and 4). The additional ORF8/ORFX was identified at the 3′ end of the genome in all bat NL63-like and 229E-like viruses characterized in this study, although it was missing in both human viruses (HCoV-229E and HCoV-NL63). The ORF8 in bat 229E-like genomes is named in analogy with the ORF8 of Ghanaian bat and dromedary 229E-like CoVs (5, 12). The ORF8 of BtKY229E-1 shared 60% protein identity with its closest relatives, while BtKY229E-8 has a shorter and highly divergent ORF8. The ORFx of NL63-like viruses shared very low identity (21 to 33% at the amino acid level). Similarly to the bat 229E-like CoVs recently discovered in Ghana (5), the S genes in our bat 229E-like CoVs have a considerably longer 5′ S1 portion (additional 185 amino acids) than HCoV-229E and alpaca and dromedary 229E viruses (12).
FIG 5.
Genome organization of two bat 229E-like viruses and three bat NL63-like viruses sampled from Kenyan bats. A unified length scale is used for all the genomes. Within each genome, the ORFs (arrow boxes) and ribosomal frameshift sites (vertical lines) are indicated at their corresponding positions.
TABLE 3.
Sequence comparisons of the Kenyan bat CoVs with HCoV-229E or HCoV-NL63
| HCoV sequence compared | Kenyan bat CoV | Genome identity (% nucleotide identity) | % Amino acid identity |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concatenated domains | ADRP | nsp5 | nsp12 | nsp13 | nsp14 | nsp15 | nsp16 | 1ab | S | ORF3/4 | E | M | N | |||
| HCoV-229E | BtKY229E-1 | 88 | 98 | 92 | 98 | 97 | 99 | 97 | 96 | 94 | 95 | 75 | 92 | 93 | 90 | 78 |
| BtKY229E-8 | 88 | 97 | 89 | 98 | 98 | 98 | 97 | 97 | 94 | 96 | 74 | 94 | 97 | 90 | 68 | |
| HCoV-NL63 | BtKYNL63-9a | 78 | 91 | 75 | 89 | 93 | 94 | 89 | 88 | 94 | 86 | 53 | 67 | 80 | 82 | 69 |
| BtKYNL63-9b | 68 | 83 | 51 | 76 | 88 | 91 | 82 | 81 | 84 | 72 | 52 | 55 | 64 | 61 | 51 | |
| BtKYNL63-15 | 68 | 84 | 51 | 76 | 88 | 91 | 82 | 81 | 87 | 72 | 49 | 55 | 62 | 58 | 52 | |
TABLE 4.
Genomic features of ORFs in Kenyan bat CoVs and their putative TRSsa
| Genomic feature | 229E-like virus |
NL63-like virus |
|||
|---|---|---|---|---|---|
| BtKY229E-1 | BtKY229E-8 | BtKYNL63-9a | BtKYNL63-9b | BtKYNL63-15 | |
| Sequence length (nt) | 27,837 | 27,666 | 28,363 | 28,677 | 28,479 |
| GC% | 38 | 39 | 39 | 43 | 43 |
| ORF1ab | |||||
| ORF size (nt) | 20,286 | 20,304 | 20,277 | 20,349 | 20,355 |
| Putative TRS | TCTCAACTAAAC(N219)AUG | TCTCAACTAAAC(N219)AUG | TCAACTAAAC(N214)AUG | CTCAACTAAAC(N215)AUG | TCTCAACTAAAC(N215)AUG |
| S gene | |||||
| ORF size (nt) | 4,095 | 4,095 | 4,119 | 4,122 | 4,134 |
| Putative TRS | TCTCAACTAAATAAAAUG | UCTCAACUAAA(4)AUG | TCAACTAAAC(N1)AUG | CTCAACTAAAUG | TCAACTAAAC(N1)AUG |
| ORF3/4 | |||||
| ORF size (nt) | 681 | 684 | 684 | 684 | 684 |
| Putative TRS | TCAACTAAAC(N37)AUG | TCAACTAAAC(N37)AUG | TCAACTAAAC(N37)AUG | TCAACTAAAC(N37)AUG | CAACUAAAC(N37)AUG |
| E gene | |||||
| ORF size (nt) | 234 | 234 | 234 | 234 | 234 |
| Putative TRS | TCTCAACTAAAC(N151)AUG | TCTTCAATGTAAC(N281)AUG | TTATAAC(N79)AUG | TCTGCTAAC(N151)AUG | TCTGATAAC(N151)AUG |
| M gene | |||||
| ORF size (nt) | 681 | 681 | 693 | 681 | 684 |
| Putative TRS | CTAAACTAAAC(N4)AUG | CTAAACTAAAC(N4)AUG | CTAAAC(N6)AUG | TCTAAACTAAAC(N4)AUG | UCUAAACUAAA(N4)AUG |
| N gene | |||||
| ORF size (nt) | 1,161 | 1,200 | 1,225 | 1,302 | 1,302 |
| Putative TRS | TTAATCTAAAC(N11)AUG | ATCTAAAC(N11)AUG | TCTAAACTAAAC(N3)AUG | CTAAACCAAAC(N4)AUG | UCUAAACUAAAC(N4)AUG |
| ORF8/ORFx | |||||
| ORF size (nt) | 288 | 198 | 287 | 291 | 270 |
| Putative TRS | UCAACUAAAAC(1)AUG | UCAACUAAAAC(4)AUG | CAAAACCUAAC(N12)AUG | TCAACTAAAC(N567)AUG | CAACUAAAC(N234)AUG |
TRS, transcription regulatory sequence.
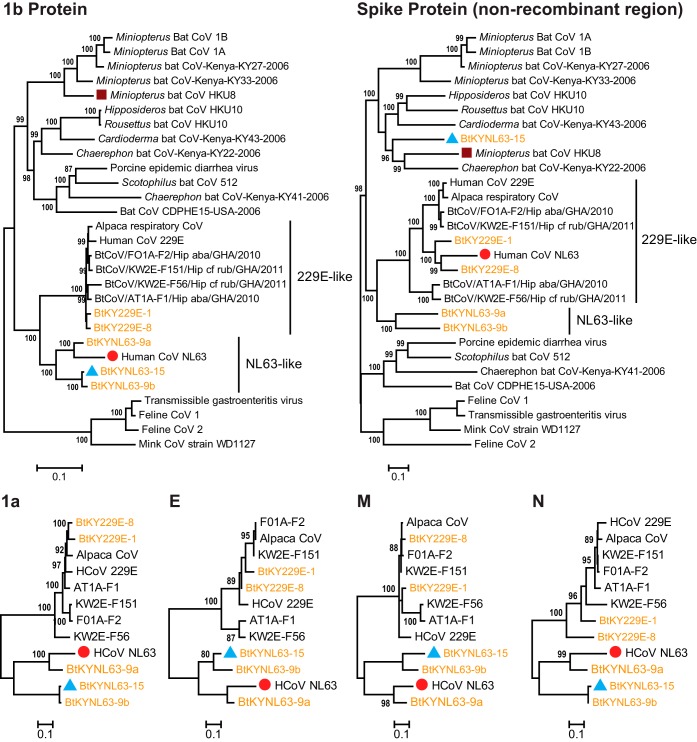
For comparison, we also included 21 genome sequences representative of the diversity in the genus Alphacoronavirus. The phylogeny based on the ORF1b protein alignment confirmed that NL63-like and 229E-like groups are monophyletic (Fig. 6). Given that each group is associated with a specific bat genus, it is likely that the ORF1b genes of the human viruses (i.e., HCoV-NL63 and HCoV-229E) were ultimately derived from Triaenops-associated CoVs and Hipposideros-associated CoVs, respectively. The relationship between Hipposideros bat CoVs and HCoV-229E was also demonstrated by Corman et al. (5) based on specimens obtained in Ghana. Compared to the viruses described in that study, the newly identified Kenyan viruses (BtKY229E-1 and BtKY229E-8) were among those more distantly related to HCoV-229E (Fig. 6 and Table 3). As for the NL63-like group, HCoV-NL63 was nested within the diversity of three lineages of Triaenops-associated CoVs, among which BtKYNL63-9a showed the closest relationship in all genome regions with the exception of the S gene (Fig. 6 and Table 3).
FIG 6.
Phylogenetic analyses of major open reading frames of NL63-like and 229E-like CoVs in the context of alphacoronaviruses revealing evidence of recombination. The names of viruses sequenced in this study are shown in orange. Three potential recombinant genomes, of HCoV-NL63, BtKYNL63-15, and HKU8, are indicated with red circles, blue triangles, and brown squares, respectively.
Strikingly, the phylogeny of the S protein suggested an entirely different evolutionary history for HCoV-NL63 compared to the rest of the genome (Fig. 6). Specifically, for all the proteins with the exception of S, HCoV-NL63 clustered with the NL63-like group. However, for the S protein, HCoV-NL63 was deeply nested within the 229E-like group, associated exclusively with viruses from Hipposideros bats, and in sequence was particularly similar to the BtKY229E-1 and BtKY229E-8 CoVs newly identified during this study (Fig. 6). Interestingly, BtKY229E-1 exhibited the closest resemblance to HCoV-NL63 in the receptor binding domain (RBD) (34), especially in the three receptor binding motifs (RBM), whereas other viruses exhibited less similarity in these regions (Fig. 7A). A phylogeny based on the RBD region confirmed our observation (Fig. 7B), although it remains uncertain whether these bat viruses utilize the same host cell receptor.
FIG 7.
Relationships between HCoV-NL63 and related viruses at the receptor binding domain. (A) Alignment of NL63-like and 229E-like viruses and related viruses at the receptor binding domain. The positions of three receptor binding motifs (RBMs) are marked with double-arrowed black lines. Residues in the NL63-CoV RBMs that directly contact the ACE2 receptor are marked with red downward arrows. (B) Phylogenetic relationships of NL63-like and 229E-like viruses at the receptor binding domain of HCoV-NL63. The tree is based on an amino acid alignment and midpoint rooted.
To further characterize this recombination event, we performed genome-scale similarity comparisons between HCoV-NL63 and related viruses (Fig. 8). The analysis confirmed the chimeric nature of the HCoV-NL63 genome, with only the spike protein gene involved in recombination via two breakpoints: one located near the 5′ end of the S gene and the other at around 200 nucleotides upstream of the 3′ end. To exclude the possibility of any artificial recombination, the breakpoint was further confirmed by RT-PCR and Sanger sequencing, using a single amplicon to cover each breakpoint. Collectively, these data show that HCoV-NL63 evolved from a recombination event between CoVs from the NL63-like and 229E-like groups.
FIG 8.
Recombination analyses of HCoV-NL63 using Simplot. Genome-scale similarity comparisons of HCoV-NL63 (query) against BtKYNL63-9a (major parental group; blue), BtKYNL63-9b (green), BtKY229E-8 (minor parental group; red), HCoV-229E (orange), BtCoV/FO1A-F2/Hip_aba/GHA/2010 (pink), and alpaca respiratory CoV (brown) were done. A full genome structure, with reference to HCoV-NL63, is shown above the similarity plot, with the positions and boundaries of the major open reading frames indicated. At the beginning of the S gene, the flat line followed by a sudden drop in similarity is due to a gap (deletion within HCoV-229E S gene) in the alignment.
In addition to HCoV-NL63, we identified a number of other recombination events between divergent CoVs involving the S gene. One example is the BtKYNL63-15 CoV newly identified here. Throughout the genome, BtKYNL63-15 showed strong similarity (79% to 99% protein identities in the ORF1ab, ORF4, and M, E, and N genes) with BtKYNL63-9b. In contrast, the genetic identity between S protein sequences of these viruses was only 53%. In the S protein phylogeny, BtKYNL63-15 did not cluster with NL63-like viruses but instead clustered with Miniopterus bat CoV HKU8 and Chaerophon bat CoV KY22 (Fig. 6). Interestingly, HKU8 itself is a recombinant in the S gene region (Fig. 6). These results suggest that the spike protein of CoVs is subject to relatively frequent recombination even between divergent viruses.
DISCUSSION
In this study, we significantly extended existing knowledge on CoV diversity, the association of CoVs with specific bat species, the relatedness between bat and human CoVs, and natural recombination events in the CoV spike (S) protein gene between viruses from different lineages.
Notably, we found that the host species poses a greater influence on CoV diversity in bats than the geographic distance, which can be explained by the ability of bats to fly (including long-distance migrations typical for some species) and disperse their pathogens over vast territories (35). A closer inspection of the Mantel correlogram suggests the presence of less-structured (homogenous; Mantel statistic r > 0) and highly structured (Mantel statistic r < 0) diversity which, strikingly, corresponds to the division between intragenus (10% ∼ 20%) and intergenus (>20%) host genetic distances (Fig. 4B). This suggests that within-genus virus transmissions occur significantly more frequently than between-genus transmissions, which is consistent with the previous observations that phylogenetic clustering is less constrained at the host species level than at the genus level (16, 36). While it is commonly accepted that host phylogeny constrains virus cross-species transmission to some extent (37), the stronger demarcation at the genus level is of particular interest. In fact, bats of different species, genera, and families frequently roost together (in caves, tree holes, and other shelters), sometimes in dense aggregations, which provides abundant opportunity for mechanical transmission of pathogens between host species. Therefore, our data suggest that distinctions between bats at the genus level might mark a threshold where the differences in cellular and immunological environments become a major challenge for a virus to switch hosts. This, in turn, will lead to the pattern of “preferential host switching” that has been observed in a number of other viruses (38).
The detection of distinctive HCoV-NL63-like and HCoV-229E-like sequences in bats sheds new light on CoV evolution. In particular, we provide strong evidence that HCoV-NL63 has a zoonotic recombinant origin. Although the majority of the HCoV-NL63 genome originates from the viruses circulating in Triaenops afer bats, its spike protein gene is derived from a 229E-like virus circulating in Hipposideros species bats. However, despite the strong signal for recombination, both putative parental strains show substantial genetic distances from human CoVs. This most likely reflects extensive postrecombination sequence divergence, which in turn suggests that the recombination event has occurred prior to the emergence of HCoV-NL63 in humans.
Most of the recombination events reported here involve breakpoints around the S gene. Indeed, similar breakpoints are also reported for SARS-CoV and SARS-like CoVs (24, 25), HCoV-OC43 (26), and a feline CoV (39), such that it is seemingly a recombination hot spot in many CoVs. It has been argued that a strong secondary structure between ORF1a and S gene may promote transcriptional pulsing, facilitating recombination (40). However, there is also evidence that this recombination hot spot does not exist under nonselective conditions (41), such that it may reflect the successful spread of beneficial recombinants rather than an elevated rate of recombination per se. This hypothesis is supported by the fact that the spike protein is intimately involved in the interaction with the host immune system.
Importantly, our results also revealed that recombination has resulted in similar S proteins in the two human viruses HCoV-NL63 and HCoV-229E, such that acquisition of a 229E-like S protein may have contributed to the emergence of NL63-like viruses in humans. However, despite this similarity of S protein sequences, these two human viruses utilize different receptors (ACE2 and aminopeptidase-N for HCoV-NL63 and HCoV-229E, respectively) to enter human cells. Within the 229E-like group, the RBD of HCoV-NL63 is more closely related to BtKY229E-8 than to HCoV-229E. The RBD of BtKY229E-8 exhibits greater similarity with that of HCoV-NL63 (Fig. 7) and is therefore more likely to be the prototype of RBD in HCoV-NL63.
Until recently, most reported recombination events in CoVs involved viruses associated with closely related host species, although recombination between highly divergent CoVs has been demonstrated experimentally (42–44). The apparent lack of interspecies recombination under natural conditions is most likely due to the insufficient collection of complete genome sequences that are truly representative of coronavirus diversity. Indeed, a number of viruses, such as HKU2, display phylogenetic incongruence across different parts of the genome (45), although the lack of one of the putative parental strains has prevented clear identification of a recombinant history.
Finally, our study provides insights into the evolutionary history of CoVs. Although it is unclear whether bats are direct ancestors of all alpha- or beta-CoVs due to the presence of nonbat CoV clades at the basal phylogenetic positions of both genera (Fig. 3), bat-borne CoVs constitute a substantial part of the diversities of alpha- or beta-CoVs. In addition, six lineages of nonbat CoVs are nested within the bat-borne clades. These likely represent independent and successful adaptations via shifts from the progenitor reservoir species (bats) to other mammals. Four well-characterized human CoVs lie within these clades. However, it is worth noting that bats may not have directly transmitted the viruses to humans. Indeed, HCoV-229E is more closely related to viruses circulating in camels than to those in bats, suggesting that camels may be intermediate hosts between bats and humans (12). Similarly, other human CoVs such as SARS-CoV and MERS-CoV all use terrestrial mammals rather than bats as intermediate hosts, which have an increased chance of contact with humans. This underlines a typical zoonotic link of bat-associated CoV to humans via terrestrial mammals.
MATERIALS AND METHODS
Sample collection.
Between 2007 and 2010, a total of 2,050 bat specimens were collected from 30 different locations in Kenya (Table S1) in collaboration with the CDC GDD regional country office in Kenya and National Museums of Kenya. The bats were captured using mist nets, hand nets, or manually. The protocol (2096FRAMULX-A3) was approved by the CDC IACUC and by Kenya Wildlife Services. Upon capture, each bat was measured, sexed, and identified to species by a trained field biologist. Subsequently, fecal and oral swabs (if possible) were collected in compliance with field protocol and were then transported on dry ice from the field to −80°C storage before further processing.
CoV RNA detection.
Each fecal and oral swab was suspended in 200 μl of a phosphate-buffered saline. Viral total nucleic acids (TNA) were extracted using the QIAamp mini viral spin kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, followed by seminested RT-PCR (SuperScript III One-Step RT-PCR kit and Platinum Taq kit; Invitrogen, San Diego, CA, USA) using primer sets designed to target the conserved genome region of alpha-, beta-, gamma-, and delta-CoVs, respectively (15). PCR products of the expected size (∼400 nucleotides) were purified by gel extraction using the QIAquick gel extraction kit (Qiagen, Valencia, CA, USA) and sequenced in both directions on an ABI Prism 3130 automated sequencer (Applied Biosystems, Foster City, CA, USA). As validation, the RT-PCR procedure was repeated for each of the CoV-positive specimens.
Bat mitochondrial gene sequencing.
Bat species were further confirmed by sequencing the host mitochondrial cytochrome b (cytB) gene in each of the CoV-positive specimens. Both the method and the primers used have been described previously, and a final 1,104-bp fragment of the cytB gene was amplified and sequenced as described previously (14, 15).
Phylogenetic analyses.
This study generated a total of 240 CoV RdRP sequences (402 bp) from Kenyan bats. These sequences were first aligned in MAFFT v7.013 (46), using amino acid sequences as a guide for the nucleotide sequence alignment. Phylogenetic trees were then inferred using the maximum likelihood (ML) method available in PhyML version 3.0 (47), assuming a general time-reversible (GTR) model with a discrete gamma distributed rate variation among sites (Γ4) and the SPR branch-swapping algorithm. To produce a more condensed data set, we clustered the highly similar sequences from the same geographic location and host species and randomly chose one or two to represent each cluster. This condensed data set was subsequently combined with 121 reference sequences representative of the genetic diversity of alpha- and beta-CoVs on a global scale taken from GenBank. ML phylogenetic trees of these final alignments were inferred using the same procedure and substitution models as described above.
Comparisons of viral genetic, geographic, and host genetic distance matrices.
To determine the relationship between viral genetic, geographic, and host genetic distances, we compiled a data set containing the Kenyan CoV samples generated in this study. The genetic distance matrices were produced from pairwise comparisons either in the form of uncorrected percentage differences or calculated from the phylogenetic trees (patristic distance) using the Patristic v1.0 program (48). The geographic distances (Euclidean distance) were calculated using the formula distance = (acos((sin(latitude1) × sin(latitude2)) + (cos(latitude1) × cos(latitude2) × cos(longitude2 − longitude1)))) × 6,371, with spatial coordinates of the samples derived from the geographic location information.
We used Mantel correlation analyses to test the extent of the correlation between these matrices (49). Both a simple Mantel's test and a partial Mantel's test were performed, and the correlation was evaluated with 10,000 permutations. To access which of the two factors—geographic or host genetic distance—best explained total variation in the virus genetic distance matrices, we performed multiple linear regression analyses on these distance matrices (50). The statistical significance of each regression was evaluated by performing 10,000 permutations. To examine whether the degree of virus genetic relatedness corresponded to the scale of geographic distance or host relatedness, we generated Mantel correlograms. In each correlogram, 10 to 12 distance classes were assigned based on an equal-frequency criterion: each class had similar numbers of pairwise comparisons. All statistical analyses were performed using the Ecodist package implemented in R 3.0.2 (51), and all statistical results were considered significant at a P of 0.05.
Full genome sequencing and sequence analyses.
Five viruses representative of the full diversity of the CoVs newly described here were selected for full genome sequencing: BtKY229E-1, BtKY229E-8, BtKYNL63-9a, BtKYNL63-9b, and BtKYNL63-15. We first sequenced a number of conserved regions throughout the genome using several seminested or nested consensus degenerate RT-PCR amplicons. These regions were then bridged using sequence-specific RT-PCR, followed by Sanger sequencing (<2 kb), or using the PacBio platform (>2 kb). The assembled consensus genome sequences from PacBio sequencing were later confirmed by sequence-specific RT-PCR and Sanger sequencing (GenBank accession numbers KY073744 to KY073748). The 5′ and 3′ genome termini were not determined due to the limited RNA remaining and were derived with PCR primers based on the conserved genome regions in alpha-CoVs.
For each complete genome sequence, potential ORF products were predicted based on the conserved core sequence, 5′-CUAAAC-3′, with a minimum length of 66 amino acids. Ribosomal frameshifts were identified based on the presence of the conserved slippery sequence, UUUAAAC. For phylogenetic analyses, the data set was first separated into six ORFs, namely, ORF1a, ORF1b, and the spike (S), envelope (E), membrane (M), and nucleoprotein (N) genes. The data set for each gene was translated into amino acid sequences and aligned using MAFFT v7.013. Phylogenetic trees were then inferred using PhyML as described above. Recombination events were first identified from the occurrence of incongruent topologies in these initial phylogenies and were then confirmed and characterized using Simplot v3.5.1 (52). In the Simplot analysis, seven sequences were analyzed, including the potential recombinant and the parental viruses, as well as an outgroup. The similarity comparisons of recombinant and other sequences were plotted using a sliding window with a size of 1,000 bp and a step size of 10 bp.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Niezgoda, Richard Franka, Amy T. Gilbert, Charles E. Rupprecht (CDC, Atlanta, GA), and Bernard Agwanda (National Museums of Kenya, Nairobi) for field sampling of bats, as well as Evelyne Mulama, Edwin Danga, Robert F. Breiman, and Joel M. Montgomery (CDC-Kenya, Nairobi) for logistical support during field expeditions.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01953-16.
REFERENCES
- 1.Weiss SR, Leibowitz JL. 2011. Coronavirus pathogenesis. Adv Virus Res 81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MJ, Lefkowitz EJ, King AM, Bamford DH, Breitbart M, Davison AJ, Ghabrial SA, Gorbalenya AE, Knowles NJ, Krell P, Lavigne R, Prangishvili D, Sanfacon H, Siddell SG, Simmonds P, Carstens EB. 2015. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2015). Arch Virol 160:1837–1850. doi: 10.1007/s00705-015-2425-z. [DOI] [PubMed] [Google Scholar]
- 3.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, da Silva Filho LV, Leroy EM, Thiel V, van der Hoek L, Poon LL, Tschapka M, Drosten C, Drexler JF. 2015. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, Muller MA, Annan A, Vallo P, Adu-Sarkodie Y, Kruppa TF, Drosten C. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis 15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EK, Lina PH, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis 19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SK, Li KS, Tsang AK, Lam CS, Ahmed S, Chen H, Chan KH, Woo PC, Yuen KY. 2013. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol 87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corman V, Ithete N, Richards L, Schoeman M, Preiser W, Drosten C, Drexler J. 2014. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ithete N, Stoffberg S, Corman V, Cottontail V, Richards L, Schoeman M, Drosten C, Drexler J, Preiser W. 2013. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis 19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, Qureshi MI, Abu-Zeid M, Zhang Y, Khiyami MA, Alharbi NS, Hajrah NH, Sabir MJ, Mutwakil MH, Kabli SA, Alsulaimany FA, Obaid AY, Zhou B, Smith DK, Holmes EC, Zhu H, Guan Y. 2016. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 12.Corman V, Eckerle I, Memish Z, Liljander A, Dijkman R, Jonsdottir H, Juma NK, Kamau E, Younan M, Al Masri M, Assiri A, Gluecks I, Musa B, Meyer B, Müller M, Hilali M, Bornstein S, Wernery U, Thiel V, Jores J, Drexler J, Drosten C. 2016. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci U S A 113:9864–9869. doi: 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, Luis AD, Peel AJ, Plowright RK, Wood JL. 2014. Bat flight and zoonotic viruses. Emerg Infect Dis 20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao Y, Tang K, Shi M, Conrardy C, Li KS, Lau SK, Anderson LJ, Tong S. 2012. Genomic characterization of seven distinct bat coronaviruses in Kenya. Virus Res 167:67–73. doi: 10.1016/j.virusres.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong S, Conrardy C, Ruone S, Kuzmin IV, Guo X, Tao Y, Niezgoda M, Haynes L, Agwanda B, Breiman RF, Anderson LJ, Rupprecht CE. 2009. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis 15:482–485. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drexler JF, Corman VM, Drosten C. 2014. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res 101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, Osterhaus AD. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A 101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fielding BC. 2011. Human coronavirus NL63: a clinically important virus? Future Microbiol 6:153–159. doi: 10.2217/fmb.10.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Rasche A, Diallo TD, Cottontail VM, Stocker A, Souza BF, Correa JI, Carneiro AJ, Franke CR, Nagy M, Metz M, Knornschild M, Kalko EK, Ghanem SJ, Morales KD, Salsamendi E, Spinola M, Herrler G, Voigt CC, Tschapka M, Drosten C, Drexler JF. 2013. Highly diversified coronaviruses in neotropical bats. J Gen Virol 94:1984–1994. doi: 10.1099/vir.0.054841-0. [DOI] [PubMed] [Google Scholar]
- 21.Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford University Press, New York, NY. [Google Scholar]
- 22.Lai MM. 1992. RNA recombination in animal and plant viruses. Microbiol Rev 56:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baric RS, Fu K, Schaad MC, Stohlman SA. 1990. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology 177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hon CC, Lam TY, Shi ZL, Drummond AJ, Yip CW, Zeng F, Lam PY, Leung FC. 2008. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J Virol 82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau SK, Li KS, Huang Y, Shek CT, Tse H, Wang M, Choi GK, Xu H, Lam CS, Guo R, Chan KH, Zheng BJ, Woo PC, Yuen KY. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol 84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau SK, Lee P, Tsang AK, Yip CC, Tse H, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. 2011. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol 85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyrc K, Dijkman R, Deng L, Jebbink MF, Ross HA, Berkhout B, van der Hoek L. 2006. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anthony SJ, Ojeda-Flores R, Rico-Chavez O, Navarrete-Macias I, Zambrana-Torrelio CM, Rostal MK, Epstein JH, Tipps T, Liang E, Sanchez-Leon M, Sotomayor-Bonilla J, Aguirre AA, Avila-Flores R, Medellin RA, Goldstein T, Suzan G, Daszak P, Lipkin WI. 2013. Coronaviruses in bats from Mexico. J Gen Virol 94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.August TA, Mathews F, Nunn MA. 2012. Alphacoronavirus detected in bats in the United Kingdom. Vector Borne Zoonotic Dis 12:530–533. doi: 10.1089/vbz.2011.0829. [DOI] [PubMed] [Google Scholar]
- 30.Balboni A, Palladini A, Bogliani G, Battilani M. 2011. Detection of a virus related to betacoronaviruses in Italian greater horseshoe bats. Epidemiol Infect 139:216–219. doi: 10.1017/S0950268810001147. [DOI] [PubMed] [Google Scholar]
- 31.Geldenhuys M, Weyer J, Nel LH, Markotter W. 2013. Coronaviruses in South African bats. Vector Borne Zoonotic Dis 13:516–519. doi: 10.1089/vbz.2012.1101. [DOI] [PubMed] [Google Scholar]
- 32.Shirato K, Maeda K, Tsuda S, Suzuki K, Watanabe S, Shimoda H, Ueda N, Iha K, Taniguchi S, Kyuwa S, Endoh D, Matsuyama S, Kurane I, Saijo M, Morikawa S, Yoshikawa Y, Akashi H, Mizutani T. 2012. Detection of bat coronaviruses from Miniopterus fuliginosus in Japan. Virus Genes 44:40–44. doi: 10.1007/s11262-011-0661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuda S, Watanabe S, Masangkay JS, Mizutani T, Alviola P, Ueda N, Iha K, Taniguchi S, Fujii H, Kato K, Horimoto T, Kyuwa S, Yoshikawa Y, Akashi H. 2012. Genomic and serological detection of bat coronavirus from bats in the Philippines. Arch Virol 157:2349–2355. doi: 10.1007/s00705-012-1410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A 106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peel AJ, Sargan DR, Baker KS, Hayman DT, Barr JA, Crameri G, Suu-Ire R, Broder CC, Lembo T, Wang LF, Fooks AR, Rossiter SJ, Wood JL, Cunningham AA. 2013. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat Commun 4:2770. doi: 10.1038/ncomms3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Muller MA, Deng H, Herrler G, Drosten C. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol 84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. 2010. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 38.Charleston MA, Robertson DL. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst Biol 51:528–535. doi: 10.1080/10635150290069940. [DOI] [PubMed] [Google Scholar]
- 39.Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol 72:4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills DR, Dobkin C, Kramer FR. 1978. Template-determined, variable rate of RNA chain elongation. Cell 15:541–550. doi: 10.1016/0092-8674(78)90022-3. [DOI] [PubMed] [Google Scholar]
- 41.Banner LR, Lai MM. 1991. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology 185:441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baric RS, Yount B, Hensley L, Peel SA, Chen W. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J Virol 71:1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, Denison MR. 2008. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters PS, Rottier PJ. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr Top Microbiol Immunol 287:133–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau SK, Woo PC, Li KS, Huang Y, Wang M, Lam CS, Xu H, Guo R, Chan KH, Zheng BJ, Yuen KY. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 48.Fourment M, Gibbs M. 2006. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol 6:1–5. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220. [PubMed] [Google Scholar]
- 50.Lichstein JW. 2007. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol 188:117–131. doi: 10.1007/s11258-006-9126-3. [DOI] [Google Scholar]
- 51.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Software 22:1–19. [Google Scholar]
- 52.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.