ABSTRACT
Latent membrane protein 1 (LMP1) is an Epstein-Barr virus (EBV)-encoded oncoprotein that is packaged into small extracellular vesicles (EVs) called exosomes. Trafficking of LMP1 into multivesicular bodies (MVBs) alters the content and function of exosomes. LMP1-modified exosomes enhance the growth, migration, and invasion of malignant cells, demonstrating the capacity to manipulate the tumor microenvironment and enhance the progression of EBV-associated cancers. Despite the growing evidence surrounding the significance of LMP1-modified exosomes in cancer, very little is understood about the mechanisms that orchestrate LMP1 incorporation into these vesicles. Recently, LMP1 was shown to be copurified with CD63, a conserved tetraspanin protein enriched in late endosomal and lysosomal compartments. Here, we demonstrate the importance of CD63 presence for exosomal packaging of LMP1. Nanoparticle tracking analysis and gradient purification revealed an increase in extracellular vesicle secretion and exosomal proteins following LMP1 expression. Immunoisolation of CD63-positive exosomes exhibited accumulation of LMP1 in this vesicle population. Functionally, CRISPR/Cas9 knockout of CD63 resulted in a reduction of LMP1-induced particle secretion. Furthermore, LMP1 packaging was severely impaired in CD63 knockout cells, concomitant with a disruption in the perinuclear localization of LMP1. Importantly, LMP1 trafficking to lipid rafts and activation of NF-κB and PI3K/Akt pathways remained intact following CD63 knockout, while mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) and noncanonical NF-κB activation were observed to be increased. These results suggest that CD63 is a critical player in LMP1 exosomal trafficking and LMP1-mediated enhancement of exosome production and may play further roles in limiting downstream LMP1 signaling.
IMPORTANCE EBV is a ubiquitous gamma herpesvirus linked to malignancies such as nasopharyngeal carcinoma, Burkitt's lymphoma, and Hodgkin's lymphoma. In the context of cancer, EBV hijacks the exosomal pathway to modulate cell-to-cell signaling by secreting viral components such as an oncoprotein, LMP1, into host cell membrane-bound EVs. Trafficking of LMP1 into exosomes is associated with increased oncogenicity of these secreted vesicles. However, we have only a limited understanding of the mechanisms surrounding exosomal cargo packaging, including viral proteins. Here, we describe a role of LMP1 in EV production that requires CD63 and provide an extensive demonstration of CD63-mediated exosomal LMP1 release that is distinct from lipid raft trafficking. Finally, we present further evidence of the role of CD63 in limiting LMP1-induced noncanonical NF-κB and ERK activation. Our findings have implications for future investigations of physiological and pathological mechanisms of exosome biogenesis, protein trafficking, and signal transduction, especially in viral-associated tumorigenesis.
KEYWORDS: Epstein-Barr virus, cell signaling, exosomes, extracellular vesicles, human herpesviruses
INTRODUCTION
Epstein-Barr virus (EBV) is a human herpesvirus (HHV-4) that infects the vast majority of the world's population (1). In healthy individuals, EBV infections can range from asymptomatic to a mild, self-limited illness. However, in immunocompromised individuals, a population that has grown in size due to modern medical treatment and the AIDS epidemic, latent infection can contribute to the development of numerous lymphoid or epithelioid malignancies (2, 3). It is also likely that genetic and environmental factors contribute to virus-associated tumorigenesis, as the prevalence of EBV-associated cancers differs by geographical region (4–6). EBV-associated cancers include, but may not be limited to, gastric carcinoma, Burkitt's lymphoma, Hodgkin's lymphoma, and nasopharyngeal carcinoma. One viral protein, latent membrane protein 1 (LMP1), likely serves as the main oncogene product in virally infected cells. LMP1 is a six-pass transmembrane protein that is detected in most EBV-associated cancers and has the ability to transform cells in vitro through the activation of intracellular signaling pathways (7–9). When present in the host cell, LMP1 acts as a mimic of CD40, a tumor necrosis factor receptor (TNFR) (8, 10), activating NF-κB, mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK), phosphatidylinositol 3-kinase (PI3K)/Akt, and c-Jun N-terminal kinase (JNK) pathways. The activation of these pathways results in upregulation of multiple genes involved with regulation of apoptosis, cell cycle progression, cell proliferation, migration, and invasion (8, 11–16). Notably, LMP1 can signal in the absence of a ligand (17) through recruitment of TNFR-associated factors (TRAFs) to interaction sites at C-terminal activation region (CTAR) domains (18, 19). Localization of LMP1 to perinuclear regions of the cell is believed to be necessary to mediate these signaling functions, independent of the transmembrane protein aggregation on the plasma membrane (20).
LMP1 has also been demonstrated to localize to internal Golgi and multivesicular body (MVB) compartments and is packaged into exosomes for release from the cell (21). Exosomes are a population of small (40 to 150 nm) endocytically derived extracellular vesicles. Extracellular vesicles (EVs) broadly encompass a variety of vesicle populations, including exosomes, microvesicles, apoptotic bodies, and viral particles (22–25). These vesicle populations reflect a diversity of sizes, densities, and intracellular origins of EVs. While microvesicles are considered larger EVs shed directly from the plasma membrane into the extracellular milieu, exosomes are produced from inward budding events on the limiting membrane of late endosomal organelles, forming intraluminal vesicles in MVBs. Similar to mechanisms of egress used by viral particles, MVBs can fuse with the plasma membrane to release exosomes into the extracellular space (22, 24). Functionally, exosomes have been revealed to play a role in cell-to-cell communication and modulation of immune responses (26–29). Thus, it is likely that packaging of LMP1 into these vesicles mediates a number of functions, including facilitation of viral replication, immunosuppression, the establishment of latency, and promotion of cell growth.
Exosomal trafficking of LMP1 has been linked to both the intra- and intercellular signaling capabilities of the viral protein. For instance, blockage of exosomal LMP1 secretion has been demonstrated to lead to downstream intracellular NF-κB overstimulation within the cell, as measured by luciferase reporter assay (30). Additionally, transfer of LMP1-containing exosomes induces the activation of PI3K/Akt and MAPK/ERK signaling pathways in naive recipient cells (31). This evidence suggests that secretion of LMP1 into exosomes plays a role in cell-to-cell communication in the context of viral infection and may contribute to the pathogenesis of EBV-associated diseases. It is increasingly evident that viruses such as EBV can hijack and utilize the host cell exosome pathway to modulate cell-to-cell signaling in the context of cancer. Expression of LMP1 has been shown to modify the protein cargo of vesicles released from cells (31–33), and in turn, LMP1-modified exosomes can increase the growth, migration, and invasion of malignant cells (34–36). Thus, in the context of EBV-associated cancers, the properties of LMP1-containing vesicles likely alter the tumor microenvironment and contribute to cancer progression.
Despite the clear biomedical significance of exosomal packaging of the viral oncoprotein, the mechanisms of LMP1 trafficking into exosomes remain obscure. Recently, LMP1 has been shown to coimmunoprecipitate with CD63, a tetraspanin protein enriched in late endosomal and lysosomal compartments (30). CD63 is also commonly enriched in the membranes of intraluminal vesicles which are secreted as exosomes from cells. We recently demonstrated that small-vesicle secretion was reduced following CD63 knockout, suggesting that CD63 plays a major role in the exosomal pathway (37). Here we demonstrate the impact of LMP1 expression on extracellular vesicle secretion and that CD63 is critical for the trafficking of LMP1 into secreted vesicles.
RESULTS
LMP1 enhances secretion of small extracellular vesicles.
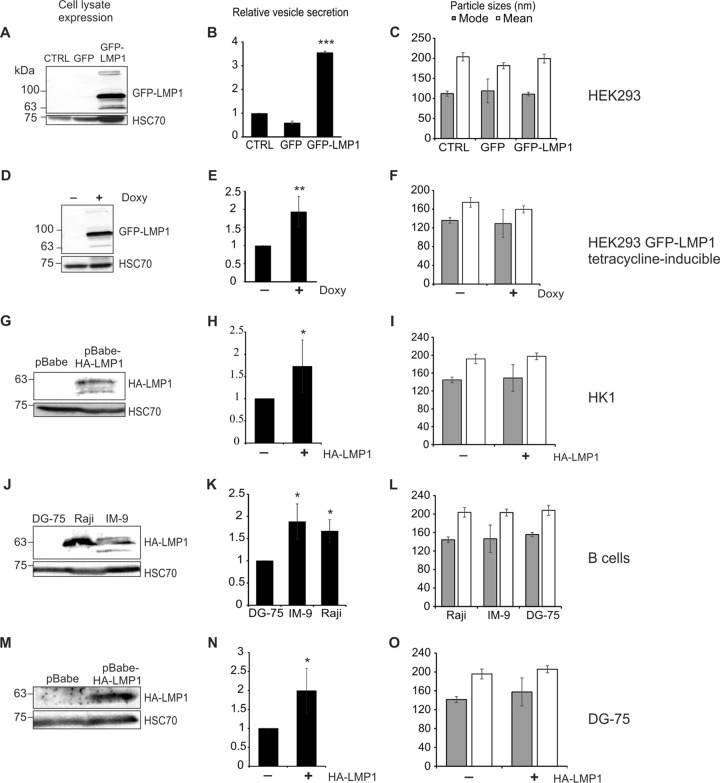
Epstein-Barr virus LMP1 is efficiently sorted into exosomes and has been suggested to modify the content of vesicles released from infected cells (21, 23, 31) In fact, early research describing LMP1-mediated protein trafficking alluded to a potential effect of the viral protein on exosome secretion quantity (38). To determine the direct impact of LMP1 expression on the quantity of vesicles released, we harvested EVs from five models of LMP1-negative and -positive cells for nanoparticle tracking analysis (NTA). In a transfected model, a green fluorescent protein (GFP) control or GFP-tagged LMP1 vector was introduced into HEK293 cells (Fig. 1A). Twenty-four hours posttransfection, EV secretion was found to be markedly increased in cells containing LMP1 (P < 0.001) (Fig. 1B). A second model using a GFP-tagged LMP1 tetracycline-inducible system was constructed in a HEK293 cell line, and cells were treated with doxycycline to activate LMP1 expression (Fig. 1D). EV quantity was similarly increased following viral protein induction (P = 0.001) (Fig. 1E). To investigate vesicle secretion in the context of lower stable expression of LMP1, vector control (pBabe) or hemagglutinin (HA)-tagged LMP1 retroviral vectors were used to transduce HK1 cells, an EBV-negative nasopharyngeal carcinoma cell line (Fig. 1G). LMP1 expression was detectable but notably lower than in the transfected or inducible models. Previous reports have shown that this level of LMP1 expression is comparable to those found in EBV-infected cell lines (39). Again, nanoparticle tracking analysis revealed an increase in vesicle secretion compared to that with control (empty pBabe vector) cells (P = 0.026) (Fig. 1H). To test the effect of LMP1 on vesicle quantity in the context of EBV infection, EV secretion was measured across an EBV-negative B cell lymphoma line (DG-75) and compared to those in EBV-positive B cell lines expressing LMP1 (IM-9 and Raji) (Fig. 1J). EV secretion was observed to be greater in Raji (P = 0.011) and IM-9 (P = 0.018) cell lines than in uninfected DG-75 cells (Fig. 1K). Because the observed differences in vesicle secretion could be due to other cellular or viral products besides LMP1 expressed in Raji or IM-9 cells, we generated DG-75 cells stably expressing LMP1 or the vector control (Fig. 1M). Introduction of LMP1 into DG-75 cells resulted in an increase in vesicle secretion (P = 0.043) (Fig. 1N), consistent with the previous cell models in this study. Particles tracked in all models revealed 100- to 200-nm mode and mean sizes, consistent with extracellular vesicle sizes (Fig. 1C, F, I, L, and O). Altogether, these data reveal that LMP1 enhances EV secretion in a variety of cellular models relevant to EBV pathogenesis.
FIG 1.
LMP1 expression increases extracellular-vesicle secretion. EVs were harvested from cell-conditioned media by the ExtraPEG method and quantitatively compared by nanoparticle tracking analysis. Immunoblot analyses of equal protein demonstrate detectable levels of cellular LMP1 compared to that in control cells in each model. Increased 100- to 200-nm particles were secreted from cells transfected with GFP-LMP1 compared to untransfected HEK293 cells or GFP-transfected cells (P < 0.001) (A to C), following induction of GFP-LMP1 expression by doxycycline in HEK293 GFP-LMP1 tetracycline-inducible cells (P = 0.001) (D to F), from cells with stable expression of pBabe-HA-LMP1 in HK1 cells compared to empty pBabe vector expression (P = 0.026) (G to I), from B cells expressing LMP1 (Raji and IM-9) compared to LMP1-negative DG-75 cells (P = 0.011 and P = 0.018) (J to L), and from DG-75 cells stably expressing the pBabe-HA-LMP1 compared to empty pBabe vector (P = 0.04) (M to O). Loading control HSC70 was probed on blots of cell lysates. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
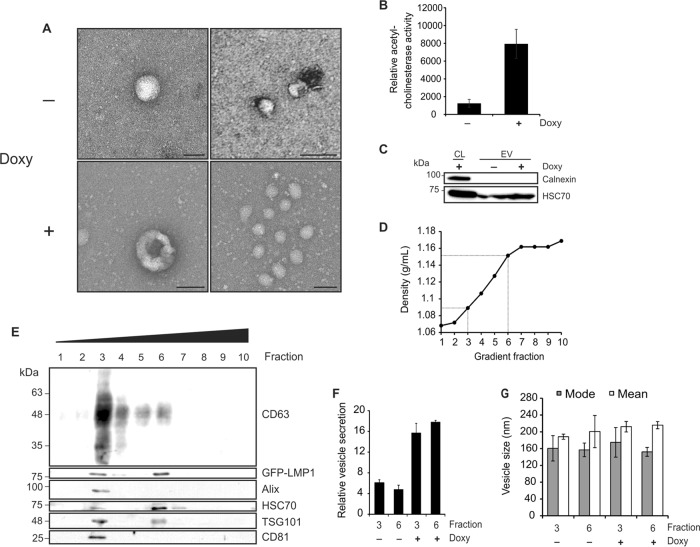
Electron microscopy of EVs harvested from HEK293 GFP-LMP1-inducible cells additionally demonstrated the presence of small EVs in enrichments (Fig. 2A). In general, vesicles ranged in size from less than 100 nm to 250 nm, supporting a mixture of EV populations harvested. Clusters of 100-nm vesicles were observed frequently in EVs induced by LMP1 expression, suggesting that particle increases seen by nanoparticle tracking analysis likely reflect an increase in small-EV secretion. Moreover, small EVs are often enriched in acetylcholinesterase, and increasing enzymatic activity can be correlated with EV number (40). Esterase activities in vesicles harvested from HEK293 GFP-LMP1-inducible cells were compared (Fig. 2B). Over a 5-fold increase in enzyme activity was observed in LMP1-induced EVs, further quantifying an increase in vesicle secretion. Of note, acetylcholinesterase has been shown to be increased in the cytoplasm of apoptotic cells and present in apoptotic bodies (41). To ensure that cellular debris and apoptotic bodies were not found in our vesicle preparations, the absence of an endoplasmic reticulum protein, calnexin, was confirmed (Fig. 2C).
FIG 2.
Exosome production is enhanced by LMP1 expression. (A) EVs harvested by ExtraPEG from HEK293 GFP-LMP1-inducible cells were examined by electron microscopy. Scale bar = 200 nm. Doxy, doxycycline. (B) Relative levels of acetylcholinesterase activity in EV samples, measured by EXOCET assay. (C) Immunoblot analysis showing the absence of calnexin protein in vesicle isolates, equal masses loaded. CL, cell lysate. (D) Densities of fractions following iodixanol gradient purification of EVs. Dotted lines denote fractions 3 and 6, where vesicular markers were abundant. (E) Immunoblots of iodixanol density gradient fractions demonstrating the presence of two distinct populations of EVs corresponding to densities of 1.089 (fraction 3) and 1.151 (fraction 6), equal volumes loaded. (F and G) Nanoparticle tracking analysis of the quantity (F) and size (G) of EVs in fractions 3 and 6 following LMP1 induction in HEK293 cells.
To further examine the enhanced secretion of small EVs, vesicles were purified on an iodixanol density gradient to separate small-EV populations, according to the method of Kowal et al. (42). Two major populations of vesicles were observed in fractions of the gradient, corresponding to densities of 1.089 and 1.151 g/ml (Fig. 2D). Vesicle populations floating to the third fraction of this gradient and enriched in CD63, CD81, and TSG101 have been reported to represent “bona fide exosomes” (42), while higher fractions may represent vesicles of plasma membrane origin. Here, we confirm the presence of this exosomal population positive for protein markers CD63, Alix, HSC70, CD81, and TSG101 (Fig. 2E). Furthermore, we demonstrate the coaccumulation of LMP1 in these exosomal vesicles. LMP1 has been previously detected in various populations of extracellular vesicles, including exosomes, as well as larger shed microvesicles (43, 44). Our data here suggest that LMP1 is present in both exosomes (fraction three) and denser EVs not enriched in Alix and CD81 (fraction six). Nanoparticle tracking of fractions three and six revealed both populations of vesicles to be increased following LMP1 expression (Fig. 2F and G). These findings suggest that LMP1 expression increases the number of exosomes secreted from HEK293 cells and may additionally enhance the production of other EVs that warrant future characterization.
LMP1 increases CD63-positive vesicle populations.
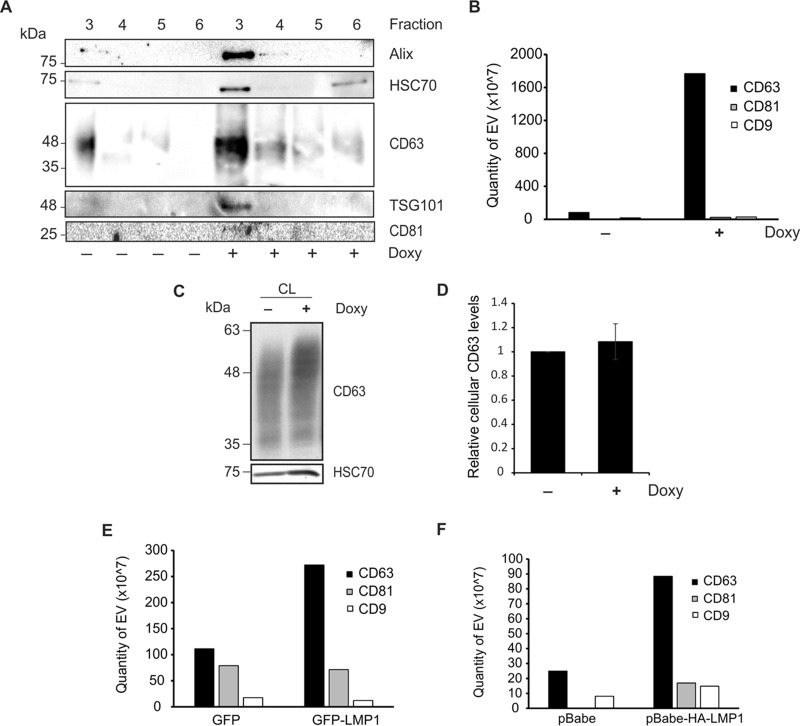
To confirm the presence of increased levels of exosomal proteins in vesicles following LMP1 induction, density gradient purified fractions 3 to 6 were compared between control and LMP1-expressing EVs (Fig. 3A). Increases in exosomal markers Alix, HSC70, CD63, and TSG101 were observed following LMP1 induction. Elevation of CD81 levels in LMP1-expressing exosomes was detectable, though not dramatic, by immunoblot analysis. LMP1 has been previously shown to associate with tetraspanin proteins for endosomal trafficking and exosome secretion (30). To more closely examine the subpopulation of exosomes increased by LMP1 expression, protein levels of common vesicular tetraspanins CD63, CD81, and CD9 were compared between control and LMP1-expressing cells by enzyme-linked immunosorbent assay (ELISA) (Fig. 3B). EVs were enriched from untreated or doxycycline-treated HEK293 GFP-LMP1 tetracycline-inducible cells. Levels of CD81- and CD9-positive vesicles were relatively unchanged between EV samples. However, levels of CD63-positive vesicles were dramatically increased following LMP1 expression, suggesting that CD63-containing vesicles account for the majority of EV secretion induced by the viral protein LMP1. Interestingly, cellular levels of CD63 were not significantly increased in LMP1-expressing cells (Fig. 3C and D), suggesting that the increase seen in CD63 secretion may be explained by more efficient sorting into the exosome pathway. To confirm the CD63-specific increase observed, EVs from GFP- or GFP-LMP1-transfected HEK293 cells (Fig. 3E) and HK1 pBabe control or pBabe-HA-LMP1-expressing cells (Fig. 3F) were analyzed by ELISA. Again, vesicles containing tetraspanin proteins CD81 and CD9 were not significantly varied in number, while levels of CD63-positive vesicles were increased in LMP1-expressing EV samples.
FIG 3.
LMP1 enhances secretion of CD63-positive vesicle subpopulations. (A) Comparison of equal volumes of gradient fractions 3 to 6 following LMP1 expression in HEK293 cells, demonstrating increased exosomal protein markers in fraction 3. (B) Equal volume of vesicle samples from HEK293 GFP-LMP1-inducible cells enriched by the ExtraPEG method were quantified by ExoELISA to measure differences in tetraspanin protein secretion. (C) Equal masses (30 μg) of cell lysate from HEK293 GFP-LMP1-inducible cells (untreated and doxycycline treated) were probed for cellular CD63. (D) Quantitation of cellular CD63 levels from three independent experiments relative to those in control cells. (E and F) Tetraspanin ExoELISAs of EVs derived from HEK293 cells transfected with GFP or GFP-LMP1 (E) and HK1 cells stably expressing a pBabe control vector or pBabe-HA-LMP1 (F).
LMP1 accumulates in CD63-positive vesicles.
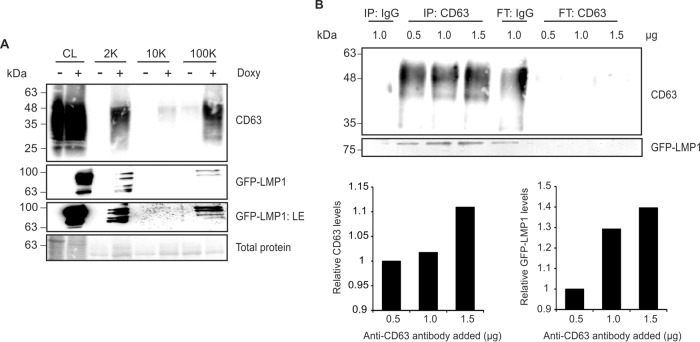
As LMP1 has been previously demonstrated to associate with CD63 within tetraspanin-enriched microdomains and in endosomal multivesicular bodies (30), we aimed to investigate whether LMP1 was present in the CD63-positive population of EVs increased following viral protein expression. Vesicles were harvested from untreated or doxycycline-treated HEK293 GFP-LMP1 tetracycline-inducible cells and separated by density using differential centrifugation. In cells treated with doxycycline, LMP1 was induced in cell lysates and found predominately in the fractions from the centrifugations at 2,000 and 100,000 × g (2K and 100K fractions), largely corresponding to the vesicle populations enriched in CD63 (Fig. 4A). Upon a longer exposure, low levels of LMP1 were detectable in the 10K fraction (GFP-LMP1:LE), consistent with small amounts of CD63 protein found in this vesicle population. As previously seen (Fig. 3), levels of CD63 in small EVs (100K pellet) were markedly augmented following LMP1 induction, along with low levels of detectable CD63 present in the 10K pellet. Notably, previous evidence has similarly demonstrated the presence of CD63 in larger vesicles isolated by a low-speed (2,000 × g) centrifugation (42). These data suggest that LMP1 accumulates in small exosome-sized vesicles and may be additionally secreted in association with CD63 in larger EV populations.
FIG 4.
LMP1 is present in small CD63-positive extracellular vesicles. (A) EVs were harvested from untreated or doxycycline-treated HEK293 GFP-LMP1 tetracycline-inducible cells by differential centrifugation. Pellets from cell lysates (equal masses) and the centrifugation steps at 2,000 (2K), 10,000 (10K), and 100,000 (100K) × g (equal volumes) were compared by immunoblot analysis for relative CD63 and LMP1 levels in each fraction. LE, immunoblot with longer exposure. (B) Immunoprecipitation was performed using equal volumes of EVs from doxycycline-treated LMP1-inducible cells by incubation with 1.0 μg of a control mouse IgG antibody or increasing amounts (0.5, 1.0, or 1.5 μg) of anti-CD63 antibody. Vesicles pulled down (IP) were compared with residual flowthrough (FT) material by loading equal volume into a gel for immunoblot analysis. Levels of CD63 and LMP1 in IP pulldowns from increasing amount of anti-CD63 antibody were quantitated.
If LMP1 is present in CD63-positive vesicles, then vesicles immunoisolated with CD63-specific antibodies should contain LMP1. To test this, vesicles were harvested from cells induced with doxycycline and were recovered by addition of anti-CD63 antibody. Bound EV populations (IP) were compared to the unbound flowthrough (FT) material (Fig. 4B). Increasing amounts of anti-CD63 antibody (0.5, 1.0, and 1.5 μg) effectively captured CD63-positive vesicles in cumulative levels. Trace amounts of CD63 were consistently pulled down with IgG antibody, indicating unavoidable nonspecificity of vesicles containing CD63 binding to the protein G beads. However, CD63 was largely present in the FT material of the IgG control. Immunoblot analysis further revealed LMP1 to be present in the CD63 pulldowns in increasing levels corresponding to accumulating CD63. LMP1 was abundant in the IgG flowthrough, though a small amount was also detected in the IgG IP, likely due to nonspecific pulldown of CD63-positive vesicles. Altogether, these data suggest that LMP1 is packaged into CD63-positive vesicles, consistent with the subpopulation of EVs increased by LMP1 expression.
LMP1-induced vesicle secretion and packaging into exosomes requires CD63.
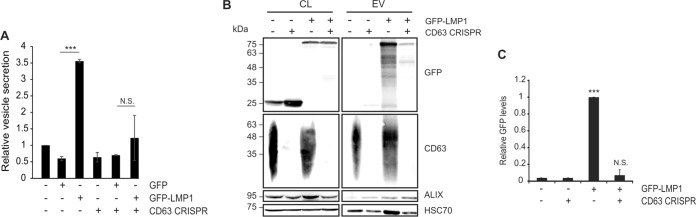
As LMP1 accumulates in vesicles containing CD63 and enhances the secretion of this population of vesicles when expressed in cells, we sought to examine the role of CD63 in exosomal LMP1 trafficking. LMP1 has been found to be copurified with CD63 under gentle cell lysis conditions that maintain tetraspanin-enriched microdomains (TEMs) (30). In the same study, short hairpin RNA (shRNA) knockdown of CD63 resulted in a decrease in exosomal LMP1 levels. However, a reduction in other exosomal protein markers was also observed, suggesting that the decrease in exosomal LMP1 may be primarily due to a global abatement in vesicle secretion. More recently, a reduction in small EV secretion following CD63 knockout was verified (37). Bearing in mind these results, we directly compared vesicle secretion from cells expressing LMP1 following complete CD63 knockout. For these experiments, HEK293 cells expressing a CRISPR/Cas9 construct targeting CD63 were used. Endogenous levels of CD63 protein were undetectable in knockout cells. Control HEK293 or CD63 knockout cells were transfected with a GFP or GFP-tagged LMP1 vector. Nanoparticle tracking analysis again revealed an increase in EV secretion following LMP1 expression in control cells (P < 0.001) that was not observed in CD63 knockout cells (P = 0.38) (Fig. 5A), suggesting that LMP1-induced vesicle secretion is dependent upon the presence of CD63.
FIG 5.
LMP1-induced EV secretion and trafficking into exosomes is CD63 dependent. (A) EVs were harvested from HEK293 control and CD63 CRISPR knockout cells transiently transfected with GFP or GFP-LMP1 and analyzed by nanoparticle tracking. Expression of LMP1 resulted in an increase in EV secretion (P < 0.001) that was abrogated by CD63 knockout (P = 0.38). (B) Immunoblot analyses of GFP- or GFP-LMP1-transfected cell lysates and EVs demonstrate reduced packaging of LMP1 into vesicles following CD63 knockout. EV markers ALIX and HSC70 were used as loading controls. (C) Normalized quantification of immunoblots from three independent experiments was calculated as protein quantity [(LMP1EV/HSC70EV)/(LMP1cell/HSC70cell)]. Statistical significance compared to values with control cells transfected with GFP is indicated as follows: ***, P < 0.001; N.S., not significant (P = 0.38).
Vesicle lysates were also examined by immunoblot analysis to evaluate LMP1 packaging into vesicles following CD63 knockout (Fig. 5B). Control and CD63 knockout cells transfected with the GFP-LMP1 construct showed similar levels of cellular GFP-LMP1, and GFP-LMP1 was successfully packaged in EVs enriched from control cells. However, in CD63 knockout cells, LMP1 packaging into EVs was reduced to the basal levels of GFP packaging. Consistent with evidence demonstrating an overall reduction in vesicle secretion, HSC70 levels in CD63 knockout EVs were also reduced compared to those in controls. To account for differences in vesicle secretion, levels of GFP-LMP1 were compared by normalization to HSC70 expression in lysates (Fig. 5C). Quantification of three independent experiments revealed a >90% decrease in LMP1 packaging in the absence of CD63. Altogether, these data demonstrate a CD63-dependent mechanism of viral LMP1 packaging into extracellular vesicles.
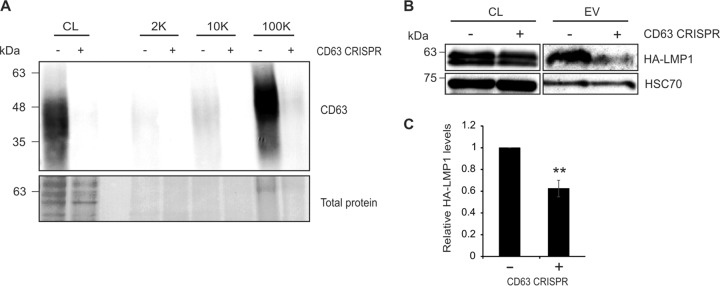
Extracellular vesicle LMP1 packaging is reduced after CD63 knockout in NPC cells.
Nasopharyngeal carcinoma (NPC) is an epithelial cancer that is almost exclusively associated with latent EBV infection. To test whether a CD63-dependent mechanism of vesicular LMP1 trafficking was present in NPC cells, endogenous CD63 was knocked out in an EBV-infected HK1 cell line. Again, vesicle lysates from 2K, 10K, and 100K fractions of control and knockout cells were compared. Immunoblot analysis revealed CD63 to be enriched in the 100K fraction of control cells (Fig. 6A), while levels were nearly undetectable in vesicles derived from cells expressing CRISPR/Cas9 directed against CD63. As the low levels of viral LMP1 were undetectable in the EBV-infected cells by immunoblotting, cells were transfected with an HA-tagged LMP1 construct to determine the efficiency of exosomal packaging in the absence of CD63. Introduced HA-LMP1 levels were similarly expressed in CD63 knockout cells, but LMP1 packaging into vesicles pelleted following a spin at 100,000 × g was significantly decreased (Fig. 6B). Quantification of three independent experiments exhibited a 40% reduction in exosomal LMP1 secretion (P = 0.001) (Fig. 6C), supporting a role for CD63 in LMP1 trafficking into vesicles originating from NPC cells.
FIG 6.
Extracellular vesicle packaging of LMP1 in nasopharyngeal carcinoma cells is mediated by CD63. (A) Cells (equal masses loaded) and vesicle lysates (2K, 10K, and 100K; equal volumes loaded) from EBV-infected HK1 or EBV-infected HK1 CD63 CRISPR cell lines were analyzed by immunoblotting for CD63 levels. Control cells exhibited enrichment of CD63 predominately in the 100K fraction. (B) Small EVs (100K fraction) were isolated from EBV-infected HK1 or EBV-infected HK1 CD63 CRISPR cells by differential centrifugation and analyzed by immunoblotting for LMP1 and EV marker HSC70. (C) Quantification of HA-LMP1 EV levels from three independent experiments. HA-LMP1 packaging into small EVs (100K fraction) was reduced following CD63 knockout (P = 0.001).
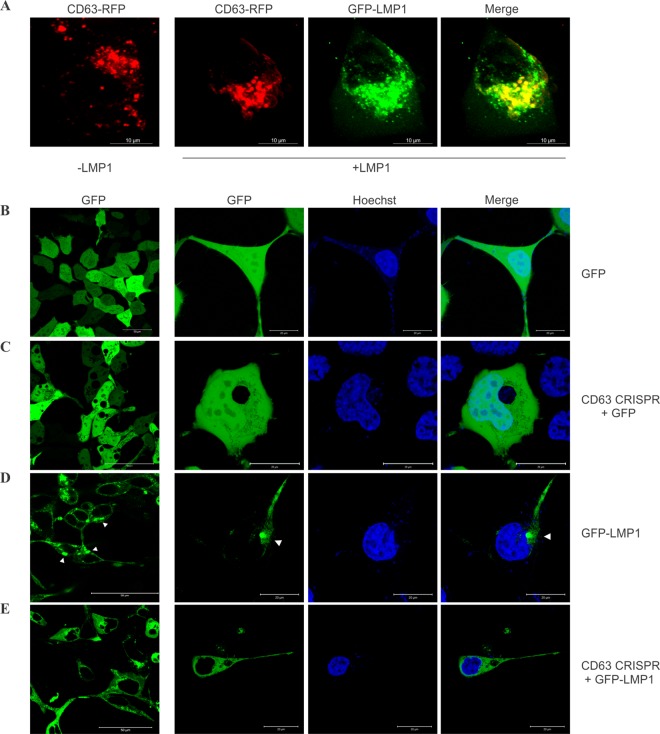
Internal perinuclear accumulation of LMP1 is disturbed following CD63 knockout.
LMP1 has been shown to colocalize with endogenous CD63 in perinuclear compartments and on the plasma membrane (30). Here, we similarly demonstrate the perinuclear colocalization of CD63 and LMP1 in HEK293 cells (Fig. 7A). As LMP1 trafficking into extracellular vesicles was disturbed by knockout of CD63 protein (Fig. 5B and C and 6B and C), we hypothesized that cellular localization of LMP1 may be altered in the absence of CD63. HEK293 cells expressing GFP or GFP-tagged LMP1 were examined by live-cell confocal microscopy following Hoechst 33342 staining to visualize nuclei. Control cells expressing GFP-LMP1 displayed a distinct punctate perinuclear signal (Fig. 7D) compared to the diffuse fluorescent signal seen in cells expressing a GFP control construct (Fig. 7B). CD63 knockout cells transfected with GFP similarly showed a diffuse pattern (Fig. 7C). However, introduction of GFP-tagged LMP1 into CD63 knockout cells displayed fluorescence that lacked the strong punctate subcellular signal seen in control cells (Fig. 7E). Based on this evidence, we conclude that LMP1 is dependent upon CD63 for proper perinuclear accumulation.
FIG 7.
Perinuclear localization of LMP1 requires CD63. (A) HEK293 cells were transfected with CD63-RFP alone or with GFP-LMP1 for live-cell confocal imaging. Scale bar = 10 μm. (B to E) HEK293 control (B and D) or CD63 CRISPR (C and E) cells were transfected with GFP (B and C) or GFP-LMP1 (D and E) and imaged by confocal microscopy. The prominent perinuclear localization of LMP1 (arrowheads) was interrupted following CD63 CRISPR knockout. The leftmost column depicts multiple cell images at a magnification of ×60. Scale bar = 50 μm. The rest of the columns show zoomed-in representative cells. Scale bar = 20 μm.
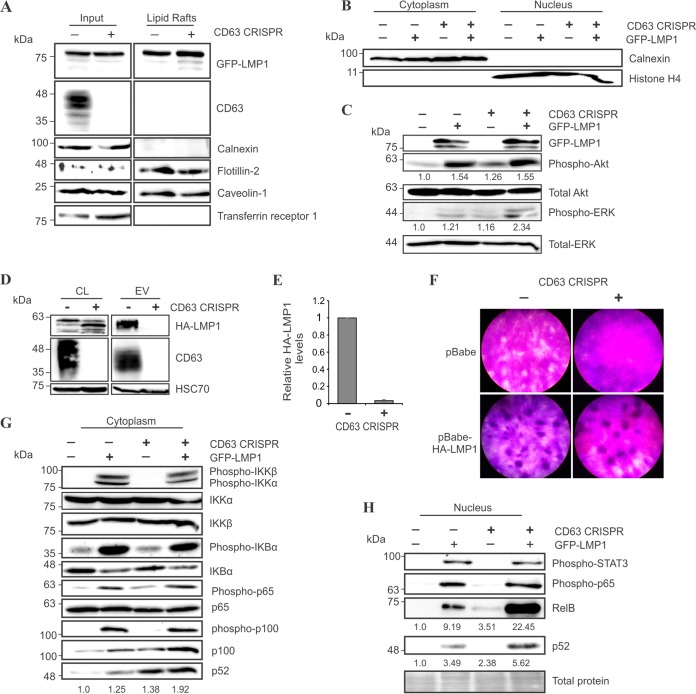
Intracellular LMP1 trafficking to lipid rafts does not require CD63.
Based on the diffuse localization of LMP1 following CD63 knockout, we aimed to investigate whether LMP1 trafficking to lipid rafts was intact. LMP1 has been previously demonstrated to accumulate in lipid raft microdomains, membrane subcompartments that are largely enriched in sphingolipids and cholesterol and serve as organizing sites for selective lipid and protein interactions (45). Localization of LMP1 to lipid rafts is believed to mediate its downstream NF-κB, PI3K/Akt, and MAPK/ERK signaling targets (32, 46). Based on their detergent-resistant biochemical properties, lipid rafts were isolated from control or CD63 knockout HEK293 cells following introduction of LMP1 (Fig. 8A). In both cell lines, raft isolates were enriched in lipid raft scaffolding proteins flotillin-2 and caveolin-1, as previously described (47). Cytoplasmic calnexin and non-lipid raft marker transferrin receptor (48) were not detected in the isolated rafts. Surprisingly, LMP1 localization to lipid rafts remained intact following CD63 knockout. These findings may be explained by our observation that CD63 was not enriched in Triton X-100-insoluble lipid raft isolates. Together, our data suggest that CD63 does not aid in LMP1 lipid raft localization and further propose that raft and exosome trafficking are distinct processes. This is supported by data describing efficient exosome packaging of an LMP1 mutant that fails to localize to lipid rafts (30).
FIG 8.
LMP1 trafficking to lipid rafts and downstream signaling do not require CD63. (A) HEK293 control and CD63 knockout cells were transfected with GFP-LMP1. Lipid rafts were biochemically isolated for immunoblot analysis of whole-cell (input) and lipid raft-associated proteins, equal masses loaded. (B) Cytoplasmic and nuclear fractions of HEK293 control and CD63 CRISPR cells transfected with GFP or GFP-LMP1 were separated and confirmed by enrichment of cytoplasmic calnexin or nuclear histone H4 protein, equal masses loaded. (C) Akt and ERK activation in cytoplasmic fractions of HEK293 and CD63 CRISPR cells following GFP-LMP1 transfection was measured, equal masses loaded. Relative levels of phospho-Akt and phospho-ERK were averaged over three independent experiments. (D) HA-LMP1 packaging in EVs (equal volumes) from Rat1 cells stably expressing a pBabe-HA-LMP1 vector following CD63 knockout. (E) Quantitation of HA-LMP1 packaging in Rat1 EVs from three independent experiments. (F) Focus formation assay was performed using Rat1 control or CD63 knockout cells transduced with an empty pBabe vector or pBabe-HA-LMP1. (G and H) Immunoblot analysis of LMP1-induced NF-κB signaling activation in cytoplasmic (G) and nuclear (H) fractions of HEK293 control and CD63 CRISPR cells, equal masses loaded. The results shown are representative of findings from multiple experiments.
CD63 limits LMP1 activation of noncanonical NF-κB signaling.
Since LMP1 signaling is thought to occur in lipid raft microdomains, we next hypothesized that LMP1-mediated signaling may similarly be unimpaired in the absence of CD63. To further assess downstream LMP1 signaling efficacy, cytoplasmic and nuclear compartments of the cell were isolated (Fig. 8B). The C-terminal activation region 1 (CTAR1) domain of LMP1 has been demonstrated to activate the PI3K/Akt and MAPK/ERK pathways necessary for transformation of rodent fibroblasts through recruitment of TRAF proteins to lipid rafts (32, 49–51). In this study, we observed detectable increases in cytoplasmic Akt and ERK activation, independent of CD63 presence within HEK293 cells (Fig. 8C). In fact, ERK activation appeared to be slightly elevated in the absence of CD63. As LMP1 transforming properties have been well characterized in Rat1 fibroblasts (52), we confirmed a similar reduction of LMP1 packaging into EVs from Rat1 fibroblasts following CD63 knockout (Fig. 8D and E). A reproducible increase in LMP1 breakdown was observed in Rat1 cell lysates in the absence of CD63-mediated vesicle secretion, possibly due to altered protein trafficking into lysosomes. To determine whether LMP1 could transform rat fibroblasts in the absence of CD63-mediated LMP1 vesicle secretion, a focus formation assay was performed using Rat1 control or CD63 knockout cells. Crystal violet staining revealed foci present in both cell lines following LMP1 introduction (Fig. 8F). Together, these findings suggest that CD63 is not necessary for LMP1-mediated Akt and ERK activation, likely due to the maintained ability of LMP1 to traffic to lipid raft domains. This evidence also implies that exosomal packaging of LMP1 is not necessary for rat fibroblast transformation.
Previously, CD63 knockdown has been demonstrated to increase NF-κB induction by LMP1 (30), as measured by a luciferase reporter assay. However, LMP1 activation of NF-κB is complex. The CTAR2 domain of LMP1 can activate canonical NF-κB signaling, while CTAR1 mediates NF-κB activation through multiple routes, including canonical, noncanonical, and atypical pathways (19, 53–55). One disadvantage of using a reporter assay to examine activation of NF-κB is that this method does not discriminate between converging signaling pathways upstream of the reporter readout. In this study, we thoroughly examined LMP1-induced activation of canonical and noncanonical NF-κB pathways in the absence of CD63. In general, NF-κB is activated by release of cytoplasmic inhibitory proteins that allows nuclear translocation of NF-κB family proteins. Through recruitment of TRAF proteins, LMP1 can canonically activate the IKK complex, resulting in phosphorylation of IκB kinase α (IKKα) and IKKβ, followed by phosphorylation of IKBα, which is targeted for proteasomal degradation (54, 56, 57). Following release from IKBα, activated NF-κB p65 protein can translocate into the nucleus for transcriptional regulation. Activation of the noncanonical pathway requires IKKα phosphorylation of p100, which is cleaved into p52 and forms dimers with NF-κB RelB for nuclear translocation (58). In this study, we observed LMP1-induced activation of the canonical NF-κB pathway to be unaffected following CD63 knockout (Fig. 8G). Interestingly, activation of noncanonical pathway proteins p100/p52 appeared increased in the absence of CD63. In nuclear fractions, phospho-STAT3 and NF-κB p65 were similarly increased by LMP1 in control and CD63 knockout cells (Fig. 8H). However, again, we observed increased levels of noncanonical NF-κB proteins RelB and p52. These data support the previous observation by Verweij et al. that NF-κB activation was increased following knockdown of CD63 (30) and further support a specific effect on the noncanonical pathway. Together, our findings suggest that a CD63-mediated mechanism of LMP1 exosomal sorting may be linked to the noncanonical NF-κB signaling activation induced by the viral protein.
DISCUSSION
In this study, we examined the impact of EBV oncoprotein LMP1 on extracellular vesicle secretion using a number of novel techniques. A myriad of evidence has demonstrated the role of EVs in manipulation of the tumor microenvironment, including altering the immune response and enhancing tumor growth, invasion, and metastasis (59, 60). Furthermore, recent clinical studies have found an increase in circulating EVs in late-stage cancer patients corresponding to disease progression (61–63). In the context of EBV-associated cancer, LMP1 has been shown to modify the content of exosomes emitted from infected cells, resulting in an upregulation of signal transduction molecules released that may have a role in invasive and metastatic processes of neoplastic cells (31–33, 64). Here, we demonstrate that expression of LMP1 in cells directly results in an augmentation of EV secretion. In cell lines expressing high levels of LMP1 (transiently transfected GFP-LMP1 or tetracycline-induced GFP-LMP1), vesicle secretion was observed to be increased by 2- to 4-fold compared to that in control cells. An HK1 cell line stably expressing lower levels of LMP1 also demonstrated an increase in vesicle secretion, though less dramatically than cell lines expressing high levels of LMP1. Furthermore, naturally infected B cells (Raji and IM-9) showed greater EV secretion than did EBV-negative DG-75 B cells, and introduction of LMP1 into DG-75 cells resulted in a corresponding increase in vesicle production. One explanation for the increase in vesicle secretion seen may be that EVs released from virally infected cells can function to enhance naive B cell growth and differentiation toward a memory B cell or dampen the immune response toward the virus (31, 33, 34). Combined with our knowledge of the role of EVs and LMP1 in cancer progression, these findings lead us to conclude that LMP1-modified EVs act as a mechanism of cell-to-cell communication that may be instrumental to virus-associated cancer growth and progression. Enhancing the production of specific EV subpopulations and the selective sorting of EV cargo may represent important mechanisms of LMP1 oncogenesis.
To gain insights into these mechanisms, we further interrogated the identity of the subpopulations of extracellular vesicles secreted following LMP1 expression. We found that LMP1 in part accumulated in a vesicle population consistent with the density and protein markers of exosomes, and we confirmed the presence of LMP1 in CD63-containing vesicles by immunoisolation of this vesicle subpopulation. We also found tetraspanin CD63 to be the major vesicular tetraspanin protein increased in these EVs, concurrent with prior evidence demonstrating complex formation between viral LMP1 and host cell CD63 (30). Strikingly, cellular levels of CD63 were not dramatically altered, supporting an increased efficiency of CD63 sorting into exosomes, possibly deviating from a lysosomal degradation pathway. LMP1 may also upregulate CD63 expression, accounting for the elevated levels detected outside the cell. In work published by Verweij et al. (30), intracellular CD63 levels were found to be decreased following LMP1 expression in an inducible BJAB cell model. The authors proposed a more efficient sorting process of CD63 into exosomes for secretion as a possible explanation for this finding. Our data demonstrated a dramatic and specific increase in vesicular levels of CD63, supporting this theory. However, in this study, we did not measure cellular CD63 levels to be decreased in HEK293 cells following LMP1 expression.
Verweij et al. have shown that exosomal secretion of LMP1 is inversely linked to downstream signaling activation of the viral protein through pathways such as NF-κB, and they hypothesized that CD63 may limit LMP1 signaling through chaperoning the viral protein into late endosomes for secretion via exosomes (30). Here, we directly demonstrate that packaging of LMP1 into exosomes is nearly abrogated following CD63 protein depletion in HEK293 and Rat1 cells and markedly reduced in EBV-infected nasopharyngeal carcinoma epithelial cells. Differences in LMP1 packaging between different cell lines may reflect diverse mechanisms of EV biogenesis, a large topic which certainly warrants future study.
In addition to reduced exosomal packaging, LMP1-induced activation of the noncanonical NF-κB pathway is enhanced in the absence of CD63. It is clear that cellular levels of LMP1 must be balanced to prevent NF-κB overstimulation and potential transformation of cells, as dysregulation of the noncanonical NF-κB pathway is associated with malignant lymphomagenesis (58). These data solicit a larger picture surrounding the circumstances of exosomal LMP1 secretion in the context of EBV infection. It is possible that host cell CD63 is recruited by the virus to chaperone LMP1 into the late endosomal pathway for secretion. Secretion of LMP1, and other viral components, allows the virus to escape proteasomal and lysosomal degradative pathways, as supported by our evidence showing increased LMP1 breakdown by Rat1 cells following CD63 knockout. Packaging into secreted membranous sacs likely facilitates cell-to-cell transmission of LMP1 and other viral factors and may serve to evade the immune system by cloaking viral components within host cell vesicles, as previously suggested (65, 66). Moreover, high levels of LMP1 are detrimental within the host cell (67, 68), and LMP1 has demonstrated roles in the regulation of autophagy and apoptosis (69, 70). Indeed, our results showing increased CD63 and LMP1 protein in larger vesicles (pelleted at 2,000 × g) likely indicate autophagic or apoptotic pathway activation, as large membrane-bound vesicles are often secreted from autophagic cells (71). Rapid turnover of LMP1 has been suggested to regulate the signaling activity within the host cell (72, 73). LMP1 secretion via exosomes may therefore serve to limit injurious downstream signaling by decreasing LMP1 levels within the cell, thus supporting intracellular virus survival.
On the other hand, interaction of CD63 with LMP1 may serve as a cellular defense mechanism to limit uncontrollable and cytotoxic intracellular signaling activation in the context of abundant LMP1. Host cell recognition of the foreign viral protein may result in dispatchment of CD63 to the viral oncoprotein to reduce intracellular levels of LMP1 by packaging it into exosomes for secretion. Exosomal secretion may therefore serve as a host cell mechanism to prevent constitutive activation of LMP1-activated signaling pathways, and CD63 specifically may be a cellular response to limit overstimulation of LMP1 oncogenic signaling. One benefit to the host of LMP1 secretion into exosomes rather than intracellular degradation could be priming noninfected immune cells for LMP1 antigen recognition (74).
Overall, it is difficult to tease apart the impetus for LMP1 trafficking into exosomes via a CD63-dependent mechanism. It is quite possible that both competing viral offense and host cell defensive mechanisms drive the interaction between the viral oncoprotein and host cell tetraspanin protein. Regardless of whether the virus or the host is driving LMP1 secretion from the cell, it is clear that LMP1-modified exosomes can impact the surrounding tumor microenvironment and contribute to viral pathogenesis in the context of EBV-associated cancers. Future research is necessary to further elucidate the mechanism of CD63-mediated trafficking of LMP1 into small EVs and the impact on both viral pathogenesis and host immunity.
Taken together, the findings presented here implicate a direct link between LMP1 expression and oncogenic vesicle release in the context of viral-associated tumorigenesis. Surprisingly, we observed that LMP1 trafficked to Triton X-100-insoluble lipid raft microdomains independent of CD63, suggesting that lipid raft compartmentalization does not mediate LMP1 secretion into exosomes. In light of our evidence showing increased noncanonical NF-κB activity in the absence of CD63-mediated exosomal secretion, we propose that exosomal sorting of LMP1 is independent of lipid raft localization and may instead represent a competing trafficking mechanism within the cell. Previous evidence has suggested that LMP1 and CD63 associate in tetraspanin-enriched microdomains (TEMs), membrane subcompartments that are enriched in EVs and biochemically susceptible to Triton X-100 detergent (75). The TEM and EV proteomes largely overlap, and trafficking to TEMs may be in part necessary for exosomal sorting (76). We thereby propose that LMP1 may modify CD63-containing TEMs to enhance vesicle production and alter their resulting cargo. We believe that LMP1 serves as an excellent model for understanding more general physiological and pathological mechanisms of exosome subpopulation formation and content selection. Future elucidation of the protein interaction network involved in LMP1- and CD63-dependent exosome trafficking will provide a greater understanding of these cellular mechanisms and offer clear therapeutic targets to combat EBV-associated cancers.
MATERIALS AND METHODS
Retrovirus production.
Retrovirus particles for transduction and stable cell generation were produced in HEK293T cells following JetPrime transfection of expression plasmids (pBabe neo, pBabe-HA-LMP1 neo, pBabe puro, or pBabe-HA-LMP1 puro) and packaging plasmids pMD2.G (Addgene; number 12259; a gift from Didier Trono) and PSPAX2 (Addgene; number 12260; a gift from Didier Trono) according to the manufacturer's instructions (Polyplus). Medium was collected at 48, 72, and 96 h posttransfection, centrifuged for 10 min at 1,000 × g, filtered through a 0.45-μm filter, and frozen at −80°C until use.
Cell culture.
HEK293 and Rat1 cells (gifts from Nancy Raab-Traub, University of North Carolina) were cultured in Dulbecco modified Eagle medium (DMEM; Lonza; 12-604Q) and HK1 (a gift from George Tsao, Hong Kong University), and B cell lines DG-75 (ATCC) and Raji and IM-9 (gifts from Nancy Raab-Traub) were grown in RPMI 1640 (Lonza; 12-702Q). Medium was supplemented with 10% fetal bovine serum (FBS; Seradigm; 1400-500), 2 mM l-glutamine (Corning; 25-005-CI), 100 IU of penicillin-streptomycin (Corning; 30-002-CI), and 100 μg/ml:0.25 μg/ml antibiotic/antimycotic (Corning; 30-002-CI). Serum used for experiments was depleted of extracellular vesicles by ultracentrifugation at 100,000 × g for 20 h and filtered through a 0.2-μm filter prior to being added to medium. For nanoparticle tracking experiments, live cells were counted with an automated cell counter (Cellometer Vision, software version 2.1.4.2; Nexcelom Biosciences) at the time of harvest by staining with 0.2% trypan blue (Sigma; T8154) in phosphate-buffered saline (PBS).
Stable HK1 cell lines expressing an empty pBabe vector or pBabe-HA-LMP1 were created by retrovirus transduction in the presence of 10 μg/ml of Polybrene (Sigma; H9268) as previously described (49, 77). Stable cell lines were selected and maintained in 2 μg/ml of puromycin (Amresco; 58-58-2)-containing medium. EBV-infected HK1 cells (a gift from George Tsao, Hong Kong University) were maintained in 1 mg/ml of G418 sulfate (Corning; 30-234-CI), further transduced with CD63 lentiCRISPRv2 plasmid as previously described (37), and then selected in 2 μg/ml of puromycin. Stable DG-75 and Rat1 cells expressing an empty pBabe vector or pBabe-HA-LMP1 were created by retrovirus transduction and maintained in 1 mg/ml of G418 sulfate as described above. Rat1 cells were further transduced with the CD63 lentiCRISPRv2 plasmid that was mutated to contain Rattus norvegicus-specific seed sequence and selected in puromycin.
Transfection.
The GFP-LMP1 plasmid was constructed by PCR amplification of LMP1 from pBabe-HA-LMP1 using primers containing HindIII and EcoRI restriction sites. The LMP1 PCR product was cloned into entry vector pENTR4-GFP-C1 (Addgene; number 17396; a gift from Eric Campeau) following ligation of DNA products digested with HindIII and EcoRI restriction enzymes (New England BioLabs [NEB]). The resulting sequenced plasmid was recombined into pQCXP CMV/TO destination vector (Addgene; number 17386; a gift from Eric Campeau) by LR recombination using Gateway LR Clonase II (Invitrogen; number 11791-020). The GFP vector was constructed by LR recombination of pENTR4-GFP-C1 with the pQCXP destination vector. The RFP-CD63 plasmid was constructed by PCR amplification of CD63 from pCT-CD63-GFP using primers containing EcoRI and BamHI cut sites. The CD63 PCR product was cloned into pENTRDsRedEx2 (a gift from Nathan Lawson; Addgene; plasmid number 22451). The resulting sequenced plasmid was recombined into pQCXP CMV/TO destination vector by LR recombination using Gateway LR Clonase II according to the manufacturer's instructions (Invitrogen). The pCDNA3.1/Zeo and pCDNA3.1/Zeo HA-LMP1 constructs were provided by Nancy Raab-Traub (University of North Carolina). HEK293 cells were transfected with plasmids using JetPRIME transfection reagent (number 114-15) according to the manufacturer's instructions (Polyplus).
Focus formation assay.
To assess LMP1-mediated transformation, Rat1 control and CD63 knockout cells were seeded in 6-well plates (0.3 × 106 cells per dish). Cells were transduced with empty pBabe or pBabe-HA-LMP1 retroviral particles with the addition of Polybrene (10-μg/ml final concentration) 24 h after seeding. Viral medium was aspirated 24 h later, and cells were grown in complete DMEM for 2 weeks. Finally, foci were stained according to the method of Alvarez et al. (78). Briefly, medium was aspirated and plates were washed twice with cold PBS before fixation with cold methanol for 10 min on ice. Methanol was removed, 0.5% crystal violet in 25% methanol was added for 5 min to stain foci, and then plates were rinsed with Milli-Q water and dried for imaging.
Generation of GFP-LMP1-inducible cells.
Cells stably expressing GFP-LMP1 under the control of a tetracycline-inducible promoter were created by first transducing HEK293 cells with lentivirus particles containing pLenti CMV TetR BLAST (Addgene; number 17492). Stable cells were selected with medium containing 10 μg/ml of blasticidin (Invivogen; ant-bl-1) and then transduced with retrovirus particles containing pQCXP GFP-LMP1. Doubly stable cells were selected with medium supplemented with blasticidin (10 μg/ml) and puromycin (2 μg/ml) for 2 weeks. LMP1 expression was induced for 24 h with the addition of doxycycline (Sigma; D3447) to a final concentration of 1 μg/ml.
Extracellular-vesicle enrichment.
EV samples for nanoparticle tracking analysis, ELISA quantification, acetylcholinesterase measurement, and immunoprecipitation assays were enriched using the ExtraPEG method, as previously described (79). EVs harvested using this method have been thoroughly characterized by immunoblotting, electron microscopy, NTA, mass spectrometry, and RNA profiling (37, 79). Briefly, cell-conditioned medium was centrifuged at 500 × g for 5 min to remove cells, followed by 2,000 × g for 30 min to remove large cellular debris in an Eppendorf 5804R using an S-4-104 rotor. A 1:1 volume of 16% (2×) polyethylene glycol (average Mn, 6000; Alfa Aesar; 25322-68-3) and 1 M sodium chloride was added to samples and incubated overnight at 4°C. Following the incubation, samples were centrifuged at 3,214 × g for 1 h (maximum speed in an S-4-104 rotor) and washed by ultracentrifugation at 100,000 × g for 70 min in a Beckman Optima MAX-E tabletop ultracentrifuge using a TLA120.2 rotor. Samples were resuspended in particle-free PBS for NTA, EXOCET, and IP analyses or in System Biosciences binding buffer (see below) for ELISA quantification.
To examine the presence of LMP1 in extracellular vesicles of various sizes, a differential centrifugation method of EV enrichment was employed. Cell-conditioned medium was centrifuged at 500 × g for 5 min (twice) and 2,000 × g for 10 min (twice) using an S-4-104 rotor, 10,000 × g for 30 min (twice) in an FA-45-630 rotor, and 100,000 × g for 70 min in a Beckman Optima XL using an SW41 Ti rotor, followed by a final ultracentrifugation wash for 70 min at 100,000 × g in a Beckman Optima MAX-E using a TLA120.2 rotor. EVs pelleted following the centrifugations at 2,000, 10,000, and 100,000 × g were lysed in strong lysis buffer (5% SDS, 10 mM EDTA, 120 mM Tris-HCl [pH 6.8], 8 M urea) or directly in 2× nonreducing Laemmli sample buffer (4% SDS, 100 mM Tris [pH 6.8], 0.4 mg/ml of bromophenol blue, 20% glycerol) for immunoblot analysis. All centrifugations were performed at 4°C.
Iodixanol density gradient.
EVs from HEK293 GFP-LMP1-inducible cells harvested by the ExtraPEG method were further purified on a density gradient, according to the method of Kowal et al. (42). Briefly, EV pellets following the ultracentrifugation step were resuspended in 1.5 ml of 0.25 M sucrose buffer (10 mM Tris [pH 7.4]). A 60% stock (wt/vol) Optiprep (Sigma, D1556) solution was added 1:1 to EV suspensions and transferred to an MLS-50 rotor tube (Beckman; 344057). Iodixanol stock was then diluted in 0.25 M sucrose-Tris buffer to make 10 and 20% iodixanol solutions, and then 1.3 ml of 20% iodixanol and 1.2 ml of 10% iodixanol solutions were carefully layered on top of EV suspensions. Gradients were centrifuged for 90 min at maximum MLS-50 rotor speed (268,000 × g) and separated by collection of 490-μl fractions from the top of the gradient. Densities were measured with a refractometer (Refracto 30PX). Individual fractions were washed with PBS and repelleted by ultracentrifugation in an SW41 Ti rotor at 100,000 × g for 2 h. Pellets were resuspended in particle-free PBS for NTA or strong urea-containing lysis buffer (see above) for immunoblot analysis.
Extraction of cytoplasmic and nuclear proteins.
Cells were grown in serum-free medium, transfected with GFP or GFP-LMP1 at 90% confluence, then washed with cold PBS after 24 h, and scraped and pelleted by centrifugation at 1,000 × g for fractionation as described previously (80). Briefly, cell pellets were lysed in a hypotonic buffer (20 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA) for 15 min on ice with the addition of protease and phosphatase inhibitor cocktails (Thermo) and 0.4 mM sodium orthovanadate. Samples were vortexed after adding NP-40 to a final concentration of 1% and then centrifuged at 1,000 × g for 10 min to pellet nuclei. The cytoplasmic supernatant was further solubilized by adding SDS to a final concentration of 1%. Crude nuclear pellets were resuspended in 1 ml of homogenization buffer (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tricine-KOH [pH 7.8]) and then mixed 1:1 with a 50% iodixanol (Optiprep) solution in a diluent buffer (150 mM KCl, 30 mM MgCl2, 120 mM Tricine-KOH [pH 7.8]). For preparation of crude nuclei, 1 ml of 35% iodixanol–diluent buffer was pipetted into the bottom of a 5-ml clear ultracentrifuge tube, followed by layering of 2 ml of 30% iodixanol. The 2 ml of nuclei in 25% iodixanol was carefully layered on top. Gradients were centrifuged for 35 min at 10,000 × g in an MLS-50 rotor with minimum acceleration/deceleration. Nuclear bands were collected at the interface of 30 to 35% iodixanol, washed in 4 ml of PBS, and repelleted at 10,000 × g for 10 min. Purified nuclear fractions were lysed in strong urea-containing lysis buffer (see above) with 2.5% β-mercaptothanol (BME) and then sonicated using a Bioruptor (Diagenode) for 5 cycles of 30 s on/off before immunoblot analysis.
Immunoblot analysis.
To prepare cell lysates, cells were washed and then scraped into cold PBS and collected by centrifugation at 1,000 × g for lysis in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl, 50 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholate). EV samples were harvested as described above. Cell and EV lysates probed for CD63 were run under nonreducing conditions. To prepare all cell lysates, and EV lysates run under reducing conditions for SDS-PAGE, additional sample buffer (5×) also containing 0.2 M dithiothreitol (DTT) and 2% BME was added to samples. Lysates were boiled for 5 min before an equal mass of cell lysate or equal volume of EV lysate was run in an SDS 10% polyacrylamide gel and subsequently transferred onto a nitrocellulose membrane (GE Healthcare). Total protein was measured by Ponceau S stain. Blots were blocked with 5% (wt/vol) nonfat dry milk powder in Tris-buffered saline with Tween 20 (TBS-T). Blots were probed with primary antibodies against the following: Alix (Q-19; Santa Cruz Biotechnology), HSC70 (B-6; Santa Cruz), TSG101 (C-2; Santa Cruz), calnexin (11397; Santa Cruz), caveolin-1 (D46G3; Cell Signaling), flotillin-2 (H-90; Santa Cruz), CD63 (TS63; Abcam), GFP (600-101-215; Rockland), HA (C29F4; Cell Signaling), LMP1 (CS1-4; Dako), histone H4 (81, 82), IKKα (11930; Cell Signaling), IKKβ (8943; Cell Signaling), phospho-IKKα/β (2697; Cell Signaling), IκBα (4814; Cell Signaling), phospho-IκBα (2859; Cell Signaling), NF-κB p65 (8242; Cell Signaling), phospho-NF-κB p65 (3033; Cell Signaling), RelB (4922; Cell Signaling), NF-κB p100/p52 (3017; Cell Signaling), phospho-NF-κB p100/p52 (4810; Cell Signaling), ERK 2 (C-14; Santa Cruz), phospho-p44/42 MAPK (9106; Cell Signaling), Akt (9272; Cell Signaling), phospho-Akt (4060; Cell Signaling), and phospho-STAT3 (9134; Cell Signaling). Blots were subsequently probed with the following horseradish peroxidase (HRP)-conjugated secondary antibodies: rabbit anti-mouse IgG (Genetex; 26728), rabbit anti-goat IgG (Genetex; 26741), goat anti-rabbit IgG (Fab fragment) (Genetex; 27171), or anti-mouse kappa light chain (H139-52.1; Abcam). Blots were imaged using an ImageQuant LAS4000 (General Electric) and processed with ImageQuant TL v8.1.0.0 software, Adobe Photoshop CS6, and CorelDraw Graphic Suite X5.
Nanoparticle tracking.
Nanoparticle tracking analysis (NTA), a technology used to quantify nanoparticle concentrations and sizes consistent with EV populations, was used to determine increases in vesicle secretion following LMP1 expression in cells. Following EV enrichment, vesicles were resuspended in particle-free PBS for quantitation using a Malvern NanoSight LM10 instrument as previously described in detail (37, 79). The camera level was set to 13, and the threshold was maintained at 3 for all samples. Videos were processed using NTA 3.1 software. The quantity of particles measured by NTA was normalized to the number of live cells counted at the time of harvest to generate a measure of the number of EVs secreted per cell. Relative vesicle secretion was obtained by normalization of EV levels to control cells in each experiment.
Transmission electron microscopy.
Following enrichment of EVs by the ExtraPEG method, pellets were resuspended in 100 μl of particle-free PBS for electron microscopy imaging. Samples were prepared as described by Lässer et al. (83) and visualized on an FEI CM120 transmission electron microscope.
ELISA.
Following EV enrichment by the ExtraPEG protocol, tetraspanin-containing vesicle subpopulations were quantitated using the System Biosciences ExoELISA kit. Equal volumes of EV samples were immobilized onto the wells of each microtiter plate—CD63 ExoELISA (EXOEL-CD63A-1), CD9 ExoELISA, (EXOEL-CD9A-1), or CD81 ExoELISA, (EXOEL-CD81A-1)—and assays were performed according to the manufacturer's instructions.
Acetylcholinesterase activity.
Exosomal acetylcholinesterase activity was measured by the EXOCET exosome quantification kit (System Biosciences) according to the manufacturer's instructions. EVs were harvested by the ExtraPEG method from HEK293 GFP-LMP1-inducible cells, with or without doxycycline (1.0 μg/ml) treatment, and equal volumes of samples were loaded into the assay.
Immunoprecipitation.
EVs from HEK293 cells expressing the inducible GFP-LMP1 construct were harvested 24 h after treatment with 1.0 μg/ml of doxycycline using the ExtraPEG protocol. An equal volume of sample was added to increasing amounts (0.5 μg, 1.0 μg, or 1.5 μg) of mouse anti-human CD63 antibody (TS63; Abcam) or 1.0 μg of mouse IgG control (12-371; Millipore) in PBS with 0.01% Tween 20 (PBS-Tween) and incubated overnight at 4°C with rotation. The next day, 100 μl of protein G magnetic beads (Thermo; number 21349) were added to sample-antibody mixtures and incubated with rotation at room temperature for 45 min. EVs bound to magnetic beads were collected and washed three times in PBS-Tween. Unbound supernatant (FT) was concentrated by ultracentrifugation for 70 min at 100,000 × g. Bead-bound EVs and FT material were lysed in 2× nonreducing Laemmli buffer.
Confocal microscopy.
HEK293 CD63 knockout and control cells were seeded into 35-mm glass well plates (Greiner Bio-One; number 627860), transfected 24 h later with 1 μg of GFP or GFP-LMP1 vectors as described above, and imaged 24 h posttransfection using a Zeiss LSM 880 microscope with a live-cell imaging chamber. Hoechst 33342 nuclear staining (5 μg/ml; Thermo Scientific; number 62249) was added 15 min prior to imaging. Confocal images were taken using 488-nm and 405-nm lasers and processed using Zen 2.1 Black software. HEK293 cells cotransfected with 1 μg of GFP-LMP1 and CD63-RFP were imaged on an Andor revolution spinning-disk laser confocal microscope. Images were obtained using the standard fluorescence (SF) 100× objective and 405-nm, 488-nm, and 561-nm lasers and analyzed in Imaris and ImageJ.
Lipid raft isolation.
HEK293 control and CD63 CRISPR cells were seeded into 150-mm plates and transfected with GFP-LMP1 for 24 h. Cell pellets (equivalent to 300 μl of packed cells) were collected as described above and frozen at −80°C prior to lysis and lipid raft extraction as previously described (32). Cells were lysed in 700 μl of 1% Triton X-100 in MNE buffer (25 mM morpholineethanesulfonic acid, 150 mM NaCl, 5 mM EDTA [pH 8.5]) and then homogenized with a tight-fit Dounce homogenizer (30 to 35 strokes). Lysates were incubated on ice for 30 min and then centrifuged at 1,000 × g for 1 min to pellet and discard nuclei. Supernatants were then mixed with 1 ml of 80% sucrose (wt/vol) in MNE buffer and transferred into 12-ml ultracentrifuge tubes (Beckman). Gradients were constructed by layering 7 ml of 30% sucrose in MNE buffer, followed by 3 ml of 5% sucrose in MNE buffer on top of the lysate-sucrose solution, and then ultracentrifuged at 188,000 × g in an SW41 Ti rotor (Beckman) for 20 h. Following the overnight centrifugation, 2 ml from the top of the gradient was discarded, and the subsequent 2-ml fractions containing lipid rafts were transferred to new ultracentrifuge tubes. PBS (10 ml) was added and mixed to lipid raft isolates, and then samples were washed by ultracentrifugation at 111,000 × g for 1 h in an SW41 Ti rotor. The remaining pellet containing the lipid rafts and associated proteins was dissolved in RIPA buffer for immunoblot analysis.
Statistical analysis.
Statistical significance of results was evaluated by Student's two-sample t test. Figures were constructed using Microsoft Excel, Adobe Photoshop CS6, and CorelDraw X5 software.
ACKNOWLEDGMENTS
We thank Marius Kostelic for his help in the generation of DG-75 cells stably expressing pBabe-HA-LMP1, Akash Gunjan for providing the anti-histone H4 antibody, and Lauren Howell for assistance in the confocal microscopy experiments performed at the FSU College of Medicine Confocal Microscopy Laboratory.
This study was supported by grants from the National Institutes of Health (RO1CA204621 and R15CA188941) awarded to D.G.M.
REFERENCES
- 1.Cohen JI. 2000. Epstein-Barr virus infection. N Engl J Med 343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 2.Young LS, Murray PG. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 3.Pattle SB, Farrell PJ. 2006. The role of Epstein-Barr virus in cancer. Expert Opin Biol Ther 6:1193–1205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 4.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. 2012. Epstein-Barr virus (EBV)-associated gastric carcinoma. Viruses 4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady G, MacArthur GJ, Farrell PJ. 2007. Epstein-Barr virus and Burkitt lymphoma. J Clin Pathol 60:1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu EA, Wu JM, Tunkel DE, Ishman SL. 2008. Nasopharyngeal carcinoma: the role of the Epstein-Barr virus. Medscape J Med 10:165. [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilger E, Kieser A, Baumann M, Hammerschmidt W. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J 17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirmeier U, Neuhierl B, Kilger E, Reisbach G, Sandberg ML, Hammerschmidt W. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res 63:2982–2989. [PubMed] [Google Scholar]
- 10.Uchida J, Yasui T, Takaoka-Shichijo Y, Muraoka M, Kulwichit W, Raab-Traub N, Kikutani H. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300–303. doi: 10.1126/science.286.5438.300. [DOI] [PubMed] [Google Scholar]
- 11.Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J, Nakano H, Mak TW, Yeh WC, Li X, Akira S, Suzuki N, Suzuki S, Mosialos G, Kieff E. 2003. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci U S A 100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliopoulos AG, Gallagher NJ, Blake SM, Dawson CW, Young LS. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem 274:16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 13.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem 278:3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 14.Eliopoulos AG, Young LS. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 15.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J 16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito N, Courtois G, Chiba A, Yamamoto N, Nitta T, Hironaka N, Rowe M, Yamaoka S. 2003. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-kappaB through distinct signaling pathways in fibroblast cell lines. J Biol Chem 278:46565–46575. doi: 10.1074/jbc.M302549200. [DOI] [PubMed] [Google Scholar]
- 17.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J 16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandberg M, Hammerschmidt W, Sugden B. 1997. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J Virol 71:4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huen DS, Henderson SA, Croom-Carter D, Rowe M. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549–560. [PubMed] [Google Scholar]
- 20.Lam N, Sugden B. 2003. LMP1, a viral relative of the TNF receptor family, signals principally from intracellular compartments. EMBO J 22:3027–3038. doi: 10.1093/emboj/cdg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flanagan J, Middeldorp J, Sculley T. 2003. Localization of the Epstein-Barr virus protein LMP 1 to exosomes. J Gen Virol 84:1871–1879. doi: 10.1099/vir.0.18944-0. [DOI] [PubMed] [Google Scholar]
- 22.Meckes DG, Raab-Traub N. 2011. Microvesicles and viral infection. J Virol 85:12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meckes DG, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N. 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meckes DG. 2015. Exosomal communication goes viral. J Virol 89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte-'t Hoen E, Cremer T, Gallo RC, Margolis LB. 2016. Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci U S A 113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Théry C. 2011. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobrie A, Colombo M, Raposo G, Théry C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 29.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. 2009. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J 23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. 2011. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J 30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meckes DG, Menaker NF, Raab-Traub N. 2013. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol 87:1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutzeit C, Nagy N, Gentile M, Lyberg K, Gumz J, Vallhov H, Puga I, Klein E, Gabrielsson S, Cerutti A, Scheynius A. 2014. Exosomes derived from Burkitt's lymphoma cell lines induce proliferation, differentiation, and class-switch recombination in B cells. J Immunol 192:5852–5862. doi: 10.4049/jimmunol.1302068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, Yoshizaki T, Pagano JS, Shackelford J. 2014. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 33:4613–4622. doi: 10.1038/onc.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. 2013. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J Virol 87:10334–10347. doi: 10.1128/JVI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurwitz SN, Conlon MM, Rider MA, Brownstein NC, Meckes DG Jr. 2016. Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J Extracell Vesicles 5:31295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceccarelli S, Visco V, Raffa S, Wakisaka N, Pagano JS, Torrisi MR. 2007. Epstein-Barr virus latent membrane protein 1 promotes concentration in multivesicular bodies of fibroblast growth factor 2 and its release through exosomes. Int J Cancer 121:1494–1506. doi: 10.1002/ijc.22844. [DOI] [PubMed] [Google Scholar]
- 39.Wasil LR, Wei L, Chang C, Lan L, Shair KH. 2015. Regulation of DNA damage signaling and cell death responses by Epstein-Barr virus latent membrane protein 1 (LMP1) and LMP2A in nasopharyngeal carcinoma cells. J Virol 89:7612–7624. doi: 10.1128/JVI.00958-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savina A, Vidal M, Colombo MI. 2002. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci 115:2505–2515. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XJ, Yang L, Zhao Q, Caen JP, He HY, Jin QH, Guo LH, Alemany M, Zhang LY, Shi YF. 2002. Induction of acetylcholinesterase expression during apoptosis in various cell types. Cell Death Differ 9:790–800. doi: 10.1038/sj.cdd.4401034. [DOI] [PubMed] [Google Scholar]
- 42.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazirabadi G, Geiger TR, Coffin WF, Martin JM. 2003. Epstein-Barr virus latent membrane protein-1 (LMP-1) and lytic LMP-1 localization in plasma membrane-derived extracellular vesicles and intracellular virions. J Gen Virol 84:1997–2008. doi: 10.1099/vir.0.19156-0. [DOI] [PubMed] [Google Scholar]
- 44.Keryer-Bibens C, Pioche-Durieu C, Villemant C, Souquère S, Nishi N, Hirashima M, Middeldorp J, Busson P. 2006. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer 6:283. doi: 10.1186/1471-2407-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 46.Yasui T, Luftig M, Soni V, Kieff E. 2004. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc Natl Acad Sci U S A 101:278–283. doi: 10.1073/pnas.2237224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salzer U, Prohaska R. 2001. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood 97:1141–1143. doi: 10.1182/blood.V97.4.1141. [DOI] [PubMed] [Google Scholar]
- 48.Harder T, Scheiffele P, Verkade P, Simons K. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol 141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mainou BA, Everly DN, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 50.Shair KH, Bendt KM, Edwards RH, Bedford EC, Nielsen JN, Raab-Traub N. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog 3:e166. doi: 10.1371/journal.ppat.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kung CP, Meckes DG, Raab-Traub N. 2011. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J Virol 85:4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell T, Sugden B. 1995. Stimulation of NF-kappa B-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol 69:2968–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kung CP, Raab-Traub N. 2010. Epstein-Barr virus latent membrane protein 1 modulates distinctive NF-kappaB pathways through C-terminus-activating region 1 to regulate epidermal growth factor receptor expression. J Virol 84:6605–6614. doi: 10.1128/JVI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci U S A 101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perkins ND. 2007. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 57.Jost PJ, Ruland J. 2007. Aberrant NF-kappaB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood 109:2700–2707. [DOI] [PubMed] [Google Scholar]
- 58.Sun SC. 2011. Non-canonical NF-κB signaling pathway. Cell Res 21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. 2010. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics 9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, Garcia JM, Dominguez G, Campos-Martin Y, Cuevas J, Peña C, Herrera M, Diaz R, Mohammed N, Bonilla F. 2012. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 51:409–418. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 62.Taylor DD, Gercel-Taylor C. 2008. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. 2015. Exosomes in cancer: small particle, big player. J Hematol Oncol 8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wasil LR, Shair KH. 2015. Epstein-Barr virus LMP1 induces focal adhesions and epithelial cell migration through effects on integrin-α5 and N-cadherin. Oncogenesis 4:e171. doi: 10.1038/oncsis.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gould SJ, Booth AM, Hildreth JE. 2003. The Trojan exosome hypothesis. Proc Natl Acad Sci U S A 100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammerschmidt W, Sugden B, Baichwal VR. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J Virol 63:2469–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Floettmann JE, Ward K, Rickinson AB, Rowe M. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29–40. doi: 10.1006/viro.1996.0452. [DOI] [PubMed] [Google Scholar]
- 69.Lee DY, Sugden B. 2008. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene 27:2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 70.Lu JJ, Chen JY, Hsu TY, Yu WC, Su IJ, Yang CS. 1996. Induction of apoptosis in epithelial cells by Epstein-Barr virus latent membrane protein 1. J Gen Virol 77(Part 8):1883–1892. [DOI] [PubMed] [Google Scholar]
- 71.Pallet N, Sirois I, Bell C, Hanafi LA, Hamelin K, Dieudé M, Rondeau C, Thibault P, Desjardins M, Hebert MJ. 2013. A comprehensive characterization of membrane vesicles released by autophagic human endothelial cells. Proteomics 13:1108–1120. doi: 10.1002/pmic.201200531. [DOI] [PubMed] [Google Scholar]
- 72.Martin J, Sugden B. 1991. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ 2:653–600. [PubMed] [Google Scholar]
- 73.Martin J, Sugden B. 1991. Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J Virol 65:3246–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qazi KR, Gehrmann U, Domange Jordö E, Karlsson MC, Gabrielsson S. 2009. Antigen-loaded exosomes alone induce Th1-type memory through a B-cell-dependent mechanism. Blood 113:2673–2683. doi: 10.1182/blood-2008-04-153536. [DOI] [PubMed] [Google Scholar]
- 75.Le Naour F, André M, Boucheix C, Rubinstein E. 2006. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6:6447–6454. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- 76.Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, Vázquez J, Yáñez-Mó M. 2013. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem 288:11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shair KH, Schnegg CI, Raab-Traub N. 2008. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res 68:6997–7005. doi: 10.1158/0008-5472.CAN-08-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alvarez A, Barisone GA, Diaz E. 2014. Focus formation: a cell-based assay to determine the oncogenic potential of a gene. J Vis Exp 2014(94):e51742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rider MA, Hurwitz SN, Meckes DG. 2016. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kung CP, Raab-Traub N. 2008. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J Virol 82:5486–5493. doi: 10.1128/JVI.00125-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gunjan A, Verreault A. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537–549. doi: 10.1016/S0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 82.Singh RK, Kabbaj MH, Paik J, Gunjan A. 2009. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lässer C, Eldh M, Lötvall J. 2012. Isolation and characterization of RNA-containing exosomes. J Vis Exp 2014:e3037. [DOI] [PMC free article] [PubMed] [Google Scholar]