ABSTRACT
Owing to a complex history of host-parasite coevolution, lentiviruses exhibit a high degree of species specificity. Given the well-documented viral archeology of human immunodeficiency virus (HIV) emergence following human exposures to simian immunodeficiency virus (SIV), an understanding of processes that promote successful cross-species lentiviral transmissions is highly relevant. We previously reported natural cross-species transmission of a subtype of feline immunodeficiency virus, puma lentivirus A (PLVA), between bobcats (Lynx rufus) and mountain lions (Puma concolor) for a small number of animals in California and Florida. In this study, we investigate host-specific selection pressures, within-host viral fitness, and inter- versus intraspecies transmission patterns among a larger collection of PLV isolates from free-ranging bobcats and mountain lions. Analyses of proviral and viral RNA levels demonstrate that PLVA fitness is severely restricted in mountain lions compared to that in bobcats. We document evidence of diversifying selection in three of six PLVA genomes from mountain lions, but we did not detect selection among 20 PLVA isolates from bobcats. These findings support the hypothesis that PLVA is a bobcat-adapted virus which is less fit in mountain lions and under intense selection pressure in the novel host. Ancestral reconstruction of transmission events reveals that intraspecific PLVA transmission has occurred among panthers (Puma concolor coryi) in Florida following the initial cross-species infection from bobcats. In contrast, interspecific transmission from bobcats to mountain lions predominates in California. These findings document outcomes of cross-species lentiviral transmission events among felids that compare to the emergence of HIV from nonhuman primates.
IMPORTANCE Cross-species transmission episodes can be singular, dead-end events or can result in viral replication and spread in the new species. The factors that determine which outcome will occur are complex, and the risk of new virus emergence is therefore difficult to predict. We used molecular techniques to evaluate the transmission, fitness, and adaptation of puma lentivirus A (PLVA) between bobcats and mountain lions in two geographic regions. Our findings illustrate that mountain lion exposure to PLVA is relatively common but does not routinely result in communicable infections in the new host. This is attributed to efficient species barriers that largely prevent lentiviral adaptation. However, the evolutionary capacity for lentiviruses to adapt to novel environments may ultimately overcome host restriction mechanisms over time and under certain ecological circumstances. This phenomenon provides a unique opportunity to examine cross-species transmission events leading to new lentiviral emergence.
KEYWORDS: bobcat, cross-species transmission, feline, feline immunodeficiency virus, mountain lion, retroviruses
INTRODUCTION
The Lentivirus genus comprises complex retroviruses with a propensity for rapid mutation and recombination that results in a high rate of viral evolution. Lentiviruses typically infect hosts in a species-specific manner, and distinct viral subtypes or clades are characteristically associated with single host species. Transmission of these host-adapted viruses to new species is uncommon (1, 2). Host restriction is attributed to several factors, including lower viral fitness in the novel host, intrinsic antiviral defense mechanisms, and/or limited contact sufficient for transmission between different host species (3–7). Notable examples of successful cross-species lentiviral infection include multiple transmissions of simian immunodeficiency viruses (SIVs) from nonhuman primates to humans, which gave rise to the various circulating subtypes of human immunodeficiency virus (HIV) (reviewed in reference 8). It is thought that a convergence of social, cultural, and behavioral factors resulted in viral transmission and subsequent adaptation, culminating in a devastating pandemic infecting an estimated 35 million people worldwide (9).
At least 11 felid species have been diagnosed with infections with lentiviruses known as feline immunodeficiency viruses (FIVs), which represent the most well-defined lentiviral group outside the SIVs (10, 11). As with other lentiviruses, FIV phylogenetic relationships support a pattern of species-specific viral evolution (12, 13). In domestic cats (Felis catus), reported morbidity and mortality vary widely, from mild or inapparent infection to a terminal AIDS-like syndrome (14–17). In nondomestic felids, infections are apparently subclinical, though reduced CD4 T-lymphocyte counts and an increased prevalence of opportunistic pathogens have been documented for some feline hosts (18–20). Experimental transmission of FIVs isolated from mountain lions (Puma concolor; also referred to as pumas, cougars, and panthers) to domestic cats resulted in productive yet avirulent infections (21). Host-mediated cytidine deamination ultimately produced defective viral genomes (21, 22), suggesting that adaptation of FIVs to new host species does not readily occur.
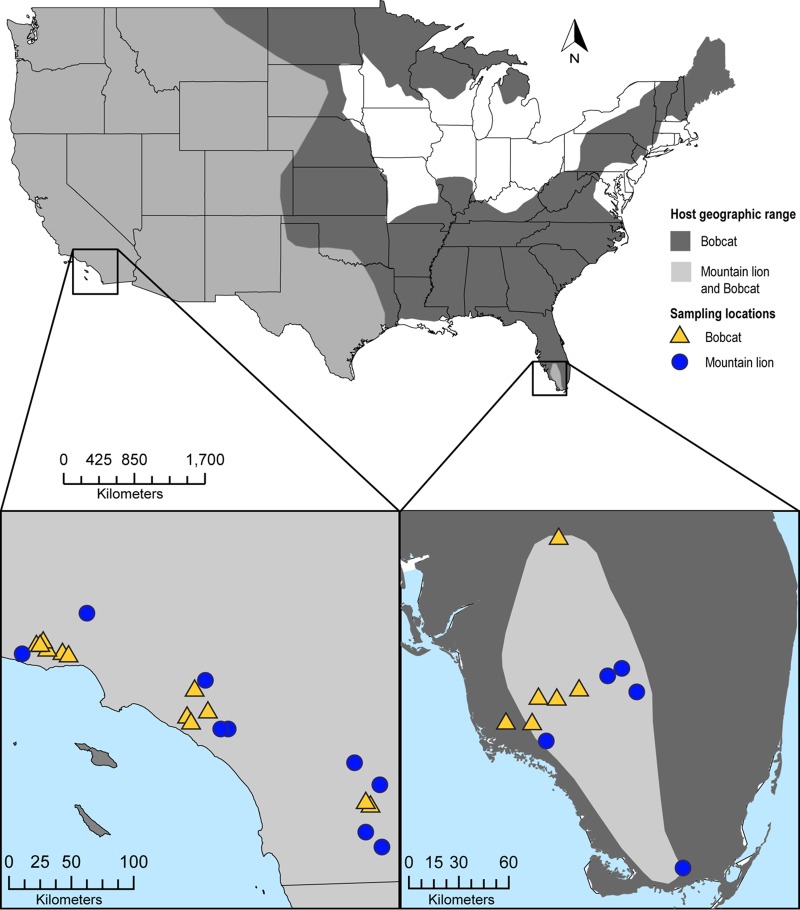
An exception to the pattern of lentiviral host specificity is puma lentivirus A (PLVA; also referred to as FIVpcoA), an FIV subtype documented to infect two different species in the wild: bobcats (Lynx rufus) and mountain lions (23). Mountain lions inhabit a geographic range from western Canada to southern Chile, while bobcats are sympatric mesopredators throughout much of North America (Fig. 1). Both species are habitat generalists but are sensitive to anthropogenic influences and have experienced regional extinctions and population subdivision due to overhunting and habitat degradation (24–27). PLVA is the only FIV that has been isolated from bobcats, and it is endemic in California and Florida but has not been identified in other geographic regions (23, 28–31). PLVA has been identified in mountain lions only for populations that are sympatric with PLVA-infected bobcats (Fig. 1), is absent throughout most of the mountain lion geographic range, and is much less common in mountain lions than a second FIV subtype, puma lentivirus B (PLVB; also referred to as FIVpcoB), in regions where the two viruses cocirculate (2, 11, 13, 23, 25, 28). In contrast to PLVA, PLVB has been shown to infect mountain lions throughout their entire geographic range and thus has likely coevolved with mountain lions since prior to their proposed recolonization of North America after the last Ice Age (10,000 to 15,000 years ago) (28, 32). The genetic distance between PLVA and PLVB is similar to that separating the other species-specific strains of FIV and is suggestive of divergent evolutionary histories in separate host species (11, 13).
FIG 1.
Host species geographic ranges and sample collection sites for PLVA isolates from bobcats and mountain lions in California and Florida. Light gray shading represents regional sympatry; the geographic range of bobcats overlaps that of mountain lions throughout the United States (http://www.icunredlist.org/).
We previously described several aspects of PLV evolution, including the roles of mutation, recombination, and natural selection in generating genetic diversity over time and space (30). Specifically, both PLVA and -B evolve under predominantly purifying selection, with high rates of synonymous mutations occurring at the nucleotide level but relatively infrequent nonsynonymous substitutions resulting in diversity in the corresponding proteins. Multiple recombination breakpoints were detected across the PLV genomes, indicating that this is an important mechanism for generating genetic diversity, which can lead to fit viral variants that differ from both parental isolates. Finally, we were able to document through viral phylogenies that PLV diversity reflects the population structure of its hosts, with large genetic distances separating geographically distinct host populations but apparent admixture within populations.
In this study, we present new findings following examination of an expanded set of samples to specifically characterize the dual-host tropism of PLVA. This report represents the first analysis of cross-species transmission, viral fitness, and viral adaptation in relation to the evolution of PLVA in the native host (bobcat) and a secondary host (mountain lion). Our results indicate that PLVA viral fitness is severely reduced in the mountain lion compared to that in the bobcat and that adaptation (episodic diversifying selection) has occurred in mountain lion PLVA isolates. In California, most mountain lion isolates have arisen from cross-species transmission from bobcats. In contrast, the phylogeny of PLVA isolates from Florida panthers (Puma concolor coryi; a regional subspecies of mountain lion) is consistent with primarily intraspecific transmission events, suggesting possible PLVA adaptation in this population. The dual-host tropism of PLVA provides a unique opportunity to understand the ecological and evolutionary factors involved in lentivirus host range expansion, analogous to the transmission of SIVs to humans leading to the emergence of HIV.
RESULTS
Host-virus phylogeny.
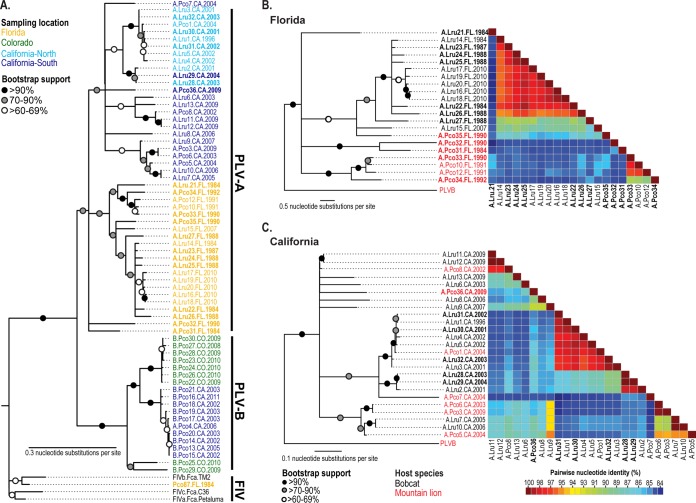
We previously evaluated PLV phylogenetic relationships in the context of broad-scale geographic patterns and gene-by-gene evolution (30). In the present analysis, we report on as yet uncharacterized traits of PLV diversity that provide insight into the evolutionary dynamics of this two-host, two-virus system. The PLVB clade contains only mountain lion isolates, and ancestral host state reconstruction of the basal node of this clade resulted in a 0.99 posterior probability of mountain lion ancestry (Fig. 2A and Fig. 3). In contrast, the PLVA clade comprises 15 mountain lion isolates and 32 bobcat isolates, and the basal node of PLVA was assigned a probability of bobcat ancestry of 0.84.
FIG 2.
PLVA and PLVB are separated by large genetic distances, and each comprises geographically associated subgroups. (A) Maximum likelihood phylogenetic tree constructed from a 474-bp region of pol. Isolates original to this study are highlighted in bold text. Nodes with <60% bootstrap support have been collapsed. Isolate names provide the following information: (i) PLV clade (clade A or B), (ii) host species (Lru, bobcat; Pco, mountain lion), (iii) animal identification number (Table 3), (iv) sampling location, and (v) sample year (1984 to 2011). Regional PLVA subtrees and pairwise identity matrices demonstrate different patterns of host-virus relationships. Isolates in Florida (B) tend to form well-supported clusters by host species, while most California mountain lion isolates (C) are more closely related to sympatric bobcat isolates than to viruses from other mountain lions.
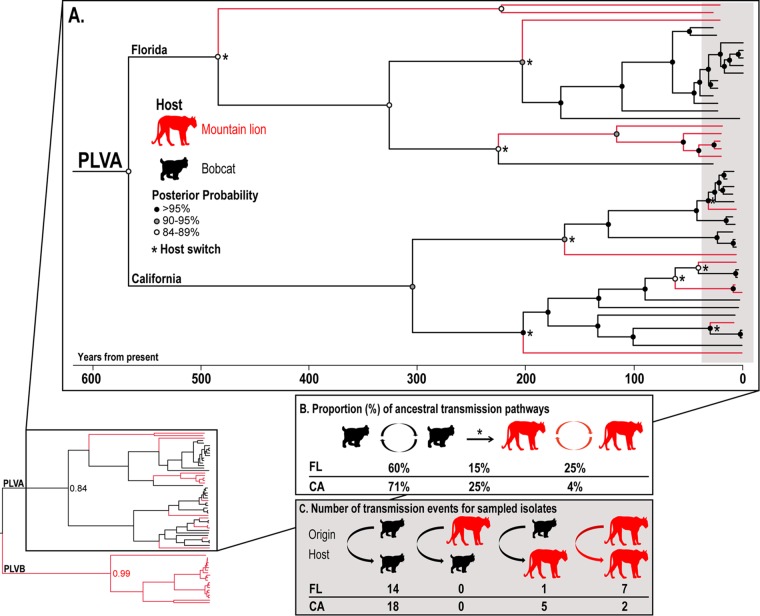
FIG 3.
An ancestral reconstruction of host states across the PLVA phylogeny depicts different patterns of intra- and interspecies transmissions in California and Florida. (A) Maximum clade credibility tree constructed from the pol sequences used for Fig. 2, depicting historic and contemporary transmission dynamics. Host state posterior probability values relevant to transmission directionality are indicated by shaded circles at nodes. Asterisks indicate predicted cross-species transmission events (3 in Florida and 6 in California). (B) The proportion of inferred host state transitions across the PLVA phylogeny depicts substantial bobcat-to-mountain lion transmission rates at each site (15% of Florida and 25% of California transmissions). Predicted mountain lion-to-mountain lion transmissions occur with far greater frequency in Florida (25%) than in California (4%). (C) The gray shaded region of panel A corresponds to host states for the sampled isolates depicted here. More sampled mountain lion isolates were predicted to arise from intrahost transmission events in Florida (7 of 8 isolates) than in California (2 of 7 isolates).
PLVA isolates form two distinct groups of viral sequences exclusively from California or Florida (Fig. 2). Samples from Florida cluster by host species: 14 of 14 bobcat and 7 of 8 panther isolates have predicted most recent common ancestors from a bobcat and a panther, respectively (Fig. 2B and 3). In California, 18 of 18 bobcat PLVA isolates arose from predicted bobcat ancestors; however, in contrast to the case in Florida, 5 of 7 California mountain lion isolates were predicted to have arisen from a most recent common ancestor from a bobcat (Fig. 2C and 3). No mountain lion-to-bobcat transmission was inferred for either population. In support of these results from the host state ancestral reconstruction analysis, pairwise identity matrices demonstrate different patterns of host-virus relationships in California and Florida (Fig. 2B and C). In Florida, the majority of panther isolates share higher pairwise identity with other panther isolates than with bobcat isolates, while in California, the most closely related isolate to most mountain lion isolates is a bobcat isolate.
One viral isolate from a Florida panther (Pco87.FL1984) is paraphyletic to all PLV isolates, with high bootstrap support for its exclusion from PLVA and PLVB (Fig. 2A). This isolate clusters with domestic cat FIV isolates and is most similar to FIVFca subtype B (92% pairwise identity) (data not shown).
Within-host fitness.
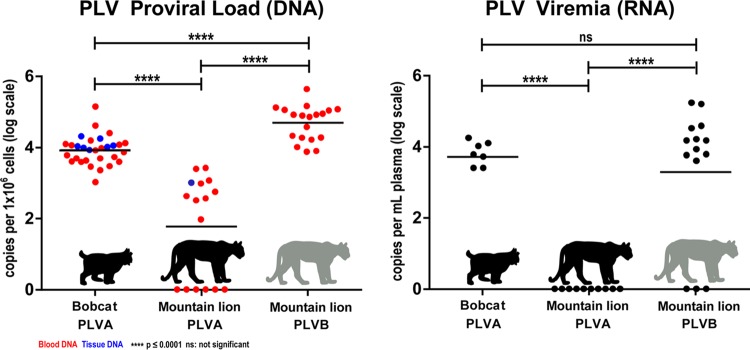
PLVA proviral loads in bobcats (mean = 103.8; standard deviation [SD] = 0.49) and PLVB proviral loads in mountain lions (mean = 104.7; SD = 0.50) were 1 to 2 orders of magnitude higher than PLVA proviral loads in mountain lions (mean = 103.0; SD = 0.93) (P < 0.0001 by analysis of variance [ANOVA]) (Fig. 4, left panel). This result was consistent for proviral loads quantified from both blood and tissue samples. A quantitative PCR (qPCR) assay did not detect PLVA provirus in 6 PLVA-infected pumas, despite amplification of integrated proviral DNA by nested PCR assays.
FIG 4.
Within-host viral fitness differs significantly among hosts and among viral clades. (Left) Individual sample proviral loads (means for triplicates) are depicted for PLVA in bobcats (n = 30), PLVA in mountain lions (n = 10), and PLVB in mountain lions (n = 18). Horizontal lines represent mean values for the host-virus relationships. The mountain lion PLVA proviral copy number was significantly lower than that for PLVA in bobcats and PLVB in mountain lions (P < 0.0001 for both). The mountain lion PLVB proviral load was significantly higher than the bobcat PLVA proviral load (P < 0.0001). No significant difference was detected between proviral loads obtained from blood (represented in red) versus tissue (represented in blue). (Right) Individual sample viremia values (means for triplicates) are depicted for PLVA in bobcats (n = 7), PLVA in mountain lions (n = 10), and PLVB in mountain lions (n = 14). PLVA viremia was below the limit of quantification for all mountain lion samples. PLVA viremia in bobcats was not significantly different (ns) from the mean PLVB viremia in mountain lions.
Analysis of the raw data documented significantly higher viremia for PLVA in bobcats and PLVB in mountain lions than for PLVA in mountain lions (Fig. 4, right panel), and there was no significant difference in viremia values for each virus in the apparent primary host. The majority of bobcat and mountain lion plasma viral loads for both PLVA and PLVB were below the lower limit of quantitation (LLOQ), and 10 of 10 mountain lions with detectable PLVA provirus had no detectable viremia.
Host-specific selection.
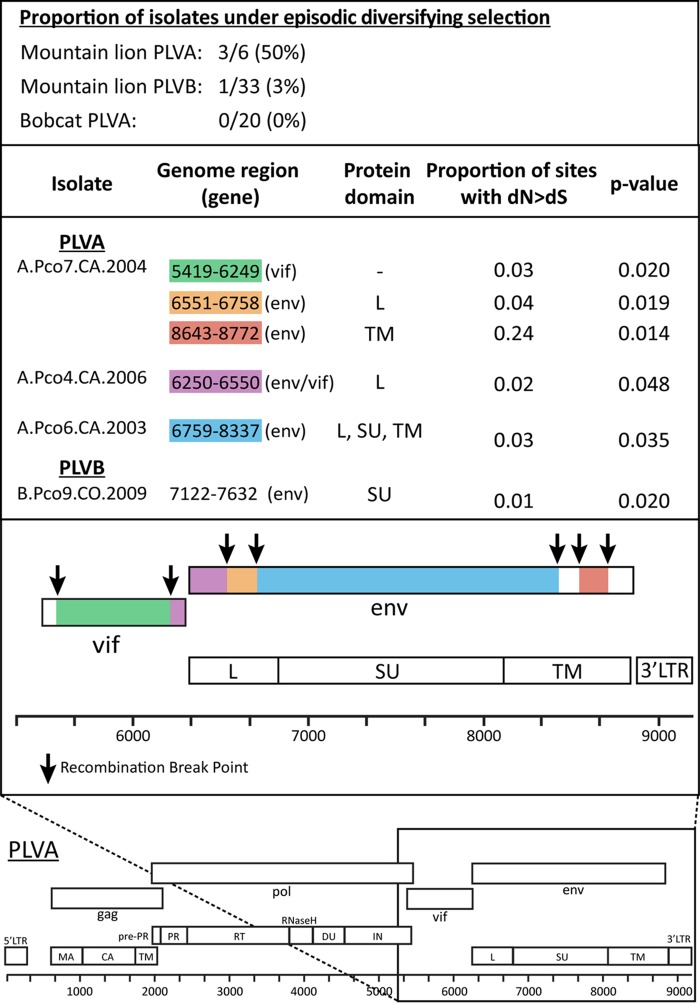
Full genome sequences of PLVA (n = 26) and PLVB (n = 33) were analyzed to detect individual isolates subject to episodic bursts of diversifying selection. Episodic diversifying selection (nonsynonymous substitution rate [dN] > synonymous substitution rate [dS]) was detected in at least one genomic region in 50% (3/6 sequences) of the PLVA sequences isolated from mountain lions (Fig. 5). Selective pressure was detected within vif and several regions of env, including those encoding the leader domain, the transmembrane domain, and the region spanning the surface and transmembrane domains. In contrast, no evidence of diversifying selection was detected in any segments of 20 PLVA genome sequences from bobcats. One of 33 PLVB genome sequences had evidence of selection in a single region of env (data not shown).
FIG 5.
The selection results are consistent with the hypothesis that PLVA is not adapted to the mountain lion and is therefore under strong pressure to evolve in this host. All nonrecombinant regions of PLVA and PLVB genomes were analyzed by branch-site REL without a priori designation of isolates as originating from a bobcat or a mountain lion. Diversifying selection was detected in 3 of 6 PLVA isolates from mountain lions, 0 of 20 bobcat PLVA isolates, and 1 of 33 PLVB isolates from mountain lions. The table at the top lists the isolates evolving under diversifying selection, the corresponding genomic regions under selection, and the proportion of sites (codons) diversifying in each region (L, leader; SU, surface; TM, transmembrane). P values shown were derived via ANOVA and corrected for multiple tests by using the Holm-Bonferroni method. The bottom panel depicts the locations of the PLVA regions identified as evolving under episodic diversifying selection. All segments under selection were at the 3′ end of the genome and included multiple loci within env and vif. Previously characterized recombination breakpoints are indicated by arrows. The full PLVA genome is outlined below for reference.
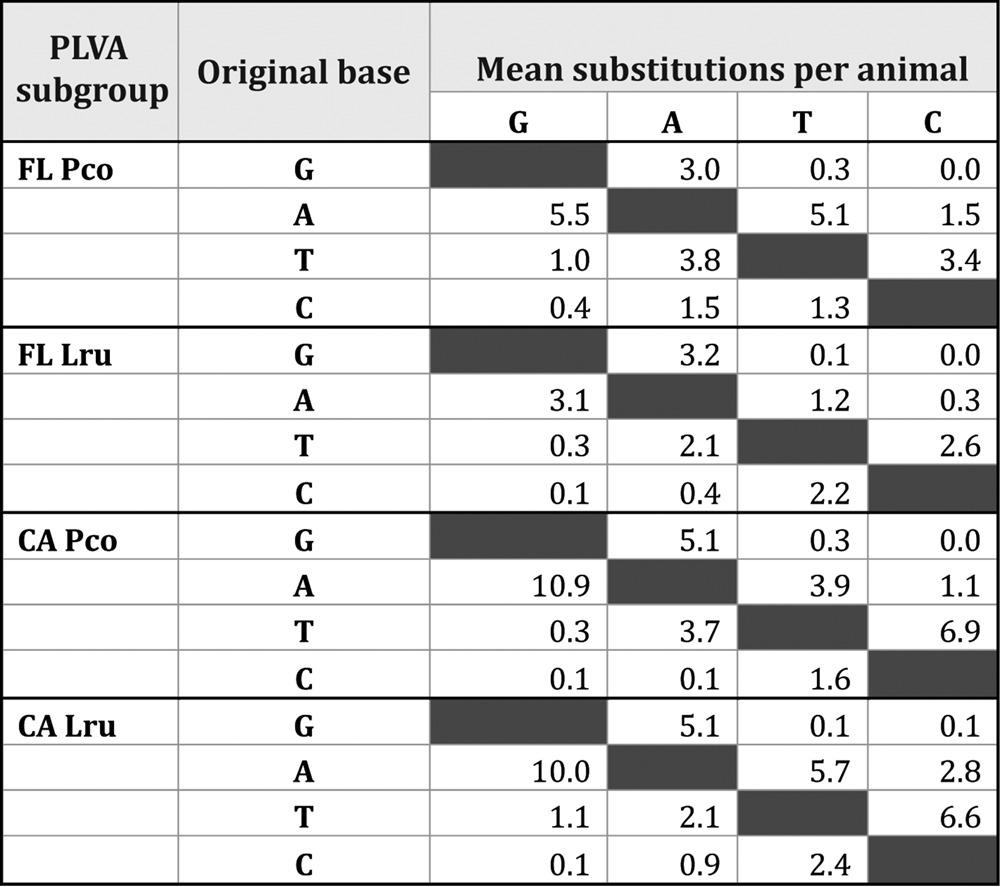
Host-specific substitution bias.
The most common type of nucleotide substitution in 3 of 4 groups of hosts (California bobcats, California mountain lions, and Florida panthers) was an A-to-G change (Table 1). G-to-A transitions were slightly more common within bobcat isolates from Florida, but we did not identify a significant bias for G-to-A substitutions in any group (Table 2).
TABLE 1.
G-to-A substitution rates across a 474-bp segment of PLVA pol do not vary by host speciesa
FL, Florida; CA, California; Pco, Puma concolor; Lru, Lynx rufus.
TABLE 2.
G/A variability in pol genes from Florida isolatesa
| Parameter | Value |
|
|---|---|---|
| Bobcat (n = 14) | Panther (n = 9) | |
| Total no. (%) of G nucleotides | 297 (26.5) | 192 (26.2) |
| Total no. (%) of A nucleotides | 820 (73.4) | 540 (73.7) |
| % other nucleotides | 0.1 | 0.1 |
| Mean %G/animal | 21.2 | 21.3 |
| Mean %A/animal | 58.6 | 60 |
G/A variability was identified at 18% of sites within a 474-bp segment of pol from Florida isolates, but both host species had the same proportions of G and A nucleotides.
DISCUSSION
Investigations of cross-species viral transmission events and “host jumps” have improved our understanding of the factors that can lead to virus emergence in new hosts (33, 34). While the specific ecological and adaptive drivers of emergence are case and virus specific, some general patterns and processes have been elucidated. Independent cross-species transmissions of host-adapted viruses often occur with little to no subsequent transmission among the new hosts (spillover events, e.g., transmission of avian influenza A viruses to humans) (35). This is because non-host-adapted viruses may have low fitness, low transmission efficiency, or both in the novel host (36). However, with low fitness in a novel environment, selective pressures acting on existing and de novo genetic diversity can increase the frequency of beneficial mutations, leading to viral adaptation. This process may be observed through the accumulation of genetic changes that differ from those present in the reservoir population (37). This process of viral adaptation leading to increased fitness and transmission efficiency may be necessary for the virus to persist in the new host (host jump, e.g., the emergence of canine parvovirus from felids) (38).
SIV spillover into humans has occurred many times, but only a small number of these resulted in sufficient viral adaptation to allow widespread human-to-human transmission in the worldwide HIV epidemic (8, 39, 40). We thus hypothesized that the jump of PLVA from bobcats into mountain lions would similarly represent a poorly adapted viral infection in a new host species, with a relatively low rate of transmission within the novel host. To test this hypothesis, we reconstructed ancestral virus-host phylogenetic relationships, measured within-host viral fitness, and analyzed patterns of viral adaptation for PLVA and PLVB isolates.
We inferred intra- and interspecies transmission dynamics by using a phylogenetic analysis with ancestral reconstruction of each host species along the phylogeny (Fig. 3). The results support our hypothesis that PLVA and PLVB have evolved with different primary hosts, as the ancestral host at the basal node of each clade was inferred with high posterior support to be the bobcat and the mountain lion, respectively. Further, bobcats were predicted to be the ancestral host state at over 85% of nodes across the PLVA phylogeny, and all cross-species transmissions occurred in a unidirectional pattern from bobcats to mountain lions.
Although similar numbers of bobcat and mountain lion PLVA isolates were evaluated from California and Florida, the phylogenetic relationships and predicted patterns of cross-species transmission differed greatly between the two sample sites. Most PLVA isolates sampled from mountain lions in California arose via cross-species transmission, while the majority of panther isolates sampled in Florida resulted from intraspecies transmission. The median posterior date estimates for the cross-species transmission events in Florida predate those for events in California, suggesting that PLVA has circulated in Florida panthers since prior to any detected cross-species transmission events in California (the original tree, annotated with dates and 95% highest posterior density intervals, is available upon request). Additionally, the branch lengths and pairwise identity values for PLVA isolates from Florida panthers demonstrate that lineages in Florida have diverged since their shared ancestry with bobcat PLVAs, a pattern that differs from that for the California population (Fig. 2). These findings are consistent with PLVA cross-species transmission from bobcats to panthers followed by viral divergence through drift or adaptation during subsequent intraspecific spread in Florida.
This contrast in transmission patterns between California and Florida provides a unique opportunity to consider ecological and host factors that may contribute to viral adaptation in a new host. The Florida panther is historically endangered and was reduced to a dwindling population of 20 to 25 individuals by the early 1990s (41). Remaining panthers were highly inbred, with significant deleterious impacts on the population, including congenital defects and an array of infections unlikely to occur to a similar degree in immunocompetent hosts (42). The small population size, small patches of suitable habitat, and isolation from other populations significantly restricted interactions among conspecifics for decades (41–43). This ecological situation increased contact rates among remnant panthers concentrated within small habitat patches (43). Such drastic shifts in density and distribution, paired with changes in social structure and decreased host genetic diversity, may underlie our observation that PLVA transmission occurred within the Florida panther cohort and was followed by ongoing genetic divergence in the new host.
Interestingly, many of the California mountain lions with PLVA also derive from small, isolated populations with genetic characteristics similar to those documented for the historic Florida panther population. Mountain lions from the Santa Ana Mountains (south of Los Angeles) and the Santa Monica Mountains (north of Los Angeles) have high average pairwise relatedness, small estimated effective population sizes, and strong evidence of past genetic bottlenecks (44, 45). While our phylogenetic data suggest that PLVA in California mountain lions is acquired primarily from bobcats versus intraspecific contacts, the condition of these small, isolated populations may similarly predispose them to chains of intraspecific infection and emergence of a mountain lion-adapted PLVA infection over time. Observations gleaned from these results support the notion that the outcome of lentiviral infection in a new host is highly circumstantial, with the lentivirus typically restricted from emerging in the new host and dependent on a complex interplay between ecological and evolutionary forces to adapt to the new species.
We detected both host-specific and clade-specific differences in viral fitness as measured by proviral loads and plasma viremia (Fig. 4). PLVA proviral loads in bobcats and PLVB proviral loads in mountain lions were significantly higher than PLVA proviral loads in mountain lions. PLVB proviral loads in mountain lions were significantly higher than PLVA proviral loads in bobcats, though viral copy numbers in both species were within a range similar to those reported for host-adapted subtypes of FIV in domestic cats (∼103 to 104 proviral copies/106 cells) (46–48). Our inability to quantitate PLVA provirus from a number of animals that were positive by conventional PCR is likely a reflection of low proviral loads and potentially reflects partial degradation of some archival samples. Alternatively, the possibility of mutations in the primer/probe region of the env gene resulting from selection and/or genetic drift cannot be excluded as a plausible cause for inconsistencies in the assay.
A second measure of fitness that may relate more directly to transmission efficiency is plasma viremia. Similar to the proviral load data, the levels of viremia for bobcat PLVA and mountain lion PLVB were higher than those for mountain lion PLVA. While many of the viremia values were below the LLOQ, precluding a robust quantitative comparison among groups, the fact that PLVA was not detectable in any of the mountain lions analyzed is interesting and suggests that, in most instances, PLVA is not highly replication competent in mountain lions, despite the fact that intraspecific transmission of PLVA in Florida panthers is strongly supported by viral phylogenies.
Given that adaptive viral evolution is known to occur in episodic bursts driven by the host immune response, we predicted that the low fitness of PLVA in mountain lions would result in detectable genetic signatures of adaptation. As a measure of adaptation, we therefore estimated which viral lineages may be evolving under episodic diversifying selection, identified as an increase in nonsynonymous substitutions relative to synonymous substitutions within short segments of viral proteins. In line with our prediction, diversifying selection was detected in segments from half of the mountain lion PLVA isolates analyzed (3 of 6 isolates) but was rare among PLVB isolates (1 of 33 isolates) and not detected among bobcat PLVA isolates (0 of 20 isolates) (Fig. 5). All diversifying lineages were evolving under episodic selection via changes in env, encoding an antigenic protein involved in the binding and entry of viral particles into host cells and acting as a target for neutralizing antibodies. Env variation has important fitness implications for other lentiviruses (49–54), and our results suggest that Env may be important for the adaptation of PLVA in mountain lions.
In two of the three mountain lion PLVA isolates under selection, viral adaptation was also detected in vif, the gene encoding the accessory protein that counteracts the innate antiviral activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3). In the absence of Vif, APOBEC3 enzymes are packaged into virions to restrict replication via deamination of cytidines during reverse transcription (55). APOBEC3 activity is detectable in proviral DNA as increased G-to-A mutation rates, which can result in eventual viral degradation. The complex interplay between Vif and APOBEC3 has been identified as an important driver of host-pathogen coevolution for a number of lentiviruses (56, 57), and G-to-A alterations have been documented for the infection of domestic cats with PLV (22). Thus, adaptation in the Vif protein of PLVA in mountain lions is not unexpected and may represent a critical evolutionary mechanism shaped by host immune system and intrinsic restriction pressures.
Given the well-documented importance of Vif adaptations to host range expansion of other lentiviruses, we surmised that APOBEC3 antiviral activity could be a factor limiting PLVA fitness in the Florida panther. However, we did not detect a bias of G-to-A substitutions in mountain lion PLVA sequences. While this limited analysis of a small segment of PLVA genomes did not provide evidence of APOBEC3-mediated cytidine deamination, it is possible that highly mutated genomes may have been eliminated from circulation.
Our findings support growing evidence that mountain lions, the apex feline carnivore in North America, are regularly exposed to a diverse array of pathogens (58). Recent reports have documented mountain lion predation on bobcats and domestic cats (59, 60), a likely mechanism by which mountain lions are exposed to pathogens of sympatric felids. The relationship between predator-prey interactions and viral transmission is not well studied and may represent a process by which predators accumulate pathogens by a mechanism that is similar to the process of bioaccumulation of environmental toxins (61). For example, feline leukemia virus has spilled over from domestic cats and caused outbreaks with high morbidity in Florida panthers on multiple occasions (62, 63). Another example is the recent discovery of a novel feline gammaherpesvirus that appears to be transmitted from bobcats to mountain lions (64). This pattern is further documented in the present study by the identification of frequent transmissions of PLV from bobcats to mountain lions and by the first recorded account of domestic cat FIV in a free-ranging Florida panther (Fig. 2A and 3). This phenomenon of predator-driven pathogen exposure warrants further study, as it may represent an as yet unappreciated ecological driver of emerging diseases.
With the notable exception of SIVs, this study represents the first robust analysis documenting contemporary, naturally occurring lentiviral cross-species transmission. Our findings confirm that PLVA and PLVB are different viral species, derived from bobcats and mountain lions, respectively. We therefore propose that PLVA be reclassified as a distinct species of FIV, designated FIVLru to indicate that the bobcat is the primary host. We further suggest that references to “puma lentivirus” be reserved for PLVB, more accurately referred to as FIVPco.
Our findings demonstrate that viral transmission from a reservoir host into a sympatric relative can occur frequently. Further, we found evidence of PLVA adaptation to mountain lions, an evolutionary process that may be influenced by both host and ecological conditions. Given the prevalence of PLVA in mountain lions that are sympatric with infected bobcats and the evidence for sustained intraspecies transmission of PLVA in relic Florida panthers, the emergence of a mountain lion-adapted PLVA with a broader geographic distribution is clearly possible.
This study provides further evidence that viral transmission from one species to another does not routinely result in intraspecific spread within the new host, but it also suggests that viral evolution may ultimately overcome host restriction mechanisms. This contribution to the current understanding of pathogen-host dynamics also highlights the susceptibility of small, stressed populations to novel viruses and sets the stage for further study of risk factors for viral emergence in apex predators.
MATERIALS AND METHODS
Sample collection and nucleic acid extraction.
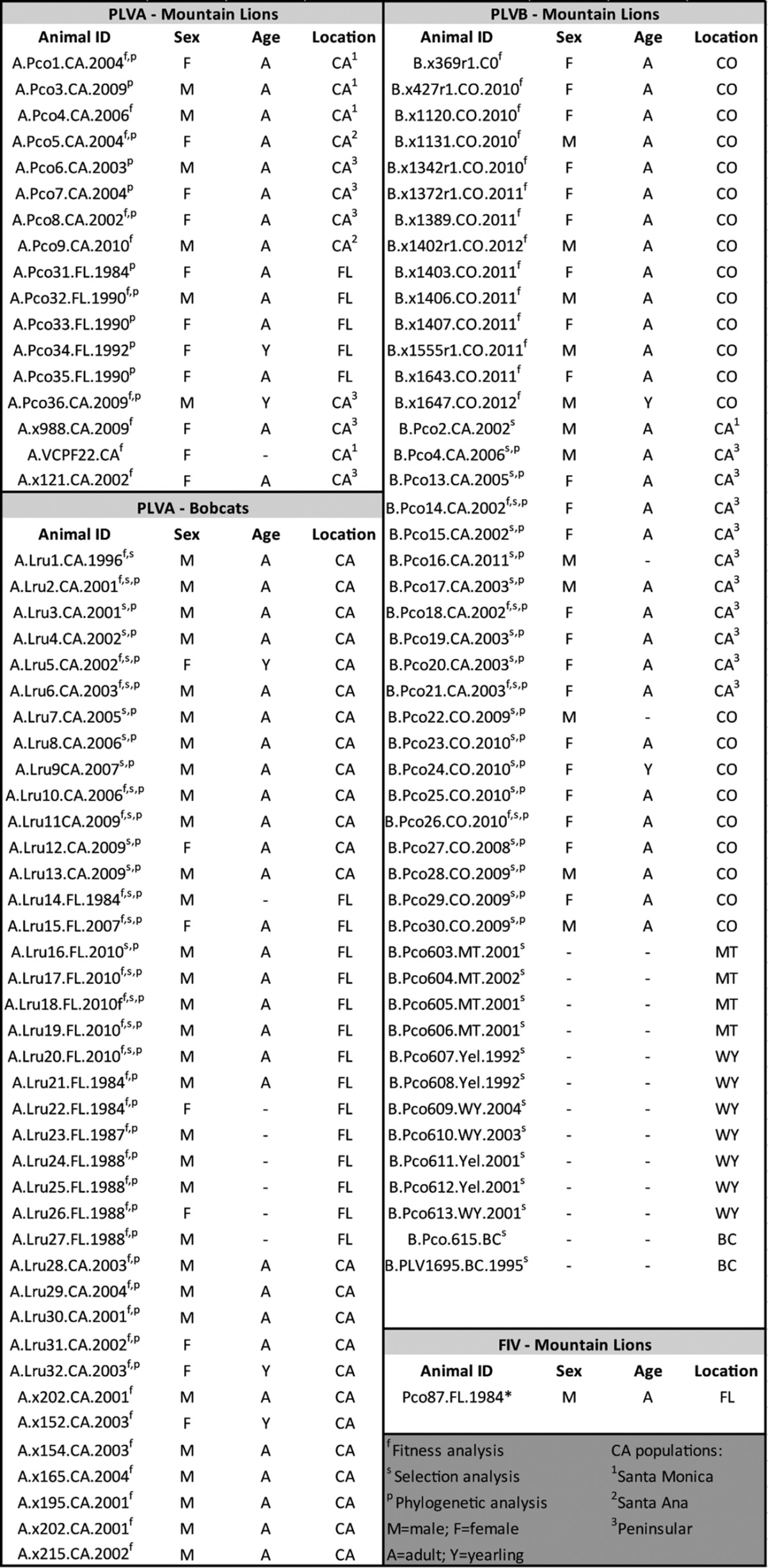
Mountain lion and bobcat samples were collected from natural populations in three locations over the following time spans: California, 1996 to 2010; Colorado, 2009 to 2013; and Florida, 1983 to 2010. Isolates yielding PLVA sequences from bobcats or mountain lions are illustrated in Fig. 1. Samples were collected from live, free-ranging animals captured using baited cage traps or scent-trained tracking hounds, as previously described (65). Animals were chemically sedated for blood collection. All animal capture and handling protocols followed approved Institutional Animal Care and Use Committee guidelines and, where applicable, local government regulations. Additional samples were opportunistically collected during routine postmortem examinations by local government authorities. Aliquots of blood and tissue samples were sent to Colorado State University for characterization as described below. Tissue samples consisted of lymphoid organs, including lymph node, spleen, and thymus samples, as well as one sample each of liver, kidney, and muscle. Table 3 provides the sex, age, location, and collection date for each sample included in this study.
TABLE 3.
Demographic information for all samples included in the present analyses
DNA was extracted from tissue, whole blood, or peripheral blood mononuclear cells (PBMCs) by using the DNeasy Blood and Tissue protocol (Qiagen Inc., Valencia, CA). Approximately 130 samples from archival collections were screened to identify the maximal number of positive samples that could be sequenced further or subjected to qPCR. Plasma or serum was available for a subset of positive samples (n = 7 bobcats and n = 24 mountain lions). RNA was extracted from these samples by use of a QIAmp viral RNA minikit (Qiagen Inc., Valencia, CA) according to the manufacturer's recommendations.
Phylogenetic relationships.
Proviral DNA sequences of the pol genes (474 bp) from 59 PLV isolates (28 PLVA isolates and 31 PLVB isolates) were obtained from GenBank (Table 4) (66). Previously described nested PCR protocols (11) were used to amplify PLVA proviral DNA from additional bobcats (n = 12) and mountain lions (n = 6). This represented approximately 20% of bobcat samples tested and 10% of available mountain lion samples, consistent with previously reported prevalences in the sampled regions (23, 30, 31). Reaction mixtures contained 100 to 1,000 ng of genomic DNA. PCR products were sequenced on an ABI 3130xl genetic analyzer (Applied Biosystems Inc., Foster City, CA) and aligned using the default parameters in MEGA6 (67). A maximum likelihood phylogenetic tree was constructed using PhyML (68) parameters in Seaview (69), based on the GTR+i model of nucleotide substitution. Cluster support was estimated with 1,000 bootstrap replicates. An HIV-1 sequence and three domestic cat FIV sequences (subtypes A, B, and C) were included as outgroups.
TABLE 4.
GenBank accession numbers for published PLV sequencesa
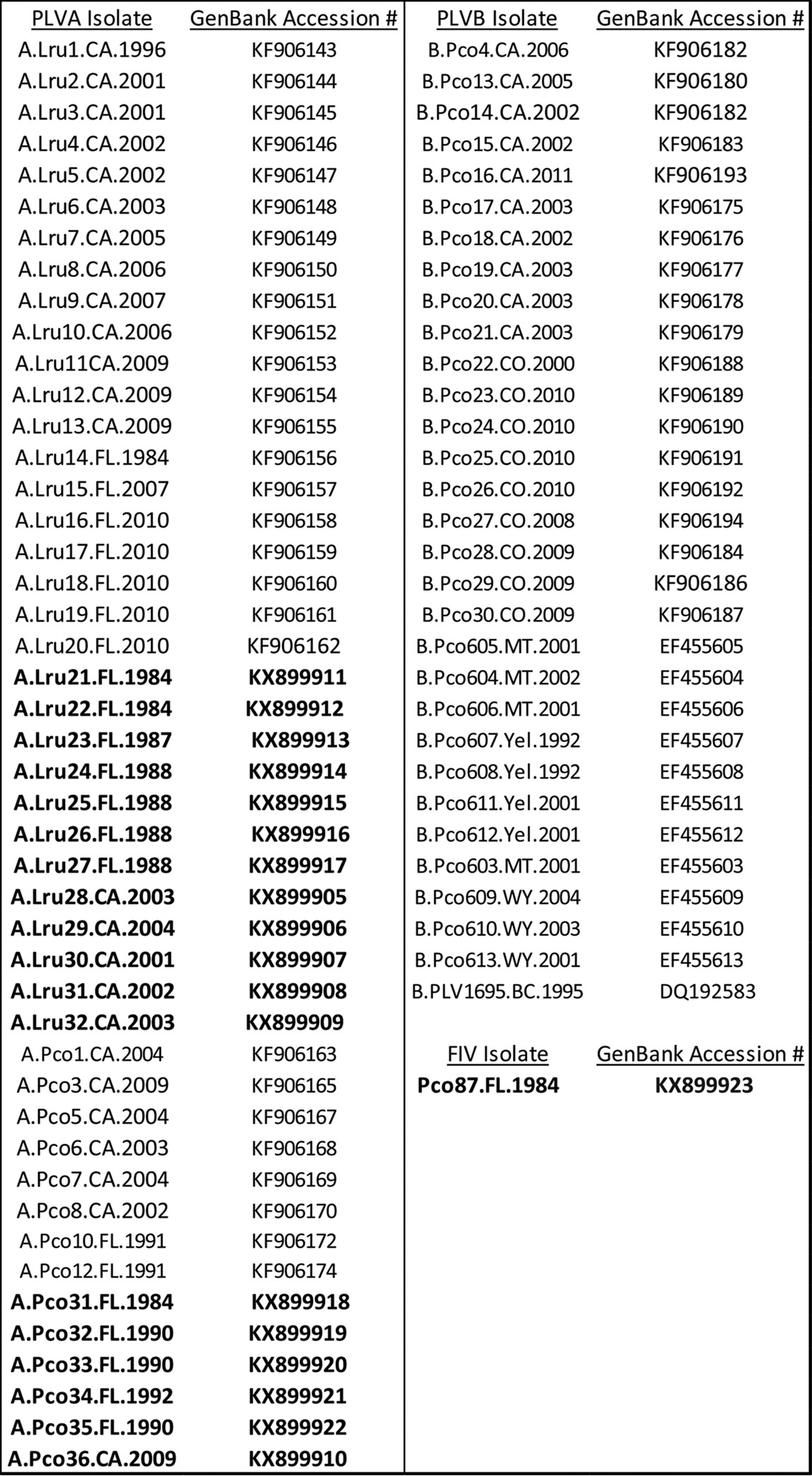
Data for new isolates obtained for this study are indicated in bold.
Two maximum likelihood phylogenetic subtrees were constructed using the above-described methods and including PLVA sequences from either Florida or California. A PLVB sequence was included in each subtree as an outgroup, and cluster support was estimated with 1,000 bootstrap replicates. We further investigated relationships between bobcat and mountain lion PLVA isolates by generating heat maps based on pairwise distance matrices, using Sequence Demarcation Tool software (70).
Within-host viral fitness.
We developed PLVA- and PLVB-specific qPCR assays to quantify proviral loads and viremia for natural infections. Thirty-four full-length PLVA genomes and 33 full-length PLVB genomes were aligned for primer design. Despite high intrasubtype genetic diversity, we identified primer binding sites in PLVA env and PLVB gag that were 100% conserved. The PLVA primers were 8083F (GCA GCC CTG ACG GTA TCC) and 8165R (GCA GTC TCC TCT GAA CAA TCC), and the PLVB primers were 643F (CTG TCT GTC ATG GGG AAT GAG T) and 773R (GTC CTG TAG CTA CCA AGG CAA).
(i) Proviral loads.
qPCRs for PLVA (n = 30 bobcats and n = 10 mountain lions) and PLVB (n = 18 mountain lions) (Table 3) were conducted under identical conditions, using 50 to 100 ng of genomic target DNA. All samples were run in triplicate, while standards and controls were run in duplicate. Reactions were conducted using SsoFast EvaGreen supermix (Bio-Rad, Hercules, CA) according to the manufacturer's recommendations, with a 20 μM (each) primer concentration and cycling conditions of 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 62°C for 10 s. Reactions were analyzed by melting curve analysis for temperatures ranging from 65°C to 95°C (increasing 0.5°C every 10 s) in order to confirm PCR product size and uniformity. All melting curves generated from PLV-infected feline DNA matched melting curves generated from plasmid standards, indicating consistent detection of virus-specific DNA. Plasmid standards of known copy number were prepared by cloning PLVA and PLVB target sequences into pCR4-TOPO by use of a TOPO TA cloning kit (Life Technologies, Carlsbad, CA). Plasmid standards giving 102 to 106 PLV copies per reaction mixture were prepared using TE buffer (10 mM Tris and 1 mM EDTA) containing 10 ng/μl DNA from FIV-negative bobcats. All qPCRs had amplification efficiencies within the acceptable range of 90 to 110% (71).
Negative-control DNA samples from specific-pathogen-free domestic cats and PLV-negative bobcats yielded negative results with both the PLVA and PLVB qPCR assays, demonstrating that the assay primers do not cross-react with feline genomic DNA. The number of PLV proviral copies per 106 cells was calculated for each sample based on the number of cell equivalents of input DNA as described previously (46). The lower limit of detection of this assay is approximately 100 proviral copies per reaction mixture.
(ii) Plasma viremia.
cDNA synthesis was performed using 10 μl of purified RNA and the following reagents: 4 μl of 5× First Strand buffer (Life Technologies, Grand Island, NY), 1 μl of deoxynucleoside triphosphates (10 mM [each]) (dNTPs; Bio-Rad), 1 μl of 0.1 M dithiothreitol (DTT; Life Technologies), 0.25 μl of 40-U/μl RNase Out (Life Technologies), 0.25 μl of 200-U/μl SuperScript II reverse transcriptase (Life Technologies), 2 μl of 300-ng/μl random primers (Life Technologies), and 1.5 μl of nuclease-free water. Samples were incubated at 42°C for 50 min followed by 95°C for 5 min. cDNA was stored at −20°C until used for testing in the qPCR assays. qPCR was performed as described above for provirus quantification. The number of PLV RNA copies per milliliter of plasma was calculated as follows: number of RNA copies per milliliter of plasma = SQ × 1/C × D × E × 1/P, where SQ is the mean starting quantity per sample, C is the cDNA volume (microliters) added per well for the qPCR assay (5 μl), D is the inverse of the dilution used for cDNA synthesis (e.g., for a 1-in-2 dilution, D equals 2), E is the elution volume (microliters) used for viral RNA extraction (60 μl), and P is the plasma volume (milliliters) used for viral RNA extraction (generally, 0.14 ml).
All calculated values for numbers of RNA copies per milliliter of plasma were rounded to two significant digits based on the precision of the method. To determine the lower limit of quantitation (LLOQ), PLV standards (diluted in TE buffer) were tested on multiple days to determine the lowest concentration that could consistently be quantified. Standard concentrations tested were 102 to 107 plasmid copies/well (10-fold dilutions), as well as values below this range (10, 20, 40, 50, 60, and 80 copies/well). The LLOQ was determined to be 102 copies/well, equivalent to 17,000 RNA copies/ml plasma.
For each virus-host relationship, mean copies of integrated provirus (proviral load) and circulating viral RNA (plasma viremia) were statistically compared by one-way analysis of variance (ANOVA) followed by Tukey's multiple-means comparison test, using Prism v. 5.0 (Graph Pad Software, La Jolla, CA).
Host-specific selection.
Previous analyses of PLVA and PLVB genomic characterizations were performed using the Datamonkey tools MEME and FEL, and results were reported on a gene-by-gene basis independent of the host of origin (30). To compare selection pressures across host species, separate PLVA (n = 26) and PLVB (n = 33) translation alignments were generated using the Muscle plug-in, with default parameters, in Geneious (72, 73). Each alignment was split into multiple nonrecombinant sections based on the recombination breakpoints identified previously (30) and was screened using branch-site random-effects likelihood analysis (branch-site REL) in Datamonkey (74). Branch-site REL does not use a priori assumptions about which viruses may be diversifying, but rather selection is allowed to vary among all lineages and across all sites. Viral lineages identified as evolving under episodic diversifying selection have a nonsynonymous substitution rate (dN) significantly greater than the synonymous substitution rate (dS) (dN > dS; corrected P value, <0.05) at a proportion of sites within any region of the genome. All full-length PLV sequences were included in these analyses (Table 3).
Host-specific substitution bias.
To further examine host-pathogen interactions, we quantified each type of nucleotide substitution within a nonrecombinant segment of the pol gene for all available PLVA isolates. Consensus sequences were generated for each host in each region (California and Florida), based on a strict (50%) majority. Mutations at polymorphic sites were summed for each isolate, and the mean numbers of each type of base change (e.g., cytosine to thymine) within each group were compared. Additionally, we screened for evidence of host-mediated cytidine deamination, under the working hypothesis that a bias for guanine-to-adenine (G-to-A) transitions could be detected in isolates of nonadapted virus in a novel host. Using the same region of pol, the mean numbers of G and A nucleotides were compared between host species in Florida.
Transmission dynamics.
Discrete trait mapping in BEAST v1.8.3 (75) was used to infer ancestral host states across the PLV pol phylogeny. Ancestral states were reconstructed at nodes and along branches, and state change counts were estimated across the entire tree (76). Cross-species transmission events were evaluated with an asymmetric Bayesian stochastic search variable selection model (77). Model parameters for the phylogeny included an HKY substitution model with estimated base frequencies. Substitution rates were estimated separately for two codon partitions (1 + 2 and 3) with a gamma distribution and four rate categories. The relative rates of mutation for both codon partitions were modeled using a lognormal prior with a standard deviation and initial value set equal to 1. The tree prior included a coalescent model with a constant population size and a random starting tree (75). Evolution over time was modeled using an uncorrelated relaxed molecular clock with a lognormal distribution of rates, an initial value of 0.1, and a standard deviation equal to 0.5. Two separate Markov chain Monte Carlo (MCMC) runs were performed for 1 × 108 generations, each sampled every 1 × 104 generations.
The log files were viewed in Tracer (78) to confirm that the models reached convergence (as indicated by effective sample sizes of >200) and to ensure that posterior parameter estimates were similar between the two independent MCMC runs. The posterior distributions were downsampled to every 2 × 104 generations and combined using Log Combiner (79) after removing the first 10% of sampled states from each file as burn-in. The posterior tree files were similarly combined, and then the maximum clade credibility tree was identified and annotated with median node heights by using Tree Annotator (75). Discrete trait transitions were categorized as intra- or interspecific transmission events based on a posterior probability support of >0.80 that a node ancestral to a given branch was for the same or different host species, respectively.
Accession number(s).
The sequences determined in this study were submitted to GenBank under the accession numbers listed in Table 4.
ACKNOWLEDGMENTS
This work was supported by NSF-EID awards 0723676 and 1413925, Morris Animal Foundation award D10ZO-415, the Merial Veterinary Summer Student Fellowship award, and NHLBI, NIH, award 5R01HL092791.
We thank the many field biologists and state and federal agency workers for invaluable assistance with obtaining sample collections. We also thank Simona Kraberger for invaluable technical support.
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
REFERENCES
- 1.Apetrei C, Robertson DL, Marx PA. 2004. The history of SIVs and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci 9:225–254. doi: 10.2741/1154. [DOI] [PubMed] [Google Scholar]
- 2.Troyer JL, VandeWoude S, Pecon-Slattery J, McIntosh C, Franklin S, Antunes A, Johnson W, O'Brien SJ. 2008. FIV cross-species transmission: an evolutionary prospective. Vet Immunol Immunopathol 123:159–166. doi: 10.1016/j.vetimm.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.VandeWoude S, Troyer J, Poss M. 2010. Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet Immunol Immunopathol 134:25–32. doi: 10.1016/j.vetimm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munk C, Hechler T, Chareza S, Lochelt M. 2010. Restriction of feline retroviruses: lessons from cat APOBEC3 cytidine deaminases and TRIM5 alpha proteins. Vet Immunol Immunopathol 134:14–24. doi: 10.1016/j.vetimm.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 6.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 7.Zielonka J, Marino D, Hofmann H, Yuhki N, Lochelt M, Munk C. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J Virol 84:7312–7324. doi: 10.1128/JVI.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.UNAIDS. 2013. Global report: UNAIDS report on the global AIDS epidemic. UNAIDS, Geneva, Switzerland. [Google Scholar]
- 10.VandeWoude S, Apetrei C. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev 19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, Brown M, Frank L, Woodroffe R, Winterbach C, Winterbach H, Hemson G, Bush M, Alexander KA, Revilla E, O'Brien SJ. 2005. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol 79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien SJ, Troyer JL, Roelke M, Marker L, Pecon-Slattery J. 2006. Plagues and adaptation: lessons from the Felidae models for SARS and AIDS. Biol Conserv 131:255–267. doi: 10.1016/j.biocon.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pecon-Slattery J, Troyer JL, Johnson WE, O'Brien SJ. 2008. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet Immunol Immunopathol 123:32–44. doi: 10.1016/j.vetimm.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beczkowski PM, Litster A, Lin TL, Mellor DJ, Willett BJ, Hosie MJ. 2015. Contrasting clinical outcomes in two cohorts of cats naturally infected with feline immunodeficiency virus (FIV). Vet Microbiol 176:50–60. doi: 10.1016/j.vetmic.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen NC, Ho EW, Brown ML, Yamamoto JK. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 16.Liem B, Dhand N, Pepper A, Barrs V, Beatty J. 2013. Clinical findings and survival in cats naturally infected with feline immunodeficiency virus. J Vet Intern Med 27:798–805. doi: 10.1111/jvim.12120. [DOI] [PubMed] [Google Scholar]
- 17.Kohmoto M, Miyazawa T, Sato E, Uetsuka K, Nishimura Y, Ikeda Y, Inada G, Doi K, Mikami T. 1998. Cats are protected against feline immunodeficiency virus infection following vaccination with a homologous AP-1 binding site-deleted mutant. Arch Virol 143:1839–1845. doi: 10.1007/s007050050422. [DOI] [PubMed] [Google Scholar]
- 18.VandeWoude S, Hageman CA, O'Brien SJ, Hoover EA. 2002. Nonpathogenic lion and puma lentiviruses impart resistance to superinfection by virulent feline immunodeficiency virus. J Acquir Immune Defic Syndr 29:1–10. doi: 10.1097/00042560-200201010-00001. [DOI] [PubMed] [Google Scholar]
- 19.Poss M, Ross H, Rodrigo A, Terwee J, VandeWoude S, Biek R. 2008. The molecular biology and evolution of feline immunodeficiency viruses of cougars. Vet Immunol Immunopathol 123:154–158. doi: 10.1016/j.vetimm.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roelke ME, Brown MA, Troyer JL, Winterbach H, Winterbach C, Hemson G, Smith D, Johnson RC, Pecon-Slattery J, Roca AL. 2009. Pathological manifestations of feline immunodeficiency virus (FIV) infection in wild African lions. Virology 390:1–12. doi: 10.1016/j.virol.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terwee JA, Yactor JK, Sondgeroth KS, Vandewoude S. 2005. Puma lentivirus is controlled in domestic cats after mucosal exposure in the absence of conventional indicators of immunity. J Virol 79:2797–2806. doi: 10.1128/JVI.79.5.2797-2806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poss M, Ross HA, Painter SL, Holley DC, Terwee JA, Vandewoude S, Rodrigo A. 2006. Feline lentivirus evolution in cross-species infection reveals extensive G-to-A mutation and selection on key residues in the viral polymerase. J Virol 80:2728–2737. doi: 10.1128/JVI.80.6.2728-2737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franklin SP, Troyer JL, Terwee JA, Lyren LM, Boyce WM, Riley SP, Roelke ME, Crooks KR, Vandewoude S. 2007. Frequent transmission of immunodeficiency viruses among bobcats and pumas. J Virol 81:10961–10969. doi: 10.1128/JVI.00997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooks KR. 2002. Relative sensitivities of mammalian carnivores to habitat fragmentation. Conserv Biol 16:488–502. doi: 10.1046/j.1523-1739.2002.00386.x. [DOI] [Google Scholar]
- 25.Lee JS, Ruell EW, Boydston EE, Lyren LM, Alonso RS, Troyer JL, Crooks KR, VandeWoude S. 2012. Gene flow and pathogen transmission among bobcats (Lynx rufus) in a fragmented urban landscape. Mol Ecol 21:1617–1631. doi: 10.1111/j.1365-294X.2012.05493.x. [DOI] [PubMed] [Google Scholar]
- 26.Riley SPD, Pollinger JP, Sauvajot RM, York EC, Bromley C, Fuller TK, Wayne RK. 2006. A southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15:1733–1741. doi: 10.1111/j.1365-294X.2006.02907.x. [DOI] [PubMed] [Google Scholar]
- 27.Riley SPD, Boyston EE, Crooks KR, Lyren LM. 2010. Bobcats (Lynx rufus), p 121–140. In Gehrt SD, Riley SPD, Cypher BL (ed), Urban carnivores—ecology, conflict, and conservation. The Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 28.Carpenter MA, Brown EW, Culver M, Johnson WE, Pecon-Slattery J, Brousset D, O'Brien SJ. 1996. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J Virol 70:6682–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmsted RA, Langley R, Roelke ME, Goeken RM, Adger-Johnson D, Goff JP, Albert JP, Packer C, Laurenson MK, Caro TM. 1992. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J Virol 66:6008–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JS, Bevins SN, Serieys LE, Vickers W, Logan KA, Aldredge M, Boydston EE, Lyren LM, McBride R, Roelke-Parker M, Pecon-Slattery J, Troyer JL, Riley SP, Boyce WM, Crooks KR, VandeWoude S. 2014. Evolution of puma lentivirus in bobcats (Lynx rufus) and mountain lions (Puma concolor) in North America. J Virol 88:7727–7737. doi: 10.1128/JVI.00473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagana DM, Lee JS, Lewis JS, Bevins SN, Carver S, Sweanor LL, McBride R, McBride C, Crooks KR, VandeWoude S. 2013. Characterization of regionally associated feline immunodeficiency virus (FIV) in bobcats (Lynx rufus). J Wildl Dis 49:718–722. doi: 10.7589/2012-10-243. [DOI] [PubMed] [Google Scholar]
- 32.Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ. 2006. The Late Miocene radiation of modern Felidae: a genetic assessment. Science 311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- 33.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mollentze N, Biek R, Streicker DG. 2014. The role of viral evolution in rabies host shifts and emergence. Curr Opin Virol 8:68–72. doi: 10.1016/j.coviro.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, De Jong MD, Yuen KY. 2005. Avian influenza A (H5N1) infection in humans. N Engl J Med 353:1374. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 36.Woolhouse MEJ, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streicker DG, Altizer SM, Velasco-Villa A, Rupprecht CE. 2012. Variable evolutionary routes to host establishment across repeated rabies virus host shifts among bats. Proc Natl Acad Sci U S A 109:19715–19720. doi: 10.1073/pnas.1203456109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shackelton LA, Parrish CR, Truyen U, Holmes EC. 2005. High rate of viral evolution associated with the emergence of canine parvoviruses. Proc Natl Acad Sci U S A 102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 40.Sharp PM, Robertson DL, Hahn BH. 1995. Cross-species transmission and recombination of AIDS viruses. Philos Trans R Soc Lond B Biol Sci 349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 41.Johnson WE, Onorato DP, Roelke ME, Land ED, Cunningham M, Belden RC, McBride R, Jansen D, Lotz M, Shindle D, Howard J, Wildt DE, Penfold LM, Hostetler JA, Oli MK, O'Brien SJ. 2010. Genetic restoration of the Florida panther. Science 329:1641–1645. doi: 10.1126/science.1192891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roelke ME, Forrester DJ, Jacobson ER, Kollias GV, Scott FW, Barr MC, Evermann JF, Pirtle EC. 1993. Seroprevalence of infectious disease agents in free-ranging Florida panthers (Felis concolor coryi). J Wildl Dis 29:36–49. doi: 10.7589/0090-3558-29.1.36. [DOI] [PubMed] [Google Scholar]
- 43.Roelke ME, Martenson JS, O'Brien SJ. 1993. The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Curr Biol 3:340–350. [DOI] [PubMed] [Google Scholar]
- 44.Ernest HB, Vickers TW, Morrison SA, Buchalski MR, Boyce WM. 2014. Fractured genetic connectivity threatens a southern California puma (Puma concolor) population. PLoS One 9:e107985. doi: 10.1371/journal.pone.0107985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernest HB, Boyce WM, Bleich VC, May B, Stiver SJ, Torres SG. 2003. Genetic structure of mountain lion (Puma concolor) populations in California. Conserv Genet 4:353–366. doi: 10.1023/A:1024069014911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.TerWee JA, Carlson JK, Sprague WS, Sondgeroth KS, Shropshire SB, Troyer JL, VandeWoude S. 2008. Prevention of immunodeficiency virus induced CD4+ T-cell depletion by prior infection with a non-pathogenic virus. Virology 377:63–70. doi: 10.1016/j.virol.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J, MacMillan M, Boegler K, Wood C, Elder JH, VandeWoude S. 2011. Pathogenicity and rapid growth kinetics of feline immunodeficiency virus are linked to 3′ elements. PLoS One 6:e24020. doi: 10.1371/journal.pone.0024020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X, Carver S, Troyer RM, Terwee JA, VandeWoude S. 2011. Prior virus exposure alters the long-term landscape of viral replication during feline lentiviral infection. Viruses 3:1891–1908. doi: 10.3390/v3101891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes EC, Zhang LQ, Simmonds P, Ludlam CA, Brown AJL. 1992. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human-immunodeficiency-virus type-1 within a single infected patient. Proc Natl Acad Sci U S A 89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huisman W, Schrauwen EJA, Rimmelzwaan GF, Osterhaus A. 2008. Intrahost evolution of envelope glycoprotein and OrfA sequences after experimental infection of cats with a molecular clone and a biological isolate of feline immunodeficiency virus. Virus Res 137:24–32. doi: 10.1016/j.virusres.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Overbaugh J, Rudensey LM. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol 66:5937–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Payne SL, Fang FD, Liu CP, Dhruva BR, Rwambo P, Issel CJ, Montelaro RC. 1987. Antigenic variation and lentivirus persistence—variations in envelope gene-sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology 161:321–331. doi: 10.1016/0042-6822(87)90124-3. [DOI] [PubMed] [Google Scholar]
- 53.Ross HA, Rodrigo AG. 2002. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol 76:11715–11720. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.da Silva J, Coetzer M, Nedellec R, Pastore C, Mosier DE. 2010. Fitness epistasis and constraints on adaptation in a human immunodeficiency virus type 1 protein region. Genetics 185:293–303. doi: 10.1534/genetics.109.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Compton AA, Hirsch VM, Emerman M. 2012. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martcheva M. 2009. Evolutionary consequences of predation for pathogens in prey. Bull Math Biol 71:819–844. doi: 10.1007/s11538-008-9383-5. [DOI] [PubMed] [Google Scholar]
- 59.Moss WE, Alldredge MW, Pauli JN. 29 November 2015. Quantifying risk and resource use for a large carnivore in an expanding urban-wildland interface. J Appl Ecol doi: 10.1111/1365-2664.12563. [DOI] [Google Scholar]
- 60.Smith JA, Wang Y, Wilmers CC. 2015. Top carnivores increase their kill rates on prey as a response to human-induced fear. Proc Biol Sci 282:20142711. doi: 10.1098/rspb.2014.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rattner BA, Lazarus RS, Elliott JE, Shore RF, van den Brink N. 2014. Adverse outcome pathway and risks of anticoagulant rodenticides to predatory wildlife. Environ Sci Technol 48:8433–8445. doi: 10.1021/es501740n. [DOI] [PubMed] [Google Scholar]
- 62.Cunningham MW, Brown MA, Shindle DB, Terrell SP, Hayes KA, Ferree BC, McBride RT, Blankenship EL, Jansen D, Citino SB, Roelke ME, Kiltie RA, Troyer JL, O'Brien SJ. 2008. Epizootiology and management of feline leukemia virus in the Florida puma. J Wildl Dis 44:537–552. doi: 10.7589/0090-3558-44.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown MA, Cunningham MW, Roca AL, Troyer JL, Johnson WE, O'Brien SJ. 2008. Genetic characterization of feline leukemia virus from Florida panthers. Emerg Infect Dis 14:252. doi: 10.3201/eid1402.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Troyer RM, Beatty JA, Stutzman-Rodriguez KR, Carver S, Lozano CC, Lee JS, Lappin MR, Riley SP, Serieys LE, Logan KA, Sweanor LL, Boyce WM, Vickers TW, McBride R, Crooks KR, Lewis JS, Cunningham MW, Rovnak J, Quackenbush SL, VandeWoude S. 2014. Novel gammaherpesviruses in North American domestic cats, bobcats, and pumas: identification, prevalence, and risk factors. J Virol 88:3914–3924. doi: 10.1128/JVI.03405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bevins SN, Carver S, Boydston EE, Lyren LM, Alldredge M, Logan KA, Riley SPD, Fisher RN, Vickers TW, Boyce W, Salman M, Lappin MR, Crooks KR, VandeWoude S. 2012. Three pathogens in sympatric populations of pumas, bobcats, and domestic cats: implications for infectious disease transmission. PLoS One 7:e31403. doi: 10.1371/journal.pone.0031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benson D, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2011. GenBank. Nucleic Acids Res 39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 69.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 70.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 72.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kosakovsky P, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rambaut A, Drummond A. 2007. Tracer, version 1.4. http://beast.bio.ed.ac.uk/Tracer.
- 79.Rambaut A, Drummond A. 2010. LogCombiner v1.5.4. MCMC output combiner. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, United Kingdom. [Google Scholar]