ABSTRACT
Epstein-Barr virus (EBV) latently infects normal B cells and contributes to the development of certain human lymphomas. Newly infected B cells support a highly transforming form (type III) of viral latency; however, long-term EBV infection in immunocompetent hosts is limited to B cells with a more restricted form of latency (type I) in which most viral gene expression is silenced by promoter DNA methylation. How EBV converts latency type is unclear, although it is known that type I latency is associated with a germinal center (GC) B cell phenotype, and type III latency with an activated B cell (ABC) phenotype. In this study, we have examined whether expression of TET2, a cellular enzyme that initiates DNA demethylation by converting 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), regulates EBV latency type in B cells. We found that TET2 expression is inhibited in normal GC cells and GC type lymphomas. In contrast, TET2 is expressed in normal naive B cells and ABC type lymphomas. We also demonstrate that GC type cell lines have increased 5mC levels and reduced 5hmC levels in comparison to those of ABC type lines. Finally, we show that TET2 promotes the ability of the EBV transcription factor EBNA2 to convert EBV-infected cells from type I to type III latency. These findings demonstrate that TET2 expression is repressed in GC cells independent of EBV infection and suggest that TET2 promotes type III EBV latency in B cells with an ABC or naive phenotype by enhancing EBNA2 activation of methylated EBV promoters.
IMPORTANCE EBV establishes several different types of viral latency in B cells. However, cellular factors that determine whether EBV enters the highly transforming type III latency, versus the more restricted type I latency, have not been well characterized. Here we show that TET2, a cellular enzyme that initiates DNA demethylation by converting 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), regulates EBV latency type in B cells by enhancing the ability of the viral transcription factor EBNA2 to activate methylated viral promoters that are expressed in type III (but not type I) latency. Furthermore, we demonstrate that (independent of EBV) TET2 is turned off in normal and malignant germinal center (GC) B cells but expressed in other B cell types. Thus, restricted TET2 expression in GC cells may promote type I EBV latency.
KEYWORDS: 5hmC, EBNA2, EBV, Epstein-Barr virus, TET2, germinal center B cell, latency
INTRODUCTION
Epstein-Barr virus (EBV) is human gammaherpesvirus that infects over 90% of the world population. EBV is the causative agent of infectious mononucleosis and is also associated with several malignancies of B cell and epithelial cell origin, including Burkitt lymphoma (BL), Hodgkin's disease, posttransplant lymphoproliferative disorder, diffuse large B cell lymphoma (DLBCL), nasopharyngeal carcinoma (NPC), and gastric carcinoma (1, 2). EBV establishes latent infection in normal B cells, in which the virus persists as an episome and is replicated once per cell cycle by the cellular DNA polymerase in conjunction with the viral EBNA1 protein (3). There are at least three different forms of EBV latency, which differ with regard to the number of viral genes expressed. Type III latency (in which all 9 EBV latency proteins are expressed) is the only latency form sufficient to transform B cells in vitro (reviewed in reference 4). However, since type III latency is highly immunogenic, it occurs in immunocompetent humans only during the initial stage of viral infection. Subsequently, EBV infection in B cells converts to a more stringent form of viral latency (type I latency), in which only the EBNA1 protein is expressed (in addition to noncoding viral encoded RNAs (reviewed in references 5 and 6). The cellular factors that regulate EBV latency type in infected B cells remain poorly understood.
Following EBV infection of B cells, the virus initially establishes type III latency. The first latent viral transcript expressed is derived from the EBV Wp promoter and is biscistronic, encoding both EBNA2 and EBNA-LP (2). EBNA2, a transcription factor, then activates expression of the EBV Cp promoter, which drives expression of all EBNA genes during type III latency (including EBNA2), and promoters for the EBV latent membrane proteins (LMPs) (1, 2). The divergent LMP1/LMP2B promoter drives expression of the LMP1 and LMP2B genes, and the LMP2A promoter drives the expression of LMP2A.
EBNA2 does not bind to DNA directly but instead activates EBNA gene transcription by interacting with the cellular transcription factor RBP-Jκ, which binds to sites in the C promoter (7). EBNA2 also interacts with RBP-Jκ to activate LMP2Ap (8). In the case of the LMP1/LMP2B promoter, EBNA2 interacts with RBP-Jκ as well as the cellular transcription factor PU.1 to activate transcription (9). In addition, many EBNA2 binding sites in the cellular genome have been shown to colocalize with binding sites for the essential B cell differentiation factor EBF1 as well as other cellular transcription factors (10).
During type I latency, Cp promoter and EBNA2 expression is turned off, and EBNA1 transcription is instead regulated by the viral Q promoter (Qp). The EBV genome becomes highly methylated during the establishment of type I latency, and stringent type I gene expression is enforced in part by CpG methylation of the viral Cp, LMP1/2B, and LMP2A promoters (11, 12). In contrast, the Cp, LMP1/LMP2B, and LMP2A promoters remain unmethylated in cells with type III latency. Treatment of cells exhibiting type I latency with demethylating agents is sufficient to convert cells to type III latency (13). Furthermore, the ability of the Cp and LMP1/LMP2B promoters to be activated by EBNA2 in transient reporter gene assays has been previously shown to be inhibited by promoter methylation (14, 15). Of note, methylation of Cp does not prevent EBNA2/RBPJκ-mediated activation but instead prevents binding of the cellular AUF1 protein to the EBNA2 enhancer site within Cp (14, 16).
Nevertheless, how methylation of the type III latency promoters is established, or reversed, during normal B cell infection is not clear. Interestingly, type I EBV latency in EBV-infected lymphomas (including Burkitt lymphomas and diffuse large B cell lymphomas) is almost always found in tumors that have a germinal center (GC) phenotype (17–19). In contrast, type III latency is almost exclusively confined to tumors with an activated B cell (ABC) phenotype. Furthermore, there is some evidence that EBV superinfection of EBV-negative DLBCLs in vitro converts these lines to a ABC phenotype, but only when the EBV infection assumes the type III latency form (18).
We hypothesized that a cellular environment unique to GC B cells may promote stringent type I EBV latency by enhancing de novo methylation, and/or decreasing demethylation, of the Cp, LMP1/LMP2B, and LMP2A promoters following EBV infection. Reversal of promoter methylation is mediated by the cellular TET family of enzymes that hydroxylate 5-methylcytosine (5mC), producing 5-hydroxymethylcytosine (5hmC) (reviewed in reference 20). 5hmC is an intermediate for DNA demethylation and is also stably maintained on the genome in some cell types, where it is commonly associated with gene activation (21).
Here we demonstrate that TET2 expression is turned off in normal GC tonsil cells, as well as in EBV-infected GC type BLs with type I latency and EBV-negative GC type DLBCLs. In contrast, we found that TET2 is expressed in normal naive tonsil B cells, in EBV-positive BLs with type III latency (which have an ABC phenotype), and in ABC type DLBCLs. Furthermore, we show that BLs and DLBCLs with an ABC phenotype have globally increased 5hmC-modified cellular DNA, and decreased 5mC-modified cellular DNA, compared to those of BLs and DLBCLs with a GC phenotype. Importantly, we demonstrate that TET2 enhances EBNA2's ability to activate the methylated form of the LMP1 promoter in reporter gene assays and increases its ability to activate LMP1 expression from the endogenous methylated viral genome in BL cells with type I latency. Finally, we show that the EBV C promoter is 5hmC modified in TET2-expressing cells with type III latency and, conversely, is highly methylated and silenced in TET2-deficient cells that maintain type I EBV latency. These results are the first to show that TET2 is downregulated in B cells with the GC phenotype, and they suggest that TET2 expression plays a critical role in determining the EBV latency type in B cells.
RESULTS
TET2 is not expressed in Burkitt lymphoma cell lines with type I latency and a GC type but is expressed in BL cells with type III latency and an ABC phenotype.
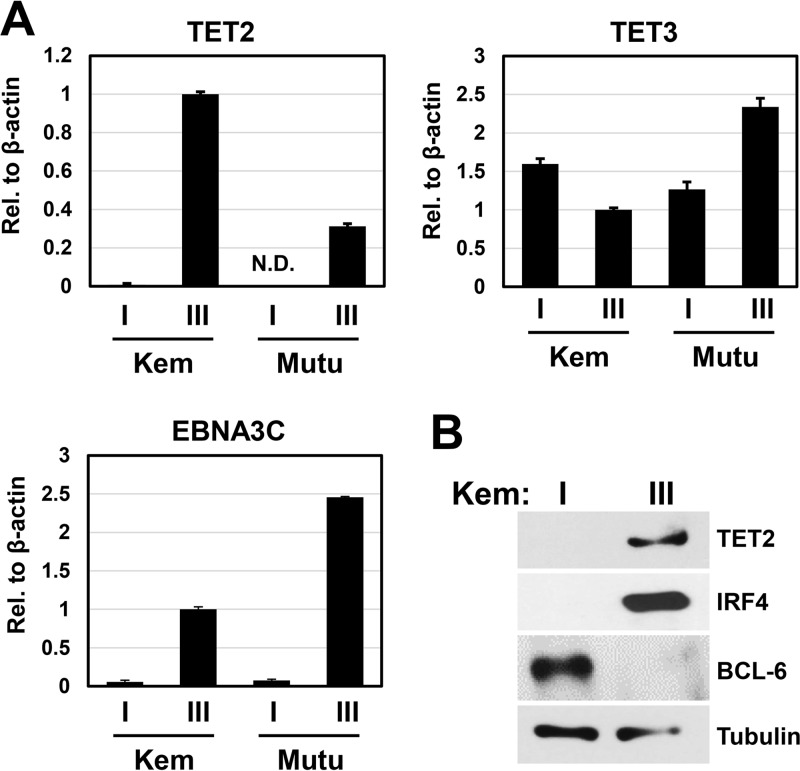
Given that EBV infection in GC type Burkitt lymphomas is associated with a restricted latency gene expression program called type I latency, in which expression of the EBV type III latency genes is silenced by promoter methylation, we explored whether cellular TET proteins might regulate EBV genome CpG methylation in BLs with type I versus type III latency. TET proteins are 5-methylcytosine (5mC) hydroxylases that initiate DNA demethylation by converting 5mC into 5-hydroxymethylcytosine (5hmC) (20). Importantly, TET2/TET3 in vivo knockout in the mouse B cell lineage was recently shown to decrease B cell maturation and inhibit the germinal center reaction (22). Therefore, levels of TET2 and TET3 expression were compared in isogenic EBV+ BL cell lines with type I versus type III latency. Interestingly, reverse transcription-quantitative PCR (RT-qPCR) analysis of the TET2 transcript indicated that EBV+ BL cell lines that had drifted to the type III EBV latency program all expressed the TET2 transcript, while the lines that maintained a type I EBV latent infection did not express TET2 (Fig. 1A). In contrast, levels of TET3 expression were similar in all cells lines (Fig. 1A). EBV latency type was determined by EBNA3C expression (Fig. 1A), which is expressed in type III but not type I latency. Immunoblot analysis of cell lines derived from the same BL tumor (Kem) that had either type I latency (Kem I) versus type III latency (Kem III) confirmed that TET2 protein is expressed in Kem III cells but not Kem I cells (Fig. 1B). Similar results were obtained with Mutu I versus Mutu III cells (data not shown). As previously reported, type I lines had a GC phenotype (BCL6+/IRF4−), whereas type III lines had an ABC phenotype (BCL6−/IRF4+) (Fig. 1B). These results suggest that EBV proteins specifically expressed in type III (but not type I) latency, and/or differences associated with the GC versus ABC phenotype, regulate TET2 expression in Burkitt lymphoma cells.
FIG 1.
TET2 is not expressed in EBV+ BL cell lines with a GC phenotype and type I latency. (A) RNA was isolated from EBV+ BL cell lines (Kem I and Mutu I) with a GC phenotype that maintain type I EBV latency, and isogenic EBV+ BL cell lines (Kem III and Mutu III) with an ABC phenotype that maintain type III EBV latency. Expression of TET2 and TET3, and the type III latency viral gene for EBNA3C, was quantified with RT-qPCR. Transcript levels were quantified relative to β-actin expression, and Kem III transcript levels relative to β-actin were set to 1. Error bars indicate +1 standard deviation calculated from triplicate samples. N.D., not detected. (B) Immunoblot analysis was performed to examine TET2 protein expression in Kem I and Kem III cells, and the B cell phenotype was analyzed by probing for IRF4 (ABC marker) and BCL-6 (GC marker). Tubulin served as a loading control.
TET2 is turned off in EBV-negative DLBCL lines with a GC phenotype but is expressed in DLBCL cell lines with the ABC phenotype.
To determine whether phenotypic differences between GC type and ABC type B cells are associated with altered TET2 expression in the absence of EBV infection, we examined the level of TET2 protein in a series of EBV-negative DLBCL cell lines that had either GC or ABC phenotypes (23). As shown in Fig. 2, TET2 protein was not expressed in four different GC type EBV-negative DLBCL lines (HT, SUDHL4, RL, and DB) but was expressed in four different EBV-negative ABC DLBCL cell lines (HBL1, OCI-Ly10, OCI-Ly3, and SUDHL2). The ABC DLBCL cell lines were confirmed to have an ABC phenotype by the presence of IRF4 expression (Fig. 2). These results suggest that TET2 is specifically turned off in B cells with a GC phenotype rather than activated by EBV infection per se.
FIG 2.
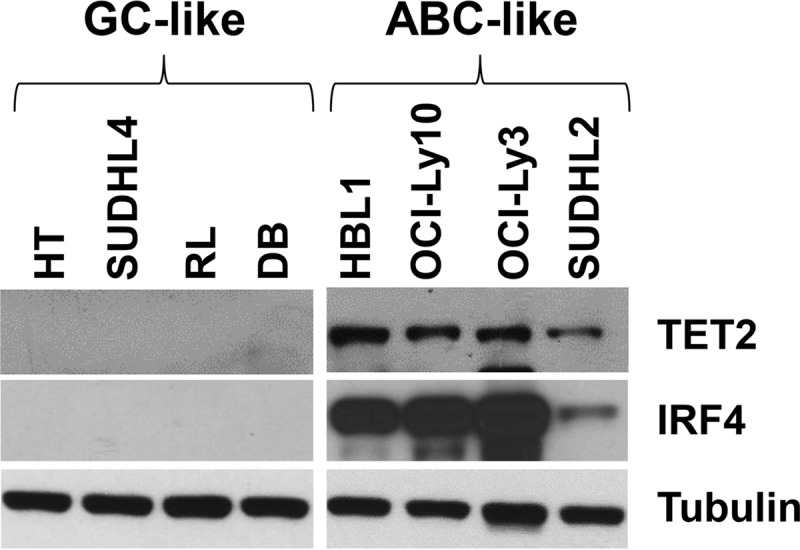
TET2 is not expressed in EBV-negative DLBCLs with a GC phenotype. Immunoblots were performed to compare TET2 protein expression in EBV-negative diffuse large B cell lymphoma (DLBCL) cell lines with a GC phenotype (HT, SUDHL4, RL, and DB) versus an ABC phenotype (HBL1, OCI-Ly10, OCI-Ly3, and SUDHL2); IRF4 served as a marker for ABC phenotype and β-actin as the loading control.
Normal human GC cells do not express TET2.
Given that DLBCL and BL cell lines with a GC phenotype express little, if any, TET2, we hypothesized that TET2 is turned off in normal human GC B cells. To examine this, naive and GC type B cells were isolated from human tonsils, and RNA was extracted from each cell type, as previously described (24). Isolated naive B cells were confirmed by fluorescence-activated cell sorter (FACS) analysis to be IgD+ CD38low CD27−, and isolated GC B cells (centroblasts) were confirmed to be CD77+ CD38high (data not shown). TET2 transcript levels (relative to glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were quantitated by RT-qPCR and were compared to those in Kem I (TET2-negative) versus Kem III (TET2-positive) cell lines.
As shown in Fig. 3, TET2 was highly expressed in primary naive tonsillar B cells (similar to the level expressed in Kem III BL cells). In contrast, little, if any, TET2 expression was observed in either primary GC tonsillar B cells or Kem I BL cells. Since TET2 has been previously demonstrated to be highly expressed in CD19+ B cells (a pan-B cell marker) (25), TET2 expression appears to be specifically inhibited in normal GC B cells, rather than activated in naive B cells and ABC type DLBCLs.
FIG 3.
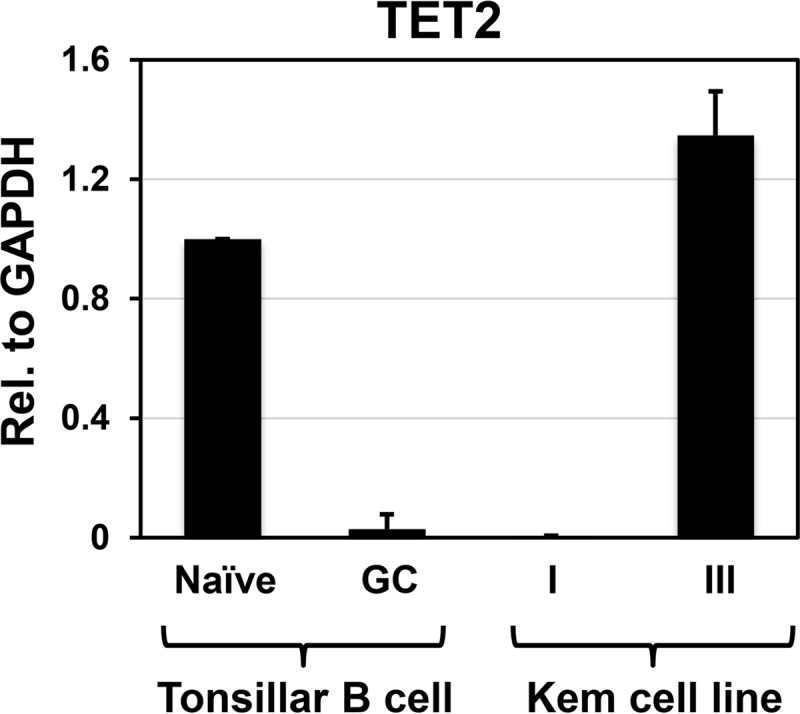
TET2 is turned off in normal human GC B cells. RNA was harvested from separated normal human tonsil naive and GC B cells, and Kem I and Kem III EBV-positive BL cells, and converted to cDNA by RT-PCR. TET2 transcript levels relative to GAPDH were determined by qPCR. The level of TET2 relative to GAPDH in naive B cells was set to 1, and error bars indicate +1 standard deviation calculated from duplicate samples.
Loss of TET2 expression correlates with globally decreased 5hmC-modified DNA, and globally increased 5mC-modified DNA, in BL and DLBCL cell lines.
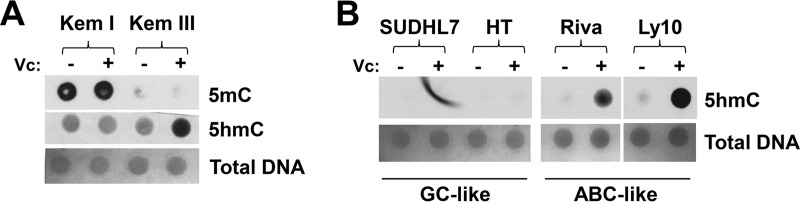
To determine if TET2 expression correlates with the amount of globally 5mC- and/or 5hmC-modified DNA, total DNA was isolated from BL and DLBCL cell lines that had been treated or not for several days with vitamin C (enhances TET enzyme activity) (26–28), and dot blots were performed to quantify global 5mC- and 5hmC-modified DNA in TET2-positive versus TET2-negative cell lines. As shown in Fig. 4A, EBV-infected BL cells with type I latency (Kem I) have globally higher levels of cytosine methylation than isogenic cells that had type III latency (Kem III). Furthermore, Kem I cells have less global 5hmC-modified DNA than Kem III cells. Similar results were obtained with Mutu I versus Mutu III cells (data not shown). Additionally, EBV-negative GC-type DLBCL cell lines have less global 5hmC-modified DNA than EBV-negative ABC-type DLBCL cell lines (Fig. 4B). These results suggest that loss of TET2 expression in normal GC B cells, as well as in GC-type BL and DLBCL lymphomas, inhibits global 5hmC modification while promoting global 5mC modification of DNA.
FIG 4.
GC B cells have less 5hmC, and more 5mC, than B cells with an ABC phenotype. Global levels of 5hmC-modified and 5mC-modified DNA in EBV+ BL cell lines with a type III EBV latency (TET2+, ABC) versus type I EBV latency (TET2−, GC) (A) or EBV-negative DLBCL cell lines with a GC-like phenotype versus ABC-like phenotype (B) were assessed using dot blotting. Cells were treated or not, as indicated, for 4 days with vitamin C (Vc), which enhances TET2 activity prior to harvesting cellular DNA. DNA was spotted on a nitrocellulose membrane (1,000 ng of DNA for 5hmC and 62.5 ng for 5mC), and equal loading of DNA was confirmed by methylene blue staining.
The EBV genome is 5hmC modified at a type III latency promoter in BL cells with an ABC phenotype.
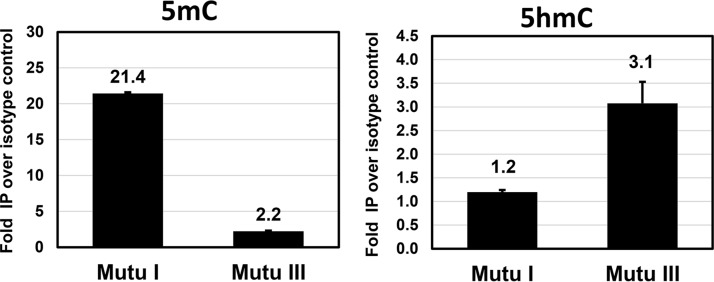
To determine if EBV promoters that are active in cells with type III, but not type I, latency can be 5hmC modified, we quantitated the amount of 5hmC- and 5mC-modified DNA on the type III Cp using the DNA immunoprecipitation (DIP) technique. Mutu I and Mutu III cells were treated for 4 days with acyclovir (to prevent lytic EBV DNA replication, which results in loss of 5hmC and 5mC on the EBV genome), and isolated total DNA was immunoprecipitated using antibodies that specifically detect 5hmC- or 5mC-modified DNA. As expected, the EBV Cp promoter is highly methylated in Mutu I cells (where the promoter is silent) (Fig. 5) and much less methylated in Mutu III cells (where the promoter is active). In contrast, the Cp had little or no 5hmC modification in Mutu I cells (in which TET2 is not expressed) but had detectable 5hmC modification in Mutu III cells (which express TET2) (Fig. 5). These data suggest that TET2 expressed in EBV-infected cell lines can target methylated type III latency promoters for 5hmC modification, which subsequently may promote demethylation and activation of these promoters.
FIG 5.
The EBV type III latency C promoter is methylated in Mutu I cells, and 5hmC modified, but not methylated, in Mutu III cells. A DIP assay was performed to quantitate 5mC and 5hmC modification of the Cp in Mutu I (TET2−, GC) versus Mutu III (TET2+, ABC) BL cells. Cells were treated for 4 days with acyclovir, and then DNA was harvested, sonicated, and immunoprecipitated with anti-5mC, anti-5hmC, or appropriate Ig isotype controls. Promoter immunoprecipitation was quantified using qPCR with primers spanning the Cp. Promoter modification is shown as fold pulldown relative to the isotype control (set to 1).
TET2 enhances EBNA2-mediated activation of the LMP1 promoter.
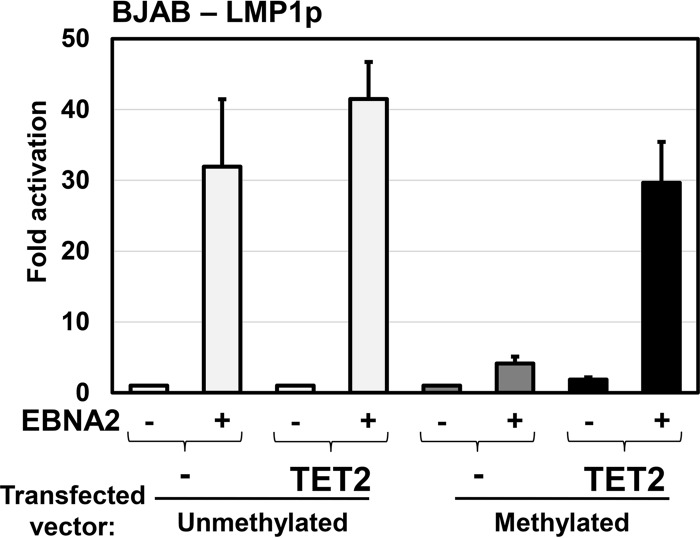
The EBV oncoprotein latent membrane protein 1 (LMP1) is expressed in cells with type III EBV latency, but not type I latency, and the activity of the LMP1 promoter has been shown to be regulated in part by CpG methylation (11, 12, 15). Since EBNA2 activates LMP1 expression, and LMP1 expression is associated with LMP1p demethylation, we explored whether TET2 enhances EBNA2-mediated activation of LMP1 when the promoter is methylated. The LMP1 ED-L1 promoter (the primary LMP1 promoter used in B cells [29]) was cloned upstream of luciferase in a reporter gene vector that lacks any CpGs motifs (pCpGL-Basic), thus preventing nonspecific silencing due to methylation of the luciferase gene itself. The LMP1p luciferase vector was methylated or mock treated in vitro and transfected into the EBV-negative B cell line BJAB, with or without an EBNA2 expression vector, in the presence or absence of a cotransfected TET2 expression vector.
As shown in Fig. 6, in the absence of cotransfected TET2, EBNA2 efficiently activated the unmethylated, but not the methylated, form of the LMP1 promoter. Cotransfected TET2 had little effect on the ability of EBNA2 to activate the unmethylated form of the LMP1 promoter. Importantly, however, coexpression of TET2 greatly increased the ability of EBNA2 to activate the methylated form of LMP1p (Fig. 6). These results suggest that TET2 is required for efficient activation of methylated LMP1 promoter by EBNA2.
FIG 6.
TET2 restores EBNA2 activation of the methylated LMP1 promoter. The ability of EBNA2 to activate the unmethylated, methylated, and 5hmC-modified forms of the EBV LMP1 promoter was analyzed using reporter gene assays. LMP1p was cloned upstream of the luciferase gene in the CpG-free vector pCpGL-basic and was methylated (dark gray) or mock treated (light gray) in vitro. The construct was transfected into EBV− BJAB cells with and without TET2, which converts 5mC into 5hmC (black), in the presence and absence of EBNA2. Luciferase assays were performed 2 days posttransfection. The fold luciferase activation is shown relative to the activity of the unmethylated vector transfected with control vectors, which is set to 1. Error bars indicate standard errors calculated from 2 replicate experiments consisting of triplicate conditions.
TET2 expression enhances EBNA2-mediated type III latency gene expression from the endogenous viral genome in cells with type I latency.
To determine if TET2 enhances the ability of EBNA2 to induce gene expression from the endogenous highly methylated viral genome in cells with type I latency, Mutu I cells were transfected with an EBNA2 expression vector in the presence and absence of a cotransfected (constitutively active) TET2 vector. EBNA2-mediated activation of the type III latency program was determined by immunoblot analysis for LMP1. As shown in Fig. 7, transfected EBNA2 alone induced a very low level of LMP1, while cotransfection of EBNA2 and TET2 greatly enhanced EBNA2-mediated LMP1 expression. These results suggest that the lack of TET2 expression in EBV-infected B cells with a GC phenotype helps to maintain EBV type I latency by suppressing the ability of low-level EBNA2 to induce type III gene transcription.
FIG 7.
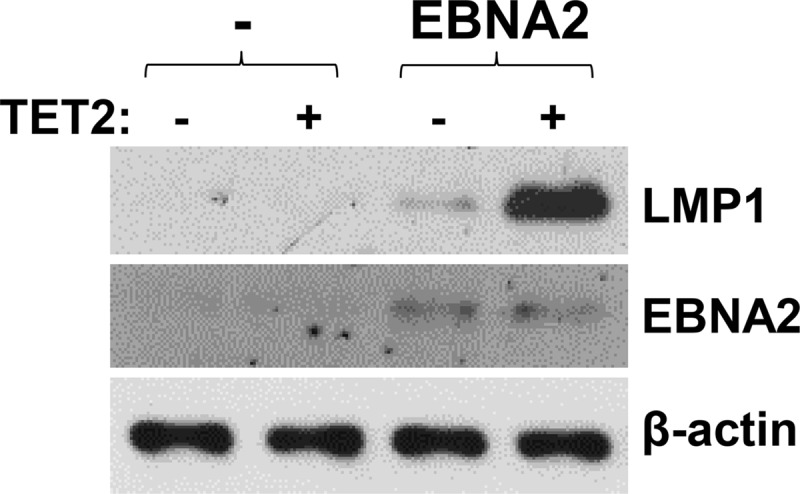
EBNA2 activation of LMP1 in an EBV+ BL cell line with a type I latency is augmented by TET2 expression. Mutu I cells were transfected with a control vector, or an EBNA2 expression vector with and without a TET2 expression vector, as indicated. Two days later, immunoblotting was performed to examine transfected EBNA2 protein expression and induced LMP1 protein expression. β-Actin served as a loading control.
DISCUSSION
EBV-infected cells can enter the most stringent form of viral latency (type I) when critical viral promoters active in type III latency are silenced via cytosine methylation (11, 12). However, exactly why these promoters become methylated in certain cells but not others is not clear. The cellular TET family of enzymes hydroxylates 5-methylcytosine (5mC), producing an intermediate for DNA demethylation, 5-hydroxymethylcytosine (5hmC) (20), and enhances gene transcription. Here we show that TET2 expression is extremely low both in normal GC cells isolated from human tonsils and in B cell lymphomas with a GC phenotype, including Burkitt lymphomas and GC type DLBCLs. Furthermore, we found that TET2 expression in EBV-infected B cells is strongly correlated with the type of viral latency and demonstrated that TET2 enhances the ability of EBNA2 to transcriptionally activate the methylated form of the LMP1 promoter. These results suggest that a lack of TET2 expression in GC cells contributes to the tendency of EBV to convert to type I latency in this cell type, and conversely, TET2 expression in ABC type B cells promotes type III latency both by reversing 5mC modification of the EBV genome and by increasing the ability of EBNA2 to activate methylated viral promoters.
Although the role of EBV latency promoter methylation in enforcing type I EBV latency has been known for some time, how such methylation is regulated remains largely unclear. Methylation of the EBV genome first becomes apparent around 2 weeks after infection of B cells (30). Regulation of viral genome methylation appears to be complex, since inactivation of single DNA methyltransferases (DNMTs) or a combination of DNMT1 and DNMT3B is not sufficient to induce Cp demethylation or activation of type III latency gene expression (31). Two different EBV latency proteins, LMP1 and LMP2A, have been reported to increase expression of cellular DNA methyltransferases (32, 33). Furthermore, the viral latency proteins EBNA3A and EBNA3C induce promoter methylation of the cellular proapoptotic Bim gene through an unknown mechanism (34). Thus, EBV-encoded proteins expressed during type III latency seem to promote EBV (and cellular) genome cytosine methylation, allowing the virus to switch to the most stringent form of viral latency and better evade the host immune response.
Nevertheless, at least in tissue culture, it is clear that certain EBV-infected Burkitt lymphoma lines with type I latency can drift into type III latency. Burkitt cells that convert to type III latency express more anti-apoptotic proteins and have a survival advantage (reviewed in reference 4) and thus tend to overgrow cells with type I latency in vitro. Such lines provide definitive evidence that type I latency, and methylation of type III latency promoters, is reversible. In addition, cells with type III latency are commonly found in immunosuppressed patients (1, 2), although in this case it cannot be ruled out that such cells are newly infected and have not yet undergone viral genome methylation. Given the ability of type I Burkitt lines to switch to type III latency, and to lose cytosine methylation of the latency III promoters, we hypothesized that the demethylating functions of cellular TET protein(s) might be involved in this phenomenon.
In this study, we have unexpectedly discovered that B cell lymphomas with a GC phenotype (regardless of whether they are EBV infected or not) express little, if any, TET2 (Fig. 1 and 2) and that, likewise, normal tonsillar GC cells express much less TET2 than normal naive B cells (Fig. 3). Furthermore, we found that lymphoma cells with a GC phenotype have globally decreased 5hmC-modified DNA, and globally increased 5mC modified DNA, in comparison to those of their ABC counterparts (Fig. 4) and that this effect is EBV independent. In support of our findings in this study, examination of the TET1, -2, and -3 transcript levels reported in a recently published transcriptome sequencing (RNA-seq) data set from four human GC DLBCL cell lines and four human ABC DLBCL cell lines (half of which were not used in this study) (35) revealed that TET2 expression was increased more than 4-fold in ABC lines compared to GC DLBCL lines. In contrast, ABC and GC DLBCLs expressed similar levels of TET3 and expressed little, if any, TET1 (35).
Since TET2 is expressed in many tissue types (36) and has been shown to be highly expressed in pooled CD19+ B cells (a pan-B cell marker) (25), our finding that TET2 is expressed very poorly (if at all) in normal tonsillar GC cells and GC type lymphomas likely results from loss of TET2 expression in GC type B cells rather than a gain of expression in ABC type B cells. TET2 plays a critical role in early stages of B cell development, since in vivo knockout of TET2/TET3 in the mouse B-cell lineage prevents demethylation of B cell-specific enhancers and key regulatory sites, such as the Igκ locus, that occurs normally beginning in early in B cell development, at the pro-B cell stage (22). Phenotypically, TET2/TET3 knockout inhibits B cell maturation and renders B cells unable to mount an immune response and form germinal centers (22). Since the master regulator of the GC phenotype, BCL-6, is a transcriptional repressor (reviewed in reference 37), it is possible that BCL-6 itself, or other transcription factors activated downstream of BCL-6, inhibits transcription of TET2 once B cells have become GC cells. As BCL-6 expression is lost when B cells exit the GC phenotype, this could explain why TET2 is expressed when BL cells convert to an ABC type. Alternatively, cellular transcription factors activated in ABCs (such as IRF4 [23]) may be required for TET2 transcription in B cells. Conversely, TET2 expression may influence B cell phenotype by activating genes involved in GC exit.
Like other groups (19), we found that type III latency in BL cells is associated with an ABC phenotype, while type I latency is associated with a GC phenotype (Fig. 1). Since the EBV EBNA2 latency protein is the key transcription factor required for activation of type III latency viral promoters, we examined how methylation of the LMP1 promoter (the major viral oncoprotein) affects its ability to be activated by EBNA2. We found that cytosine methylation of the LMP1 promoter greatly decreases the ability of EBNA2 to activate the promoter in reporter gene assays and that cotransfection with a constitutively active TET2 vector reverses this effect (Fig. 6). Furthermore, TET2 greatly enhances the ability of EBNA2 to activate expression of the methylated LMP1 promoter from the endogenous viral genome in BL cells with type I latency (Fig. 7). Thus, a lack of TET2 expression in GC cells may promote type I latency both by increasing methylation of viral promoters and by attenuating EBNA2's ability to activate methylated viral promoters.
Interestingly, EBV superinfection of GC-type DLBCLs in vitro converts the cells to an ABC phenotype in lines where EBV establishes type III latency, but not in lines where EBV establishes type I latency, and both LMP1 and EBNA2 contribute to loss of the GC phenotype (18). Thus, EBV proteins expressed during type III latency may stabilize this form of latency by inducing loss of the GC phenotype, with concomitant activation of TET2 expression. Although silencing type III protein expression in EBV-infected B cells normally involves methylation of type III latency promoters, a subset of Burkitt lymphomas has been found to have EBNA2 deletions that convert the virus to more stringent form of latency termed Wp restricted, even in an ABC environment (38); furthermore, some of these EBNA2-deleted tumors have been shown to express IRF4 and thus do not have a strict GC phenotype (38). Thus, deletion of the EBNA2 gene serves as another mechanism by which the type III latency program could be shut down in BLs.
EBNA2 does not bind directly to DNA but is instead tethered to promoters via its interactions with cellular transcription factors, including RBP-Jκ, PU.1, and EBF1 (7, 9, 10). Both PU.1 and EBF1 binding motifs in the LMP1 promoter contribute to its activation by EBNA2 (9, 10). Intriguingly, both PU.1 and EBF-1 were also recently shown to directly interact with TET2, but not the other TET enzymes (21). PU.1 targets TET2 to PU.1 sites in cellular promoters of genes required for osteoclast differentiation, and this interaction results in the promoters becoming 5hmC modified, and then demethylated, during osteoclast differentiation (39). Although the PU.1 DNA binding consensus motif does not contain a CpG motif, the interaction between TET2 and PU.1 results in demethylation of surrounding CpGs that inhibit promoter transcription when methylated (39). Thus, it is likely that the PU.1/EBNA2 complex bound to the LMP1p induces activation of the promoter by recruiting TET2 and inducing demethylation of the promoter. Similarly, the TET2/EBF1 interaction is thought to inhibit methylation of promoters, since TET2 and EBF1 directly interact and colocalize at EBF1 binding sites that are commonly located within promoters that become hypermethylated in cancers with inactive TET function due to IDH1/2 mutations (40). Importantly, the previously reported findings showing that PU.1 and EBF1 specifically interact with TET2 but not other TET proteins help to explain the unique requirement for TET2 (versus TET1/TET3) in EBV-infected B cells for efficient EBNA2 transcriptional function.
It will be important to confirm whether loss of TET2 expression and 5hmC modification is an important part of normal GC cell biology. Somatic hypermutation and affinity maturation of antibodies (which specifically occur in GC cells) are associated with immunoglobulin gene mutation and demethylation via deamination of cytosine into uracil, and perhaps via deamination of 5mC into thymine, by activation-induced deaminase (AID) (41, 42). Demethylation may also be mediated by the activation of TET2 expression, which converts 5mC into 5hmC and the further oxidized forms 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) (20). 5caC-G base pairs were recently shown to be targeted more efficiently for repair back to C-G by thymine-DNA glycosylase (TDG) than T-G mismatches (43). Possibly, the TET2 function interferes and/or competes with the AID function, and hence TET2 must be turned off in GC cells. Finally, it is also possible that TET2 is specifically turned off in malignant GC-type lymphomas and that loss of TET2 plays a critical “driver” role in these lymphomas. Although PU.1 is required at the first stage of B lymphoid development, and its expression level determines whether lymphoid progenitors differentiate into the myeloid or B cell lineage (44), PU.1 also functions as a tumor suppressor during later stages of B cell development and is often turned off in certain B cell malignancies (45, 46). Given the previous findings that TET2 is required for efficient PU.1 activation of genes (39), loss of TET2 expression in B cell malignancies may serve as another mechanism to inhibit PU.1 activity.
Importantly, TET2 loss has recently been implicated in the development of EBV+ nasopharyngeal carcinoma (NPC) (47). EBV exclusively, lytically infects more differentiated normal epithelial cells (48–53), and thus, the viral genome remains highly unmethylated (11). However, in the case of NPC, EBV maintains a latent infection and the viral genome becomes highly methylated (54). We recently showed that TET2 5hmC modifies the EBV genome to promote lytic gene expression in epithelial cells and that NPC tumors lose TET2 expression, which may enhance oncogenesis (47). Furthermore, TET2 was recently found to be a resistance factor of EBV-induced DNA methylation in gastric carcinoma cells, demonstrating that TET2 may have an important role in protection against EBV+ tumor development (55). Thus, regulation of TET2 activity may play critical roles in the development of both epithelial and B cell malignancies associated with EBV.
MATERIALS AND METHODS
Cell lines and culture.
All cell lines were maintained in RPMI 1640 growth medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Invitrogen) at 37°C with 5% CO2 and 100% humidity. Mutu I and Kem I (gifts from Alan Rickinson [Birmingham, England] and Jeff Sample [Penn State]) are EBV+ Burkitt lymphoma cell lines that maintain a type I EBV latency. Mutu III and Kem III (also gifts from Alan Rickinson and Jeff Sample) originated from Mutu I and Kem I cell lines, respectively, but the EBV infection has drifted to type III latency in culture. EBV-negative human diffuse large B cell lymphoma (DLBCL) cell lines with a germinal center (GC)-like phenotype (HT, SUDHL4, RL, DB, and SUDHL7) and activated B cell (ABC)-like phenotype (HBL1, OCI-Ly10, OCI-Ly3, SUDHL2, and RIVA) were acquired from Louis Staudt (NCI) via Lixin Rui (University of Wisconsin-Madison). BJAB (a kind gift from Janet Mertz, University of Wisconsin-Madison) is an EBV-negative B cell lymphoma line thought to be derived from an EBV-negative BL tumor (56).
Plasmids and cloning.
Plasmid DNA was purified with columns using the Qiagen midi/maxiprep kit according to the manufacturer's instructions. pSG5 was acquired from Stratagene. SG5-EBNA2 (a gift from Paul Ling, Baylor College of Medicine) contains the EBNA2 gene under the control of the simian virus 40 (SV40) promoter. pcDNA3.1(+) was obtained from Invitrogen. pcDNA3-FLAG-TET2 (a gift from Yi Zhang, Howard Hughes Medical Institute and Harvard Medical School) contains the FLAG-tagged catalytic domain of the mouse Tet2 coding sequence under the control of cytomegalovirus (CMV) promoter (57).
pCpGL-basic (a gift from Micheal Rehli, Universitätsklinikum Regensburg) is a CpG-free, promoterless luciferase reporter gene vector that was constructed as previously described (58). The proximal LMP1 ED-L1 promoter was PCR amplified using the EBV B95.8 genome as the template with the primers 5′-GCTACTAGTTCATGACACTCGCACAGCCCACACC-3′ and 5′-CTGAGATCTCAGTGTGTCAGGAGCAAGGCAGTTG-3′ and cloned upstream of the luciferase gene in pCpGL-basic using the SpeI and BglII restriction sites.
Isolation of primary tonsillar naive and GC B cells.
Isolation of GC B cells was done described before (59). Briefly, magnetic cell separation with the MidiMACS system (Miltenyi Biotec) was used to isolate naive B cells and GC B cells from human tonsils according to the protocol published by Klein et al. (24). The purity of separated B cell subpopulations was confirmed with FACScan analysis (Becton Dickinson) with the following surface markers: naive B cells were IgD+ CD38low CD27− and GC B cells (centroblasts) were CD77+ CD38high.
RNA isolation and RT-qPCR.
RNA was isolated from the EBV-positive BL cell lines Mutu I, Mutu III, Kem I, and Kem III and primary tonsillar naive and GC B cells using the RNeasy Plus minikit (Qiagen) according to the manufacturer's instructions. Equal amounts of RNA derived from EBV-positive BL cell lines were DNase treated, and cDNA was made using the Improm-II reverse transcription (RT) system (Promega). Equal amounts of primary tonsillar B cell RNA were reversed transcribed with a first-strand cDNA synthesis kit (Invitrogen). Relative transcript levels were quantified with iTaq universal SYBR green (Bio-Rad) using quantitative PCR (qPCR) analysis with primers for the following: TET2 (5′-AAAGATGAAGGTCCTTTTTATACCC-3′ and 5′-TTTCAACTTCTGTCCAAACCTT-3′), TET3 (5′-CAGCAGCCGAGAAGAAGAAG-3′ and 5′-GGACAATCCACCCTTCAGAG-3′), EBNA3C (5′-GGGCTGTCAAGCAATCGCAC-3′ and 5′-GTGGTGCATTCCACGGGTAA-3′), β-actin (5′-GCCGGGACCTGACTGACTAC-3′ and 5′-TTCTCCTTAATGTCACGCACGAT-3′), and GAPDH (5′-TGAAGGTCGGAGTCAACGGATTG-3′ and 5′-GCCATGGAATTTGCCATGCCATGGGTGG-3′).
Immunoblotting.
Immunoblotting was performed as previously described (60). Cells were harvested in Sumo lysis buffer with 1× proteasome inhibitor cocktail (Roche). Lysate protein concentration was determined using the Sumo protein assay (Bio-Rad), and equal amounts of protein were loaded on 6%, 10%, or 4 to 20% gradient (Bio-Rad) sodium dodecyl sulfate (SDS)-polyacrylamide gels. Proteins were resolved and transferred to nitrocellulose membranes, which were blocked in a phosphate-buffered saline solution with 5% milk and 0.1% Tween 20. The membranes were then incubated with primary antibody diluted in the blocking solution. The following antibodies were used: anti-β-actin AC-15 (Sigma; A5441; 1:5,000), anti-BCL-6 (Santa Cruz; 0.N.26; 1:200), anti-EBNA2 PE2 (Abcam; ab90543; 1:250), anti-IRF4 (Santa Cruz; sc-6059; 1:200), anti-LMP1 S12 (Kerafast; ETU001; 1:5000), anti-TET2 (Proteintech; 21207-1-AP; 1:1,000), and anti-tubulin (Sigma; T5168; 1:2,000). The secondary antibodies used were horseradish peroxidase (HRP)-goat anti-mouse (Pierce; 1:5,000) and HRP-goat anti-rabbit (Pierce; 1:10,000).
Dot blots.
Cells were pretreated or not with vitamin C (50 μg/μl) daily for 4 days prior to DNA isolation. DNA was isolated using the GenElute mammalian genomic DNA miniprep kit (Sigma) according to the manufacturer's instructions. NaOH was added at a final concentration of 0.2 M to 2 μg of DNA. Samples were incubated at 95°C for 5 min, followed by immediate incubation on ice to denature the DNA. Ammonium acetate was added to a final concentration of 0.8 M to neutralize the NaOH. A 0.22-μm nitrocellulose membrane was presoaked in 2× SSC (0.3 M NaCl, 30 mM sodium citrate [pH 7.0]), and DNA was then spotted onto the membrane through a dot blot apparatus. One thousand nanograms of DNA was loaded to probe for 5hmC, and 62.5 ng was loaded to probe for 5mC. The DNA was cross-linked to the membrane with UV irradiation, and the membrane with incubated with blocking solution (Tris-buffered saline solution with 5% milk and 0.1% Tween 20) and then incubated with primary antibody diluted in the blocking solution. The following antibodies were used: anti-5-methylcytosine 33D3 (Active Motif; 39649; 1:1,000) and anti-5-hydroxymethylcytosine (Active Motif; 39769; 1:10,000). The secondary antibodies used were HRP-goat anti-mouse (Pierce; 1:2,000) and HRP-goat anti-rabbit (Pierce; 1:2,000). Equal loading of DNA was determined by staining with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2).
DNA immunoprecipitation (DIP).
Mutu I and Mutu III were treated with acyclovir (100 μg/ml) for 4 days, and DNA was isolated using the GenElute mammalian genomic DNA miniprep kit (Sigma) according to the manufacturer's instructions. DNA (10 μg in 500 μl of Tris-EDTA [TE]) was sonicated to an average size of 300 bp and denatured by heating at 95°C for 10 min followed by rapid chilling on ice. Fifty microliters (10%) was reserved to be used as input, and then 50 μl of 10× IP buffer (100 mM sodium phosphate buffer [pH 7.0], 1.4 M NaCl, 0.5% Triton X-100) was added to the sample. The samples were precleared with protein A/G beads for 2 h and separated into four conditions of equal volume (one for each antibody and isotype control). Samples were immunoprecipitated overnight with the following antibodies or isotype controls: 1 μl of anti-5hmC (Active Motif; 39769, rabbit), 1 μg of rabbit isotype IgG control (Cell Signaling; 39769), 1 μg of anti-5mC 33D3 (Active Motif; 39649; mouse), and 1 μg of mouse isotype IgG control (Santa Cruz). The antibody bound DNA was captured with protein A/G beads for 2 h at 4°C, washed 5 times with 1× IP buffer, eluted overnight at 55°C in elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5% SDS, 0.25 mg/ml of proteinase K), and cleaned using a Qiagen gel extraction kit. qPCR was used to determine the presence and relative amount of specific DNA fragments modified by 5hmC and 5mC.
DIP qPCR.
The relative amount of immunoprecipitated DNA fragments from DIP assays was measured using qPCR analysis with SYBR green (Bio-Rad) according to the manufacturer's instructions. Samples were analyzed using an ABI Prism 7900 real-time PCR system (Applied Biosystems). The C promoter was amplified with the following primer set: 5′-CCTAGGCCAGCCAGAGATAAT-3′ and 5′-AGATAGCACTCGACGCACTG-3′. A dilution series of each input sample was utilized to create a standard curve of the threshold cycle (CT) value for each dilution. Percent input bound of each sample was calculated with the determined standard curve. Each condition and input dilution was loaded in triplicate.
In vitro methylation of reporter gene constructs.
The EBV LMP1 ED-L1 promoter inserted into the pCpGL-basic reporter gene construct was methylated or mock treated in vitro using CpG methyltransferase M.SssI (New England BioLabs [NEB]) according the manufacturer's instructions. The DNA was cleaned by phenol-chloroform extraction and salt precipitation. Promoter CpG methylation was verified by enzymatic digestion with two restriction enzymes (NEB) that share the same cut site: HpaII (methylation inhibits digestion) and MspI (cuts regardless of methylation status).
Reporter gene assays.
BJAB cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions with 0.3 μg of LMP1p-ED-L1-pCpGL reporter vector, 0.5 μg of SG5-EBNA2, 0.5 μg of TET2 or pcDNA3.1(+), and up to 1.6 μg of SG5. Cells were harvested in 1× reporter lysis buffer (Promega) 2 days posttransfection and subjected to one freeze-thaw cycle. Luciferase activity was quantified using a BD monolight 3010 (BD biosciences) with luciferase assay reagent (Promega). Each luciferase assay condition was performed in triplicate on two separate days.
Nucleofection.
Mutu I cells were transfected with 1 μg of EBNA2 or SG5 and 1 μg of TET2 or pcDNA3.1(+) using an Amaxa nucleofector device (Lonza) according to the Raji cell protocol (Lonza). Briefly, 3 × 106 actively growing cells per condition were resuspended in buffer V (Lonza), aliquoted in cuvettes with 2 μg of DNA, and nucleofected using program G-016. Cells were harvested for immunoblot analysis 48 h postnucleofection.
ACKNOWLEDGMENTS
We thank Alan Rickinson, Jeff Sample, and Janet Mertz for cell lines, Yi Zhang for the TET2 expression vector, Paul Ling for the EBNA2 expression vector, Michael Rehli for the pCpGL-basic luciferase vector, and members of the Kenney, Johannsen, Rui, and Mertz laboratories for suggestions and discussions.
This work was supported by HHS | NIH | National Cancer Institute (NCI) grants 4P01CA022443-39 and R01-CA174462-04 and by HHS | NIH | National Institute of Dental and Craniofacial Research (NIDCR) grant 4R01DE023939-04.
REFERENCES
- 1.Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700. In Knipe, DM, Howley, PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Kieff E, Rickinson AB. 2007. Epstein-Barr virus and its replication, p 2603–2654. In Knipe, DM, Howley, PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Yates JL, Guan N. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol 65:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price AM, Luftig MA. 2014. Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation. Adv Virus Res 88:279–313. doi: 10.1016/B978-0-12-800098-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen JI. 2005. Clinical aspects of Epstein-Barr virus infection, p 35–54. In Robertson E. (ed), Epstein-Barr virus. Caister Academic Press, Norfolk, England. [Google Scholar]
- 6.Thorley-Lawson DA. 2005. EBV persistence and latent infection in vivo, p 309–357. In Robertson E. (ed), Epstein-Barr virus. Caister Academic Press, Norfolk, England. [Google Scholar]
- 7.Henkel T, Ling PD, Hayward SD, Peterson MG. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 8.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. 1994. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J 13:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman SR. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J Virol 69:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B, Zou J, Wang H, Johannsen E, Peng C, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, Aster JC, Bernstein BE, Kieff E. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A 108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez AF, Rosales C, Lopez-Nieva P, Graña O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, Pino I, Javierre B, Carmona FJ, Acquadro F, Steenbergen RDM, Snijders PJF, Meijer CJ, Pineau P, Dejean A, Lloveras B, Capella G, Quer J, Buti M, Esteban J- I, Allende H, Rodriguez-Frias F, Castellsague X, Minarovits J, Ponce J, Capello D, Gaidano G, Cigudosa JC, Gomez-Lopez G, Pisano DG, Valencia A, Piris MA, Bosch FX, Cahir-McFarland E, Kieff E, Esteller M. 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res 19:438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minarovits J. 2006. Epigenotypes of latent herpesvirus genomes. Curr Top Microbiol Immunol 310:61–80. [DOI] [PubMed] [Google Scholar]
- 13.Masucci MG, Contreras-Salazar B, Ragnar E, Falk K, Minarovits J, Ernberg I, Klein G. 1989. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt's lymphoma line Rael. J Virol 63:3135–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson KD, Hayward SD, Ling PD, Samid D, Ambinder RF. 1995. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol Cell Biol 15:6150–6159. doi: 10.1128/MCB.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minarovits J, Hu LF, Minarovits-Kormuta S, Klein G, Ernberg I. 1994. Sequence-specific methylation inhibits the activity of the Epstein-Barr virus LMP 1 and BCR2 enhancer-promoter regions. Virology 200:661–667. doi: 10.1006/viro.1994.1229. [DOI] [PubMed] [Google Scholar]
- 16.Fuentes-Pananá EM, Peng R, Brewer G, Tan J, Ling PD. 2000. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J Virol 74:8166–8175. doi: 10.1128/JVI.74.17.8166-8175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone A, Gaidano G, Gloghini A, Pastore C, Saglio G, Tirelli U, Dalla-Favera R, Falini B. 1997. BCL-6 protein expression in AIDS-related non-Hodgkin's lymphomas: inverse relationship with Epstein-Barr virus-encoded latent membrane protein-1 expression. Am J Pathol 150:155–165. [PMC free article] [PubMed] [Google Scholar]
- 18.Boccellato F, Anastasiadou E, Rosato P, Kempkes B, Frati L, Faggioni A, Trivedi P. 2007. EBNA2 interferes with the germinal center phenotype by downregulating BCL6 and TCL1 in non-Hodgkin's lymphoma cells. J Virol 81:2274–2282. doi: 10.1128/JVI.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trivedi P, Spinsanti P, Cuomo L, Volpe M, Takada K, Frati L, Faggioni A. 2001. Differential regulation of Epstein-Barr virus (EBV) latent gene expression in Burkitt lymphoma cells infected with a recombinant EBV strain. J Virol 75:4929–4935. doi: 10.1128/JVI.75.10.4929-4935.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastor WA, Aravind L, Rao A. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delatte B, Deplus R, Fuks F. 2014. Playing TETris with DNA modifications. EMBO J 33:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlanski S, Labi V, Reizel Y, Spiro A, Lichtenstein M, Levin-Klein R, Koralov SB, Skversky Y, Rajewsky K, Cedar H, Bergman Y. 2016. Tissue-specific DNA demethylation is required for proper B-cell differentiation and function. Proc Natl Acad Sci U S A 113:5018–5023. doi: 10.1073/pnas.1604365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaffer AL, Rosenwald A, Staudt LM. 2002. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol 2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- 24.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. 2003. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A 100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RAP, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. 2009. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 26.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minor EA, Court BL, Young JI, Wang G. 2013. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem 288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin R, Mao S-Q, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, Jiao Y, Li C, Liu S, Wu D, Gu W, Yang Y-G, Xu G-L, Wang H. 2013. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 29.Sadler RH, Raab-Traub N. 1995. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J Virol 69:4577–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci U S A 107:850–855. doi: 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes DJ, Marendy EM, Dickerson CA, Yetming KD, Sample CE, Sample JT. 2012. Contributions of CTCF and DNA methyltransferases DNMT1 and DNMT3B to Epstein-Barr virus restricted latency. J Virol 86:1034–1045. doi: 10.1128/JVI.05923-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai C-N, Tsai C-L, Tse K-P, Chang H-Y, Chang Y-S. 2002. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc Natl Acad Sci U S A 99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K, Fukayama M. 2009. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res 69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 34.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. 2009. Epstein-Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog 5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardee J, Ouyang Z, Zhang Y, Kundaje A, Lacroute P, Snyder M. 2013. STAT3 targets suggest mechanisms of aggressive tumorigenesis in diffuse large B-cell lymphoma. G3 (Bethesda) 3:2173–2185. doi: 10.1534/g3.113.007674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. 2012. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res 22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basso K, Dalla-Favera R. 2010. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol 105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 38.Kelly GL, Milner AE, Baldwin GS, Bell AI, Rickinson AB. 2006. Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells. Proc Natl Acad Sci U S A 103:14935–14940. doi: 10.1073/pnas.0509988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Rica L, Rodríguez-Ubreva J, García M, Islam AB, Urquiza JM, Hernando H, Christensen J, Helin K, Gómez-Vaquero C, Ballestar E. 2013. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol 14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilhamon P, Eskandarpour M, Halai D, Wilson GA, Feber A, Teschendorff AE, Gomez V, Hergovich A, Tirabosco R, Fernanda Amary M, Baumhoer D, Jundt G, Ross MT, Flanagan AM, Beck S. 2013. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun 4:2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jolly CJ, Neuberger MS. 2001. Somatic hypermutation of immunoglobulin κ transgenes: association of mutability with demethylation. Immunol Cell Biol 79:18–22. doi: 10.1046/j.1440-1711.2001.00968.x. [DOI] [PubMed] [Google Scholar]
- 42.Bhutani N, Burns DM, Blau HM. 2011. DNA demethylation dynamics. Cell 146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu G-L, Luo C, Jiang H, He C. 2012. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramírez J, Lukin K, Hagman J. 2010. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol 22:177–184. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein TL, Plass C, Tenen DG. 2006. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 46.Jundt F, Kley K, Anagnostopoulos I, Schulze Pröbsting K, Greiner A, Mathas S, Scheidereit C, Wirth T, Stein H, Dörken B. 2002. Loss of PU.1 expression is associated with defective immunoglobulin transcription in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease. Blood 99:3060–3062. doi: 10.1182/blood.V99.8.3060. [DOI] [PubMed] [Google Scholar]
- 47.Wille CK, Nawandar DM, Henning AN, Ma S, Oetting KM, Lee D, Lambert P, Johannsen EC, Kenney SC. 2015. 5-Hydroxymethylation of the EBV genome regulates the latent to lytic switch. Proc Natl Acad Sci U S A 112:E7257–E7265. doi: 10.1073/pnas.1513432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenspan JS, Greenspan D, Lennette ET, Abrams DI, Conant MA, Petersen V, Freese UK. 1985. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N Engl J Med 313:1564–1571. doi: 10.1056/NEJM198512193132502. [DOI] [PubMed] [Google Scholar]
- 49.Niedobitek G, Young LS, Lau R, Brooks L, Greenspan D, Greenspan JS, Rickinson AB. 1991. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J Gen Virol 72(Part 12):3035–3046. [DOI] [PubMed] [Google Scholar]
- 50.Gilligan K, Rajadurai P, Resnick L, Raab-Traub N. 1990. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc Natl Acad Sci U S A 87:8790–8794. doi: 10.1073/pnas.87.22.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walling DM, Flaitz CM, Nichols CM, Hudnall SD, Adler-Storthz K. 2001. Persistent productive Epstein-Barr virus replication in normal epithelial cells in vivo. J Infect Dis 184:1499–1507. doi: 10.1086/323992. [DOI] [PubMed] [Google Scholar]
- 52.Tsang CM, Yip YL, Lo KW, Deng W, To KF, Hau PM, Lau VMY, Takada K, Lui VWY, Lung ML, Chen H, Zeng M, Middeldorp JM, Cheung AL-M, Tsao SW. 2012. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc Natl Acad Sci U S A 109:E3473–E3482. doi: 10.1073/pnas.1202637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Temple RM, Zhu J, Budgeon L, Christensen ND, Meyers C, Sample CE. 2014. Efficient replication of Epstein-Barr virus in stratified epithelium in vitro. Proc Natl Acad Sci U S A 111:16544–16549. doi: 10.1073/pnas.1400818111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol 12:431–441. doi: 10.1016/S1044579X0200086X. [DOI] [PubMed] [Google Scholar]
- 55.Namba-Fukuyo H, Funata S, Matsusaka K, Fukuyo M, Rahmutulla B, Mano Y, Fukayama M, Aburatani H, Kaneda A. 5 November 2016. TET2 functions as a resistance factor against DNA methylation acquisition during Epstein-Barr virus infection. Oncotarget doi: 10.18632/oncotarget.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein G, Lindahl T, Jondal M, Leibold W, Menézes J, Nilsson K, Sundström C. 1974. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc Natl Acad Sci U S A 71:3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klug M, Rehli M. 2006. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 59.Chiorazzi M, Rui L, Yang Y, Ceribelli M, Tishbi N, Maurer CW, Ranuncolo SM, Zhao H, Xu W, Chan W-CC, Jaffe ES, Gascoyne RD, Campo E, Rosenwald A, Ott G, Delabie J, Rimsza LM, Shaham S, Staudt LM. 2013. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc Natl Acad Sci U S A 110:3943–3948. doi: 10.1073/pnas.1217271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhende PM, Seaman WT, Delecluse H-J, Kenney SC. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet 36:1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]