Abstract
This perspective considers the use of oral cell DNA adducts, together with exposure and genetic information, to potentially identify those cigarette smokers at highest risk for lung cancer, so that appropriate preventive measures could be initiated at a relatively young age before too much damage has been done. There are now well established and validated analytical methods for the quantitation of urinary and serum metabolites of tobacco smoke toxicants and carcinogens. These metabolites provide a profile of exposure and in some cases lung cancer risk. But they do not yield information on the critical DNA damage parameter that leads to mutations in cancer growth control genes such as KRAS and TP53. Studies demonstrate a correlation between changes in the oral cavity and lung in cigarette smokers, due to the field effect of tobacco smoke. Oral cell DNA is readily obtained in contrast to DNA samples from the lung. Studies in which oral cell DNA and salivary DNA have been analyzed for specific DNA adducts are reviewed; some of the adducts identified have also been previously reported in lung DNA from smokers. The multiple challenges of developing a panel of oral cell DNA adducts that could be routinely quantified by mass spectrometry are discussed.
Introduction
The tobacco control community has made great strides in the past several decades to de-normalize the use of tobacco products, especially combusted ones such as cigarettes, cigars, and pipes. Smoking has become, in part, a socioeconomic phenomenon, so most North American and Western European readers of this perspective will never encounter cigarette smokers in indoor environments and probably rarely in the outdoors during their normal daily routine. They may not realize that in spite of this remarkable progress there are still about 40 million adult smokers in the U.S. and an astounding one billion in the world.1,2 It is likely that many of these smokers are generally aware of the hazards of cigarette smoking but they cannot break their habit because nicotine, the principal alkaloid of the tobacco plant, is highly addictive.3
Cigarette smoking is the major cause of lung cancer, a disease that is generally detected too late for effective curative therapy. While some remarkable advances in lung cancer therapy have been achieved, the overall 5 year survival rate varies from 4 - 17% depending on stage and regional differences, and median survival following diagnosis is typically measured in months.4 Cigarette smoking causes 90% of lung cancer in the U.S., where 158,080 deaths were expected in 2016, making it the leading cause of cancer death in both men and women.5 Worldwide, there were 1,589,000 deaths from lung cancer in 2012, an average of about 3 per minute.6 Cigarette smoking caused 80% of this worldwide death toll in males and 50% in females.7 This is an immense public health crisis but it receives relatively little attention because it is so common.
While cigarette smoking is clearly the major cause of lung cancer, only 11% of female and 24% of male lifetime smokers will get lung cancer by age 85 or greater, and this relatively small percentage is not due to competing causes of death from smoking.8 The major goal of the research approach discussed in this perspective is to identify individuals who are highly susceptible to the carcinogenic effects of tobacco products. These individuals would be candidates for intensive lung cancer surveillance and screening, increasing the probability of detection of a tumor at an early stage. When lung cancer is detected in its early stages, surgical resection can achieve far higher 5 year survival rates, 50-70%, than when it is detected at later stages. In this perspective, we are not proposing methods for early detection of tumors such as the identification in exhaled breath condensate of specific proteins which are characteristic of lung tumors,9 but rather identification of susceptible individuals. While there are already algorithms relating various parameters to lung cancer susceptibility, they are mostly retrospective in nature, with pack-years of cigarette smoking being a major prognostic factor.10-15 Thus, these algorithms are typically applied to subjects who are older, when the process may be more advanced.16-20 Our ultimate goal is to develop a risk model that is prospective in nature. Overall, there would be a greater probability of success if one identifies high risk individuals early in the carcinogenic process. Even if this were effective in only 10% of tobacco users, the outcome could be prevention of more than 15,000 lung cancer deaths per year in the U.S. alone and massive savings in terms of dollars not spent on therapy.
Urinary and Serum Metabolites as Biomarkers of Lung Cancer Risk
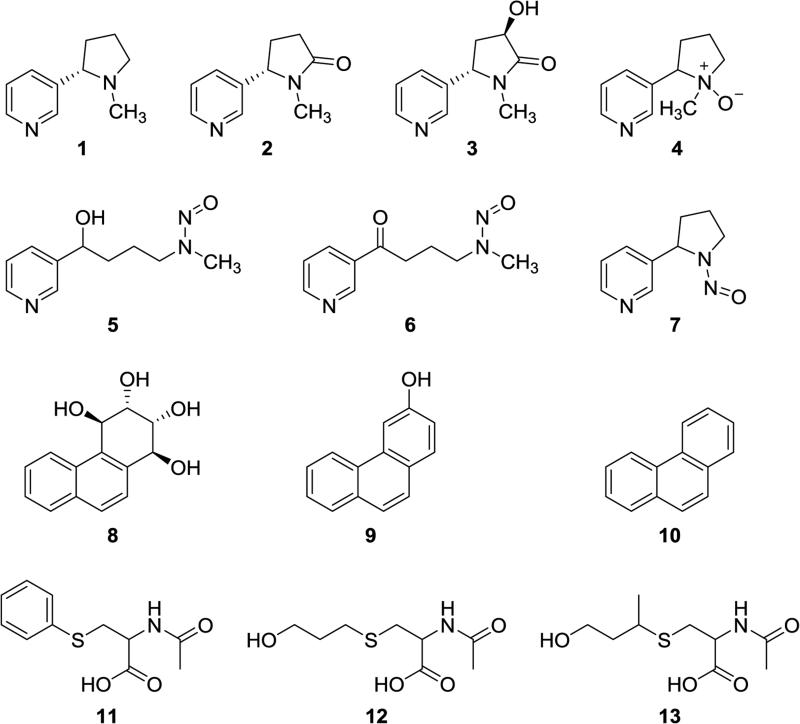
In our ongoing research to date, we have focused on several biomarkers of tobacco smoke toxicant and carcinogen uptake as quantified by the relevant parent substances and metabolites detected in urine.21 Thus, we and others have developed and applied analytically validated methods for urinary substances such as total nicotine equivalents (the sum of nicotine (1), cotinine (2), 3′-hydroxycotinine (3) and their glucuronides, as well as nicotine-N-oxide (4)); total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL, 5), a metabolite of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK, 6); total N′-nitrosonornicotine (NNN, 7), an important tobacco-specific carcinogen; r-1,t-2,3,c-4-tetrahydroxy-1,2,3,4-tetrahydrophenanthrene (PheT, 8) and 3-hydroxyphenanthrene (9), metabolites of the representative polycyclic aromatic hydrocarbon phenanthrene (10); S-phenylmercapturic acid (SPMA, 11), a metabolite of the carcinogen benzene; 3-hydroxypropylmercapturic acid (HPMA, 12), a metabolite of acrolein; and 3-hydroxy-1-methylpropylmercapturic acid (HMPMA, 13), a metabolite of crotonaldehyde (see Figure 1 for structures).22-25 Massive amounts of data demonstrate the involvement of these classes of compounds in cancer induction by tobacco smoke. 3,8 Some advantages and disadvantages of selected urinary biomarkers are summarized in Table 1.
Figure 1.
Structures of compounds discussed in the text with respect to quantitation of parent compounds and metabolites in the urine of people who use tobacco products.
Table 1.
A panel of tobacco smoke carcinogen and toxicant metabolites quantified in urine.
| Urinary Metabolite or Parent Compound | Biological Effect(s) Represented | Advantages | Risk Biomarker in Smokers? | Disadvantages | References |
|---|---|---|---|---|---|
| Total nicotine equivalents | Toxicity/addiction | Measures nearly 90% of the nicotine dose; relatively specific to tobacco use | Yes (Shanghai and Singapore cohorts) | Not a direct measure of DNA damage | 73 |
| Total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) | Lung cancer | Good measure of lung carcinogen dose (NNK); specific to tobacco use | Yes (Shanghai and Singapore cohorts; PLCO – serum) | Not a direct measure of DNA damage | 65,74-76 |
| Phenanthrene tetraol (PheT) | Cancer | Good measure of polycyclic aromatic hydrocarbon (PAH) dose plus metabolic activation | Yes (Shanghai cohort) | Not a direct measure of DNA damage | 65,75 |
| 3-Hydroxyphenanthrene (3-OH-Phe) | Cancer | Good measure of PAH exposure | Not known | Not a direct measure of DNA damage or metabolic activation | 77 |
| S-Phenylmercapturic acid (SPMA) | Cancer | Good measure of benzene exposure | No | Confounded by GST-T1 status | 78,79 |
| 3-Hydroxypropylmercapturic acid (3-HPMA) | Toxicity, cancer(?) | Good measure of acrolein exposure | No | Not a direct measure of DNA damage | 66 |
| 3-Hydroxy-1-methylpropylmercapturic acid (HMPMA) | Toxicity, cancer(?) | Good measure of crotonaldehyde exposure | No | Not a direct measure of DNA damage | 66 |
| Monohydroxybutylmercapturic acid (MHBMA) | Cancer | Good measure of 1,3-butadiene exposure | Not known | Confounded by GST-T1 status | 78,80 |
| F2-Isoprostanes (8-iso-PGF2α) | Oxidative damage | Accepted measure of oxidative damage | Not known | Not a direct measure of DNA damage | 81 |
| Prostaglandin E2 metabolite (PGEM) | Inflammation | Biomarker of inflammation | Not known | Not a direct measure of DNA damage | 82 |
We have collaborated with epidemiologists to evaluate the relationship of these urinary metabolites to cancer, as determined in prospective epidemiology studies. These studies collect and store bio-samples such as blood or urine from large numbers of healthy subjects, then follow the subjects for decades until sufficient numbers of cancer cases occur for statistical analysis. Samples from the cases and matched controls without cancer are recovered from freezers and analyzed for specific biomarkers. This is the most powerful epidemiologic study design because the subjects are recruited and interviewed when healthy, so the eventual occurrence of disease has no bearing on their biomarker levels. The results of these studies have been reviewed.26,27 In summary, statistically significant relationships of urinary total cotinine (2 plus its glucuronide), total NNAL (5), and PheT (8) with lung cancer risk, and total NNN (7) with esophageal cancer risk were observed in prospective epidemiological studies of male smokers in Shanghai. Urinary total cotinine and total NNAL were related to lung cancer risk in a study of male and female smokers in Singapore, and total NNAL in serum was related to lung cancer risk in a study of male and female smokers in the U.S.26,27 Levels of urinary SPMA (11), HPMA (12), and HMPMA (13) were not independently related to lung cancer (after correction for cotinine levels) in the Shanghai study. These results support the use of total cotinine, total NNAL, and PheT as possible biomarkers of lung cancer risk, but the relationships uncovered to date, when examined in the context of receiver operating characteristics analysis, are not strong enough to support their independent application as predictive biomarkers.
Specific DNA Adducts as Biomarkers of Lung Cancer Risk
The urinary and serum metabolites discussed above are mainly biomarkers of exposure and do not inform us about the critical parameter - DNA adduct levels. Thus, an individual with high exposure but excellent detoxification or DNA repair capacity will not have relatively high DNA adduct levels and would be misclassified as having high risk based on exposure measurements only. Conversely, an individual with relatively low exposure but high levels of metabolic activation or low DNA repair capacity would be classified as low risk based on exposure biomarkers only. Therefore, the next logical step in the development of a predictive biomarker formula is to quantify DNA adducts. In this perspective, we discuss DNA adducts in human oral cells, quantified by mass spectrometry, as potentially useful biomarkers for lung cancer risk. Our main hypothesis is that these DNA adduct measurements in combination with certain exposure biomarkers as well as selected genetic information will produce a predictive algorithm for susceptibility to lung cancer in smokers.
The oral mucosa is the mucous membrane lining the inside of the mouth. Oral mucosa cells are an excellent source of material for evaluating DNA adducts and molecular changes potentially related to cancer.28-31 Collection of oral mucosa cells is relatively simple, in contrast to bronchial brushings and sputum, which can be difficult, expensive, and impractical to obtain. Multiple studies have demonstrated similarities in molecular changes between oral mucosa cells and bronchial cells obtained from the same individuals, particularly in smokers, consistent with the field carcinogenesis concept of lung and upper aerodigestive tract cancer.32-38 In one large study, a strong association was found between promoter methylation patterns of the p16 and FHIT genes in oral tissues and bronchial brush specimens from smokers.32 In another study, similarities were found in gene expression changes in the oral and bronchial mucosa, and smoking altered the expression of numerous genes compared to observations in never smokers.35 A third study demonstrated that smoking-induced gene expression changes in the bronchial airway were reflected both in the nasal and buccal epithelium.34 Collectively, these results support the use of oral mucosa cells as a surrogate for DNA adduct formation in the lung.38,39
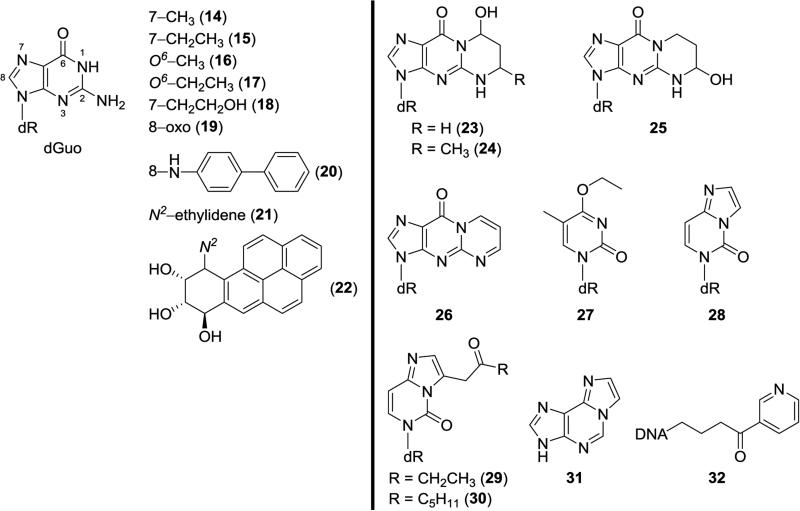
Why not quantify DNA adducts directly in lung tissue? As noted above, bronchial brushings and sputum can be difficult to obtain. However, there have been many studies of DNA adduct levels in lung tissue from smokers, mainly obtained during surgery for lung cancer or other diseases. These studies have been reviewed.40-43 Multiple investigations using non-specific DNA adduct detection methods such as 32P-postlabelling and immunoassay demonstrate higher levels of DNA adducts of unknown structure in lung tissue from smokers than from non-smokers. The characterization of the DNA adducts frequently seen in 32P-postlabelling studies, and often elevated in smokers’ lung tissue, typically described as “a diagonal radioactive zone”, remains a challenge. This material was originally thought to be derived from polycyclic aromatic hydrocarbons, but studies by Arif et al do not support that assignment.44 Relatively few structurally specific DNA adducts have been characterized in lung tissue.45 These are summarized in Figure 2, the content of which is from a review published in 2012;42 there have been no new characterizations since then. Detected adducts include 7- and O6-alkylguanines such as 14-17, aromatic-substituted guanines such as 20 and 22, and a variety of cyclic and etheno-type adducts such as 23-26 and 28-31, as well as the HPB releasing adducts 32. Most of the DNA adducts illustrated in Figure 2 were detected in relatively small studies, and some of them such as the benzo[a]pyrene adduct 22 were frequently not detected.46 None have been unequivocally related to lung cancer risk. Even the sources and identities of some of these DNA adducts are in many cases unclear. For example, the simple 7-alkylguanine, O6-alkylguanine, and O4-ethylthymidine adducts could result from direct acting methylating or ethylating agents in cigarette smoke, which are mainly uncharacterized, or from the very small amounts of dialkylnitrosamines present in smoke.
Figure 2.
Structures of DNA adducts reported in human lung tissue, using structure-specific methods (adapted from 42). Mono-substituted dGuo adducts are to the left of the vertical line, and all others to the right. The following DNA adducts have also been detected in DNA from human oral tissue or saliva: 15, 19, 20, 21, 23, 24, 27, 28, 31, and 32.
DNA Adducts in Human Oral Tissue and Saliva
Earlier studies on DNA adducts in oral tissue, detected exclusively by non-specific immunoassay and 32P-postlabelling assays, have been reviewed.28 More recent studies which used chemically specific assays, mainly but not exclusively based on mass spectrometry, are summarized here.
Borthakur et al quantified 8-oxo-dGuo (19, Figure 2) in buccal mucosa cells as a marker of oxidative damage. Subjects were non-smokers who reported being healthy and not drinking alcohol excessively. The subjects gave blood and buccal cells at baseline and after 3 and 7 days. The participants rinsed their mouth with distilled water before collection of buccal cells using a soft bristle toothbrush. Levels of 8-oxo-dGuo were quantified by HPLC with electrochemical detection. The results demonstrated that 8-oxo-dGuo/dGuo levels were relatively stable over a 7 day period and were 3-4 times higher in buccal cell DNA (30-40 adducts per 106 dGuo) than in leukocyte DNA.31
Bessette et al47 screened for DNA adducts in buccal cell DNA of cigarette smokers using data-dependent constant neutral loss MS3 in which the neutral loss of the deoxyribose moiety in the MS/MS scan triggers the acquisition of MS3 product ion spectra of the aglycone. The study was initiated by spiking buccal cell DNA samples with several authentic DNA adducts derived from acrolein, 4-hydroxynonenal, 4-aminobiphenyl, and benzo[a]pyrene, and from metabolically activated forms of the heterocyclic aromatic amines 2-amino-3,8-dimethyl-3H-imidazo[4,5-f]quinoxaline (methyl-IQx), 2-amino-α-carboline, and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), at a level of 5 adducts per 108 bases. All of the adducts spiked into the buccal cell DNA were clearly detected. However, in unspiked buccal cell DNA samples, only adducts 23 and 25 (Figure 2), derived from acrolein, were unequivocally detected. The MS response for these adducts in the unspiked samples were 10-50 times greater than the response of the other adducts. The levels of the acrolein adducts were estimated to be above 5 adducts per 107 DNA bases. These levels were quite similar to those first reported more than 10 years earlier in gingival tissue of cigarette smokers using 32P-postlabelling.48 That study provided convincing evidence based on HPLC analysis for the presence in gingival tissue of smokers of acrolein adduct 23 and both stereoisomers of the crotonaldehyde adduct 24 (Figure 2). Levels of all of these adducts were significantly higher in smokers than in non-smokers. Further studies by this group have investigated the use of monoclonal antibodies and immunohistochemistry for analysis of the acrolein-DNA adducts in human oral cells.49,50
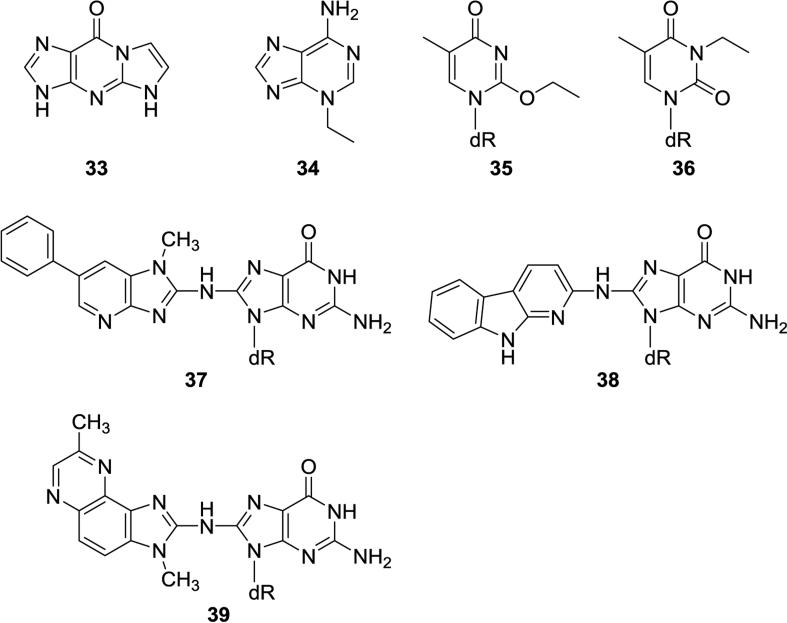
Further evidence for the presence of DNA adducts of acrolein and crotonaldehyde in oral samples was provided by Chen and Lin who used a validated nanoflow-LC-nanospray ionization tandem MS method to quantify these adducts in 27 human salivary DNA samples from healthy volunteers.51 They found mean adduct levels of 104 ± 50 adducts/108 nucleotides for acrolein adducts (consisting mostly of 23, Figure 2) and 7.5 adducts/108 nucleotides for 24. They also used this method to quantify the etheno adducts 28 and 31 (Figure 2) as well as 1,N2- ethenodGuo (33, Figure 3), with average levels of 99 ± 50, 72 ± 49, and 391 ± 198 adducts/108 nucleotides, respectively. These three adducts were highly correlated with each other suggesting a common source, most likely lipid peroxidation.
Figure 3.
Structures of DNA adducts reported in human saliva, but not in human lung.
The Chen group also used nanoflow-LC-nanospray ionization tandem MS to quantify several other DNA adducts in human saliva.52,53 Neutral thermal hydrolysis of salivary DNA released 3-Et-Ade (34, Figure 3) and 7-Et-Gua (15, Figure 2). The mean levels of 3-Et-Ade in 15 smokers and 15 non-smokers were 12.6 ± 7.0 and 9.7 ± 5.3 per 108 normal nucleotides, respectively, while those of 7-Et-Gua were 14.1 ± 8.2 and 3.8 ± 2.8 per 108 normal nucleotides.52 Further studies focused on thymidine adducts. Starting with 50 μg of DNA isolated from 3.5 ml of saliva, they were able to detect O2-Et-dThd (35, Figure 3), N3-Et-dThd (36, Figure 3), and O4-Et-dThd (27, Figure 2) in saliva DNA samples from 20 smokers, with levels ranging from 4-5 adducts per 108 normal nucleotides, while these adducts were non-detectable in saliva samples from non-smokers.53
Saliva samples from 37 human volunteers on unrestricted diets were analyzed for DNA adducts of heterocyclic aromatic amines and related compounds by LC-MS/MSn by Bessette et al.54 The dGuo-C8 adducts of PhIP (37, Figure 3), 2-amino-α-carboline (38, Figure 3), MeIQx (39, Figure 3) and 4-aminobiphenyl (20, Figure 2) were characterized and quantified using consecutive reaction monitoring. The PhIP-DNA adducts were detected most frequently, in saliva samples from 13 of 29 ever-smokers, and in 2 of 8 samples from never-smokers. Levels of these adducts ranged from 1 to 9 per 108 DNA bases.
Alcohol consumption is an established risk factor for cancer of the upper aerodigestive tract, including oral cavity cancer. Acetaldehyde, the primary metabolite of alcohol, is considered to be crucial in DNA damage by alcohol and is regarded by the International Agency for Research on Cancer as “carcinogenic to humans” when associated with the consumption of alcoholic beverages.55 The major DNA adduct of acetaldehyde is N2-ethylidene-dGuo (21, Figure 2), which can be quantified by LC-MS/MS as N2-ethyl-dGuo in DNA that has been treated with NaBH3CN. Our group quantified levels of this adduct in oral cells collected by mouthwash at various time points after consumption by non-smokers of increasing alcohol doses, administered as vodka to reach target blood alcohol concentrations of 0.03-0.07%.56 Levels of N2-ethylidene-dGuo in oral cell DNA increased as much as 100-fold from baseline within 4h after each dose for all subjects and in a dose-responsive manner, reaching levels as high as 1 adduct per 105 nucleotides. A time dependent increase and decrease in levels of this adduct in oral cells was clearly observed after all three alcohol doses. Levels of the adduct also increased in lymphocytes and granulocytes of the subjects but there was substantial intra-individual variability which obscured clear elucidation of alcohol's effects.57 Acetaldehyde exposure from alcohol consumption is expected to greatly exceed that from smoking, but further studies are needed on the potential interaction of drinking and smoking on oral cell DNA adduct levels.
The tobacco-specific nitrosamines NNK (6, Figure 1) and NNN (7, Figure 1) undergo metabolic α-hydroxylation reactions catalyzed by cytochrome P450 enzymes yielding a pyridyloxobutylating intermediate which reacts with cellular DNA to produce a variety of characterized adducts with dGuo, dThd, and dCyd.58 Treatment of this DNA with acid causes hydrolysis of several pyridyloxobutyl adducts (see 32, Figure 2) with the release of 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB). Thus, HPB-releasing DNA adducts are a measure of pyridyloxobutylation of DNA by NNK, NNN, and possibly other tobacco-specific compounds. Our group quantified HPB-releasing DNA adducts in oral cells collected by buccal brushing and by mouthwash.59 In smokers’ samples collected by mouthwash, the levels of HPB-releasing DNA adducts averaged 12.0 pmol HPB/mg DNA (4 adducts per 106 nucleotides), and they were detected in 20 of the 28 samples with quantifiable yield of DNA. Samples were also collected by buccal brushing and the levels of adducts correlated with those collected by mouthwash, and averaged 44.7 pmol HPB/mg DNA (15 adducts per 106 nucleotides) in smokers. HPB-releasing DNA adducts in non-smoker samples were mostly undetectable.
Overall, there are no coordinated studies of DNA adducts in human oral tissue and saliva vs. levels of the same adducts in human lung. Figure 2 demonstrates that there are 10 DNA adducts that have been detected in both human oral tissue or saliva and human lung, but never from the same humans. Figure 3 demonstrates that there are 7 adducts that have been detected in human saliva but have not been reported in human lung. It would most likely be impractical to carry out a study in which adducts from lung tissue and oral tissue or saliva from the same human were analyzed. Therefore, it will be necessary in future studies to depend on the assumption, supported by the studies discussed in the previous section, that genetic alterations in oral tissue reflect those in the lung because both are in the field affected by cigarette smoke. The relationship of salivary DNA adducts to adduct levels in the lung may be more complex.
Developing a Research Approach
The development of a panel of oral cell DNA adducts that might predict lung cancer susceptibility is indeed challenging. The first challenge is the selection of DNA adducts to be quantified. Considering what is known about tobacco smoke carcinogenesis, it would be prudent to include adducts derived from a variety of different types of carcinogens and relevant enhancing processes. Tobacco-specific nitrosamines, polycyclic aromatic hydrocarbons, and volatiles such as formaldehyde, acetaldehyde, 1,3-butadiene, and acrolein are representative of major classes of carcinogens and toxicants in cigarette smoke. Inflammation and oxidative damage are also likely to play a significant role in lung carcinogenesis by tobacco smoke. Thus, as an initial approach, one could consider a panel of DNA adducts consisting of HPB-releasing DNA adducts (32, Figure 2), benzo[a]pyrene-DNA adducts (22, Figure 2) or their hydrolysis products (such as benzo[a]pyrene tetraols), formaldehyde-DNA adducts, 1,3-butadiene-DNA adducts, acrolein-DNA adducts (23 and 25, Figure 2), 8-oxo-dGuo (19, Figure 2), and etheno-dGuo (33, Figure 3). Among these, only the HPB-releasing DNA adducts, acrolein-DNA adducts, and 8-oxo-dGuo have been identified and quantified in human oral tissues. Formaldehyde-DNA adducts have been quantified in human leukocyte DNA,60 benzo[a]pyrene-DNA adducts have been occasionally quantified in human lung DNA,46,61 and etheno-dGuo has been measured in human salivary DNA.51 Considering that only 1 -10 μg of oral cell DNA would be available from a typical collection, the quantitation of multiple adducts, some of which are typically found at levels of 1 per 108 nucleotides, represents a major analytical chemistry challenge. It is now realistic to consider addressing this challenge because contemporary nanoflow-LC -nanoelectrospray ionization techniques coupled with state of the art mass spectrometers can achieve detection limits in the low amol, or even high zmol range, sufficient for this type of analysis, although it still may require multiple collections per individual.
Each method must be fully validated for accuracy, precision, linearity, ruggedness, and freedom from artifacts. Another variable is the site and method of sample collection from the oral cavity. It may also be necessary to explore the contribution of oral microorganisms to DNA adduct formation. When each method has been validated, and shown not to result from the possible contribution of adducted bacterial DNA, it would be tested for inclusion in the panel. The first test would be comparison of DNA adduct levels from smokers versus non-smokers. If the DNA adduct is not elevated in smokers, or if there is not a sufficient range of values in smokers, it would not likely be a valuable addition to the panel. Typically, an initial evaluation of this type requires at least 30 samples from smokers and 30 from non-smokers, which could then be confirmed in larger studies.59,60,62 The second test would be longitudinal or temporal stability of the DNA adduct level. This is important because the adduct levels are proposed to reflect the balance between DNA adduct formation and repair. If this is the case, it would be expected that the DNA adduct level would remain relatively constant over time, if the cigarette smoker maintains his/her habit. Studies of this type have been performed for urinary metabolites, and it would be important to carry out similar studies for the oral cell DNA adducts. For example, we showed that the intra-class correlation coefficient for urinary total NNAL was 76% in 70 smokers sampled every other month for one year indicating relative stability of these measurements in a given individual.63 When these tests of the oral cell DNA adducts would be completed, the panel could be applied in prospective epidemiology studies to test its ability in risk prediction. Prospective epidemiology studies of lung cancer with collected oral cells include the Shanghai Cohort study, the Multi-Ethnic Cohort study, the Wayne State University and Karmanos Cancer Institute study, the University of Toronto study, the Los Angeles population-based case-control study, and the UCSF Northern California Lung Cancer study.64 These studies also collect extensive data on factors such as diet and alcohol consumption which would be included in the final statistical model.
While the development of a panel of oral cell DNA adducts is clearly challenging, the same could have been said about the panel of urinary metabolites summarized in Table 1. In practice, these biomarkers as well as a number of additional ones are now routinely measured in large studies of tobacco smoke toxicant and carcinogen exposure. The Population Assessment of Tobacco and Health (PATH) study, jointly sponsored by the National Institutes of Health and the Food and Drug Administration, follows 46,000 people, some of whom are tobacco users, some of whom are not, for at least 3 years. The PATH study thus generates many thousands of urine samples which are being analyzed in the laboratories of the Centers for Disease Control and Prevention using mass spectrometry-based robotically driven analytical systems. Our own relatively small laboratory is able to quantify thousands of urinary biomarker samples from smokers using high throughput technology coupled to mass spectrometry, and starting with small amounts of urine, typically 1 – 2 ml.65,66 These activities would have been incomprehensible in the early part of this century.
Exposure parameters and oral cell DNA adducts would likely only be part of an ultimate model for susceptibility to lung cancer, and it is recognized that these could be affected by polymorphisms in carcinogen metabolizing genes such as glutathione-S-transferases or UDP-glucuronosyl transferases. The model would also need to include genotyping data for CYP2A6, the highly polymorphic major enzyme involved in nicotine metabolism, with low activity forms affecting smoking behavior, nicotine uptake, and lung cancer risk.67 It would likely also include genotyping for variation in the α5 nicotinic cholinergic receptor subunit gene (CHRNA5) associated with nicotine dependence.64 The association with quantity smoked has been reported for rs16969968 and rs1051730, which are correlated genetic variants. These variants are also associated with lung cancer and COPD,64 and a haplotype rs588765-rs16969968 is also significantly associated with lung cancer (2-3 fold increased risk).68 Some studies have examined the use of these and related SNPs to assess efficacy in smoking cessation.69-71 Additional findings could be included as they evolve in genomics and epigenetics studies. For example, hypomethylation of CpG sites in smoking-related genes such as the AHRR gene has been shown to be associated with future lung cancer in some prospective epidemiology studies.72
Summary
This perspective highlights the potential use of oral cell DNA adducts to enhance the predictive power of a proposed model to identify, at a relatively young age, those individuals highly susceptible to lung cancer. Oral cell DNA is easily obtained and is part of the physiologic field affected by tobacco smoke, which includes the lung and upper aerodigestive tract, all of which are highly susceptible to tobacco induced cancer. Oral cell DNA adduct levels would complement the existing panel of urinary tobacco carcinogen and toxicant metabolites which provide excellent exposure data, but only limited information on metabolic activation. Oral cell DNA adducts could potentially identify those individuals who have relatively high DNA adduct loads and consequently are more likely to harbor mutated KRAS and TP53 genes, the most commonly mutated genes in smoking associated lung cancer.
Acknowledgements
I thank Bob Carlson and Adam Zarth for editorial assistance.
Funding
This research is supported by grants CA-81301 and CA-138338 from the U.S. National Cancer Institute.
Abbreviations
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
N′-nitrosonornicotine
- PheT
r-1,t-2,3,c-4-tetrahydroxy-1,2,3,4-tetrahydrophenanthrene
- SPMA
S-phenylmercapturic acid
- HPMA
3-hydroxypropylmercapturic acid
- HMPMA
3-hydroxy-1-methylpropylmercapturic acid
- methyl-IQx
2-amino-3,8-dimethyl-3H-imidazo[4,5-f]quinoxaline
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- HPB
4-hydroxy-1-(3-pyridyl)-1-butanone
- CHRNA5
α5 nicotinic cholinergic receptor subunit gene
Reference List
- 1.Eriksen M, Mackay J, Schluger N, Islami Gomeshtapeh F, Drope J. The Tobacco Atlas. 5th Edition. American Cancer Society and World Lung Foundation; Atlanta, GA.: 2015. [Google Scholar]
- 2.Jamal A, Homa DM, O'Connor E, Babb SD, Caraballo RS, Singh T, Hu SS, King BA. Current cigarette smoking among adults - United States, 2005-2014. MMWR Morb. Mortal. Wkly. Rep. 2015;64:1233–1240. doi: 10.15585/mmwr.mm6444a2. [DOI] [PubMed] [Google Scholar]
- 3.United States Department of Health and Human Services . A Report of the Surgeon General. U.S. Dept. of Health and Human Services, Centers for Disease Control and Prevention, Coordinating Center for Health Promotion, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; Atlanta, GA.: 2014. The Health Consequences of Smoking: 50 Years of Progress. [Google Scholar]
- 4.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr., Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2016 doi: 10.1016/S0140-6736(16)30958-8. doi: 10.1016/S0140-6736(16)30958-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.Stewart BW, Wild CP. World Cancer Report 2014. IARC; Lyon, FR.: 2014. [Google Scholar]
- 7.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 8.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, FR.: 2004. Tobacco Smoke and Involuntary Smoking. pp. 174–176. [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes SA, Haefliger S, Harris B, Pavlakis N, Clarke SJ, Molloy MP, Howell VM. Exhaled breath condensate for lung cancer protein analysis: a review of methods and biomarkers. J Breath. Res. 2016;10:034001. doi: 10.1088/1752-7155/10/3/034001. [DOI] [PubMed] [Google Scholar]
- 10.Tammemagi CM, Pinsky PF, Caporaso NE, Kvale PA, Hocking WG, Church TR, Riley TL, Commins J, Oken MM, Berg CD, Prorok PC. Lung cancer risk prediction: prostate, lung, colorectal and ovarian cancer screening trial models and validation. J. Natl. Cancer Inst. 2011;103:1058–1068. doi: 10.1093/jnci/djr173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etzel CJ, Bach PB. Estimating individual risk for lung cancer. Semin. Respir. Crit Care Med. 2011;32:3–9. doi: 10.1055/s-0031-1272864. [DOI] [PubMed] [Google Scholar]
- 12.Spitz MR, Etzel CJ, Dong Q, Amos CI, Wei Q, Wu X, Hong WK. An expanded risk prediction model for lung cancer. Cancer Prev. Res. 2008;1:250–254. doi: 10.1158/1940-6207.CAPR-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassidy A, Myles JP, van Tongeren M, Page RD, Liloglou T, Duffy SW, Field JK. The LLP risk model: an individual risk prediction model for lung cancer. Br. J. Cancer. 2008;98:270–276. doi: 10.1038/sj.bjc.6604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bach PB, Kattan MW, Thornquist MD, Kris MG, Tate RC, Barnett MJ, Hsieh LJ, Begg CB. Variations in lung cancer risk among smokers. J. Natl. Cancer Inst. 2003;95:470–478. doi: 10.1093/jnci/95.6.470. [DOI] [PubMed] [Google Scholar]
- 15.Cronin KA, Gail MH, Zou Z, Bach PB, Virtamo J, Albanes D. Validation of a model of lung cancer risk prediction among smokers. J. Natl. Cancer Inst. 2006;98:637–640. doi: 10.1093/jnci/djj163. [DOI] [PubMed] [Google Scholar]
- 16.Weissfeld JL, Lin Y, Lin HM, Kurland BF, Wilson DO, Fuhrman CR, Pennathur A, Romkes M, Nukui T, Yuan JM, Siegfried JM, Diergaarde B. Lung cancer risk prediction using common SNPs located in GWAS-identified susceptibility regions. J Thorac. Oncol. 2015;10:1538–1545. doi: 10.1097/JTO.0000000000000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, Xiao L, Huang S, Chen L, Guo Z, Lu J, Yuan K. MicroRNA-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS One. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li K, Husing A, Sookthai D, Bergmann M, Boeing H, Becker N, Kaaks R. Selecting high-risk individuals for lung cancer screening: a prospective evaluation of existing risk models and eligibility criteria in the German EPIC cohort. Cancer Prev Res (Phila) 2015;8:777–785. doi: 10.1158/1940-6207.CAPR-14-0424. [DOI] [PubMed] [Google Scholar]
- 19.Baldwin DR. Prediction of risk of lung cancer in populations and in pulmonary nodules: Significant progress to drive changes in paradigms. Lung Cancer. 2015;89:1–3. doi: 10.1016/j.lungcan.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Marcus MW, Raji OY, Field JK. Lung cancer screening: identifying the high risk cohort. J Thorac. Dis. 2015;7:S156–S162. doi: 10.3978/j.issn.2072-1439.2015.04.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht SS, Yuan J-M, Hatsukami DK. Applying tobacco carcinogen and toxicant biomarkers in product regulation and cancer prevention. Chem. Res. Toxicol. 2010;23:1001–1008. doi: 10.1021/tx100056m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SE, Park S-SL, Thompson EF, Wilkens LR, Patel Y, Stram DO, Le Marchand L. Nicotine N-glucurionidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35:2526–2533. doi: 10.1093/carcin/bgu191. PMCID: PMCID: PMC4216060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SL, Carmella SG, Ming X, Stram DO, Le Marchand L, Hecht SS. Variation in levels of the lung carcinogen NNAL and its glucuronides in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. Cancer Epidemiol. Biomarkers Prev. 2015;24:561–569. doi: 10.1158/1055-9965.EPI-14-1054. PMCID: PMCID: PMC Journal- In Process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SL, Carmella SG, Chen M, Patel Y, Stram DO, Haiman CA, LeMarchand L, Hecht SS. Mercpaturic acids derived from the toxicants acrolein and crotonaldehyde in the urine of cigarettes smokers from five ethnic groups with differing risks for lung cancer. PLoS One. 2015;10:e0124841. doi: 10.1371/journal.pone.0124841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel YM, Park SL, Carmella SG, Paiano V, Olvera N, Stram DO, Haiman CA, Le Marchand L, Hecht SS. Metabolites of the polycyclic aromatic hydrocarbon phenanthrene in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. PLoS. One. 2016;11:e0156203. doi: 10.1371/journal.pone.0156203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan JM, Butler LM, Stepanov I, Hecht SS. Urinary tobacco smoke-constituent biomarkers for assessing risk of lung cancer. Cancer Res. 2014;74:401–411. doi: 10.1158/0008-5472.CAN-13-3178. PMCID: PMCID: PMC4066207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht SS, Murphy SE, Stepanov I, Nelson HH, Yuan J-M. Tobacco smoke biomarkers and cancer risk among male smokers in the Shanghai Cohort Study. Cancer Lett. 2012;334:34–38. doi: 10.1016/j.canlet.2012.07.016. PMCID: PMC3648613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proia NK, Paszkiewicz GM, Nasca MA, Franke GE, Pauly JL. Smoking and smokeless tobacco-associated human buccal cell mutations and their association with oral cancer--a review. Cancer Epidemiol. Biomarkers Prev. 2006;15:1061–1077. doi: 10.1158/1055-9965.EPI-05-0983. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, Bender PK, Beck JC, Le Marchand L, Lum A, Alavanja M, Hayes RB, Rutter J, Buetow K, Brinton LA, Rothman N. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol. Biomarkers Prev. 2001;10:687–696. [PubMed] [Google Scholar]
- 30.Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, Orden RA, Chen J, Goldman R, Shields PG. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29:1170–1177. doi: 10.1093/carcin/bgn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borthakur G, Butryee C, Stacewicz-Sapuntzakis M, Bowen PE. Exfoliated buccal mucosa cells as a source of DNA to study oxidative stress. Cancer Epidemiol. Biomarkers Prev. 2008;17:212–219. doi: 10.1158/1055-9965.EPI-07-0706. [DOI] [PubMed] [Google Scholar]
- 32.Bhutani M, Pathak AK, Fan YH, Liu DD, Lee JJ, Tang H, Kurie JM, Morice RC, Kim ES, Hong WK, Mao L. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prev Res (Phila) 2008;1:39–44. doi: 10.1158/1940-6207.CAPR-08-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar SV, Jain R, Mokhiber K, Venezia A, Sheehan A, Spivack SD. Exfoliated buccal and microdissected lung cell expression of antioxidant enzymes. Cancer Detect Prev. 2005;29:552–561. doi: 10.1016/j.cdp.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, Liu G, Dumas YM, Zhang X, Brody JS, Lenburg ME, Spira A. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle JO, Gumus ZH, Kacker A, Choksi VL, Bocker JM, Zhou XK, Yantiss RK, Hughes DB, Du B, Judson BL, Subbaramaiah K, Dannenberg AJ. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res (Phila) 2010;3:266–278. doi: 10.1158/1940-6207.CAPR-09-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy HK, Subramanian H, Damania D, Hensing TA, Rom WN, Pass HI, Ray D, Rogers JD, Bogojevic A, Shah M, Kuzniar T, Pradhan P, Backman V. Optical detection of buccal epithelial nanoarchitectural alterations in patients harboring lung cancer: implications for screening. Cancer Res. 2010;70:7748–7754. doi: 10.1158/0008-5472.CAN-10-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teschendorff AE, Yang Z, Wong A, Pipinikas CP, Jiao Y, Jones A, Anjum S, Hardy R, Salvesen HB, Thirlwell C, Janes SM, Kuh D, Widschwendter M. Correlation of smoking-associated DNA methylation changes in buccal cells with DNA methylation changes in epithelial cancer. JAMA Oncol. 2015;1:476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 38.Sidransky D. The oral cavity as a molecular mirror of lung carcinogenesis. Cancer Prev Res (Phila Pa) 2008;1:12–14. doi: 10.1158/1940-6207.CAPR-08-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spira A. Upper airway gene expression in smokers: the mouth as a “window to the soul” of lung carcinogenesis? Cancer Prev Res. (Phila) 2010;3:255–258. doi: 10.1158/1940-6207.CAPR-10-0013. [DOI] [PubMed] [Google Scholar]
- 40.Phillips DH, Venitt S. DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. Int J Cancer. 2012;131:2733–2753. doi: 10.1002/ijc.27827. [DOI] [PubMed] [Google Scholar]
- 41.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 42.Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer. 2012;131:2724–2732. doi: 10.1002/ijc.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigoryeva ES, Kokova DA, Gratchev AN, Cherdyntsev ES, Buldakov MA, Kzhyshkowska JG, Cherdyntseva NV. Smoking-related DNA adducts as potential diagnostic markers of lung cancer: new perspectives. Exp. Oncol. 2015;37:5–12. [PubMed] [Google Scholar]
- 44.Arif JM, Dresler C, Clapper ML, Gairola CG, Srinivasan C, Lubet RA, Gupta RC. Lung DNA adducts detected in human smokers are unrelated to typical polyaromatic carcinogens. Chem. Res. Toxicol. 2006;19:295–299. doi: 10.1021/tx0502443. [DOI] [PubMed] [Google Scholar]
- 45.United States Department of Health and Human Services . How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. U.S. Department of Health and Human Services; Washington, D.C.: 2010. pp. 242–244. Ch. 5. [Google Scholar]
- 46.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. High-performance liquid chromatography electrospray ionization tandem mass spectrometry for the detection and quantitation of benzo[a]pyrene-DNA adducts. Chem Res Toxicol. 2005;18:1306–1315. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 47.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal. Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nath RG, Ocando JE, Guttenplan JB, Chung FL. 1,N2-Propanodeoxyguanosine adducts: Potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 1998;58:581–584. [PubMed] [Google Scholar]
- 49.Greenspan EJ, Lee H, Dyba M, Pan J, Mekambi K, Johnson T, Blancato J, Mueller S, Berry DL, Chung FL. High-throughput, quantitative analysis of acrolein-derived DNA adducts in human oral cells by immunohistochemistry. J Histochem. Cytochem. 2012;60:844–853. doi: 10.1369/0022155412459759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan J, Awoyemi B, Xuan Z, Vohra P, Wang HT, Dyba M, Greenspan E, Fu Y, Creswell K, Zhang L, Berry D, Tang MS, Chung FL. Detection of acrolein-derived cyclic DNA adducts in human cells by monoclonal antibodies. Chem. Res. Toxicol. 2012;25:2788–2795. doi: 10.1021/tx3004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen HJ, Lin WP. Quantitative analysis of multiple exocyclic DNA adducts in human salivary DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal. Chem. 2011;83:8543–8551. doi: 10.1021/ac201874d. [DOI] [PubMed] [Google Scholar]
- 52.Chen HJ, Lin CR. Noninvasive measurement of smoking-associated N(3)-ethyladenine and N(7)-ethylguanine in human salivary DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Toxicol. Lett. 2014;225:27–33. doi: 10.1016/j.toxlet.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 53.Chen HJ, Lee CR. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol. Lett. 2014;224:101–107. doi: 10.1016/j.toxlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem. Res. Toxicol. 2010;23:1234–1244. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 100E. IARC; Lyon, FR.: 2012. Personal Habits and Indoor Combustions. pp. 373–499. [PMC free article] [PubMed] [Google Scholar]
- 56.Balbo S, Meng L, Bliss RL, Jensen JA, Hatsukami DK, Hecht SS. Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol. Biomarkers Prev. 2012;21:601–608. doi: 10.1158/1055-9965.EPI-11-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balbo S, Meng L, Bliss RL, Jensen JA, Hatsukami DK, Hecht SS. Time course of DNA adduct formation in peripheral blood granulocytes and lymphocytes after drinking alcohol. Mutagenesis. 2012;27:485–490. doi: 10.1093/mutage/ges008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stepanov I, Muzic J, Le CT, Sebero E, Villalta P, Ma B, Jensen J, Hatsukami DK, Hecht SS. Analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB)-releasing DNA adducts in human exfoliated oral mucosa cells by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2013;26:37–45. doi: 10.1021/tx300282k. PMCID: PMC3631465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang M, Cheng G, Balbo S, Carmella SG, Villalta PW, Hecht SS. Clear differences in levels of a formaldehyde-DNA adduct in leukocytes of smokers and non-smokers. Cancer Res. 2009;69:7170–7174. doi: 10.1158/0008-5472.CAN-09-1571. PMCID: doi:10.1158/0008-5472.CAN-09-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers' lung: comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–1977. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Balbo S, Wang M, Hecht SS. Analysis of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human leukocyte DNA from smokers and nonsmokers. Chem. Res. Toxicol. 2011;24:119–124. doi: 10.1021/tx100321y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Church TR, Anderson KE, Le C, Zhang Y, Kampa DM, Benoit AR, Yoder AR, Carmella SG, Hecht SS. Temporal stability of urinary and plasma biomarkers of tobacco smoke exposure among cigarette smokers. Biomarkers. 2010;15:345–352. doi: 10.3109/13547501003753881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen LS, Hung RJ, Baker T, Horton A, Culverhouse R, Saccone N, Cheng I, Deng B, Han Y, Hansen HM, Horsman J, Kim C, Lutz S, Rosenberger A, Aben KK, Andrew AS, Breslau N, Chang SC, Dieffenbach AK, Dienemann H, Frederiksen B, Han J, Hatsukami DK, Johnson EO, Pande M, Wrensch MR, McLaughlin J, Skaug V, van der Heijden HF, Wampfler J, Wenzlaff A, Woll P, Zienolddiny S, Bickeboller H, Brenner H, Duell EJ, Haugen A, Heinrich J, Hokanson JE, Hunter DJ, Kiemeney LA, Lazarus P, Le Marchand L, Liu G, Mayordomo J, Risch A, Schwartz AG, Teare D, Wu X, Wiencke JK, Yang P, Zhang ZF, Spitz MR, Kraft P, Amos CI, Bierut LJ. CHRNA5 risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis--a meta-analysis. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv100. doi: 10.1093/jnci/djv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carmella SG, Ming X, Olvera N, Brookmeyer C, Yoder A, Hecht SS. High throughput liquid and gas chromatography-tandem mass spectrometry assays for tobacco-specific nitrosamine and polycyclic aromatic hydrocarbon metabolites associated with lung cancer in smokers. Chem Res Toxicol. 2013;26:1209–1217. doi: 10.1021/tx400121n. PMCID: PMCID: PMC3803150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carmella SG, Chen M, Zarth A, Hecht SS. High throughput liquid chromatography-tandem mass spectrometry assay for mercapturic acids of acrolein and crotonaldehyde in cigarette smokers' urine. J. Chromatog. B. 2013;935:36–40. doi: 10.1016/j.jchromb.2013.07.004. PMCID: PMCID: PMC3925436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan J-Y, Nelson HH, Butler LM, Carmella SG, Wang R, Kuriger-Laber J, Adams-Haduch J, Hecht SS, Gao Y-T, Murphy SE. Genetic determinants of cytochrome P450 2A6 and biomarkers of tobacco smoke exposure in releation to risk of lung cancer development in the Shanghai Cohort Study. Int. J. Cancer. 2015;138:2161–2171. doi: 10.1002/ijc.29963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji X, Gui J, Han Y, Brennan P, Li Y, McKay J, Caporaso N, Bertazzi PA, Landi MT, Amos CI. The role of haplotype in 15q25.1 locus in lung cancer risk: Results of scanning chromosome 15. Carcinogenesis. 2015;36:1275–1273. doi: 10.1093/carcin/bgv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen LS, Baker TB, Jorenby D, Piper M, Saccone N, Johnson E, Breslau N, Hatsukami D, Carney RM, Bierut LJ. Genetic variation (CHRNA5), medication (combination nicotine replacement therapy vs. varenicline), and smoking cessation. Drug Alcohol Depend. 2015;154:278–282. doi: 10.1016/j.drugalcdep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bierut LJ, Johnson EO, Saccone NL. A glimpse into the future - Personalized medicine for smoking cessation. Neuropharmacology. 2014;76(Pt B):592–599. doi: 10.1016/j.neuropharm.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu AZ, Zhou Q, Cox LS, David SP, Ahluwalia JS, Benowitz NL, Tyndale RF. Association of CHRNA5-A3-B4 SNP rs2036527 with smoking cessation therapy response in African-American smokers. Clin Pharmacol Ther. 2014;96:256–265. doi: 10.1038/clpt.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fasanelli F, Baglietto L, Ponzi E, Guida F, Campanella G, Johansson M, Grankvist K, Johansson M, Assumma MB, Naccarati A, Chadeau-Hyam M, Ala U, Faltus C, Kaaks R, Risch A, De Stavola B, Hodge A, Giles GG, Southey MC, Relton CL, Haycock PC, Lund E, Polidoro S, Sandanger TM, Severi G, Vineis P. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun. 2015;6:10192. doi: 10.1038/ncomms10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hukkanen J, Jacob P, III, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. PMCID: doi:10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 74.Yuan JM, Koh WP, Murphy SE, Fan Y, Wang R, Carmella SG, Han S, Wickham K, Gao YT, Yu MC, Hecht SS. Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;69:2990–2995. doi: 10.1158/0008-5472.CAN-08-4330. PMCID: PMC2664854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan J-M, Gao Y-T, Murphy SE, Carmella SG, Wang R, Zhong Y, Moy KA, Davis AB, Tao L, Chen M, Han S, Nelson HH, Yu MC, Hecht SS. Urinary levels of cigarette smoke constituent metabolites are prospectively associated with lung cancer development in smokers. Cancer Res. 2011;71:6749–6757. doi: 10.1158/0008-5472.CAN-11-0209. PMCID: PMC3392910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Church TR, Anderson KE, Caporaso NE, Geisser MS, Le C, Zhang Y, Benoit AR, Carmella SG, Hecht SS. A prospectively measured serum biomarker for a tobacco-specific carcinogen and lung cancer in smokers. Cancer Epidemiol. Biomarkers Prev. 2009;18:260–266. doi: 10.1158/1055-9965.EPI-08-0718. PMCID: PMC3513324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carmella SG, Chen M, Yagi H, Jerina DM, Hecht SS. Analysis of phenanthrols in human urine by gas chromatography-mass spectrometry: potential use in carcinogen metabolite phenotyping. Cancer Epidemiol. Biomarkers Prev. 2004;13:2167–2174. [PubMed] [Google Scholar]
- 78.Carmella SG, Chen M, Han S, Briggs A, Jensen J, Hatsukami DK, Hecht SS. Effects of smoking cessation on eight urinary tobacco carcinogen and toxicant biomarkers. Chem. Res. Toxicol. 2009;22:734–741. doi: 10.1021/tx800479s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haiman CA, Patel YM, Stram DO, Carmella SG, Chen M, Wilkens LR, Le Marchand L, Hecht SS. Benzene uptake and glutathione S-transferase T1 status as determinants of S-phenylmercapturic acid in cigarette smokers in the multiethnic cohort. PLoS. One. 2016;11:e0150641. doi: 10.1371/journal.pone.0150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park SL, Kotapati S, Wilkens LR, Tiirikainen M, Murphy SE, Tretyakova N, Le Marchand L. 1,3-Butadiene exposure and metabolism among Japanese American, Native Hawaiian, and white smokers. Cancer Epidemiol. Biomarkers Prev. 2014;23:2240–2249. doi: 10.1158/1055-9965.EPI-14-0492. PMCID: PMCID: PMC4220266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milne GL, Yin H, Hardy KD, Davies SS, Roberts LJ. Isoprostane generation and function. Chem Rev. 2011;111:5973–5996. doi: 10.1021/cr200160h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, DuBois RN. Urinary PGE-M: a promising cancer biomarker. Cancer Prev Res (Phila) 2013;6:507–510. doi: 10.1158/1940-6207.CAPR-13-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]