Abstract
We report here the main characteristics of ‘Gorbachella massiliensis’ GD7T gen. nov., sp. nov., ‘Fenollaria timonensis’ GD5T sp. nov., ‘Intestinimonas timonensis’ GD4T sp. nov., and ‘Collinsella ihuae’ sp. nov. GD8T isolated from one fresh stool of a French volunteer. We used a bacterial culturomics approach combined with taxono-genomics.
Keywords: Collinsella ihuae, fenollaria timonensis, gorbachella massiliensis, gut microbiota, intestinimonas timonensis’
In our study concerning the intolerant oxygen species from the human gut microbiota, we isolated four bacteria in 2015 using a bacterial culturomics approach. These bacteria could not be identified by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS; http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database (last access 01/30/17)) on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [1], [2]. These species were isolated from the same fresh stool from a healthy volunteer. The individual has signed informed consent and the study has been validated by the Ethics Committee of the IFR48 Federative Research Institute under the number 09-022.
Strain GD7T was isolated from a dilution of the fresh sample. The species was grown after 48 h on Columbia agar supplemented with 5% sheep blood at 37°C under strict anaerobic conditions. The colonies appeared translucent and rough, non-haemolytic, non-motile, nonspore-forming, and 1 mm size. The cells were Gram-negative, rod-shaped. The strain did not show catalase or oxidase activity. The 16S rRNA gene was sequenced using fD1-rP2 primers as described previously using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) [3]. The strain GD7T had a 16S rRNA gene sequence identity of 93.4% with Subdoligranulum variabile strain BI 114T (NR_028997), the phylogenetically closest species with standing in nomenclature (Fig. 1). This similarity <98.65% leads us to putatively classify GD7T as a new member in the Ruminococcaceae family of Firmicutes [4]. Therefore we propose the creation of the new genus ‘Gorbachella’ (Gor.ba.chel'la. NL gen fem, in honour of the microbiologist Sherwood Gorbach of the Tufts University School of Medicine, Boston, MA, USA). GD7T is the type strain of the species Gorbachella massiliensis (ma.ssi.li.en'sis L. adj. fem to Massilia, the Latin name of Marseille, France, where this strain was isolated).
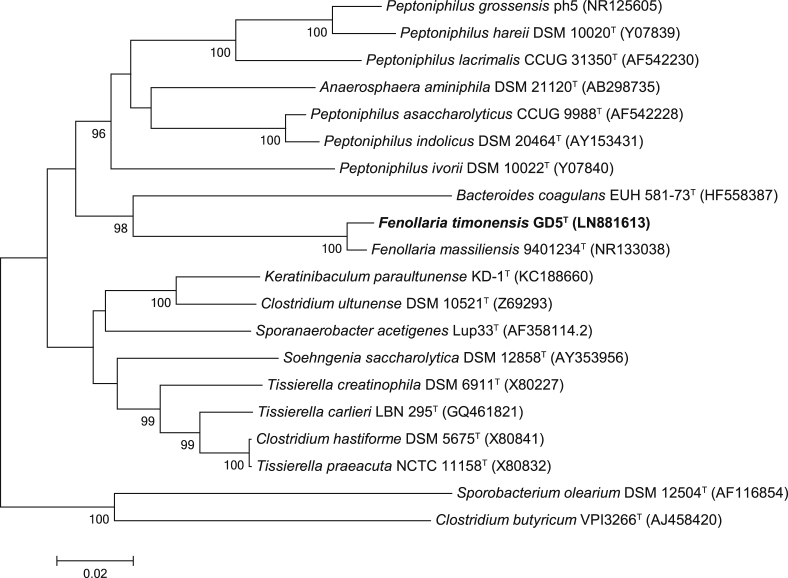
Fig. 1.
Phylogenetic tree showing the position of ‘Intestinimonas timonensis’ GD4T and ‘Gorbachella massiliensis’ GD7T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained with Kimura two-parameter models using the maximum-likelihood method within the MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. Only bootstrap values >95% are displayed. The scale bar indicates a 2% nucleotide sequence divergence.
Strain GD5T was isolated from the fresh sample after 48 h anaerobic growth on Columbia agar supplemented with 5% sheep blood at 37°C. The colonies appeared to be translucent, rough, non-haemolytic, motile, non-spore-forming, and 1 mm in size. The cells were rod-shaped with Gram-negative staining. Oxidase and catalase activities were negative. Strain GD5T showed 97.4% sequence homology with the 16S RNA of Fenollaria massiliensis strain 9401234T (NR_133038) (Fig. 2) [6]. So we propose to classify GD5T as a new species within the genus Fenollaria in the phylum Firmicutes [4]. GD5T is the type strain of the species ‘Fenollaria timonensis’ (ti.mo.nen'sis L. adj. fem to Timone, the name of the main hospital of Marseille, France, where this strain was isolated).
Fig. 2.
Phylogenetic tree showing the position of ‘Fenollaria timonensis’ GD5T relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were made as described for Fig. 1.
Strain GD4T was isolated after 48 h anaerobic growth on Columbia agar supplemented with 5% sheep blood and 5% rumen fluid at 37°C. The colonies appeared translucent, rough, non-haemolytic, motile, non-spore-forming, and 1 mm in size. The cells were Gram-negative. Catalase and oxidase activities were negative. Strain GD4T presented a sequence identity of 97.08% with 16S rRNA sequence of Intestinimonas butyriciproducens DSM 26588T (NR_118554), the closest species with a valid name (Fig. 1). We propose to putatively classify GD4T as a new member of the genus Intestinimonas in the phylum Firmicutes [4]. GD4T is the type strain of the species ‘Intestinimonas timonensis’ (ti.mo.nen'sis L. adj. fem to Timone, the name of the main hospital of Marseille, France, where this strain was isolated).
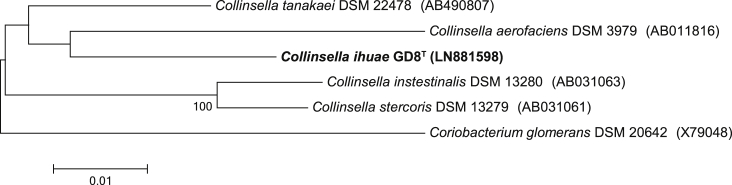
‘Collinsella ihuae’ strain GD8T was isolated after 48 h of anaerobic growth on Columbia agar supplemented with 5% sheep blood and 5% rumen fluid at 37°C. Colonies appeared as microcolonies rough, non-haemolytic, motile, non-spore-forming, and 0.5 mm in size. The cells were rod-shaped with Gram-positive staining. Catalase and oxidase activities were negative. The 16S rRNA sequence of the strain GD8T presented an identity of 96.2% with the 16S rRNA sequence of Collinsella tanakaei strain JCM 16071 (NR_113273), the closest phylogenetic species with nomenclature (Fig. 3) [5]. Then we propose the creation of the new species ‘Collinsella ihuae’ within the phylum Actinobacteria [4]. GD8T is the type strain of the species ‘Collinsella ihuae’ (i.hu.ae L. adj. fem to Institut Hospitalo-Universitaire (IHU), the name of the name of the laboratory (Marseille, France) where this strain was isolated).
Fig. 3.
Phylogenetic tree showing the position of ‘Collinsella ihuae’ GD8T relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were made as described for Fig. 1. The scale bar indicates a 1% nucleotide sequence divergence.
MALDI-TOF-MS spectra accession numbers. The MALDI-TOF-MS spectra of these species are available at http://mediterranee-infection.com/article.php? laref=256&titre=urms-database. Last access 7 December 2016.
Nucleotide sequence accession number. The 16S r RNA gene sequences were deposited in GenBank under accession numbers: ‘Gorbachella massiliensis’ GD7T (LN870316), ‘Fenollaria timonensis’ GD5T (LN881613), ‘Intestinimonas timonensis’ GD4T (LN870298) and ‘Collinsella ihuae’ GD8T (LN881598).
Deposit in a culture collection. The strains were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under the following numbers: ‘Gorbachella massiliensis’ GD7T (P2021), ‘Fenollaria timonensis’ GD5T (P2133), ‘Intestinimonas timonensis’ GD4T (P2010) and ‘Collinsella ihuae’ GD8T (P2019).
Transparency declaration
The authors have no conflicts of interest to declare.
Funding
This work was funded by the Fondation Mediterrannée-Infection.
References
- 1.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 5.Pagnier I., Croce O., Robert C., Raoult D., La Scola B. Non-contiguous finished genome sequence and description of Fenollaria massiliensis gen. nov., sp. nov., a new genus of anaerobic bacterium. Stand Genomic Sci. 2014;9:704–717. doi: 10.4056/sigs.3957647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagai F., Watanabe Y., Morotomi M. Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol. 2010;60:2639–2646. doi: 10.1099/ijs.0.017533-0. [DOI] [PubMed] [Google Scholar]