ABSTRACT
Primary human cytomegalovirus (HCMV) infection usually goes unnoticed, causing mild or no symptoms in immunocompetent individuals. However, some rare severe clinical cases have been reported without investigation of host immune responses or viral virulence. In the present study, we investigate for the first time phenotypic and functional features, together with gene expression profiles in immunocompetent adults experiencing a severe primary HCMV infection. Twenty primary HCMV-infected patients (PHIP) were enrolled, as well as 26 HCMV-seronegative and 39 HCMV-seropositive healthy controls. PHIP had extensive lymphocytosis marked by massive expansion of natural killer (NK) and T cell compartments. Interestingly, PHIP mounted efficient innate and adaptive immune responses with a deep HCMV imprint, revealed mainly by the expansion of NKG2C+ NK cells, CD16+ Vδ2(−) γδ T cells, and conventional HCMV-specific CD8+ T cells. The main effector lymphocytes were activated and displayed an early immune phenotype that developed toward a more mature differentiated status. We suggest that both massive lymphocytosis and excessive lymphocyte activation could contribute to massive cytokine production, known to mediate tissue damage observed in PHIP. Taken together, these findings bring new insights into the comprehensive understanding of immune mechanisms involved during primary HCMV infection in immunocompetent individuals.
IMPORTANCE HCMV-specific immune responses have been extensively documented in immunocompromised patients and during in utero acquisition. While it usually goes unnoticed, some rare severe clinical cases of primary HCMV infection have been reported in immunocompetent patients. However, host immune responses or HCMV virulence in these patients has not so far been investigated. In the present study, we show massive expansion of NK and T cell compartments during the symptomatic stage of acute HCMV infection. The patients mounted efficient innate and adaptive immune responses with a deep HCMV imprint. The massive lymphocytosis could be the result of nonadapted or uncontrolled immune responses limiting the effectiveness of the specific responses mounted. Both massive lymphocytosis and excessive lymphocyte activation could contribute to massive cytokine production, known to mediate tissue damage. Furthermore, we cannot exclude a delayed immune response caused by immune escape established by HCMV strains.
KEYWORDS: HCMV, NK cells, T lymphocytes, adaptive immunity, innate immunity
INTRODUCTION
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus subfamily and is prevalent worldwide. Following primary infection, HCMV establishes latency, thereby exerting a lifelong influence on the host immune system (1). With the exception of a mononucleosis-like syndrome, it is generally believed that HCMV does not cause disease in immunocompetent individuals. However, in a context of immunosuppression or acquisition in early life, HCMV infection is associated with high mortality and morbidity rates (2). The host immune response features described following HCMV primary infection or reactivation have mostly been described in the context of hematopoietic stem cell or solid-organ transplantation and acquisition during fetal development or in early life.
Following primary infection, HCMV replicates and disseminates concomitantly with the development of a robust immune response involving natural killer (NK) cells, neutralizing antibodies, γδ T cells, and a high frequency of HCMV-specific CD4+ and CD8+ T cells. Coordinated innate and adaptive immune responses lead to control of viral replication and to resolution of infection. NK cells constitute the first line of defense against viral infections and are preponderant in the early control of viral infections. This is supported by the increased susceptibility to herpesvirus infections, including HCMV, in NK cell-deficient individuals (3, 4). NK cell functions are tightly tuned by numerous inhibitory and activating cell surface molecules. Many inhibitory receptors display specificity for HLA class I molecules, allowing the discrimination of self from nonself by NK cells. They include killer cell immunoglobulin-like receptors (KIRs), immunoglobulin-like transcript 2 (ILT-2), and the heterodimer CD94-NKG2A. Among inhibitory KIRs, KIR2DL2 and KIR2DL3 receptors recognize HLA-C allotypes with an asparagine at position 80 (the C1 group), whereas KIR2DL1 receptors recognize HLA-C allotypes with a lysine at position 80 (the C2 group). ILT-2 is able to recognize a broader spectrum of HLA class I molecules. CD94-NKG2A and its activating CD94-NKG2C counterpart recognize HLA-E molecules presenting leader peptides derived from classical HLA class I molecules. These receptors are involved in the detection of “missing self,” which occurs when HLA class I molecule expression is downregulated following viral infection or tumor induction (5). Activating receptors also modulate NK cell functions and include the activating natural cytotoxic receptors (NCR) (NKp30, NKp44, Nkp46, and NKp80), DNAM-1, NKG2D, and CD161. Inhibitory and activating receptor expression is strongly associated with the differentiation and maturation status of NK cells. Thus, KIR2D-positive (KIR2D+) NKG2C+ NK cells expressing the late marker of differentiation CD57 define the late mature NK cell subset (6), which has been shown to expand during HCMV infection (7–10). Another major functional property of NK cells is their ability to perform antibody-dependent cell-mediated cytotoxicity (ADCC) via the recognition of the Fc fragment of opsonic antibodies by CD16. The diversity of NK cell receptors displaying stochastic expression, together with the modulation of their expression during the course of NK cell differentiation, defines the diversity of NK cell subsets found in the NK cell compartment.
The role of the innate-like γδ T cell compartment during HCMV infection has been clearly demonstrated in healthy HCMV-seropositive subjects (11) and during acute infection in pregnant women (12), in children (13), and in transplant recipients (14–16). HCMV has also been associated with expansion of the Vδ2(−) γδ T cell subset (15–18). Similar to NK cells, T cells are particularly important in defense against HCMV infection. This includes robust CD4+ and CD8+ T cell responses targeting a broad range of HCMV proteins during acute episodes, followed by their commitment to a late mature phenotype, a feature found in latently infected individuals.
HCMV counteracts host immune defenses by eliciting numerous sophisticated immune evasion mechanisms (19) to escape CD8+ T and NK cell recognition. In most cases, despite the numerous immune evasion strategies employed by HCMV, host immune responses mounted against primary HCMV infection are effective at controlling CMV replication and dissemination. An extensive literature on clinical cases has reported severe symptoms associated with primary HCMV infection in immunocompetent individuals without investigations of host immune responses or HCMV virulence (20, 21). As both host and viral polymorphisms may contribute to severe symptomatic HCMV infection, we focused our study on the cellular immune responses in a rare cohort of immunocompetent patients undergoing severe symptomatic primary HCMV infection. For this, we performed a broad phenotypic and functional analysis of the main lymphocyte subsets and characterized the gene expression profiles of these patients. This study should provide new insights into a comprehensive range of immune responses in immunocompetent hosts undergoing acute primary HCMV infection.
RESULTS
Significant increase of all lymphocyte compartments in PHIP.
The rare occurrence of symptomatic primary HCMV infections raises the question of the roles of both host and HCMV polymorphisms. In this study, we investigated the immune responses of 20 symptomatic primary HCMV-infected patients (PHIP) (Table 1), in comparison to HCMV-negative (HCMV−) (n = 26) and HCMV-positive (HCMV+) (n = 39) healthy individuals. All PHIP presented lymphocytosis (10.3 × 109 ± 2.2 × 109 white blood cells [WBC]/liter; n = 18) (Table 1) with an average about 10-fold more than that observed in healthy blood donors (1 × 109 ± 0.1 × 109 WBC/liter; n = 57), except for patients P3 and P5 (<4 WBC/liter). Of note, despite the time delay of around 80 days after the onset of symptom appearance, two patients (P10 and P14) presented a high number of WBC/liter and maintained a CMV load. Based on CD3 and CD56 expression, we investigated the frequencies of all 4 populations corresponding to CD3− CD56+ NK cells, CD56− and CD56+ CD3+ T cells, and CD3− CD56− B cells (Fig. 1A). While the CD3− CD56− cell and CD19+ cell frequencies are significantly lower in PHIP than in HCMV− and HCMV+ individuals (P < 0.0001 and P = 0.001, respectively), the absolute number of CD19+ B cells was higher in PHIP than in HCMV− (P = 0.008) and HCMV+ (P = 0.003) healthy individuals (Fig. 1A). HCMV− and HCMV+ individuals and PHIP displayed similar frequencies of total NK cells (Fig. 1A and B). In accordance with the substantial lymphocytosis observed in PHIP, the absolute number of NK cells in PHIP was also significantly higher than in HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals (Fig. 1B), except for 3 PHIP (P3, P5, and P11) who presented a low NK cell frequency and/or a low number of WBC per liter.
TABLE 1.
Clinical and biological characteristics of the PHIP cohorta
| Patient | Age (yr) | Sexb | Time from onset (days) | HCMV IgM (IU/ml) | HCMV IgG (IU/ml) | Avidity (%) | CMV viral load (log10 IU/ml) | WBCc (109/liter) | Lyd (%) | Activated Ly (%) | Plateletse (109/liter) | ASTf (μkat/liter) | ALTg (μkat/liter) | Fever > 38.5°C > 7 days | Lymphadenopathy, hepatomegaly, splenomegaly | Myalgia, arthralgia | Asthenia | Other clinical symptoms or disease |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 47 | M | 14 | 2.9 | Positive | 8 | 3.6 | 16.13 | 293 | 1 | 0.97 | Yes | Yes | Pneumonia | ||||
| P2 | 57 | F | 2.4 | Yes | Yes | |||||||||||||
| P3 | 22 | F | 10 | 4.8 | 199 | 10 | 5.7 | 2.3 | 38 | 6 | Aggregates | 1.16 | 1.39 | Yes | Diarrhea | |||
| P4 | 27 | M | 18 | 4.3 | >240 | 24 | 4.5 | 15.34 | 47 | 12 | Aggregates | 3.55 | 4.93 | Yes | Diarrhea, vomiting | |||
| P5 | 25 | F | 21 | 3.3 | 146 | 6 | Negative | 3.59 | 40.5 | 2.5 | 157 | 0.58 | 0.39 | Yes | Yes | Yes | ||
| P6 | 38 | M | 24 | Negative | 8 | Yes | Yes | Yes | Yes | |||||||||
| P7 | 67 | M | 27 | 3 | 147 | 7 | 3.7 | 18.63 | 42 | 18 | 249 | 1.58 | 0.87 | Yes | Diarrhea, vomiting, pulmonary embolism | |||
| P8 | 31 | M | 27 | 1.5 | 135 | 16 | Negative | 7.28 | 45 | 1 | 225 | 1.83 | 2.02 | Yes | ||||
| P9 | 41 | M | 19 | 2.1 | 137 | 4.8 | 9.52 | 57 | 6 | 130 | 1.64 | 3.05 | Yes | Yes | ||||
| P10 | 27 | F | 80 | 2.4 | 93 | 10 | 3.4 | 12.07 | 56 | 4 | 230 | 0.76 | 0.63 | Yes | Yes | Yes | Abdominal pain | |
| P11 | 53 | M | 14 | 0.3 | 132 | 14 | <2.5 | 6.27 | 38 | 4 | 106 | 1.9 | 1.65 | Yes | Yes | |||
| P12 | 59 | F | 1.9 | 147 | 4 | 9.75 | 59.5 | 4 | 193 | 3.11 | 4.64 | Yes | Yes | |||||
| P13 | 29 | M | 11 | 7.2 | 50.4 | 3.4 | 13.52 | 39.5 | 1 | 498 | 0.98 | 1.2 | Yes | Yes | Yes | |||
| P14 | 30 | F | 86 | 44 | 39.8 | 4.2 | 4.56 | 233 | Yes | Yes | Abdominal pain, hepatitis | |||||||
| P15 | 26 | F | 30 | <12 | 93.3 | <5 | 4.5 | 8.76 | 46 | 6 | 126 | 0.85 | 1.17 | Yes | Yes | Yes | ||
| P16 | 24 | F | 10 | 62.1 | Positive | 4 | 9.23 | 60 | 6 | 227 | 1.17 | 1.5 | Diarrhea, vomiting, meningoradiculitis | |||||
| P17 | 47 | M | 34 | 20.5 | 59.4 | <5 | 4.3 | 8.11 | 35 | 6 | Aggregates | 1.6 | 1.93 | Yes | Yes | Yes | ||
| P18 | 33 | M | 13 | 48.7 | >150 | 12 | 5.4 | 13.42 | 41 | 8 | 245 | 1.58 | 4.9 | Yes | Yes | Yes | Yes | |
| P19 | 43 | M | 15 | 55.8 | 97.1 | 3.2 | 13.96 | 59 | 14 | 219 | 2.3 | 3.64 | Yes | Yes | Cough | |||
| P20 | 38 | M | 24 | 30 | >140 | 6.2 | 13.26 | 40 | 8.5 | 87 | 2.59 | 4.25 | Yes | Yes | Yes | Portal vein thrombosis, macrophage activation syndrome |
Two successive generations of serologic tests were used for the confirmation of primary CMV infection. For P1 to P13, CMV serology was performed using Liaison CMV IgG (positive, >0.6 IU/ml), Liaison CMV IgM (positive, >30 arbitrary units [AU]/ml), and Vidas CMV avidity test (infection, <3 months if <0.20 to >3 months if >80%); for P13 to P20, CMV serology was performed using Liaison CMV IgG II (positive, >14 IU/ml), Liaison CMV IgM II (positive, >22 AU/ml), and Liaison CMV avidity test (recent infection if <15%; >3 months if >25%). Primary CMV infection diagnosis was confirmed by IgG seroconversion between 2 successive samples or positive IgM plus a low avidity index. For P2 and P6, the serologic diagnosis was performed in another laboratory with different tests, but the serologic criteria were the same.
M, male; F, female.
WBC (white blood cells), n = 4 × 109 to 10 × 109/liter.
Ly, lymphocytes.
Platelets, n = 150 × 109 to 400 × 109/liter.
AST, alanine transaminase; n < 0.5 μkat/liter.
ALT, aspartate transaminase; n < 0.5 μkat/liter.
FIG 1.
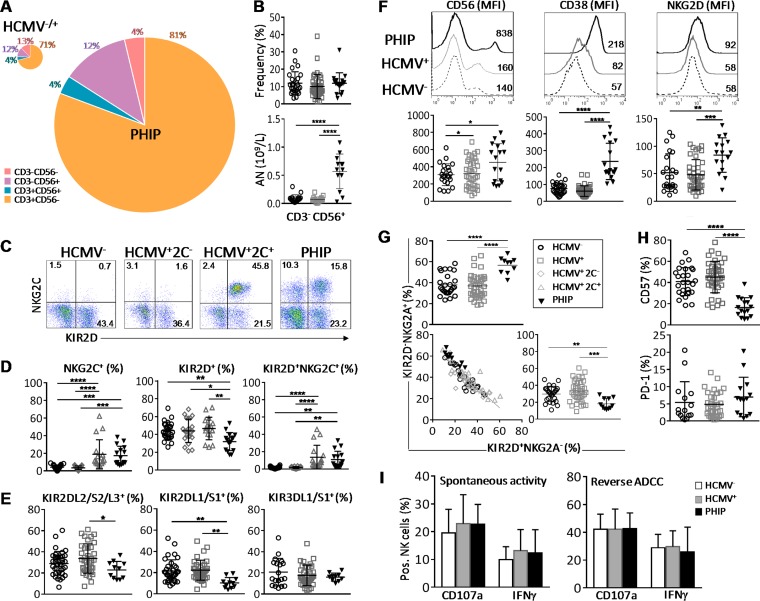
Early expansion in PHIP of activated and responsive NK cells that displayed not fully mature NKG2C, NKG2Ahi, KIR2Dlo, and CD57lo phenotypes. (A) Patterns of cell composition following CD3 and CD56 expression in HCMV−/+ individuals and PHIP. We summed CD3− CD56−, CD3− CD56+, CD3+ CD56+, and CD3+ CD56− cell subsets, weighting them according to their frequencies, as indicated. The size of the pie chart is proportional to the absolute number of total lymphocytes. (B) Scatter plots representing the percentages and the absolute numbers (AN) of CD3− CD56+ NK cells assessed by flow cytometry in HCMV− (n = 26) or HCMV+ (n = 39) individuals and PHIP (n = 17). (C) Representative density plots of CD3− CD56+ NK cells expressing KIR2D and NKG2C in HCMV−, HCMV+ 2C+, and HCMV+ 2C− individuals and PHIP. (D) Frequencies of total NKG2C+, KIR2D+, and KIR2D+ NKG2C+ NK cells for 26 HCMV−, 22 HCMV+ 2C−, and 17 HCMV+ 2C+ individuals and 16 PHIP. The results are represented as means ± standard errors of the mean (SEM). (E) Frequencies of KIR2DL2/S2/L3+ and KIR2DL1/S1+ NK cells for 34 HCMV− and 36 HCMV+ individuals and 11 PHIP. (F) CD56, CD38, and NKG2D expression (mean fluorescence intensity [MFI]) on NK cells for representative HCMV− and HCMV+ individuals and PHIP. The scatter plots represent CD56 (n = 26 HCMV−; n = 39 HCMV+; n = 17 PHIP), CD38 (n = 26 HCMV−; n = 34 HCMV+; n = 17 PHIP), and NKG2D (n = 23 HCMV−; n = 39 HCMV+; n = 16 PHIP) ΔMFI on NK cells from 26 HCMV− and 34 HCMV+ individuals and 17 PHIP after deletion of MFI obtained with isotype control (ΔMFI). (G) Scatter plots of KIR2D− NKG2A+ and KIR2D+ NKG2A− NK frequencies and correlation between the populations observed in all studied groups. (H) Scatter plots of CD57 (n = 26 HCMV−; n = 39 HCMV+; n = 16 PHIP) and PD-1 (n = 15 HCMV−; n = 32 HCMV+; n = 14 PHIP) NK cells. (I) Degranulation and IFN-γ secretion capacities of NK cells from 6 HCMV−, 14 HCMV+, and 5 PHIP PBMCs assessed by flow cytometry after PBMC coculture with medium alone, 721.221 cells for spontaneous-activity measurement, or P815 cells for reverse ADCC measurement. Statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) between more than two groups was determined using one-way ANOVA. (C) Spearman's rank correlation coefficient was calculated when a significant P value was observed (P < 0.05).
Early NKG2C NK cell expansion in PHIP.
To further investigate how HCMV shapes the NK cell compartment, we analyzed the distribution of the main NK cell subsets implicated in HCMV control. Peripheral blood NKG2C+ NK cell expansion has been described as the hallmark of HCMV infection (7–9). Therefore, we classified HCMV+ healthy individuals according to the presence (HCMV+ 2C+) or absence (HCMV+ 2C−) of NKG2C+ NK cells (Fig. 1C). Interestingly, significant NKG2C+ NK cell expansion was observed in PHIP compared to HCMV− (P = 0.0002) and HCMV+ 2C− (P = 0.0001) individuals and similar to that of HCMV+ 2C+ healthy individuals (Fig. 1D). PHIP displayed a decreased frequency of KIR2D+ NK cells compared to HCMV− (P = 0.006), HCMV+ 2C− (P = 0.01), and HCMV+ 2C+ (P = 0.002) individuals (Fig. 1D). The frequency of KIR2D+ NKG2C+ NK cell subsets in PHIP was similar to that in HCMV+ 2C+ healthy individuals and significantly higher than that observed in HCMV− (P = 0.002) and HCMV+ 2C− healthy individuals (P = 0.002) (Fig. 1D). We observed a reduced frequency of KIR2DL2/S2/L3 NK cells in PHIP compared to HCMV+ individuals (P = 0.01) (Fig. 1E). The frequency of the KIR2DL1/S1 compartment was also decreased in PHIP compared to HCMV− (P = 0.003) and HCMV+ (P = 0.001) individuals (Fig. 1E). These features were independent of the HLA-C environment (data not shown). PHIP displayed a frequency of KIR3DL1/3DS1+ NK cells similar to that in HCMV− and HCMV+ individuals (Fig. 1E).
Moreover, based on recent observations showing increased susceptibility to developing symptomatic primary HCMV infection in immunocompetent adults carrying an AA haplotype or the HLA-Bw4T allele (22), all PHIP studied were KIR and HLA typed (data not shown). We observed a lower proportion of HLA-Bw4I in PHIP (29%) than in HCMV− individuals (61%; P = 0.01; odds ratio [OR] = 0.1) and an increased proportion of HLA-Bw4T in PHIP (53%) in contrast to HCMV+ 2C− individuals (18%; P = 0.039; OR = 5.06). Taken together, these results demonstrate massive proliferation of NK cell compartments during symptomatic primary infection in immunocompetent adults, with early expression of NKG2C and delayed acquisition of KIR receptors that is not related to a particular KIR genotype or HLA environment.
Activation, partial differentiation, and functional potential of NK cells in PHIP.
To extend the investigation of NK cell responses occurring in PHIP, we evaluated their activation levels and the expression of NK cell-related markers. PHIP NK cells were highly activated, as illustrated by the strong, increased expression of CD56 and the CD38 activation marker at the surfaces of NK cells compared to HCMV− (P < 0.0001 for CD56 and CD38) and HCMV+ (P < 0.0001 for CD56 and CD38) individuals (Fig. 1F). Similarly, the expression level of the activating receptor NKG2D was increased at the NK cell surface in PHIP compared to HCMV− (P = 0.005) and HCMV+ (P = 0.0005) individuals (Fig. 1F). Interestingly, the frequencies of NK cells expressing the early activation marker CD69 were similar in the three groups (data not shown), suggesting that NK cells from PHIP had already completed the first step of activation. We observed an increased frequency of KIR2D− NKG2A+ NK cells (P < 0.0001) correlated with a decreased frequency of KIR2D+ NKG2A− NK cells in PHIP compared to HCMV− (P = 0.009) and HCMV+ (P = 0.0003) individuals (Fig. 2G). On the other hand, a lower frequency of CD57+ NK cells was observed in PHIP than in HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals (Fig. 1H) and no increased frequency of PD-1+ NK cells in PHIP. Other markers investigated, such as DNAM-1, NKp30, NKp44, NKp46, CD16, CD161, 2B4, ILT-2, granzyme A (GzmA), and perforin, showed no significant differences between the studied groups (data not shown).
FIG 2.
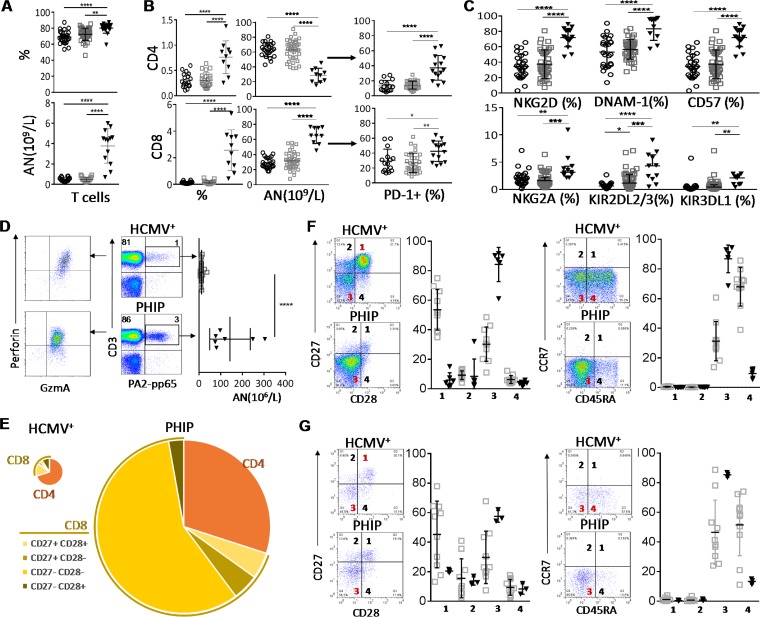
Early differentiation of CD3+ CD56− T cells in PHIP. (A) Frequency (n = 26 HCMV−; n = 39 HCMV+; n = 17 PHIP) and absolute number (n = 18 HCMV−; n = 23 HCMV+; n = 13 PHIP) of CD3+ CD56− T lymphocytes. (B) Absolute numbers (n = 18 HCMV−; n = 23 HCMV+; n = 10 PHIP) and frequencies (n = 24/25 HCMV−; n = 39 HCMV+; n = 10 PHIP) of CD4+ and CD8+ T cells. Frequencies of PD-1+ among CD4+ and CD8+ T cells (n = 15 HCMV−; n = 32 HCMV+; n = 14 PHIP). (C) AN and frequencies of NKG2D+, DNAM-1+, CD57+, NKG2A+, KIR2DL2/3+, and KIR3DL1+ T cells. Frequency results are shown as means ± SEM. (D) Density plots illustrating HLA-A2/pp65 pentamer-stained CD3+ T lymphocytes from representative HCMV+ individuals and PHIP and representative patterns of granzyme A and perforin expression in HLA-A2/pp65-specific cells. The scatter plots represent the absolute number of HLA-A2-pp65-specific CD3+ T cells from HCMV+ healthy individuals (n = 13) and PHIP (n = 6). (E) Patterns of CD4 and CD8 cell composition following CD27 and CD28 expression for CD8+ T cells in HCMV+ individuals and PHIP. We summed all cell subsets, weighting them according to their frequencies, as indicated. The size of the pie chart is proportional to the absolute number of total T lymphocytes. (F and G) Density plots illustrating the pattern of CD27/CD28 and CD45RA/CCR7 expression and corresponding scatter plots of frequencies observed in CD8+ T cells (F) and HLA-A2–pp65-specific T cells (G) from HCMV+ individuals (n = 10) and PHIP (n = 6). Means and SEM are shown. Statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) between more than two groups was determined using one-way ANOVA and between two groups using an unpaired t test.
We next assessed by flow cytometry the functional potential of NK cells from PHIP in terms of cytotoxicity (determined by expression of CD107a, a marker of degranulation) and gamma interferon (IFN-γ) production using an in vitro model for spontaneous effector functions and for reverse ADCC (Fig. 1I). NK cells from PHIP exhibited degranulation potential, as well as IFN-γ production, similar to those of HCMV− and HCMV+ healthy individuals (Fig. 1I). Taken together, our findings suggest that although NK cells from PHIP do not appear to be fully mature, as shown by increased proportions of NKG2Ahi, KIR2Dlo, and CD57lo NK subsets, they are activated and functional effectors.
Early differentiation of CD3+ CD56− T cells in PHIP.
T lymphocyte repertoire modulation has been extensively documented during HCMV infection. Our phenotypic analysis of the CD3+ CD56− T cell subset revealed a significantly higher frequency of CD3+ CD56− T cells in PHIP than in HCMV− (P < 0.0001) and HCMV+ (P = 0.0017) individuals (Fig. 2A). The absolute numbers of CD3+ CD56− T cells (Fig. 2A) and CD4+ and CD8+ T cells (Fig. 2B) were also significantly higher in PHIP than in HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals. We observed strong contraction of the CD4+ T lymphocyte frequency in PHIP compared to that in HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals and increased frequency of CD8+ T lymphocytes compared to that in HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals (Fig. 2B). This could be explained by the mobilization of the latter compartment due to the ongoing infection. We also observed an expansion of PD-1+ CD4+ and CD8+ T lymphocytes in PHIP compared to HCMV− (P < 0.0001 and P = 0. 04, respectively) and to HCMV+ (P < 0.0001 and P = 0.002, respectively) individuals (Fig. 2B). The frequencies of T lymphocytes expressing common markers with NK cells, such as NKG2D, DNAM-1, and CD57, were increased in PHIP compared to HCMV− (P < 0.0001, P < 0.0001, and P < 0.0001, respectively) and HCMV+ (P < 0.0001, P < 0.0001, and P < 0.0001, respectively) healthy individuals (Fig. 2C). Moreover, the frequencies of T cells expressing inhibitory NK receptors, like NKG2A, KIR2DL2/3, and KIR3DL1, were significantly increased in PHIP compared to HCMV− (P = 0.002, P < 0.0001, and P = 0.005, respectively) and HCMV+ (P = 0.0002, P = 0.0002, and P = 0.001, respectively) healthy individuals (Fig. 2C).
We further focused on HCMV-specific CD8+ T cells from HLA-A2+ PHIP using an HLA-A2/pp65 pentamer. While the proportion of HCMV-specific CD8+ T cells was not significantly increased in PHIP compared to HCMV+ healthy individuals, the absolute number was significantly higher (P < 0.0001) (Fig. 2D). HCMV-specific CD8+ T cells from PHIP expressed high levels of granzyme A and perforin (Fig. 2D). Based on CD27 and CD28 expression, we observed major differences in the distribution of all CD8+ T cell subsets in PHIP compared to HCMV+ individuals (Fig. 2E). Indeed, CD8+ T cells (Fig. 2F) and HCMV-specific CD8+ T cells (Fig. 2G) from PHIP were mainly characterized by a CD27lo/− CD28− CD45RAlo/− CCR7− phenotype (Fig. 2E). Altogether, our observations suggest that HCMV infection in PHIP leads to substantial proliferation of T lymphocytes and the establishment of an adaptive immune response with CD27lo CD28− CD45RAlo CCR7− GzmAhi CD8+ T cells displaying early effector features.
HCMV signature on the γδ T cell compartment.
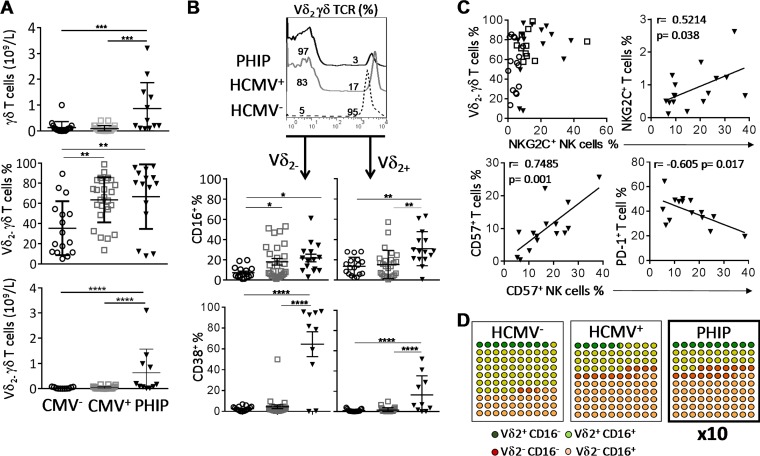
Numerous studies have implicated the γδ T cell compartment during the course of HCMV infection (11, 14–16). As in HCMV infection in kidney transplant recipients (15) or congenital infection (13), we observed a significant expansion of γδ T cells in PHIP compared to HCMV− (P = 0.0007) and HCMV+ (P = 0.0002) healthy individuals (Fig. 3A), with a preferential proliferation of Vδ2(−) γδ T cells in PHIP compared to HCMV− and HCMV+ individuals observed from the frequencies (P < 0.004 and P < 0.005, respectively) and the absolute numbers (P < 0.0001 and P < 0.0001, respectively) (Fig. 3A). Of note, for 3 PHIP (P6, P9, and P11), we did not observe an expansion of Vδ2(−) γδ T cells (Fig. 3A). We observed an increased proportion of Vδ2(−) γδ T cells expressing CD16, at various levels, in HCMV+ individuals (P < 0.05) and PHIP (P < 0.05) compared to HCMV− individuals (Fig. 3B). Interestingly, the three patients for whom a weak Vδ2(−) γδ T cell frequency was detected displayed the lowest CD16+ Vδ2− γδ T cell frequencies. Within the Vδ2(+) γδ T cell compartment, an increased frequency of CD16-expressing cells was observed only in PHIP compared to HCMV− (P < 0.002) and HCMV+ (P < 0.002) individuals (Fig. 3B). As previously described in a cohort of primary HCMV-infected pregnant women (17), both Vδ2(−) γδ T cell and Vδ2(+) γδ T cell subsets displayed higher CD38+ cell frequencies in PHIP (mean = 64.4 ± 11.9 for Vδ2(−) γδ T cells; mean = 16.4 ± 5.9 for Vδ2(+) γδ T cells) than in HCMV− (P < 0.001) or HCMV+ (P < 0.001) individuals, with an emphasis on the Vδ2(−) γδ T cell subset (Fig. 3B). The two patients for whom CD38+ Vδ2(−) γδ T cell frequency was very low (P6 and P11) also showed the lowest CD16 expression on Vδ2(−) γδ T subsets. Finally, we noticed an interesting feature when comparing Vδ2(−) γδ T cell expansion and the proportion of NKG2C+ NK cells in healthy HCMV+ individuals or in PHIP (Fig. 3C). While Vδ2(−) γδ T cell expansion was not systematic in HCMV+ NKG2C− individuals, we found, on the contrary, that all HCMV+ 2C+ healthy individuals displayed Vδ2(−) γδ T cell expansion (Fig. 3C). PHIP presented the same profile as the latter group, except for the two out of three patients (P6 and P11) who did not expand Vδ2(−) γδ T cells. Interestingly, significant positive correlations were observed when we assessed NKG2C+ T cell and NKG2C+ NK cell frequencies (P = 0.038) (Fig. 3C), as well as for CD57+ T cell and CD57+ NK cell frequencies (P = 0.001) (Fig. 3C). We also observed a significant correlation between increased frequencies of CD57+ NK cells and the diminution of PD-1+ T cell frequencies (P = 0.017) (Fig. 3C). This could reflect the diminution of T cell activation while NK cells move into a differentiation process. These results confirm that primary HCMV infection drives γδ T cell proliferation and mobilization, as illustrated by the expansion of the activated Vδ2(−) γδ T subset (Fig. 3D). Moreover, acute HCMV infection leads to coordinated changes in innate and acquired lymphocyte compartments.
FIG 3.
HCMV signature on γδ T cell compartment. (A) Absolute numbers of γδ T cells and frequencies and absolute numbers of Vδ2(−) γδ T cells in 17 HCMV− and 27 HCMV+ individuals and 15 PHIP. (B) Representative histogram illustrating γδ T cells stained with Vδ2 γδ TCR monoclonal antibody. The scatter plots represent CD16+ (n = 16 HCMV−; n = 26 HCMV+; n = 15 PHIP) and CD38+ (n = 17 HCMV−; n = 26 HCMV+; n = 10 PHIP) frequencies observed in Vδ2(+) and Vδ2(−) γδ T cell subsets. (C) Qualitative representation of NKG2C+ NK cell frequency as a function of the corresponding Vδ2(−) γδ T cell frequency for each individual for 16 HCMV+ 2C−, 11 HCMV+ 2C+, and 13 PHIP. The correlation graphs represent NKG2C+ NK cell and NKG2C+ T cell frequencies, CD57+ NK cell and CD57+ T cell frequencies, and CD57+ NK cell and PD-1+ T cell frequencies in 15 PHIP. (D) Ten-by-10 dot plot representations of Vδ2(−) and Vδ2(+) γδ T cells expressing or not CD16 in all studied groups. Means and SEM are shown. Statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) between more than two groups was determined using one-way ANOVA. (C) Spearman's rank correlation coefficient was calculated when a significant P value was observed (P < 0.05).
Activation of CD3+ CD56+ T cells in PHIP.
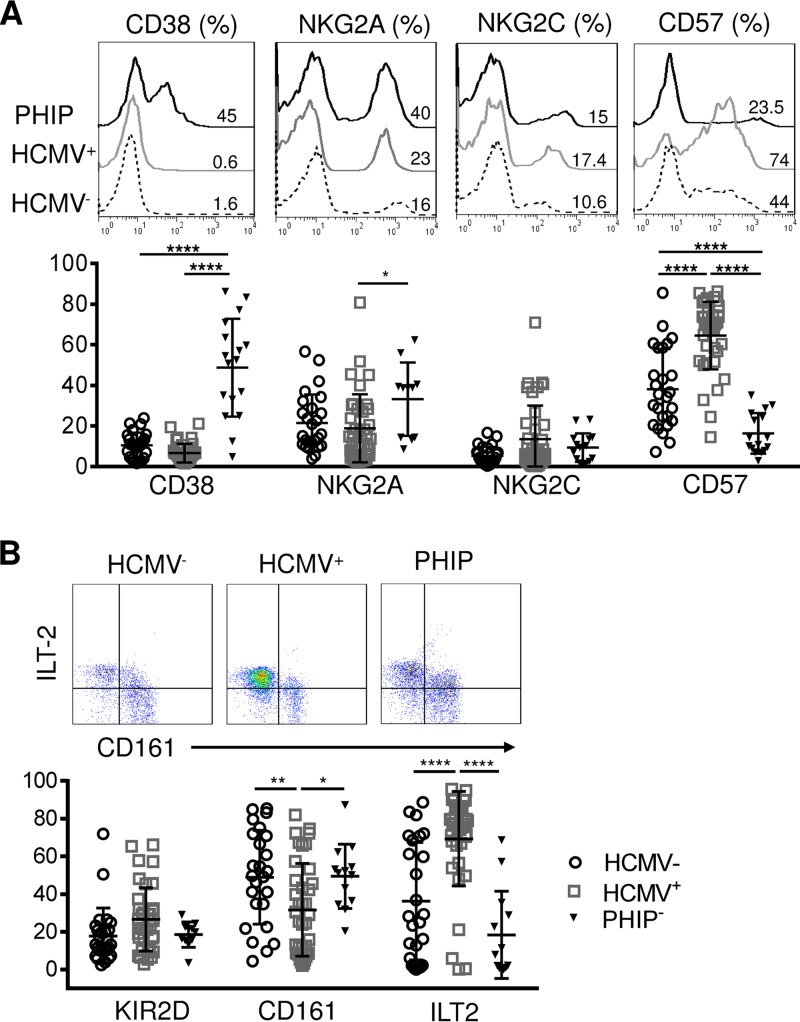
Several studies have described CD56+ T cell implication in HCMV infection (23–26). Similar to NK and conventional cells and γδ T lymphocytes in PHIP, CD56+ T cells appear to be activated, as shown by the strong increase in CD38-expressing CD56+ T cells in PHIP compared to HCMV− (P < 0.0001) and HCMV+ (P < 0.0001) individuals (Fig. 4A). NKG2A+ CD56+ T cell frequencies were significantly higher in PHIP than in HCMV+ individuals (P = 0.03), with no significant change for NKG2C+ CD56+ T cell frequencies. PHIP displayed reduced CD57+ CD56+ T cell frequencies compared to HCMV− individuals (P < 0.0001) (Fig. 4A). Interestingly, we found the highest frequencies of CD57+ CD56+ T cells in HCMV+ individuals compared to HCMV− (P < 0.0001) individuals and PHIP (P < 0.0001) (Fig. 2A). CD161 and ILT-2 appeared to be similarly expressed in PHIP and HCMV− healthy controls, and the KIR2D frequencies were similar in the three groups (Fig. 2B). However, HCMV+ individuals were characterized by a significantly lower frequency of CD161+ CD56+ T cells than PHIP (P < 0.0001) or HCMV− individuals (P = 0.008) and a significantly higher frequency of ILT-2+ CD56+ T cells than PHIP (P < 0.0001) and HCMV− individuals (P < 0.0001) (Fig. 2B), as previously observed in children (27).
FIG 4.
Phenotypic characterization of CD3+ CD56+ T cells. (A) Representative histograms illustrating CD38, NKG2A, NKG2C, and CD57 expression on CD3+ CD56+ T lymphocytes. The scatter plots represent the frequencies of CD38 (n = 26 HCMV−; n = 39 HCMV+; n = 10 PHIP), NKG2A (n = 26 HCMV−; n = 38 HCMV+; n = 17 PHIP), NKG2C (n = 25 HCMV−; n = 39 HCMV+; n = 14 PHIP), and CD57 (n = 26 HCMV−; n = 39 HCMV+; n = 16 PHIP) CD3+ CD56+ T lymphocytes. (B) Representative density plots illustrating CD161 and ILT-2 expression on CD56+ T lymphocytes (n = 26 HCMV−; n = 37 HCMV+; n = 11 PHIP). The scatter plots represent the frequencies of KIR2D, CD161, and ILT-2 on CD56+ T lymphocytes in HCMV− and HCMV+ individuals and PHIP. Means and SEM are shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Dynamics of the immune responses in PHIP.
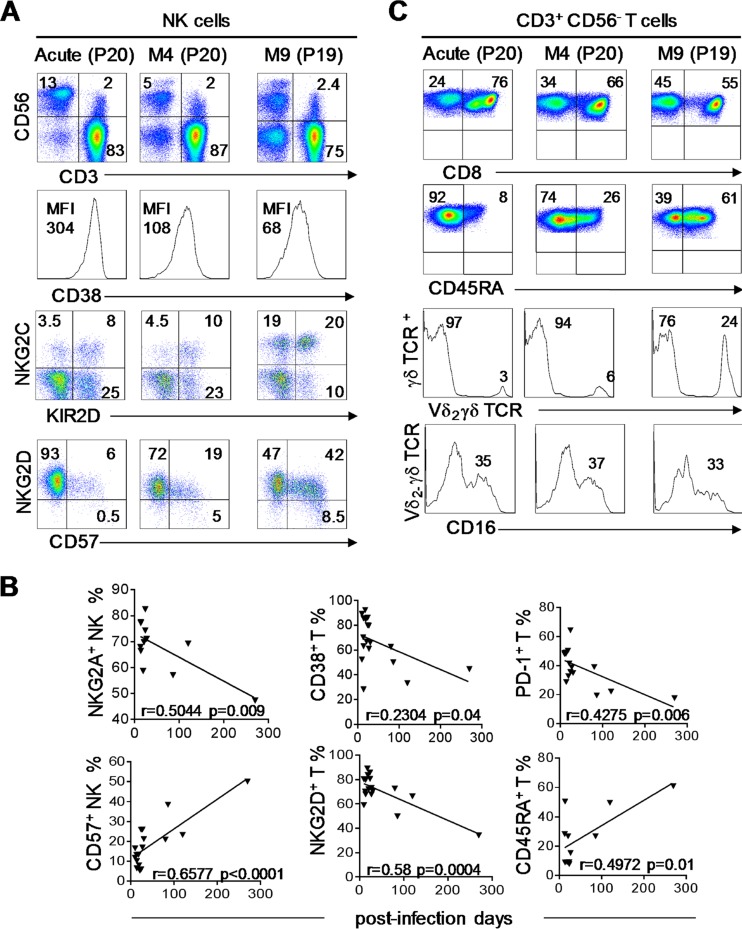
We investigated the immune response evolution kinetics from the onset of the disease to the resolution of infection in PHIP from a qualitative standpoint. As PHIP were sampled at different times after the onset of symptoms, we followed the evolution of the different cell subsets from phenotypic markers on all lymphocyte populations at the time after appearance of the first symptoms. We first noticed the recovery of normal CD56 surface expression levels on NK cells, with a distinction between CD56bright and CD56dim cells (Fig. 5A). In patient 20, we observed a decrease in the total NK cell frequency 4 months after the onset of disease. NK cells also displayed reduced expression of CD38 (Fig. 5A) on T lymphocytes (data not shown) as early as 4 months after the onset of infection, testifying to the dampened activation level. NKG2C NK cell expansion persisted for several months, and KIR2D+ NK cells tended to increase, mostly in association with NKG2C (Fig. 5A). Finally, while NKG2D+ NK cell frequencies slightly decreased (Fig. 5A) in accordance with the lower activated phenotypic level, the CD57+ NK cell frequency increased from the 4th month (Fig. 5A), matching levels observed in healthy individuals. These results reflect an evolution of the NK cell maturation status during the course of HCMV infection, from the onset of the disease to its resolution, as observed for NKG2A and CD57 on NK cells (Fig. 5B). For CD56− T lymphocytes, we observed in the late phenotype an increased frequency of CD4+ T cells (data not shown) and a decrease in CD8+ T cells (Fig. 5C). We observed the persistence of the Vδ2(−) γδ T cell subset with no change in CD16-expressing Vδ2(−) γδ T cell frequencies (Fig. 5C). Interestingly, the proportion of CD45RA+ T cells increased (Fig. 5C and D) and PD-1+ T cell frequencies decreased from the 4th month after infection onset (Fig. 5D). NKG2D expression, which is particularly high on activated T cells at the onset of HCMV infection, decreased with time, as well as NKG2D+ and CD38+ T cell frequencies (Fig. 5D). These results show commitment of T lymphocytes to differentiation.
FIG 5.
Dynamics of NK, γδ, and T cell subsets following primary HCMV infection. (A) Density plots illustrating all lymphocyte frequencies using CD3/CD56 marker combinations at different time points: acute infection (P20), month 4 (M4) (P20), and month 9 (M9) (P19). The histograms represent CD38 expression on NK cells (CD3− CD56+) at all 3 kinetic points (acute, M4, and M9), and the density plots illustrate KIR2D+, NKG2C+, CD57+, and NKG2D+ NK cells. (B) Correlation graphs representing NKG2A+ NK cell; CD57+ NK cell; and CD38+, NKG2D+, PD-1+, and CD45RA+ T cell frequencies and postinfection days for 17 PHIP. Spearman's rank correlation coefficient was calculated when a significant P value was observed (P < 0.05). (C) Density plots representing CD8+ and CD45RA+ αβ T lymphocytes at all 3 kinetic points (acute, M4, and M9). The histograms represent Vδ2(−) T lymphocytes and CD16+ γδ T cells among Vδ2(−) T lymphocytes at all 3 kinetic points.
Profound genetic modulation induced by HCMV in PHIP.
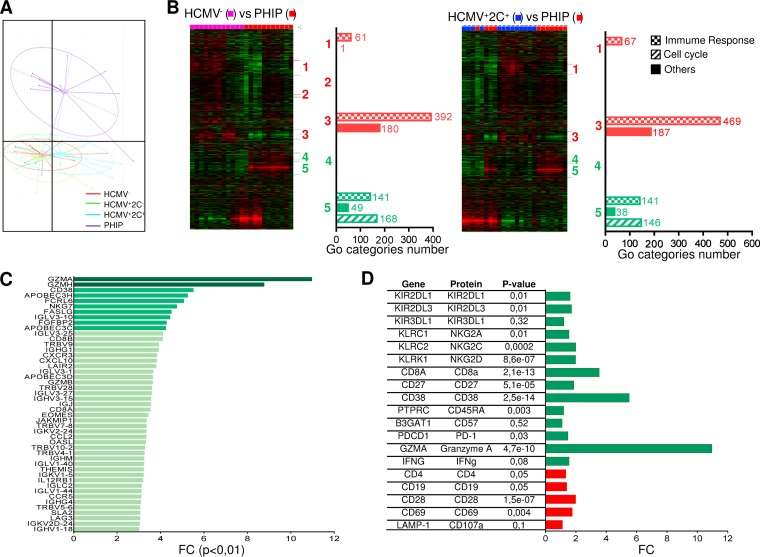
Flow cytometry analysis is useful to investigate different populations using characterized phenotypic markers but presents some limitations in investigating a wide range of cellular molecules whose expression can be modulated by HCMV infection. To further investigate the immune repertoire modulation during primary HCMV infection, we performed transcriptional analysis. We discriminated HCMV+ individuals according to the presence (HCMV+ 2C+) or absence (HCMV+ 2C−) of NKG2C+ NK cell expansion. The strong homogeneity of lymphocyte phenotype profiles of PHIP and the limited cell availability prompted us to perform the analysis of total peripheral blood mononuclear cells (PBMCs) with strong representation of conventional T lymphocytes (mean = 80.1% ± 1.5%). Principal-component analysis (PCA) demonstrated superimposed genetic profiles of HCMV-seronegative or -seropositive healthy individuals (Fig. 6A). On the other hand, the distinct genetic profile of PHIP highlighted the genetic modulations occurring during acute infection. Gene expression profiles were first subjected to unsupervised hierarchical clustering in order to highlight gene sets with similar differential expression. While no clusters were detected between control groups (data not shown), heat maps revealed five clusters of differentially expressed genes (DEG) between HCMV− or HCMV+ 2C− healthy individuals (data not shown) and PHIP and four clusters between HCMV+ 2C+ healthy individuals and PHIP (Fig. 6B). Submission to GOminer (http://discover.nci.nih.gov/gominer) to assess biological significance showed clusters 1 to 5 were identical in HCMV−, HCMV+ 2C−, and HCMV+ 2C+ healthy individuals and PHIP, except for cluster 2, which was absent in the HCMV+ 2C+ healthy individuals versus PHIP comparison. The most differentially modulated Gene Ontology (GO) categories (number of genes per GO category, >5; enrichment, >1.5; false-discovery rate [FDR], <0.01) were then classified according to immune response, cell cycle, or other cell functions (Fig. 6B). From this first analysis, we showed that PHIP present an activated gene expression profile mainly related to immune responses, corroborating the previous cellular results.
FIG 6.
Profound genetic modulation induced by HCMV in PHIP. (A) PCA illustrating genetic-profile variability in two-dimensional scale after quality control and data normalization. Each dot in the plot represents a studied individual (HCMV−, n = 13; HCMV+ 2C−, n = 12; HCMV+ 2C+, n = 12; and PHIP, n = 11). (B) Heat maps generated after hierarchical clustering using Cluster 3.0 and Treeview for HCMV− (n = 13) versus PHIP (n = 11) and HCMV+ 2C+ (n = 12) versus PHIP data set comparisons. Downregulated clusters in PHIP are shown in red, and upregulated clusters in PHIP are indicated in green. The bars indicate the numbers of GO categories belonging to the immune response, cell cycle, and other cell functions in each cluster for each data set comparison after submission to GOminer software to assess biological significance. GO categories were selected with a cutoff of enrichment of >1.5, an FDR of <0.01, and >5 changed genes. (C) FCs for all upregulated genes with an FC of >3 involved in immune response in PHIP compared to an HCMV− healthy individual gene expression profile (FC of >10, dark green; FC of >4, green; 3 < FC > 4, light green). (D) FCs for DEG between HCMV− healthy individuals and PHIP for whom protein expression was assessed by flow cytometry. Upregulated and downregulated genes are indicated in green and red, respectively. The P value observed for each gene is indicated.
A second analysis was performed to compare PHIP transcription profiles to those of all the control groups in order to individually detect differentially expressed genes. Approximately 2,000 to 3,000 genes were differentially regulated between PHIP and healthy individuals. In order to manage the data sets, we applied a cutoff of 3-fold change (FC) with a corrected P value of <0.01 (Fig. 6C). Genes encoding chemokines involved in lymphocyte adhesion and migration, including CXCL10 and its receptor CXCR3, CCL2, and CCR5, were upregulated in PHIP. Transcription of the β1 subunit of the interleukin 12 (IL-12) receptor was also increased in PHIP. Moreover, genes encoding proteins displaying antiviral properties, such as OASL and APOBEC3C, -D, and -H, were upregulated in PHIP. Among transcripts encoding T cell regulation proteins, the transcription factor gene EOMES, which drives specific CD8+ effector T cells during viral infection, was highly upregulated in PHIP. THEMIS, which is specific to T cell lineages and is required for T cell receptor (TCR) signal regulation, was also significantly increased. We also observed higher levels of JAKMIP1 transcripts in PHIP, in which the protein Jakmip1 is highly expressed in antigen (Ag)-experienced T cells (28). Moreover, we observed elevated GZMA, GZMB, and GZMH transcripts in PHIP, in accordance with the activated lymphocyte status and the high intracellular expression of granzyme A in HLA-A2–pp65-specific CD8+ T lymphocytes. These data support an efficient lytic phenotype of HCMV-specific T lymphocytes in PHIP. On the other hand, several PHIP-upregulated genes supported inhibitory regulation following T lymphocyte activation and their commitment to cell exhaustion. Finally, most of the phenotypic markers evaluated, including NKG2C, NKG2A, CD27, CD38, CD45RA, PD-1, granzyme A, CD4, CD19, and CD28, could be detected as differentially expressed at the transcriptional level in PHIP compared to HCMV− transcripts (Fig. 6D). Similar results were observed in comparing PHIP to HCMV+ 2C− and HCMV+ 2C+ healthy individuals (data not shown). Gene expression profiling of circulating PBMCs provided a global overview of differential gene expression between HCMV-free or latently HCMV-infected individuals and PHIP.
DISCUSSION
Extensive studies have reported strong immune effector mobilization during HCMV infection and its consequent imprint on the immune repertoire. However, these reports were restricted to primary infection or reactivation in immature or immunodeficient individuals (20). Here, we provide the first characterization of cellular and gene expression profiles of innate and adaptive immune repertoire modulations occurring in symptomatic primary HCMV infection in immunocompetent adults. PHIP presented a broad range of clinical features, from the viral syndrome to tissue injuries. Ideally, immunocompetent adults undergoing asymptomatic primary HCMV infection would have been better for comparison than latently infected individuals, but they are hardly manageable, by definition. The first observation in PHIP was the massive lymphocytosis (NK, conventional, and γδ T lymphocytes) associated or not with a CMV load and maintained around day 80 after the onset of symptom appearance for two patients. Interestingly, Jayasooriya et al. have recently reported the case of one asymptomatic Epstein-Barr virus (EBV)-infected Gambian child who carried a virus load equivalent to that seen in acute infectious mononucleosis (AIM) patients but who lacked significant expansion of global T cell numbers despite substantial activation of EBV-specific CD8+ T cells (29). In our study, PHIP mounted HCMV-specific T lymphocytes at frequencies comparable to those previously observed in primary HCMV-infected pregnant women and newborns (30–33). We cannot exclude the hypothesis, suggested in AIM, of the development of heterologous immunity, where an existing response to an epitope encoded by a previously encountered pathogen cross-reacts with another from CMV, amplifying the pool of T cells responsive to CMV challenge and potentially inducing an exaggerated response.
NK cells have been described as potent effectors during HCMV infection, as illustrated by the mature NKG2C+ CD57+ KIR2D+ NK cell subset expansion during HCMV infection in healthy individuals or in transplant recipients (7–9). Although NKG2C NK cell expansion has been described in other viral infections, i.e., hantavirus (34), chikungunya virus (35), and HIV (36), these patients were always HCMV seropositive. We detected the early expansion of NKG2C+ NK cells in PHIP, while KIR2D coexpression was delayed (Fig. 7). Several studies have reported an association between the KIR and HLA genotype and the development of CMV infection following hematopoietic stem cell (37) and solid-organ (38, 39) transplantation. Moreover, a recent study (22) showed increased susceptibility to developing symptomatic primary CMV infection in immunocompetent adults carrying an AA haplotype or the HLA-Bw4T allele. However, by comparing KIR and HLA genotypes of the HCMV− and HCMV+ control groups, we could not identify a specific combination. The numbers of activating or inhibitory KIR genes, as well as the frequency of each KIR gene, were similar between groups. In the same way, no singular HLA class I profile could be highlighted, in terms of either the presence or the absence of cognate KIR ligands. In parallel, we were able to distinguish the highly activated status of NK cells in PHIP, illustrated by their strong cell surface expression of CD38. Although increased in PHIP, variable levels of NKG2D expression could be explained by the time between the onset of disease and when patients were sampled. These observations, together with the high frequency of NKG2A+ NK cells and the low proportion of CD57+ NK cells, suggest that NK cells from PHIP are undergoing differentiation but are not yet fully mature. Indeed, when comparing NK cell phenotypes from a PHIP from the onset of disease to 4 months later, KIR2D receptors and CD57 expression tended to rise (Fig. 7). Interestingly, this mirrors a feature recently described in acute primary EBV infection (40), where early differentiated NKG2A+ KIR− NK cells were predominant during AIM while CD57 expression was delayed and increased at the same time as expansion of the NKG2A− KIR+ NK cell subset several months after AIM. Furthermore, functional assays performed to investigate the degranulation and IFN-γ production capacities of NK cells demonstrated similar potentials in PHIP and the control groups.
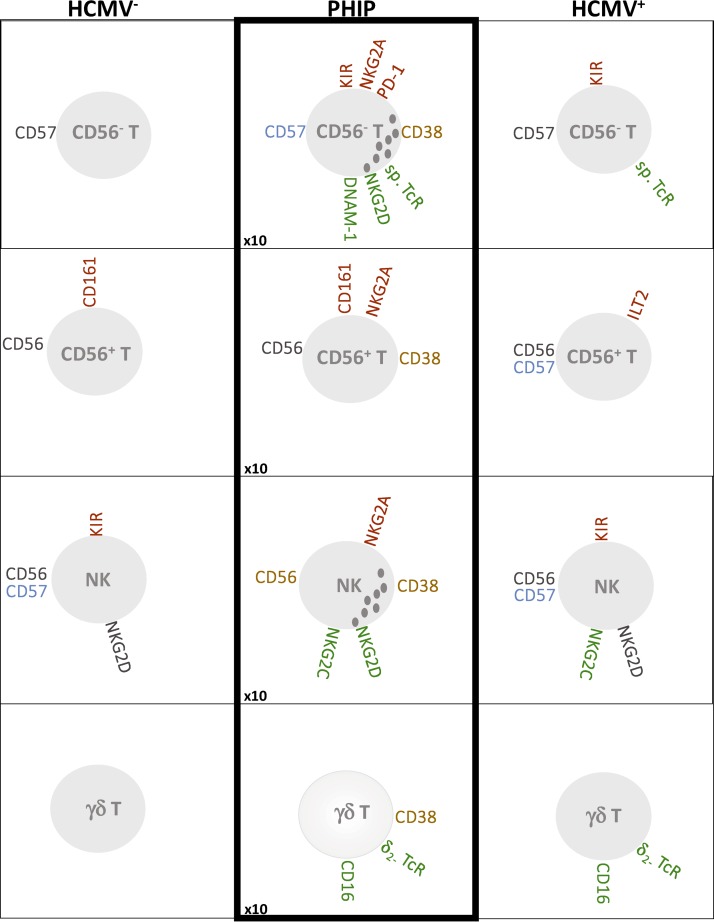
FIG 7.
Schematic representation of main phenotypic modulations on T cell (CD56−, CD56+, and γδ) and NK cell compartments. Inhibitory receptors are indicated in red, activating receptors in green, receptors with increased MFI in blue, and markers associated with activated status in orange. The increased ratio of absolute lymphocyte counts in PHIP is indicated at the bottom left.
Analysis of the γδ T cell compartment revealed expansion of a Vδ2(−) γδ T cell subset in HCMV+ healthy individuals and PHIP, as previously observed in immature or immunodeficient individuals (11–15, 18). Because these cells were activated in PHIP, especially Vδ2(−) γδ CD16+ T cells, and were not significantly detected in latent HCMV+ donors compared to HCMV− individuals, we hypothesize that CD16 expression follows γδ T cell activation, as suggested by Couzi et al. (18). Finally, we observed joint NKG2C+ NK and Vδ2(−) γδ T cell expansion, corresponding to the imprint of HCMV on immune responses in PHIP (Fig. 7).
Major changes were also observed in T lymphocyte phenotypes in PHIP, including lymphocytosis accompanied by an inversion of the CD4/CD8 ratio. This was supported by the gene expression profile analysis, where numerous cell cycle genes were upregulated. The decreased frequency of CD45RA+ T cells and increased proportions of PD-1+ T cells are consistent with immune mobilization. PHIP mount an HCMV-specific CD8+ T cell response with an effector phenotype characterized by a CD45RAlo CCR7− CD27lo CD28− GzmAhi profile with a high transcription level of EOMES, in accordance with the recent thorough characterization of all specific T cell subsets in HCMV+ individuals (41). In addition, several genes involved in T cell regulation were upregulated in PHIP. The upregulation of CXCL10, CXCR3, CCR5, CCL2, and IL-12RB1 suggests their ability to migrate to inflammatory sites, including HCMV-infected endothelial cells from blood vessels (42), and to interact with other immune effectors. Furthermore, it has been recently reported that IL-12 produced by monocytes is essential to control HCMV-driven NKG2C+ NK cell expansion (43). In addition, we observed a strong increase in APOBEC3C, -D, and -H transcripts in PHIP, suggesting a role for these enzymes in HCMV replication and the activation of antiviral immunity. APOBEC3 has been reported to be a restriction factor for some DNA viruses, including human herpesviruses, such as herpes simplex virus 1 (HSV-1) and EBV (44). Interestingly, the CD56+ T cell phenotype displayed similar features in the HCMV− control group and PHIP by comparison to latently HCMV-infected individuals, suggesting maturation in progress (Fig. 7). An interesting finding was the observation in PHIP of downregulation of CD1D (data not shown), a nonclassical HLA class I molecule that recognizes NKT cell TCR. HCMV has been shown to downregulate CD1d at the transcriptional level, thereby acting as an immune escape mechanism (45). This extensive study shows that during the symptomatic stage of acute HCMV infection, PHIP mounted appropriate cellular immune responses. However, the excessive activation could be caused by a lack of normal downregulation of activated lymphocytes and macrophages. Of note, all the studied patients presented lymphocyte activation, and P20 developed a macrophage activation syndrome. Thus, we cannot exclude the dysfunction of a particular subset, such as monocytes or macrophages, not investigated in this cohort. Moreover, the symptomatic HCMV infection was associated with massive lymphocytosis that could be the result of nonadapted or uncontrolled immune responses limiting the effectiveness of specific mounted responses. It is also possible that the lymphocytosis was associated with excessive cytokine production by macrophages, NK cells, and T lymphocytes, which is thought to be a primary mediator of tissue damage. We cannot exclude the possibility that the appearance of symptoms could be also the consequence of a delayed immune response caused by HCMV strains particularly involved in immune escape. Although the immune evasion mechanisms employed by HCMV in vitro are very well documented, their effectiveness during primary infection in vivo is not absolutely clear and perhaps may be better seen as viral functions that allow the pathogen to initially overcome host immune responses and thus create a window for the virus to replicate efficiently and disseminate to different organs. This hypothesis relies on several studies (46) suggesting HCMV virulence variation is due to genome diversity. A more thorough characterization of molecular HCMV strains isolated from PHIP samples is in progress and will allow evaluation of the immune escape mechanisms resulting from specific viral genetic polymorphisms.
MATERIALS AND METHODS
Study groups.
The HCMV serological status of healthy volunteers was assessed using the chemiluminescent immunoassay Liaison CMV IgG II (Diasorin, Saluggia, Italy). Twenty-six HCMV− and 39 HCMV+ individuals were included in the study.
Twenty adult patients (mean age, 38 ± 13 years; 12 men and 8 women) diagnosed with acute HCMV primary infection at the Nantes University Hospital were prospectively enrolled between 2009 and 2015 (Table 1). EDTA blood samples were collected during the acute phase of HCMV infection. HCMV serology was performed using the Liaison CMV IgG, Liaison CMV IgM, and Liaison CMV IgG avidity tests. Additional evidence of active HCMV replication was examined using an in-house real-time HCMV PCR in whole blood, adapted from the method of Bressollette-Bodin et al. (47).
Symptomatic primary HCMV infection was defined as a mononucleosis-like syndrome with fever, malaise, lymphocytosis (>4.0 × 109 lymphocytes/ml), and/or the presence of activated lymphocytes, combined with serology compatible with HCMV primary infection (positive HCMV IgM and/or IgG with low avidity) (Table 1). All the patients displayed general symptoms, such as fever, arthralgia/myalgia, asthenia, headache, and hepatosplenomegaly. Six patients displayed organ disease (two pneumonia, one meningoradiculopathy, one hepatitis, and two vascular thrombosis). Two out of 20 patients (P16 and P20) received curative therapy using intravenous (i.v.) ganciclovir (5 mg/kg of body weight twice daily) or oral (p.o.) valganciclovir (900 mg twice daily). HCMV viremia was determined in 17/20 patients and was positive in 14/17.
All the patients were tested for EBV serology (anti-EBNA IgG, anti-VCA IgG, and anti-VCA IgM) and showed a profile of prior immunization (positive anti-IgG EBNA and VCA). They were also tested for HIV-1/2, and hepatitis B virus (HBV). Some of the patients were also tested for herpes simplex virus (HSV), hepatitis A virus (HAV), and hepatitis C virus (HCV) serology. The combination of positive CMV IgG, positive IgM, low avidity, and positive PCR was used for confirmation of primary HCMV infection.
Ethics statement.
Healthy volunteers were recruited at the Blood Transfusion Center (Etablissement Français du Sang [EFS], Nantes, France), and informed written consent was obtained for all the blood donors. Adult patients diagnosed with acute HCMV primary infection were enrolled after giving informed written consent in accordance with the Declaration of Helsinki (CNIL declaration no. 1779686).
PBMC and cell lines.
PBMCs were isolated from citrate-phosphate-dextrose blood samples from healthy volunteers and PHIP, using Ficoll-Hypaque density gradient centrifugation (Lymphoprep; Axis-Shield, Oslo, Norway) and cryopreserved. The HLA class I-deficient lymphoblastoid cell line 721.221, referred to here as 221 cells (ATCC CRL-1855), and the murine mastocytoma cell line P815 (ATCC TIB-64) were used to determine the degranulation potential and IFN-γ production of NK cells. Both the 221 and P815 cell lines were cultured in RPMI 1640 medium (Life Technologies, Paisley, United Kingdom) containing glutamine (Life Technologies) and penicillin-streptomycin (Life Technologies) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Life Technologies) or human serum (EFS, Nantes) for the P815 cell line. Mycoplasma tests performed by PCR were negative for the two cell lines.
KIR and HLA typing.
Genomic DNA was extracted from PBMCs using a classical salting-out method (48). High-resolution typing for HLA-A, HLA-B, and HLA-C loci was carried out on all studied individuals with a sequence-based typing kit (Abbott Molecular Park, IL). All DNAs were typed for KIR2DL1, -2DL2, -2DL3, -2DL4, -2DL5A/B, -3DL1, -3DL2, -3DL3, -2DS1, -2DS2, -2DS3, -2DS4/1D, -2DS5, and -3DS1 using a KIR multiplex with PCR with sequence-specific primer (PCR-SSP) method, as previously described (49). KIR genotypes were determined based on the presence or absence of activating KIR, KIR AA genotype presenting only KIR2DS4 as the activating KIR, and KIR B+ genotype presenting several activating KIRs. KIR ligands C1, C2, Bw4, and Bw6 were defined based on allelic HLA class I typing.
NK cell phenotypic analysis by flow cytometry.
The NK cell phenotype was determined by four-color flow cytometry using the following monoclonal Abs (MAbs) against CD3 (SK7), CD56 (NCAM16.2), CD57 (HNK-1), CD69 (FN50), NKG2D (ID11), CD161 (DX12), DNAM-1 (DX11), ILT-2 (GH1/75), CD8 (HIT8a; BD Biosciences, San Jose, CA, USA), PD-1 (J105), CD38 (HB7; eBioscience), KIR2DL1 (143211; R&D Systems, Minneapolis, MN, USA), NKG2A (Z199), KIR2DL1/S1 (EB6), KIR2DL2/S2/2DL3 (GL183; Beckman Coulter, Brea, CA, USA), NKG2C (134591; R&D Systems), and KIR2DL1/L2/L3/2DS1/2 (1A6) (50). Data were collected using a FACSCalibur (BD Biosciences) and analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
CD107a mobilization assay and IFN-γ secretion assay detected by flow cytometry.
Cytotoxic and cytokine functions, respectively, were measured via CD107a (LAMP-1) and IFN-γ production by NK cells in two 5-hour functional assays to assess spontaneous lysis and ADCC functions. PBMCs were preincubated with fluorescein isothiocyanate (FITC)-conjugated anti-CD107a (H4A3; BD Biosciences) and then cocultured (effector/target ratio, 1:1) with HLA-I-defective 721.221 cells or medium (negative control) to assess NK cell degranulation according to spontaneous lysis function. The Fc receptor-positive P815 cell line was preincubated with anti-CD16 (3G8; Beckman Coulter) or IgG1 (MOPC-21; BD Biosciences) control and then cocultured (effector/target ratio, 1:1) with CD107a-preincubated PBMCs to assess the reverse ADCC function. After 1 h of incubation, brefeldin A (Sigma-Aldrich, Saint Louis, MO, USA) was added at 10 mg/ml to block trans-Golgi transport and allow the intracellular accumulation of IFN-γ. PBMC surfaces were stained and then permeabilized before intracellular staining with phycoerythrin (PE)-conjugated anti-IFN-γ (B27; BD Biosciences).
αβ T cell phenotypic analysis by flow cytometry.
The αβ T lymphocyte phenotype was determined by four-color flow cytometry using MAbs against CD3, CD4 (RPA-T4), CD8, CD56, CD57, granzyme A (CB9), perforin (γG9) (BD Biosciences), CD27 (O323; Biolegend, San Diego, CA, USA), CD45RA (MEM-56; Invitrogen, Carlsbad, CA, USA), and CD28 (CD28.2; eBioscience, San Diego, CA, USA). Common markers with NK cells as activating receptors (CD38, CD69, NKG2C, NKG2D, and DNAM-1) and inhibitory receptors (KIR2DL2/3, KIR3DL1, NKG2A, CD161, and PD-1) were investigated on both CD56− and CD56+ T cells. HLA-A*02+ HCMV+ healthy individuals and patients were selected to assess the HCMV-specific CD8+ T cell phenotype. PBMCs were stained with HLA-A*02/HCMVpp65495–504 pentamer-PE (ProImmune, Oxford, United Kingdom).
γδ T cell phenotypic analysis by flow cytometry.
The γδ T lymphocyte surface phenotype was determined by four-color flow cytometry using MAbs against CD3, CD16 (NKP15; BD Biosciences), CD38, γδ TCR (B1.1; eBioscience), and γδ TCR Vδ2 (B6; BD Biosciences).
RNA extraction. (i) Microarrays.
Total RNA from 13 HCMV−, 12 HCMV+ NKG2C−, and 12 HCMV+ NKG2C+ healthy individuals and 11 PHIP was extracted from 1 million PBMCs using the NucleoSpin RNA XS kit (Macherey-Nagel, Düren, Germany). RNA quality and quantity were checked with a NanoDrop 2000 UV-spectrophotometer (ThermoScientific, Waltham, MA, USA) and with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA preamplification, labeling, and hybridization on a HuGene 2.1 ST 24 array (Affymetrix, Santa Clara, CA, USA) were performed with 100 ng total RNA according to the manufacturer's instructions.
(ii) Microarray analysis and heat map generation.
Samples from 12 HCMV−, 12 HCMV+ 2C−, and 13 HCMV+ 2C+ individuals and 11 PHIP were included in the transcriptional study. Raw data were quality control checked using R statistical software (http://www.r-project.org). Expression data were normalized according to the robust multiarray average (RMA) method based on a quantile normalization algorithm. Then, the data were filtered to remove probes with low intensity values. Thus, 28,402 probes out of 53,617 were filtered for the analysis. Unsupervised hierarchical clustering was computed on median-gene-centered and log-transformed data using average linkage and uncentered correlation distances with the Cluster 3.0 program (51). The biological significance of each cluster was determined using GOminer software. GO categories with enrichment of >1.5, an FDR of <0.01, and >5 changed genes were selected for further analysis. A differential analysis was then performed in order to highlight the most DEG. For that, differentially expressed probes were determined using the R Limma package (Bioconductor) based on a Bayesian model. P values were then corrected by the Benjamini-Hochberg method in order to control the false-positive rate. A probe was considered differentially expressed when its corrected P value was <0.05. These sets of genes were then annotated for biological significance using GOminer.
Statistics.
All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, La Jolla, CA). One-way or two-way analysis of variance (ANOVA) tests were used to compare multiple groups. Student's t test was performed for comparisons between two groups. The χ2 test was used to analyze 2-by-2 contingency tables for frequency comparisons. Spearman's rank correlation coefficients were calculated and indicated when a significant P value was observed. P values of <0.05 were considered statistically significant.
Accession number(s).
Raw microarray data were deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE81246.
ACKNOWLEDGMENTS
We are grateful to all the blood donors and patients for participating in this study. We thank Zakia Djaoud (Department of Structural Biology and Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA, USA) for review, Nicolas Dulphy (Université Paris Diderot, Sorbonne Paris Cité, Institut Universitaire d'Hématologie, Paris, France) for advice on transcriptomic analysis, and Britt House (France) for help in editing of the manuscript. We are grateful to the Genomics and Bioinformatics Core Facility of Nantes (Biogenouest) for technical support.
We declare no competing financial interests.
This work was financially supported by the Etablissement Français du Sang (EFS)/Pays de la Loire and by grants from EFS (2014-11), Région des Pays de la Loire (ARMINA Project), Leucémie Espoir Atlantique Famille (LEAF), and Nantes Atlantique Greffe de Moelle Osseuse (NAGMO). R.R. is a Ph.D. student supported by Région des Pays de la Loire (ARMINA Project).
REFERENCES
- 1.van de Berg PJEJ, van Stijn A, Ten Berge IJM, van Lier RAW. 2008. A fingerprint left by cytomegalovirus infection in the human T cell compartment. J Clin Virol 41:213–217. doi: 10.1016/j.jcv.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Sissons JGP, Wills MR. 2015. How understanding immunology contributes to managing CMV disease in immunosuppressed patients: now and in future. Med Microbiol Immunol 204:307–316. doi: 10.1007/s00430-015-0415-0. [DOI] [PubMed] [Google Scholar]
- 3.Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 4.Orange JS. 2002. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect 4:1545–1558. doi: 10.1016/S1286-4579(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 5.Saunders PM, Vivian JP, O'Connor GM, Sullivan LC, Pymm P, Rossjohn J, Brooks AG. 2015. A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol Rev 267:148–166. doi: 10.1111/imr.12319. [DOI] [PubMed] [Google Scholar]
- 6.Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Björklund AT, Flodström-Tullberg M, Michaëlsson J, Rottenberg ME, Guzmán CA, Ljunggren H-G, Malmberg K-J. 2010. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 7.Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Vergès S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang S-M, Norris PJ, Nixon DF, Lanier LL. 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A 108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Vergès S, Lanier LL, Weisdorf D, Miller JS. 2012. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119:2665–2674. doi: 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Béziat V, Liu LL, Malmberg J-A, Ivarsson MA, Sohlberg E, Björklund AT, Retière C, Sverremark-Ekström E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaëlsson J, Ljunggren H-G, Malmberg K-J. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitard V, Roumanes D, Lafarge X, Couzi L, Garrigue I, Lafon M-E, Merville P, Moreau J-F, Déchanet-Merville J. 2008. Long-term expansion of effector/memory Vdelta2-gammadelta T cells is a specific blood signature of CMV infection. Blood 112:1317–1324. doi: 10.1182/blood-2008-01-136713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornara C, Lilleri D, Revello MG, Furione M, Zavattoni M, Lenta E, Gerna G. 2011. Kinetics of effector functions and phenotype of virus-specific and γδ T lymphocytes in primary human cytomegalovirus infection during pregnancy. J Clin Immunol 31:1054–1064. doi: 10.1007/s10875-011-9577-8. [DOI] [PubMed] [Google Scholar]
- 13.Vermijlen D, Brouwer M, Donner C, Liesnard C, Tackoen M, Van Rysselberge M, Twité N, Goldman M, Marchant A, Willems F. 2010. Human cytomegalovirus elicits fetal gammadelta T cell responses in utero. J Exp Med 207:807–821. doi: 10.1084/jem.20090348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Déchanet J, Merville P, Lim A, Retière C, Pitard V, Lafarge X, Michelson S, Méric C, Hallet MM, Kourilsky P, Potaux L, Bonneville M, Moreau JF. 1999. Implication of gammadelta T cells in the human immune response to cytomegalovirus. J Clin Invest 103:1437–1449. doi: 10.1172/JCI5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Déchanet J, Merville P, Bergé F, Bone-Mane G, Taupin JL, Michel P, Joly P, Bonneville M, Potaux L, Moreau JF. 1999. Major expansion of gammadelta T lymphocytes following cytomegalovirus infection in kidney allograft recipients. J Infect Dis 179:1–8. doi: 10.1086/314568. [DOI] [PubMed] [Google Scholar]
- 16.Lafarge X, Merville P, Cazin MC, Bergé F, Potaux L, Moreau JF, Déchanet-Merville J. 2001. Cytomegalovirus infection in transplant recipients resolves when circulating gammadelta T lymphocytes expand, suggesting a protective antiviral role. J Infect Dis 184:533–541. doi: 10.1086/322843. [DOI] [PubMed] [Google Scholar]
- 17.Roux A, Mourin G, Larsen M, Fastenackels S, Urrutia A, Gorochov G, Autran B, Donner C, Sidi D, Sibony-Prat J, Marchant A, Stern M, Sauce D, Appay V. 2013. Differential impact of age and cytomegalovirus infection on the γδ T cell compartment. J Immunol 191:1300–1306. doi: 10.4049/jimmunol.1202940. [DOI] [PubMed] [Google Scholar]
- 18.Couzi L, Pitard V, Sicard X, Garrigue I, Hawchar O, Merville P, Moreau J-F, Déchanet-Merville J. 2012. Antibody-dependent anti-cytomegalovirus activity of human γδ T cells expressing CD16 (FcγRIIIa). Blood 119:1418–1427. doi: 10.1182/blood-2011-06-363655. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson GWG, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang ECY, Griffin CA, Davison AJ. 2008. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rafailidis PI, Mourtzoukou EG, Varbobitis IC, Falagas ME. 2008. Severe cytomegalovirus infection in apparently immunocompetent patients: a systematic review. Virol J 5:47. doi: 10.1186/1743-422X-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancini D, Faddy HM, Flower R, Hogan C. 2014. Cytomegalovirus disease in immunocompetent adults. Med J Aust 201:578–580. doi: 10.5694/mja14.00183. [DOI] [PubMed] [Google Scholar]
- 22.Di Bona D, Scafidi V, Plaia A, Colomba C, Nuzzo D, Occhino C, Tuttolomondo A, Giammanco G, De Grazia S, Montalto G, Duro G, Cippitelli M, Caruso C. 2014. HLA and killer cell immunoglobulin-like receptors influence the natural course of CMV infection. J Infect Dis 210:1083–1089. doi: 10.1093/infdis/jiu226. [DOI] [PubMed] [Google Scholar]
- 23.Chan WK, Rujkijyanont P, Neale G, Yang J, Bari R, Das Gupta N, Holladay M, Rooney B, Leung W. 2013. Multiplex and genome-wide analyses reveal distinctive properties of KIR+ and CD56+ T cells in human blood. J Immunol 191:1625–1636. doi: 10.4049/jimmunol.1300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almehmadi M, Hammad A, Heyworth S, Moberly J, Middleton D, Hopkins MJ, Hart IJ, Christmas SE. 2015. CD56+ T cells are increased in kidney transplant patients following cytomegalovirus infection. Transpl Infect Dis 17:518–526. doi: 10.1111/tid.12405. [DOI] [PubMed] [Google Scholar]
- 25.Almehmadi M, Flanagan BF, Khan N, Alomar S, Christmas SE. 2014. Increased numbers and functional activity of CD56+ T cells in healthy cytomegalovirus positive subjects. Immunology 142:258–268. doi: 10.1111/imm.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis ZB, Cooley SA, Cichocki F, Felices M, Wangen R, Luo X, DeFor TE, Bryceson YT, Diamond DJ, Brunstein C, Blazar BR, Wagner JE, Weisdorf DJ, Horowitz A, Guethlein LA, Parham P, Verneris MR, Miller JS. 2015. Adaptive natural killer cell and killer cell immunoglobulin-like receptor-expressing T cell responses are induced by cytomegalovirus and are associated with protection against cytomegalovirus reactivation after allogeneic donor hematopoietic cell transplantation. Biol Blood Marrow Transplant 21:1653–1662. doi: 10.1016/j.bbmt.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monsiváis-Urenda A, Noyola-Cherpitel D, Hernández-Salinas A, García-Sepúlveda C, Romo N, Baranda L, López-Botet M, González-Amaro R. 2010. Influence of human cytomegalovirus infection on the NK cell receptor repertoire in children. Eur J Immunol 40:1418–1427. doi: 10.1002/eji.200939898. [DOI] [PubMed] [Google Scholar]
- 28.Libri V, Schulte D, van Stijn A, Ragimbeau J, Rogge L, Pellegrini S. 2008. Jakmip1 is expressed upon T cell differentiation and has an inhibitory function in cytotoxic T lymphocytes. J Immunol 181:5847–5856. doi: 10.4049/jimmunol.181.9.5847. [DOI] [PubMed] [Google Scholar]
- 29.Jayasooriya S, de Silva TI, Njie-Jobe J, Sanyang C, Leese AM, Bell AI, McAulay KA, Yanchun P, Long HM, Dong T, Whittle HC, Rickinson AB, Rowland-Jones SL, Hislop AD, Flanagan KL. 2015. Early virological and immunological events in asymptomatic Epstein-Barr virus infection in African children. PLoS Pathog 11:e1004746. doi: 10.1371/journal.ppat.1004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchant A, Appay V, van der Sande M, Dulphy N, Liesnard C, Kidd M, Kaye S, Ojuola O, Gillespie GMA, Vargas Cuero AL, Cerundolo V, Callan M, McAdam KPWJ, Rowland-Jones SL, Donner C, McMichael AJ, Whittle H. 2003. Mature CD8+ T lymphocyte response to viral infection during fetal life. J Clin Invest 111:1747–1755. doi: 10.1172/JCI200317470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miles DJC, van der Sande M, Jeffries D, Kaye S, Ojuola O, Sanneh M, Cox M, Palmero MS, Touray ES, Waight P, Rowland-Jones S, Whittle H, Marchant A. 2008. Maintenance of large subpopulations of differentiated CD8 T-cells two years after cytomegalovirus infection in Gambian infants. PLoS One 3:e2905. doi: 10.1371/journal.pone.0002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miles DJC, van der Sande M, Jeffries D, Kaye S, Ismaili J, Ojuola O, Sanneh M, Touray ES, Waight P, Rowland-Jones S, Whittle H, Marchant A. 2007. Cytomegalovirus infection in Gambian infants leads to profound CD8 T-cell differentiation. J Virol 81:5766–5776. doi: 10.1128/JVI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huygens A, Lecomte S, Tackoen M, Olislagers V, Delmarcelle Y, Burny W, Van Rysselberge M, Liesnard C, Larsen M, Appay V, Donner C, Marchant A. 2015. Functional exhaustion limits CD4+ and CD8+ T-cell responses to congenital cytomegalovirus infection. J Infect Dis 212:484–494. doi: 10.1093/infdis/jiv071. [DOI] [PubMed] [Google Scholar]
- 34.Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, Michaëlsson J, Malmberg K-J, Klingström J, Ahlm C, Ljunggren H-G. 2011. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med 208:13–21. doi: 10.1084/jem.20100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petitdemange C, Becquart P, Wauquier N, Béziat V, Debré P, Leroy EM, Vieillard V. 2011. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog 7:e1002268. doi: 10.1371/journal.ppat.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, Moretta A, Mavilio D. 2010. Chronic HIV-1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS 24:27–34. doi: 10.1097/QAD.0b013e3283328d1f. [DOI] [PubMed] [Google Scholar]
- 37.Cook M, Briggs D, Craddock C, Mahendra P, Milligan D, Fegan C, Darbyshire P, Lawson S, Boxall E, Moss P. 2006. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 107:1230–1232. [DOI] [PubMed] [Google Scholar]
- 38.Hadaya K, de Rham C, Bandelier C, Bandelier C, Ferrari-Lacraz S, Jendly S, Berney T, Buhler L, Kaiser L, Seebach JD, Tiercy JM, Martin PY, Villard J. 2008. Natural killer cell receptor repertoire and their ligands, and the risk of CMV infection after kidney transplantation. Am J Transplant 8:2674–2683. doi: 10.1111/j.1600-6143.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 39.Stern M, Elsässer H, Hönger G, Steiger J, Schaub S, Hess C. 2008. The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. Am J Transplant 8:1312–1317. doi: 10.1111/j.1600-6143.2008.02242.x. [DOI] [PubMed] [Google Scholar]
- 40.Azzi T, Lünemann A, Murer A, Ueda S, Béziat V, Malmberg K-J, Staubli G, Gysin C, Berger C, Münz C, Chijioke O, Nadal D. 2014. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124:2533–2543. doi: 10.1182/blood-2014-01-553024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Aalderen MC, Remmerswaal EBM, Verstegen NJM, Hombrink P, ten Brinke A, Pircher H, Kootstra NA, ten Berge IJM, van Lier RAW. 2015. Infection history determines the differentiation state of human CD8+ T cells. J Virol 89:5110–5123. doi: 10.1128/JVI.03478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vieira Braga FA, Hertoghs KML, van Lier RAW, van Gisbergen KPJM. 2015. Molecular characterization of HCMV-specific immune responses: parallels between CD8(+) T cells, CD4(+) T cells, and NK cells. Eur J Immunol 45:2433–2445. doi: 10.1002/eji.201545495. [DOI] [PubMed] [Google Scholar]
- 43.Rölle A, Pollmann J, Ewen E-M, Le VTK, Halenius A, Hengel H, Cerwenka A. 2014. IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J Clin Invest 124:5305–5316. doi: 10.1172/JCI77440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moris A, Murray S, Cardinaud S. 2014. AID and APOBECs span the gap between innate and adaptive immunity. Front Microbiol 5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Rho SB, Lee JY, Bae J, Park SH, Lee SJ, Lee SY, Ahn C, Kim JY, Chun T. 2013. Human cytomegalovirus (HCMV) US2 protein interacts with human CD1d (hCD1d) and down-regulates invariant NKT (iNKT) cell activity. Mol Cells 36:455–464. doi: 10.1007/s10059-013-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijmons S, Thys K, Mbong Ngwese M, Van Damme E, Dvorak J, Van Loock M, Li G, Tachezy R, Busson L, Aerssens J, Van Ranst M, Maes P. 2015. High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J Virol 89:7673–7695. doi: 10.1128/JVI.00578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bressollette-Bodin C, Coste-Burel M, Besse B, André-Garnier E, Ferre V, Imbert-Marcille B-M. 2009. Cellular normalization of viral DNA loads on whole blood improves the clinical management of cytomegalovirus or Epstein Barr virus infections in the setting of pre-emptive therapy. J Med Virol 81:90–98. doi: 10.1002/jmv.21334. [DOI] [PubMed] [Google Scholar]
- 48.Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun JY, Gaidulis L, Miller MM, Goto RM, Rodriguez R, Forman SJ, Senitzer D. 2004. Development of a multiplex PCR-SSP method for killer-cell immunoglobulin-like receptor genotyping. Tissue Antigens 64:462–468. doi: 10.1111/j.1399-0039.2004.00303.x. [DOI] [PubMed] [Google Scholar]
- 50.David G, Morvan M, Gagne K, Kerdudou N, Willem C, Devys A, Bonneville M, Folléa G, Bignon J-D, Retière C. 2009. Discrimination between the main activating and inhibitory killer cell immunoglobulin-like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology 128:172–184. doi: 10.1111/j.1365-2567.2009.03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]