Abstract
Nonalcoholic fatty liver disease (NAFLD) is associated with an increased risk of cardiovascular disease. Because the liver is the major source of circulatory proteins, it is not surprising that hepatic disease could lead to alterations in the plasma proteome, which are therein implicated in atherosclerosis. The current study used low-density lipoprotein receptor-deficient (LDLR−/−) mice to examine the impact of Western diet (WD)-induced NAFLD on plasma proteome homeostasis. Using a 2H2O-metabolic labeling method, we found that a WD led to a proinflammatory distribution of circulatory proteins analyzed in apoB-depleted plasma, which was attributed to an increased production. The fractional turnover rates of short-lived proteins that are implicated in stress-response, lipid metabolism, and transport functions were significantly increased with WD (P < 0.05). Pathway analyses revealed that alterations in plasma proteome dynamics were related to the suppression of hepatic PPARα, which was confirmed based on reduced gene and protein expression of PPARα in mice fed a WD. These changes were associated with ~4-fold increase (P < 0.0001) in the proinflammatory property of apoB-depleted plasma. In conclusion, the proteome dynamics method reveals proinflammatory remodeling of the plasma proteome relevant to liver disease. The approach used herein may provide a useful metric of in vivo liver function and better enable studies of novel therapies surrounding NAFLD and other diseases.
Keywords: NAFLD, atherosclerosis, HDL, heavy water, proteome dynamics
Graphical Abstract
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the United States, affecting one-third of the adult population, and is associated with a high rate of cardiovascular disease (CVD)-related mortality.1 Although altered lipid metabolism leading to the hepatic accumulation of triglycerides (steatosis) initiates NAFLD, hepatic oxidative stress and inflammation play central roles in disease progression to nonalcoholic steatohepatitis (NASH).2 Dyslipidemia and inflammation are also involved in the pathogenesis of atherosclerosis. Notably, hepatic inflammation is an independent predictor of cardiovascular diseases (CVDs) in patients with NASH.3 Mechanistically, ectopic fat deposition in the liver triggers hepatic oxidative stress associated with increased expression of proinflammatory mediators that initiate an acute phase response (APR) and systematic inflammation.4 However, it is unknown whether the inflamed liver is involved in the etiology of atherosclerosis through the increased secretion of pro-atherogenic and proinflammatory proteins into circulation.
Plasma proteins, including those associated with lipoproteins, are important in the APR5 and the maintenance of endothelial homeostasis.6 These circulatory proteins have multiple functions including reverse cholesterol transport, antioxidant, anti-inflammatory, and endothelial-vasoprotective activities that help to guard against CVD.6–9 In addition, several liver-secreted proteins are involved in complement activation and regulation of proteolysis and thrombosis implicated in CVD.7 However, sustained inflammation and oxidative stress in conditions like NAFLD may result in atherogenic changes of the plasma proteome composition.10–13 Indeed, the severity of NAFLD and the disease progression to cirrhosis are associated with the atherogenic lipoprotein profile that is implicated in CVD risk.14
Recent studies have demonstrated that label-free, iTRAQ-based and parallel reaction- and selected reaction-monitoring proteomics approaches can be used to characterize alterations in the plasma proteome, including high-density lipoprotein (HDL) proteome composition.15–18 Importantly, several plasma protein biomarkers of NAFLD and CVD have been identified.19–21 Although the static measurements of protein abundance are important, they do not allow an understanding of the mechanism for these changes. For instance, increased levels of a protein could be related to an accelerated rate of synthesis and/or a slower clearance from circulation. Thus, understanding the cause of proteome remodeling in pathological conditions necessitates quantification of plasma protein turnover. One could hypothesize that measurements of dynamic changes in liver-secreted proteins could also facilitate our understanding of the underlying mechanisms that contribute to altered lipid trafficking, the development of NAFLD, and its association with atherosclerosis.
Quantitative assessment of the dynamics of individual proteins has been belated by the lack of simple, and high throughput, nonradioactive methods. Recently, a deuterium oxide (2H2O or D2O)-based metabolic labeling approach coupled with high-resolution liquid chromatography-tandem mass spectrometry (LC–MS/MS) was developed for studies of proteome kinetics.22–26 The stable (nonradioactive) isotope 2H2O is easily administered in drinking water. The tracer rapidly distributes in total body water and transfers 2H from 2H2O to amino acids, which are incorporated into proteins at a rate that reflects their synthesis.24 Specifically, the fractional synthesis rate (FSR), i.e., the fraction of a protein that is newly made, is calculated from the time course of 2H enrichment in unique peptides. The absolute production rate can be calculated once the FSR and pool size of a protein are known.
Herein, we have applied our 2H2O-based flux measurements in tandem with a quantitative proteomics method to assess the effect of diet-induced NAFLD on secretome dynamics. Concurrent measurements of 2H enrichment and the abundance of tryptic peptides yielded estimates of both fractional turnover and production rates of the circulatory proteins in vivo. Studies were performed in a well-established LDLR−/− mouse model of NAFLD and atherosclerosis via feeding of a high fat diet containing cholesterol (a Western-type diet, WD), which results in hepatic steatosis, inflammation, oxidative stress, and atherosclerotic lesion formation.27 Shotgun proteomics was used to monitor relative changes in protein abundance and kinetics in apoB-depleted plasma proteins. Lastly, pathway analysis was used to determine if WD-induced alterations in the dynamics of plasma proteins could be involved in NAFLD-associated atherosclerosis.
EXPERIMENTAL SECTION
Materials
HPLC-grade solvents for reversed-phase liquid chromatography and sample preparation were purchased from Fluka (Milwaukee, WI). All other chemicals were from Sigma-Aldrich (St. Louis, MO). The labeled peptide VAPL(13C6)GAEL-(13C6)QESAR for quantification of mouse apoAI was synthesized as described.28 Mouse apoAII was quantified based on the ratio of endogenous THEQLTPLVR to the heavy labeled synthetic peptide THEQL(2H10)TPLVR (597.342+/602.342+) as an internal standard. THEQL(2H10)-TPLVR was synthesized using a solid-phase method in the Cleveland Clinic Molecular Biotechnology Core on a 396 52 Peptide Synthesizer (Advanced ChemTech, Louisville, Kentucky). Stable isotope-labeled l-[2H10]-Fmoc-leucine was coupled in the peptide sequence to give a molecular mass shift of 10 Da from the unlabeled endogenous peptide. The molecular weight of the purified peptide was verified by ESI-MS and was found to yield [M + 2H]2+ ion with the expected 602.34 m/z and an isotopic purity of 97%. The stock solution of labeled peptides VAPL(6C13)GAEL(6C13)QESAR and THEQL(2H10)TPLVR were standardized using an HPLC-UV-based amino acid analysis. The working solutions were made at concentrations of 10 and 2 µM for apoAI and apoAII, respectively (pH 3). These solutions were divided into 0.05 mL fractions and stored at −80 °C until use.
Animal Studies
All animal procedures were approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic and were performed in accordance with NIH guidelines. LDLR−/−mice used in this study were bred and housed in the animal care facility of Cleveland Clinic. The animals were accommodated with a 12:12 h light-dark cycle with chow diet (20% kcal from protein, 70% kcal from carbohydrate, and 10% kcal from fat; Harlan Teklad) and water. Eight-to-ten week old male mice (n = 10/group) fed either standard chow diet (SD) or a Western diet (WD; 20% kcal from protein, 35% kcal from carbohydrate, and 45% kcal from fat with 0.24% cholesterol added; Research Diets, Inc.) for 12 weeks. Food intake was measured twice a week, and body weight was recorded weekly. For both the relative abundance of plasma proteins and the inflammatory property of plasma to be assessed, at the end of the diet experiment, six mice from each group were euthanized. Blood was collected through cardiac puncture, and plasma was separated immediately and saved at −80 °C until the analysis.
2H-Labeling Experiment
At the beginning of the 12th week of the diet experiment, the 2H2O-based turnover study was conducted in individual mice (4 mice/group). Small blood samples (60–70 µL, which corresponds to less than 5% of total blood volume in mice) were collected at different time points. For minimizing the stress in mice related to multiple blood draws, the baseline plasma samples were taken 2 days before the metabolic labeling experiment. The animals received a loading dose (intra-peritoneal injection) of 2H2O (20 µL of 99.9% 2H-labeled saline/g of body weight) and then allowed free access to drinking water that was enriched with 5% 2H2O and food. Additional blood samples (60–70 µL) were taken from the saphenous vein at 4 h, 8 h, 1 day, and 3 day after the bolus dose of 2H2O; samples were collected in potassium EDTA-containing tubes. The animals were euthanized seven days after 2H2O exposure, and the terminal EDTA-preserved blood was collected through cardiac puncture. Plasma was separated immediately and saved at −80 °C until the analysis.
Total Body Water Enrichment
2H enrichment of total body water (TBW) was measured using a modification of the acetone exchange method.29 This method is based on the isotopic exchange between the hydrogen atoms of water and of acetone in alkaline medium (pH 12–13).
ApoB-Depleted Plasma
Plasma contains a complex mixture of diverse proteins with 12 orders of dynamic range in the protein concentrations;30 therefore, comprehensive plasma proteome analyses require prefractionation, including depletion of high abundant proteins. Because NAFLD is an inflammatory state and is associated with increased APR that may affect many abundant plasma proteins, including albumin, the highly abundant proteins in this study were not depleted. Instead, to increase the coverage of plasma proteins, we used a different strategy based on apoB depletion. Recently, using an isotope dilution method, we demonstrated that a WD results in more than a 3-fold increase in plasma apoB in LDLR−/− mice.27 The presence of highly abundant apoB associated with lipids complicates in-solution tryptic digestion due to its hydrophobicity.31 ApoB depletion with magnesium chloride/dextran sulfate allowed reproducible tryptic digestion and quantification of other plasma proteins. Although this approach does not allow quantification of low-abundance circulatory proteins, it enables simultaneous quantification of HDL-cholesterol (a known risk factor of CVD)27,28 and HDL-associated proteins. ApoB-depleted plasma samples were prepared from 25 µL of plasma using a magnesium chloride/ dextran sulfate reagent (Stanbio Laboratory, Boerne, TX).28
Tryptic Digestion of Proteins
The supernatant containing the apoB-depleted plasma was recovered and used for the proteome analysis. After precipitation of proteins with 1 mL of cold acetone (−20 °C), the pellets from apoB-depleted plasma were dried under a nitrogen gas flow. The residue was then dissolved in 200 µL of 50 mM ammonium bicarbonate solution, and 15 µL of 10% sodium deoxycholate was added to facilitate the solubility of lipid-associated large proteins.32 The fraction of the above protein solution corresponding to 20 µg of protein (based on bicinchoninic acid (BCA) protein assay) was taken from each sample followed by reducing with 2.5 µL of 0.1 M dithiothreitol (DTT) at 60 °C for 30 min. The sample was then alkylated with 2.5 µL of 0.2 M iodoacetamide at room temperature for 30 min. The excess of iodoacetamide was reacted with DTT. Tryptic digestion was then performed by the addition of 10 µL of 0.1 µg/µL of Promega sequencing grade modified porcine trypsin (20 µg of lyophilized trypsin in 200 µL of 50 mM ammonium bicarbonate solution). The samples were incubated at 37 °C overnight. The tryptic digestion was stopped, and the deoxycholate was precipitated by adding 4 µL of 20% formic acid. The sample was centrifuged at 14000g for 15 min, and the supernatant was transferred to a new 1.5 eppendorf ml tube; the pellet was discarded. Protein digests were cleaned up through solid phase extraction using Pierce Pepclean C18 spin columns.
Protein Characterization by HPLC–MS/MS
Chromatographic separation of the protein digest was performed on a Dionex ultimate 3000 UHPLC (Thermo Fisher Scientific) with an Acclaim PepMap100 Nano-Trap Column (20 mm × 100 µm, 5 µm C18, Thermo Scientific) followed by an Acclaim PepMap RSLC analytical column (150 mm × 75 µm, 2 µm C18, 100 Å, Thermo Scientific) using mobile phase A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). A 135 min stepwise gradient started with 5% of mobile phase B. After 5 min of desalting, mobile phase B was linearly increased to 40% in 105 min and held at 40% for 2 min. Mobile phase B was then ramped to 80% in 5 min. Subsequently, mobile phase B was decreased from 80 to 5% in 3 min and equilibrated for 15 min at 5% of B.
Tandem mass spectra were recorded on a Thermo Orbitrap Elite-LTQ hybrid mass spectrometer (Thermo Electron Corp.) operated in positive ion mode. The peptides were infused at a flow rate of 300 nL/min via a silica noncoated PicoTip emitter (FS360-20-10-N-20-C15, New Objective Inc.) at a voltage of 1.9 kV. The inlet capillary temperature was maintained at 200 °C. A data-dependent method was used for all of the samples. Briefly, a full profile MS scan at 60000 resolution (at 400 m/z) between 300 and 2000 m/z was performed on the Orbitrap FT instrument using a precursor isolation window of 3 m/z at 35% relative collision energy. The charge state screening was not enabled, and the dynamic exclusion option was enabled after three repeated acquisitions within a 20 s duration with each exclusion lasting 90 s.
Protein Identification
For the identification of proteins, the mass spectrometry data were analyzed using all CID spectra collected in the experiment. Peak lists were generated using Thermo Proteome Discoverer v1.3 software (Thermo Electron Corp.). The data were searched using the MASCOT v2.3 (Matrix Science, London, UK) against the National Center for Biotechnology Information Mus musculus reference sequence database (ftp://ftp.ncbi.nih.gov/refseq/) released on December 20th, 2011 containing 30438 entries. The search was performed using carbamidomethyl as a fixed modification of cysteine, oxidation as a variable modification of methionine, and allowing two missed cleavages. The mass tolerances for the precursor and product ions were 10 ppm and 1.2 Da, respectively. For peptide identification, a MASCOT score of greater than 35 was applied as the cutoff threshold along with 95% identification confidence. BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used as needed to confirm the uniqueness of identified peptides.33,34
Scaffold software (V4.3.2; Proteome Software Inc., Portland, Oregon, USA) was used to validate the accuracy of MS/MS and database search-based peptide and protein identification and also used to perform label-free relative protein quantification. Peptide identifications were established based on higher than 95% probability by the Peptide Prophet algorithm.35 Protein identifications were accepted if they could achieve greater than 99% probability and a false discovery rate (FDR) of less than 1% and contained at least two identified unique peptides. The Protein Prophet algorithm-based statistical method was used to assess the protein probabilities.36
Proteome Dynamics with 2H2O
Turnover Rate Constant and Half-Life Calculation
To determine whether the changes in plasma protein levels were due to their degradation or production rates, we applied the 2H2O-metabolic labeling approach to assess the kinetics of individual proteins. For this purpose, we followed a protocol similar to the one in our previous studies.22,28 The protein turnover is calculated based on 2H enrichment of tryptic peptides.23 The rate of 2H labeling of a proteolytic peptide of a protein represents the fractional turnover rate (or degradation rate) of that protein as long as the peptide is unique to the protein. Therefore, we can use the 2H enrichment values of a unique peptide to calculate the turnover rate constant of a protein. In addition, we used the turnover rate constant and relative protein abundance to calculate the production rate of a protein (see below).
To calculate the turnover rate constants, we used custom-made software (https://ispace.utmb.edu/users/rgsadygo/Proteomics/HeavyWater) that works with the data obtained from the mass spectrometer to list all the proteins and integrates the corresponding extracted ion chromatograms of a peptide.25 This software allows high-throughput analysis and quantification of 2H labeling of all analyzed peptides. First, the software reads all peptide IDs from MASCOT-generated .mzid files and organizes all proteins and their peptides across all time points. Second, the software locates the peptide precursor ions from high resolution full scan spectra and integrates peak areas for each isotopomer. In the output file, Mi represents the area of peak associated with ith isotopomer. Lastly, the software removes peptide IDs that have chromatographic and isobaric overlaps with other signals and spectra of low quality (low signal-to-noise ratio). It is important to note that peptides with more than 5% variability in the isotope ratio of the monoisotopic ion in the baseline sample and peptides that cannot be assigned to a unique protein are excluded. The isotope distribution was computed based on high-resolution full scan spectra, and the molar percent distribution of the isotopomeric peaks were calculated. Fractional turnover rates were determined by assuming a one-compartment model and then fitting a time course of normalized net labeling of a peptide [Epeptide(t)] into the equation25
| (1) |
where Ess is asymptotic steady-state labeling and k is the rate constant. Simulated asymptotic steady-state isotope ratios for an enriched peptide were also calculated based on total body water enrichment and the asymptotic number of deuterium incorporated into the peptide (N), estimated by integrating the labeling of intracellular free amino acids that make up a peptide of all amino acids from that peptide.22,23 This information was used as an additional parameter for elimination of outlier peptides for the kinetic calculations.
At the steady state, when the pool size of a protein does not change, the rate constant represents both the fractional synthesis rate (FSR) and fractional catabolic rate (FCR) such that k = FSR = FCR. The data from multiple peptides were aggregated to calculate the average rate constant and half-life (t1/2 = ln2/k) of a protein. Analysis of multiple unique peptide fragments within proteins of interest and rigorous exclusion criteria for the outlier peptides were applied across experimental groups.
The fractional turnover rate of proteins with longer half-life (>40 days), including hemoglobin and paraoxonase 1 (PON1), was calculated based on the precursor/product relationship assuming that 2H2O is the precursor and a proteolytic peptide is the product23
| (2) |
where slope is the rate of the increase in 2H labeling of a peptide during 2H2O administration and Ewater is the steady state enrichment of total body water. N is the asymptotic number of deuterium incorporated into a peptide.
Label-Free Quantitative Proteomics
The label-free quantifications were performed on six individual animals from each group. For the relative protein level measurements, the same amount of total protein from each sample was digested with trypsin and analyzed by LC–MS/MS. The initial concentration of total protein in apoB-depleted plasma was measured using BCA protein assay kit. Therefore, the results of these measurements represent relative levels of proteins in apoB-depleted plasma and reflect a WD-induced remodeling of proteome composition rather than the absolute concentrations of individual proteins.
Differential protein expression was assessed using the semiquantitative label-free spectral counting (SC) method using the Scaffold software package.37 The relative protein abundance in a sample is proportional to the number of MS/ MS spectra that matches the peptides of that protein. Therefore, SC of each protein/total SC of the sample gives the relative protein abundance profile of the sample. Scaffold calculates a normalized spectral abundance factor (NSAF) to assess the differences between groups: NSAF = SAFi/∑SAFi, where the spectral abundance factor (SAF) for each protein represents SC/protein length ratio.38 This normalization eliminates the differences of total protein amount between samples and allows one to compare the relative abundance of proteins within a sample and across samples. To account for the shared peptides among multiple proteins, the Scaffold program adapted the strategy previously described by Zhang and colleagues.38 Briefly, this approach distributes the spectral counts from shared peptides based on the number of unique spectral counts that results in accurate quantification. Detailed information on the label-free quantification and validation of the label-free quantitative proteomics results can be found in the Supporting Methods and Table S1.
Relative Production Rate Calculation
To assess the effect of WD-induced NAFLD on the production rate of plasma proteins, we also calculated the relative production rate (RPR) of each individual protein. For this purpose, we applied the same rational used for calculating the absolute production rate of a protein, i.e., absolute production rate (g kg−1 h−1) = k·pool size. Because in this study we measured only relative abundance but not the absolute quantity of each protein, protein levels in the WD group were normalized relative to those in the SD group. Thus, the RPR (expressed as relative protein amount per hour) represents the relative flux of each protein in the WD relative to SD group, and it was calculated as the product of the relative fractional turnover rate (k) and the abundances based on label-free quantification
| (3) |
Thus, the combination of label-free quantification with heavy water-based proteome dynamics approach allows direct comparison of pool sizes, half-lives, and production rates of individual proteins on a proteome-wide scale.
Gene Expression Analyses
Total RNA was isolated from the mouse liver tissue using an RNeasy Mini Kit (Qiagen Inc.). One microgram of total RNA was reverse transcribed using the commercial Advantage RT-for-PCR Kit (Clontech Laboratories, Inc.). Real-time PCR amplification was performed using Brilliant SYBR Green QPCR Master Mix (Agilent Technologies, Inc.) and gene-specific primers in an Mx3000p PCR machine (Agilent Technologies, Inc.) in duplicate. The sequences of the primers were as follows: β-actin forward: 5′-CTT TGC AGC TCC TTC GTT GC-3′, reverse: 5′-ACG ATG GAG GGG AAT ACA GC-3′; PPARα forward: CTTCCCAAAGCTCCTTCAAAAA, reverse: CTGCGCATGCTCCGTG. The primers used for real-time PCR analysis were synthesized by Integrated DNA Technologies, Inc. The relative amount of target mRNA was determined using the cycle threshold (Ct) method by normalizing target mRNA Ct values to that of β-actin. Fold induction ratios were calculated relative to basal conditions for each group using the formula 2−ΔΔCt.
Western Blot Analysis
Proteins for each sample (20 µg) were resolved on 4–20% precast gradient SDS-PAGE gels (Life Technologies) and then electro-transferred to polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies overnight at 4 °C followed by horseradish peroxidase-labeled second antibody (Santa Cruz Biotechnology) labeling for 1 h at room temperature. Primary antibodies used were anti-Actin (Santa Cruz Biotechnology) and anti-PPARα (Affinity Bioreagents). Finally, detection procedures were performed using an ECL Plus Western Blotting Detection Kit (Amersham).
Pro-Inflammatory Index of Plasma
To assess the functional consequence of WD-induced changes on the dynamics and remodeling of plasma proteome, we also measured proinflammatory function of plasma. A modification of the cell-free assay developed by Navab and colleagues39 was used to quantify the proinflammatory index of apoB-depleted plasma.40 This assay measures the antioxidant capacity of plasma proteins to prevent Cu2+-induced oxidative stress. Briefly, oxidation in apoB-depleted plasma was initiated with Cu2+ and rates of total oxidation quantified with 2′,7′-dichlorodihydrofluorescein (DCFH) in a microtiter plate at 37 °C. Fluorescent emission with 530 nm wavelength was measured after serial excitation at 485 nm.
To determine whether proinflammatory remodeling of plasma proteome was related to myeloperoxidase (MPO), a pro-oxidant neutrophil-derived heme protein, we also quantified the peroxidase MPO activity in apoB-depleted plasma. The circulatory MPO is mechanistically linked to oxidative alterations in plasma proteins and is also involved in the pathogenesis of both NAFLD41 and atherosclerosis.11 The peroxidase activity of MPO in plasma was measured by a spectrophotometer with 3,3′5,5′-tetramethylbenzidine (TMB) as a substrate. In this assay, MPO catalyzes oxidation of TMB to 3,3′5,5′-tetramethylbenzidine diimine, a blue-colored product with light absorption at 650 nm. Thus, the proinflammatory index and MPO activity assays reflect the ability of plasma to cope with or propagate the oxidative stress, respectively, and thereby characterize the functional consequences of the altered plasma proteome dynamics.
Data Presentation and Statistical Analysis
Results are presented as mean ± standard deviation (SD). Label-free protein abundance quantification and inflammatory index of apoB-depleted plasma assays are based on six mice from each group. Differences in biochemical parameters were assessed using a 2-tailed t-test. P < 0.05 was considered significant. Data on 2H2O metabolic-labeling experiments are shown as the excess labeling in a given mouse experiment. To analyze data from time course experiments, we constructed a relative 2H excess curve for several (n = 3–6) peptides of each protein. All six time points in each mouse (n = 4/group) were used to model the rate constants. The calculated 95% confidence intervals for the rate constants were used as the criteria for determining the differences when we compared the 2H enrichment profiles.
Multidimensional Scaling (MDS)
MDS is an ordination method used to visualize data sets with high order dimensions. This approach scales down the selected attributes (in our case, identified individual protein abundances), and a plot is generated with R-project (version 3.1.2) function cmdscale from the stats package, which calculates classical multidimensional scaling of a data matrix. Calculating Euclidian distances across all NSAF values, a dissimilarity matrix was generated. The distances, reduced to a two-dimensional space, were used to generate a plot, where samples are organized by their similarity (dissimilarity).
Gene Ontology Analysis
These Gene Ontology (GO) terms included proteins with k values analyzed in this study. The k values are organized by these GO terms, and cumulative k values were calculated as long as a protein has that GO term annotation. Special emphasis was placed on seven GO annotations, i.e., catalytic activity (GO:0003824), lipid metabolic process (GO:0006629), transport (GO:0006810), inflammatory response (GO:0006954), immune response (GO:0006955), response to stress (GO:0006950), and proteolysis (GO:0006508), which are known to be affected in atherosclerosis. Functional annotation of proteins identified in plasma was processed using Biomart R package.42 A t-test was used to calculate significant differences at cumulative k values.
Clustering Analysis
Unsupervised hierarchical clustering of samples and proteins was used to generate a heatmap of proteins and sample dendrograms. The values are log transformed for depiction purposes in the heat maps. R-project (version 3.1.2.) hclust function with Ward-D2 method was used to generate a dendrogram plot. The height of the tree branch connecting end nodes (proteins or samples) in these plots is proportional to the value of the intergroup dissimilarity between its children.
Network Analysis
The kinetic data from proteome dynamics experiments were further analyzed by Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com). IPA queried both interaction networks and canonical pathways.
Canonical pathway analysis identified the pathways from the IPA library of canonical pathways that were significant to a data set. The statistical assessment of network enrichment and significance was calculated by IPAs implementation of Fisher’s Exact test (right-tailed), and pathways are ranked by the P-values generated from this test. This test measures the likelihood that the association between proteins measured in our experiments and a pathway is due to random chance. The smaller P-value means that it is less likely that the association is due to random chance. The networks and pathways were annotated with k and RPR values to depict changes across networks and pathways. The networks are annotated with functional terms by the IPA.
Data Accession
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004270.
RESULTS
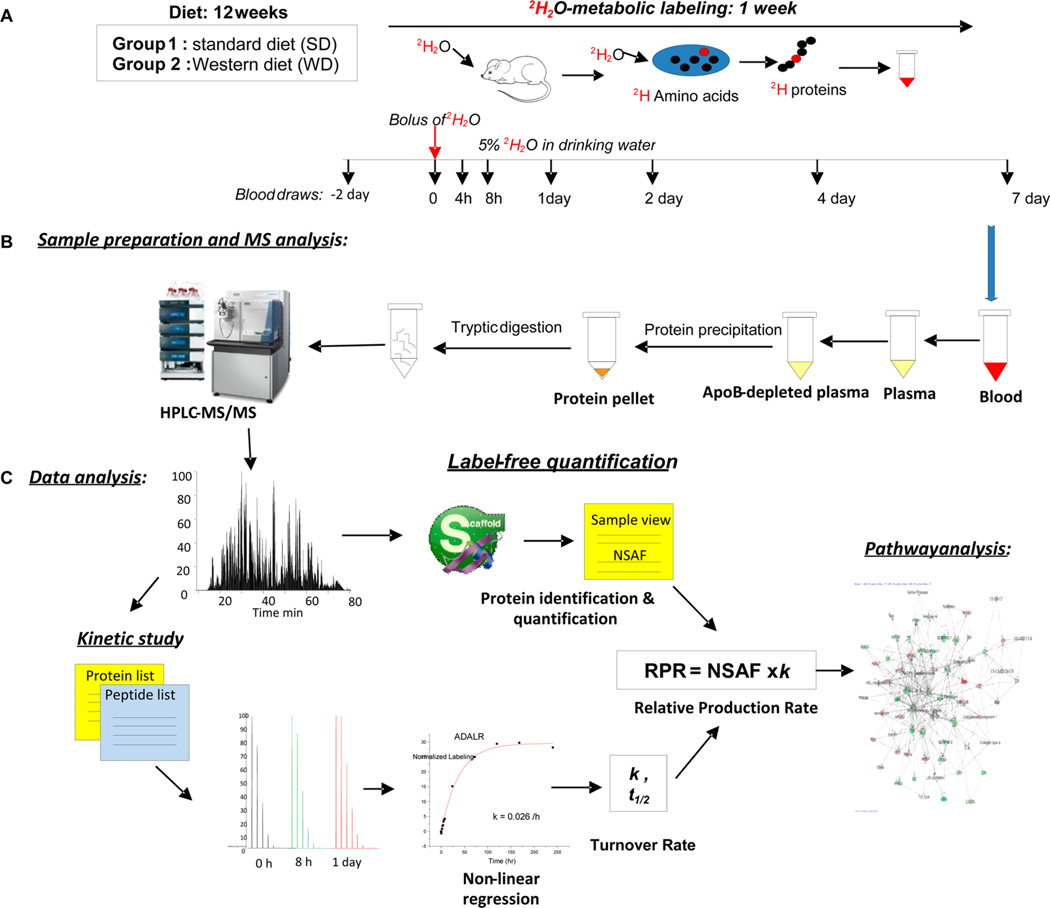
Recently, we have shown that LDLR−/− mice on a WD develop obesity as reflected by an ~65% increase in body weight, which is associated with dyslipidemia (increased plasma triglycerides and LDL cholesterol and reduced HDL cholesterol), hyper-glycemia, and hyperinsulinemia with confirmed insulin resistance. The presences of fatty liver was determined via liver histology and biochemical triglyceride assay (3.63 ± 0.39 in SD vs 8.35 ± 1.74 mg/g of liver wet weight in WD, P = 0.01).27 Importantly, the WD caused hepatocyte disarray, lobular inflammation, and mild centrilobular fibrosis, all markers of NAFLD. In addition, a WD led to severe atherosclerosis in these animals as determined from the staining of aortic root lesions.27 Conventional differential proteomics has been used to identify potential protein biomarkers of CVD. However, the static measurements provide no information on the cause of altered protein levels. In addition, protein homeostasis may not be associated with a measurable change in protein expression. In contrast, small changes in protein dynamics could be measured using tracer-based flux studies. To explore the association between NAFLD and atherosclerosis, we used the 2H2O metabolic-labeling approach to examine the effect of a WD on the dynamics of secreted plasma proteins, the assumption being that the majority of circulating proteins are derived from the liver. The experimental design for the study is presented in Figure 1. Attention was directed toward the dynamics of plasma proteins that are involved in reverse cholesterol transport, thrombosis, APR, inflammation, oxidative stress, and lipid metabolism because each is implicated in CVD.
Figure 1.
Experimental design and workflow for the proteome dynamics study. (A) At the beginning of the 12th week of the diet experiment, mice were given a bolus injection of pure 2H2O followed by the administration of 5% 2H2O in drinking water. Small blood samples were taken at different time points up to 1 week. (B) Proteins from apoB-depleted plasma were digested in solution and analyzed by LC–MS/MS. (C) Protein abundances were assessed by label-free protein quantification using Scaffold software, and protein kinetics were analyzed based on peptide isotopomer distribution using custom-made software. Networks of protein fluxes were analyzed using Gene Ontology (GO) and Ingenuity Pathway Analysis (IPA).
Differential Proteomics
In a pilot study, we used spectral counting-based label-free quantitative proteomics to assess the effect of WD on apoB-depleted plasma proteome remodeling. Proteome analyses did not detect apoB in these samples, confirming complete removal of apoB-containing particles. Overall, 175 proteins were identified in at least one animal from both groups with the mass range from 9 kDa (protein FAMc) to 582 kDa (hydrocephalus-inducing protein). A total of 157 and 171 of these proteins were present in the SD and WD groups, respectively, and 153 proteins were common to both groups (Figure S1 and Tables S1a and S1b). These proteins were represented by 2199 and 2119 unique peptides in SD and WD groups, respectively, and 1707 of these peptides were identified in both groups. Interestingly, four proteins were specific to the SD group and 18 to the WD group. The proteins that were present only in the WD group are involved in normal cardiovascular development (semaphorin-3C), lipid metabolism (diacylglycerol kinase gamma isoform X3), and degenerative changes in aortic walls (myosin-11 isoform 1) (Table S2). In addition, several proteins released by cell damage, including glyceraldehyde-3-phosphate dehydrogenase and carbonic anhy-drase, were found only in samples from WD-fed animals. The proteins that were absent in the WD group and only detected in the SD group are characterized with protease inhibition (serine protease inhibitor A3M (SERPIN A3M) and platelet biogenesis functions (neurobeachin-like protein 2 (NBEAL-2)).
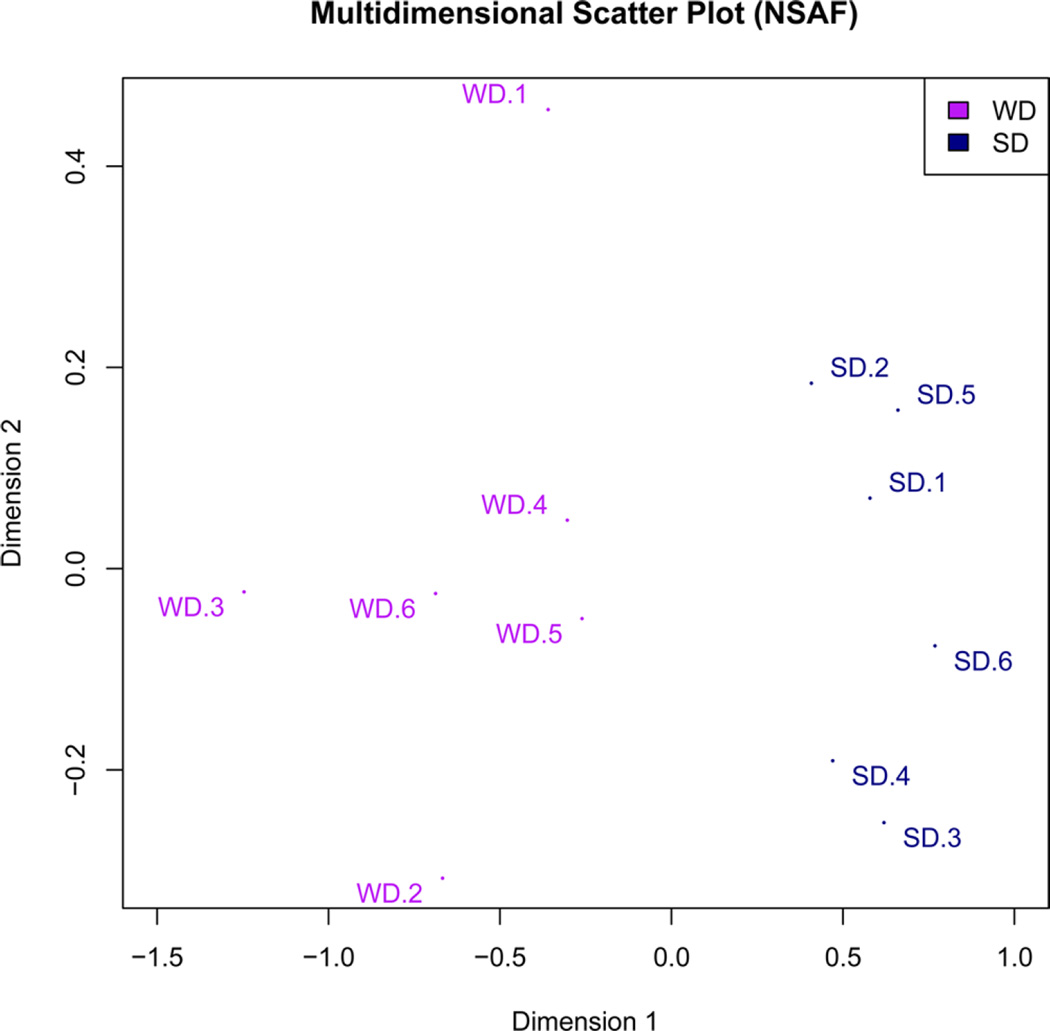
Eighty-nine of the identified proteins were present in all 12 mice (six mice/group). Most of these proteins, i.e., 69/89, were known to be associated with HDL, as previously described by other groups based on different HDL isolation methods.43 The remaining fraction consisted of water-soluble, nonlipoprotein-associated proteins. To assess the effect of WD-induced NAFLD on apoB-depleted plasma proteome composition, we interrogated these proteins using the label-free quantification approach (Tables S3a and S3b). For the results to be compared, all WD protein measurements were normalized to the SD. Because of bidirectional changes in proteome composition, the WD did not significantly affect the total protein levels in apoB-depleted plasma (26.7 ± 1.1 in SD vs 31.2 ± 6.1 mg/µL in WD, P = 0.2). However, the multidimensional scaling plot of a data matrix (Figure 2) indicated that samples from the SD and WD groups were clustered separately and split into two distinct groups, clearly demonstrating the WD-induced alteration in the distribution of proteins in apoB-depleted plasma.
Figure 2.
Effect of a WD on apoB-depleted plasma protein composition. After 12 weeks of either SD or WD, mice were euthanized, and plasma proteins were analyzed using the label-free protein quantification method (n = 6/group). A multidimensional scaling (MDS) plot of a data matrix was applied to the normalized spectral abundance factor (NSAF) values of analyzed proteins to depict the level of similarity of individual samples in the data set in two dimensions. The proximity of samples in this figure indicates the similarity between two NSAF profiles. The two dimensions after scaling the data are shown in the x and y axes. The origin corresponds to the mean of all samples, and the samples are arranged based on their similarities. On the basis of the MDS, SD and WD samples were separated into two distinct groups across the first dimension.
Comparative analysis of the normalized spectral counts revealed that 38 proteins were differentially expressed with a minimum of 1.2-fold changes (P < 0.05) in WD compared to that in SD (Figure S2): 21 proteins were upregulated and 17 proteins were downregulated due to the WD. Overall, multiple proteins involved in lipid metabolism, blood coagulation, inflammation, and stress response were markedly increased, whereas proteins with antioxidant and proteolytic inhibitory functions were significantly reduced following WD feeding. In addition, a WD significantly altered the levels of multiple acutephase reaction proteins that are known to be affected by inflammation and injury. As a part of the innate immune system, positive acute phase response proteins are increased, whereas proteins with negative acute phase response are decreased to exert a negative feedback reaction to inflammation. Next, for the accurate assessment of these changes, we applied more conservative criteria, i.e., fold change and P values that take into account the differences in spectral abundances based on a wide dynamic range of plasma proteins. On the basis of these criteria, only 18 proteins passed the fold change and significance thresholds defined in the method section (Supporting Information). Interestingly, the coagulation factor X, a key protein in the coagulation cascade, displayed a WD-induced 3.6-fold increase (P = 0.0002). We also observed an ~3-fold increase in hemoglobin, which was associated with an ~2.5-fold increase in haptoglobin (P = 0.05), an APR protein that binds hemoglobin and inhibits its oxidative activity. Likewise, the WD also significantly increased the relative abundance of the positive APR, including fibronectin (~3-fold, P = 0.009), which is known to increase during inflammation. Furthermore, ~2-fold increases were observed in the immunoglobulin J chain (P = 0.009). On the other hand, the levels of albumin, a negative APR protein that is known to decrease in response to stress, were clearly downregulated in the WD group (22% reduction, P = 0.0004); however, as the most abundant plasma protein, it did not pass the 1.3-fold change threshold. Similarly, WD-induced suppression was observed in the contents of paraoxonase 1 (PON1), the major antioxidant protein, which was reduced 33% (P = 0.005), but it also did not pass our fold change threshold.
Several remarkable changes were detected in plasma proteins that are involved in the regulation of proteolysis. In particular, inter-α-trypsin inhibitor heavy chain 2 (ITIH2) and serine protease inhibitor A3K (SERPINA3K) were significantly reduced in the WD group (P = 0.001 and 0.022, respectively). In contrast, several proteins within the complement pathway involved in innate and accrued immunity, including the complement factor B with the major function of alternative complement pathway initiation, and C5, C6, and C9, the essential components of the terminal complement pathway, were markedly increased due to WD feeding, although they did not pass our significance threshold. This was associated with decreased levels of C3 (P = 0.001), the key precursor in the alternative complement pathway, suggesting its increased utilization in an alternative complement pathway (Table 1).
Table 1.
Relative Abundances (NSAF), Fractional Turnover Rate (k), and Relative Production Rates (RPR) of Plasma Proteins
| NSAF ratio |
fractional turnover rate (%/h) |
RPR ratio |
||||
|---|---|---|---|---|---|---|
| accession | name of protein | molecular function | WD/ SD |
SD | WD | WD/ SD |
| Complement/Immune Response | ||||||
| 218156289 | complement factor B isoform 1 precursor | serine-type endopeptidase | 1.59 | 10.79 ± 1.07 | 11.29 ± 1.89 | 1.66c |
| 568966398 | complement factor D isoform X1 | serine-type endopeptidase | 0.54 | 3.72 ± 0.03 | 3.22 ± 0.03b | 0.46c |
| 109627652 | complement factor H precursor | C3b binding | 0.81 | 4.43 ± 0.22 | 5.57 ± 0.28b | 1.02 |
| 568921715 | complement factor I isoform X3 | complement activation | 0.88 | 5.21 ± 1.04 | 6.22 ± 0.72 | 1.05 |
| 114326449 | properdin precursor (factor P) | complement activation | 1.26 | 2.11 ± 0.26 | 0.71 ± 0.07b | 0.43c |
| 126518317 | complement C3 precursor | endopeptidase inhibitor | 0.67a | 11.92 ± 1.63 | 11.91 ± 1.69 | 0.67c |
| 6754164 | complement C5 preproprotein | endopeptidase inhibitor | 1.81 | 4.02 ± 0.45 | 3.96 ± 0.80 | 1.79c |
| 161086891 | complement component 6 precursor | immune response | 1.72 | 5.52 ± 0.57 | 6.53 ± 0.43b | 2.03c |
| 345110552 | complement component C7 precursor | immune response | 0.76 | 1.49 ± 0.16 | 1.67 ± 0.05 | 0.85 |
| 22122667 | complement component C8 alpha chain isoform 1 precursor |
complement activation | 1.51 | 4.90 ± 0.22 | 4.19 ± 0.22 | 1.55 |
| 15375312 | complement component C9 precursor | complement activation | 1.81 | 6.59 ± 0.73 | 7.60 ± 0.49b | 2.09c |
| 568986620 | clusterin isoform X2 (ApoJ) | misfolded protein binding | 1.04 | 12.40 ± 1.25 | 14.63 ± 1.63b | 1.23 |
| Antioxidants | ||||||
| 261823995 | serum paraoxonase/arylesterase 1 precursor | antioxidant | 0.67 | 1.54 ± 0.20 | 1.44 ± 0.23 | 0.63c |
| 15011841 | glutathione peroxidase 3 precursor | antioxidant | 0.65 | 2.23 ± 0.24 | 2.50 ± 0.53 | 0.73 |
| Lipid Metabolism | ||||||
| 160333304 | apolipoprotein A-I preproprotein | cholesterol transporter | 0.68a | 4.64 ± 0.42 | 4.92 ± 0.45 | 0.72c |
| 157951676 | apolipoprotein A-II precursor | cholesterol transporter | 1.18 | 5.58 ± 0.62 | 4.12 ± 0.32b | 0.87 |
| 244791354 | phosphatidylcholine-sterol acyltransferase | phosphatidylcholine-sterol O- acyltransferas |
1.03 | 5.76 ± 0.80 | 5.08 ± 0.46 | 1.36 |
| 568915857 | phospholipid transfer protein isoform X1 | lipid binding | 1.96 | 2.90 ± 0.51 | 3.63 ± 1.14 | 2.46c |
| 158517825 | apolipoprotein C-I precursor | fatty acid binding | 1.51 | 6.92 ± 0.95 | 7.84 ± 0.81 | 1.34 |
| 577019552 | apolipoprotein C-III isoform a | lipid binding | 0.84 | 6.59 ± 0.06 | 6.80 ± 0.99 | 0.87 |
| 110347473 | apolipoprotein A-IV precursor | antioxidant, cholesterol transporter | 1.21 | 8.34 ± 0.79 | 12.10 ± 0.71b | 1.75c |
| 163644329 | apolipoprotein E precursor | antioxidant, cholesterol transporter | 1.59 | 11.81 ± 1.09 | 13.25 ± 0.70b | 1.79c |
| 75677437 | apolipoprotein D precursor | cholesterol binding | 0.73 | 2.64 ± 0.13 | 2.79 ± 0.01 | 0.77 |
| 163310765 | serum albumin precursor | DNA binding | 0.78 | 0.80 ± 0.21 | 1.29 ± 0.36b | 1.25c |
| 7305599 | transthyretin precursor | hormone binding | 0.54a | 6.64 ± 0.10 | 6.02 ± 0.49 | 0.49c |
| Vitamin Transport | ||||||
| 125347464 | afamin precursor | vitamin E binding | 0.79 | 1.54 ± 0.23 | 3.63 ± 0.42b | 1.87c |
| 51172612 | vitamin D-binding protein precursor | actin binding | 0.83 | 6.95 ± 1.16 | 11.57 ± 0.92b | 1.38c |
| 33859612 | retinol-binding protein 4 isoform 1 | retinol binding | 1.13 | 11.19 ± 1.33 | 17.63 ± 0.75 | 1.79 |
| Acute Phase Response | ||||||
| 76881807 | alpha-1-antitrypsin 1–2 precursor | serine-type endopeptidase inhibitor | 0.71a | 3.72 ± 0.65 | 3.07 ± 0.37b | 0.59c |
| 7304875 | alpha-2-HS-glycoprotein isoform 1 (Fetuin A) | cysteine-type endopeptidase inhibitor | 0.83 | 6.71 ± 0.78 | 7.21 ± 0.35 | 0.89 |
| 6755398 | serum amyloid A-4 protein precursor | acute-phase response | 0.50 | 6.55 ± 0.44 | 5.82 ± 0.22b | 0.45c |
| 110347564 | ceruloplasmin isoform b precursor | copper ion binding | 0.93 | 5.57 ± 0.65 | 6.41 ± 0.75 | 1.07 |
| 449083336 | fibronectin isoform b precursor | peptidase activator | 2.74a | 4.16 ± 0.55 | 4.12 ± 0.48 | 2.74c |
| 6753798 | prothrombin precursor | thrombospondin receptor | 0.87 | 5.27 ± 0.54 | 8.53 ± 1.39 | 1.41c |
| 156231029 | kininogen 2 isoform 2 precursor | serine-type endopeptidase inhibitor | 0.39a | 4.12 | 10.97 ± 1.64 | 0.41 |
| 160358829 | hemopexin precursor | metal ion binding | 1.24 | 2.99 ± 0.38 | 4.62 ± 0.40b | 1.92c |
| 8850219 | haptoglobin precursor | hemoglobin binding | 2.45 | 8.50 ± 1.11 | 23.04 ± 2.25b | 13.27c |
| Immune Response | ||||||
| 568999092 | H-2 class I histocompatibility antigen, Q10 alpha chain isoform X1 |
peptide antigen binding | 0.77 | 2.31 ± 0.09 | 5.66 ± 0.68b | 1.89 |
| 160415217 | zinc-alpha-2-glycoprotein precursor | peptide antigen binding | 1.31 | 5.84 ± 0.14 | 8.16 ± 0.16b | 1.83c |
| 170172530 | immunoglobulin J chain precursor | antigen binding | 1.93a | 5.15 ± 0.58 | 1.64 ± 0.01b | 0.61 |
| Proteolysis | ||||||
| 568910406 | antithrombin-III isoform X1 | serine-type endopeptidase inhibitor | 0.52a | 4.53 ± 0.11 | 5.71 ± 0.21b | 0.66 |
| 257471003 | plasminogen precursor | serine-type endopeptidase | 0.81 | 6.60 ± 0.39 | 6.50 ± 0.30 | 0.80c |
| 236465805 | plasma kallikrein precursor | serine-type endopeptidase | 1.11 | 2.49 ± 0.25 | 2.52 ± 0.42 | 1.12 |
| Proteolysis Inhibition | ||||||
| 124249351 | interalpha-trypsin inhibitor heavy chain H1 precursor |
serine-type endopeptidase inhibitor | 0.62 | 5.45 ± 0.40 | 4.77 ± 0.53 | 0.55c |
| 226874935 | interalpha-trypsin inhibitor heavy chain H2 precursor |
serine-type endopeptidase inhibitor | 0.38a | 3.90 ± 0.31 | 4.21 ± 0.58 | 0.41c |
| Proteolysis Inhibition | ||||||
| 159110717 | interalpha-trypsin inhibitor heavy chain H3 precursor |
serine-type endopeptidase inhibitor | 1.45 | 5.86 ± 1.26 | 6.82 ± 0.11 | 1.69 |
| 148747546 | serine protease inhibitor A3K precursor | serine-type endopeptidase inhibitor | 0.76a | 3.33 ± 0.54 | 3.50 ± 0.33 | 0.80 |
| 130503301 | serine protease inhibitor A3N precursor | serine-type endopeptidase inhibitor | 1.15 | 1.97 ± 0.32 | 2.87 ± 0.57b | 1.39 |
| 31982171 | murinoglobulin-1 precursor | serine-type endopeptidase inhibitor | 0.94 | 2.67 ± 0.52 | 2.11 ± 0.31 | 0.75c |
| Metal Binding | ||||||
| 20330802 | serotransferrin precursor | ferric iron binding | 0.80 | 2.61 ± 0.35 | 2.62 ± 0.37 | 0.80c |
| 17647499 | hemoglobin subunit beta-2 | heme binding | 2.44a | 0.13 ± 0.01 | 0.21 ± 0.02b | 3.85 |
| 226958456 | histidine-rich glycoprotein precursor | cysteine-type endopeptidase inhibitor |
1.28 | 2.49 ± 0.27 | 2.37 ± 0.38 | 1.21 |
| Hemostasis | ||||||
| 110347469 | alpha-2-macroglobulin precursor | endopeptidase inhibitor | 0.83a | 2.18 ± 0.29 | 2.21 ± 0.28 | 0.84c |
| 110625994 | coagulation factor X isoform 2 preproprotein | calcium ion binding | 3.58a | 5.65 ± 0.78 | 5.17 ± 0.86 | 3.27c |
| Other Plasma Proteins | ||||||
| 111378397 | phosphatidylinositol-glycan-specific phospholipase D precursor |
phospholipase D | 0.78 | 3.42 ± 0.28 | 3.06 ± 0.06 | 0.70c |
| 28916693 | gelsolin isoform 1 precursor | metal ion binding | 0.79 | 6.52 ± 1.03 | 4.58 ± 0.73b | 0.55c |
| 16418335 | leucine-rich alpha-2-glycoprotein precursor | transforming growth factor beta receptor binding |
0.99 | 4.07 ± 0.10 | 6.57 ± 0.30b | 1.59 |
Significant difference in NSAF (P < 0.05).
Significant difference in k (P < 0.05).
Significant difference in RPR (P < 0.05).
Our label-free quantification also demonstrated that the WD results in a 33% decrease in apoAI levels, the major HDL protein involved in HDL assembly and reverse cholesterol transport (P = 0.0004). In contrast, WD did not have any significant effect on apoAII levels, the second most abundant protein of HDL (Table 1). To validate the label-free quantification results, we quantified the absolute amounts of apoAI and apoAII. Similar to relative spectral count results, the isotope dilution method revealed that the WD significantly reduced the absolute amounts of apoAI (Figure S3A and B) (P = 0.02) without any significant effect on apoAII (Figure S3B and D). This also agrees with the results based on reconstructed spectral areas under the full scan ion chromatogram curves of analyzed unique peptides of apoAI and apoAII, respectively (Figure S3E and F). Furthermore, consistent with the spectral counting results, quantification of the areas under the extracted ion chromatograms of selected unique peptides determined that a WD significantly altered the levels of C3 (Figure S3G and I) and C9 (Figure S3H and J). Thus, two independent validation methods confirmed that WD profoundly altered the distribution of proteins in apoB-depleted plasma.
In Vivo Plasma Proteome Dynamics in Mice
To understand the cause of WD-induced alterations in plasma proteins, we used the heavy water-based metabolic labeling approach to assess proteome dynamics (Figure 1). Briefly, at the start of the 12th week of the diet experiment, the total body water in each mouse was enriched with a bolus load of heavy water (20 µL/kg body weight, 99.99% isotopic and chemical purity) and was maintained for 7 days using 5% (v/v) heavy water in drinking water. The time course of 2H incorporation into peptides was quantified to assess the kinetics of apoB-depleted plasma proteins. This protocol results in 2.7-3% body water enrichment,27 and no adverse effects on food intake or growth were observed due to heavy water administration. In our previous studies, we confirmed that heavy water rapidly (~10–20 min) equilibrates with the total body water and stays at the steady state provided that mice are given 2H-labeled drinking water.25 Because intracellular amino acids also reach the steady-state labeling quickly (~20 min), the temporal change in 2H labeling of a protein will represent its turnover rate.
The incorporation of 2H-labeled amino acids into newly synthesized protein results in a shift in the isotope distributions of tryptic peptides. Specifically, there is a gradual increase of heavy isotopomers (M1…Mn) and a decrease of monoisotopic isotopomer (M0) (Figure 1). The high-resolution Orbitrap MS allows analyses of the isotopic distribution of virtually all peptides with good peak quality and enables measurements of global proteome dynamics. To process the data, we used specialized software that extracts the mass isotopomer distribution of peptides.25
The mass isotopic distributions of all unique peptides were analyzed; the turnover rate was calculated using the regression analysis of time-course labeling of analyzed peptides. Because each unique peptide represents their protein of origin, the results from all unique peptides were aggregated to derive the averaged turnover rate of a protein. The high sensitivity of our assay allowed measuring the dynamics of plasma proteins in individual mice through the collection of small blood samples (~60–70 µL) at different time points.27,28 This approach, in addition to reducing the cost of the study, also improves the accuracy of the assay by minimizing the biological variability in each kinetic curve.
Effect of a WD-Induced NAFLD on Plasma Proteome Dynamics
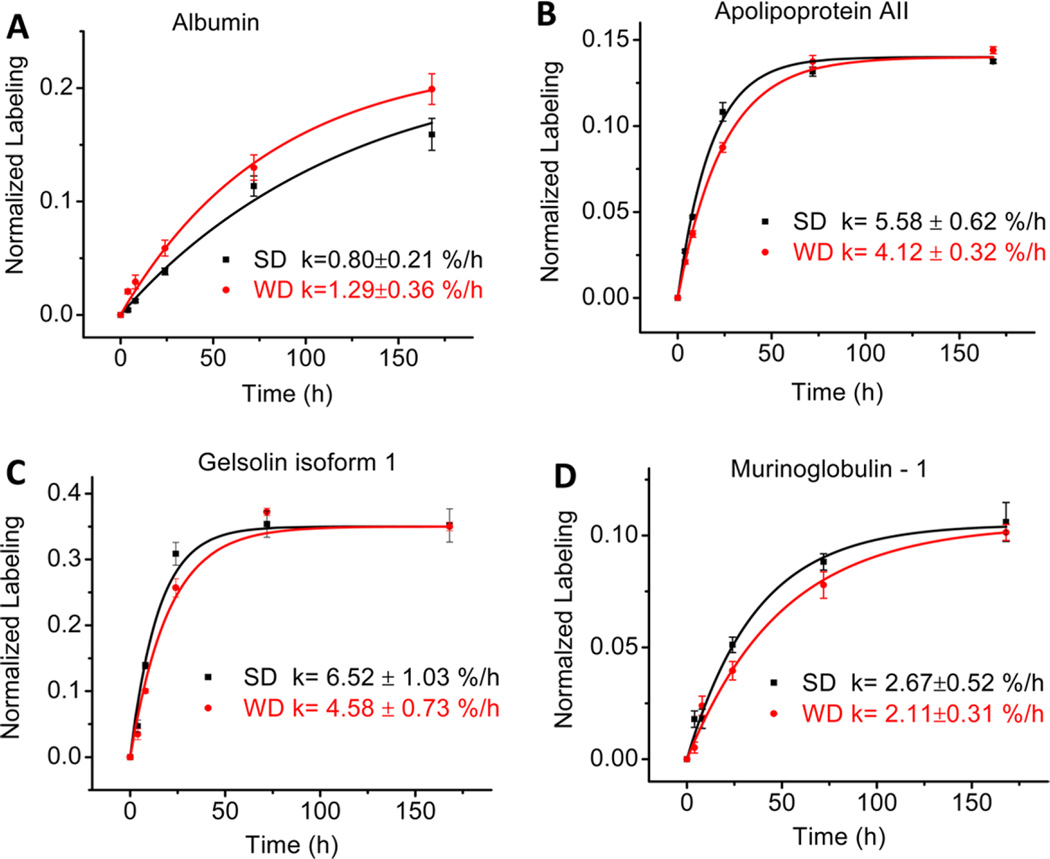
Eighty-nine proteins were identified in each time point of apoB-depleted plasma samples from animals in both the SD and WD groups. Because of the stringent criteria for quantification, i.e., at least two unique peptides with moderately higher abundances, good chromatographic characteristics, and the presence at all six time points in both the SD and WD groups, we could accurately quantify the kinetics of 57 proteins (Table 1). The proteome dynamics study revealed that a WD-induced remodeling of the plasma proteome was associated with the altered turnover of several individual proteins. The kinetic analysis of the individual plasma proteins demonstrated that WD feeding caused more than a 20% change in the half-lives of 35 out of the 57 analyzed proteins. The WD had distinct effects on the turnover rates of proteins with different functions. Interestingly, 46% of all analyzed proteins showed an increased fractional turnover rate (i.e., a reduced half-life) due to the WD. In particular, the WD resulted in a significant (≥15%) reduction of half-lives of several proteins involved in lipid metabolism, and proteins with antioxidant, stress response, and vitamin transport functions. For instance, the WD lead to an increase in the turnover of negative APR protein albumin (0.80 ± 0.21 vs 1.29 ± 0.36%/h, P < 0.0001) (Figure 3A). Although the WD significantly increased the turnover of most of the analyzed proteins, the turnover rates of some proteins were reduced and others remained unchanged. In particular, the WD significantly increased the turnover of positive APR protein haptoglobin (23.04 ± 2.25 vs 8.50 ± 1.11%/h, P < 0.001) (Table 1), whereas it reduced the turnover of apoAII (5.6 ± 0.6 vs 4.1 ± 0.3%/h, P < 0.002) (Figure 3B). Similarly, a reduced turnover was observed for gelsolin (6.5 ± 1.0 vs 4.6 ± 0.7%/h, P = 0.01) (Figure 3C) and other plasma proteins involved in inflammation and immune response (Table 1). Interestingly, the proteome dynamics also revealed a robust influence of WD on altered turnover of proteins involved in cholesterol metabolism, including apoAII, apoE, and apoAIV, although only a modest effect was observed on the differential expression of these proteins (Figure S2A). However, the WD did not have any significant effect on the turnover of murinoglobulin-1 (Figure 3D), PON1, apoAI, or other proteins (Table 1).
Figure 3.
Effect of Western Diet (WD) on turnover rates of selected plasma proteins in LDLR-deficient mice assessed with the 2H2O metabolic-labeling technique. Intraperitoneal bolus loading followed by free access to drinking water enriched with 2H2O (5%) led to a steady state body water labeling of ~2.9%. Time course enrichment of 2H incorporation into peptide LGEYGFQNAILVR from albumin (A), peptide THEQLTPLVR from apoAII (B), peptide DSQEEEKTEALTSAK from gelsolin (C), and peptide YTYGKPVPGHVK from murinoglobulin-1 (D). Western diet, square symbols; standard diet, circle symbols. Data show mean ± SD with n = 4 per group.
Consistent with the label-free quantitative proteomics results, the heavy water-based metabolic-labeling experiment demonstrated that the turnover rates of several key proteins of the complement pathway were significantly affected due to the WD. For example, more than 30% increase in turnover rates constant of complement factor H (P < 0.05), a component of the alternative complement pathway. Similar, but more modest (i.e., 20%), increases in the turnover were observed for C6 and C9, components of the terminal complement pathway, clusterin, involved in apoptosis, and transcriptional regulation of inflammation (all P < 0.05). In addition, despite the fact that the relative abundance of properdin (Factor P), the only known positive regulator of complement activation that stabilizes C3 convertases and amplifies the alternative pathway, did not change significantly, its flux was increased by ~3-fold (P < 0.05). This finding highlights the importance of the flux measurements as they can detect the changes that would otherwise be dismissed based on the static measurements of protein abundance.
Relative Production Rates
Simultaneous measurements of the relative expression of a protein’s abundance and its fractional turnover in the same experiment enabled us to calculate the relative production rates (RPR) of plasma proteins. Because the fractional turnover and quantitation were performed on 57 proteins, RPR was calculated for only these proteins (Table 1, last column). The RPR results enhance the differences between the two groups and clearly illustrate the effect of WD on plasma proteome flux. Overall, the calculated RPR revealed that the flux of more than half of the analyzed proteins were altered due to a WD as several proteins clearly displayed >1.5-fold increase in their RPR (P < 0.005). For example, consistent with the label-free quantification and turnover rate results, the production rates of the key components of the complement pathway including complement factor B, complements C5, C6, C8, and C9 were increased more than 50%. Similarly, the WD led to increased production rates of apoAIV and phospholipid transfer protein (PLTP) of 1.8- and 2.5-fold (P < 0.005), respectively. ApoAIV is involved in the regulation of appetite and satiety in rodents, whereas PLTP is responsible for the remodeling of lipid composition in HDL, which are lipoprotein particles that prevent atherosclerosis. It has been shown that an increased content of apoAIV and PLTP are associated with dysfunctional HDL and atherosclerosis. Mice deficient in PLTP are protected from atherosclerosis,44 whereas HDL from mice overexpressing PLTP has reduced cholesterol efflux capacity, and these mice develop greater atherosclerotic lesions compared to those of control mice.45 Several proteins had a >1.5-fold decrease in their RPR. For instance, PON1, properdin, transheretin, and gelsolin were among those proteins with reduced RPR (all P < 0.05).
To better understand the cause of plasma proteome remodeling, the heat maps of abundance, fractional turnover, and RPR differences were created for the same set of proteins. For the WD-induced alterations to be visualized, all WD protein measurements were normalized to the SD values. These results demonstrated that the changes in the abundance of proteins affect innate immunity, including those involved in the complement pathway, are directly related to their turnover (Table 1 and Figure 4). In particular, reduced expression of proteins involved in proteolytic inhibition was associated with increased fractional turnover and decreased RPR of these proteins. Similarly, the WD-mediated reduction in factor D, a protein involved in alternative complement pathway inhibition, was associated with decreased production of this protein.
Figure 4.
Heatmap differences of protein abundance (NSAF), turnover rate (k), and relative production rate (RPR). The analyses are based on 57 proteins with calculated k and RPR values. The image is organized by sorting the proteins by NSAF ratios. The ratios are log scaled, and a ratio of one is represented with white. Increased values in WD are shown in red, and increased values in SD are shown in blue. The color bar on the left shows the functional annotation of the proteins [UniprotKB]. Because of altered k values, it is not necessarily the case that abundant proteins also have high RPR values.
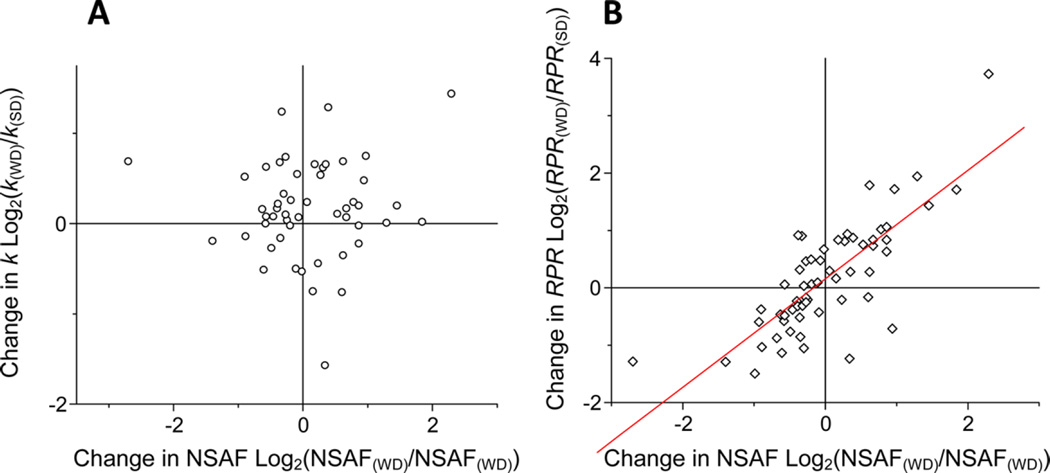
The functional grouping of the analyzed proteins demonstrated that proteins with inflammatory functions had significantly higher RPR in the WD group compared to that in the SD group. In contrast, consistent with the important role in preventing atherosclerosis, WD-induced depletion of apoAI was associated with 28% reduction of the RPR of this protein. WD also dramatically affected the dynamics of plasma proteins with antioxidant activities (Figure 4 and Table 1). Consistent with these results, the regression analysis revealed that the relative abundances of proteins were significantly correlated with their RPR (Figure 5A), but not with their turnover (Figure 5B), suggesting that the changes in protein abundance were driven mainly by their production rates.
Figure 5.
Relationships between the WD-induced change in the abundance and the change in turnover of plasma proteins. Distribution of the changes in both turnover rate (k) and abundance of each protein (A). Distribution of change in RPR and the abundance (B). The data are derived from Table 1 (means ± SD). There were no correlations between the changes in protein abundance and the changes in turnover rate (k). The changes in the abundance of proteins were directly related to the changes in RPR (r = 0.78, P < 0.0001) (B).
Identification of the Pathways Associated with the WD-Induced Alterations in Plasma Proteome Dynamics
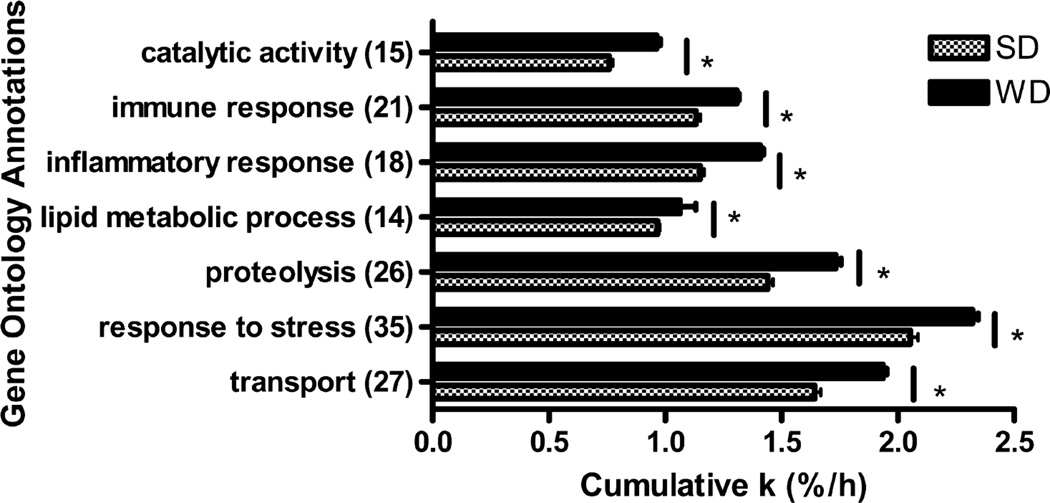
To understand the biological relevance of altered plasma proteome dynamics in the context of NAFLD and associated atherosclerosis, we further analyzed the changes in protein turnover using Gene Ontology (GO) and Ingenuity Pathway Analyses (IPA). First, we compared proteins based on their functions. The functional grouping revealed that the bulk of the proteins with altered dynamics fall into functional categories that are associated with pathways involved in atherosclerosis, including proteins with catalytic activity, immune and inflammatory response, proteolysis, stress response, and transport (all P < 0.01) (Figure 6). These alterations were consistent with the changes observed in the abundances of proteins from these GO groups (not shown).
Figure 6.
Differences in turnover rate (k) across Gene Ontology terms are shown. The analyses are based on 57 proteins with calculated turnover rates. The k values of proteins annotated with these GO terms were added to create cumulative k values for each term. The data are derived from Table 1. The cumulative turnover rate values increased in WD compared to SD significantly across all annotations (P < 0.01).
As expected, the IPA core network analysis based on the turnover results demonstrated that HDL is the major hub connecting all proteins with WD-induced altered dynamics. The functional analysis revealed that the WD significantly increased the turnover rate constants of proteins implicated in lipid metabolism, immune response, and CVD functions (Figure S4), suggesting that the WD-induced NAFLD is associated with hepatic acute phase reaction leading to proatherogenic and proinflammatory alterations of plasma proteins.
Next, we applied IPA to the proteome dynamics results to identify the canonical pathways that were significantly affected by the WD. There were multiple pathways that scored significantly (Fisher’s exact test, P < 0.05), but only three pathways, i.e., LXR/RXR, Coagulation System, and Complement System, had a pattern of activation/inhibition based on the z-score (≠0). Consistent with the results demonstrating increased flux of proteins in the terminal complement pathway (C5, C6, C8, and C9), IPA analysis suggested that WD-induced activation of the complement cascade was in part due to the activation of the alternative complement pathway (complement factors B and D, properdin, and C3) (Figure S5). A similar induction of the alternative complement pathway was previously shown based on liver gene expression in LDLR−/−mice fed a WD.46 Taken together, both GO and IPA analysis confirmed that WD-induced NAFLD alters plasma proteome dynamics in a manner that is consistent with having negative implications in CVD.
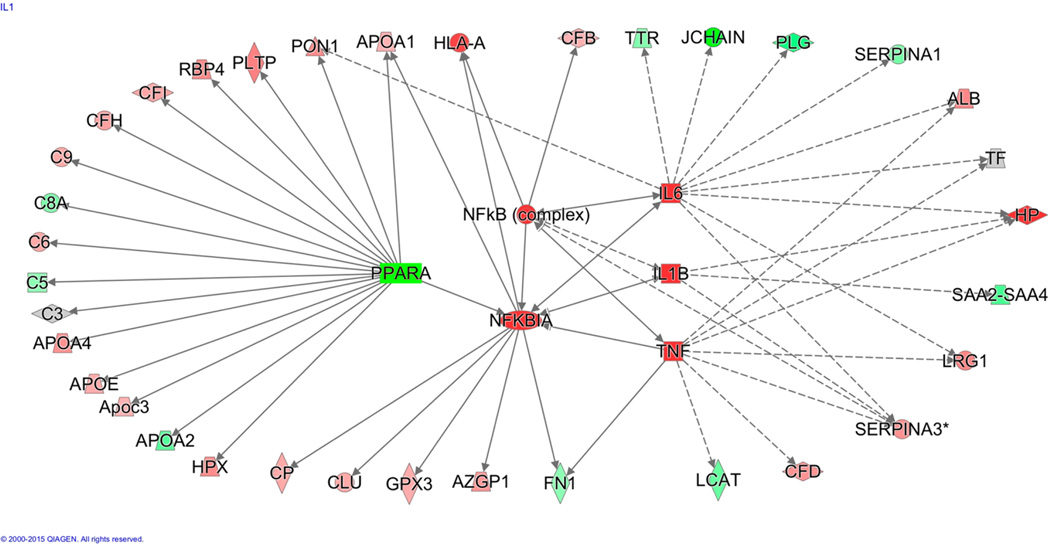
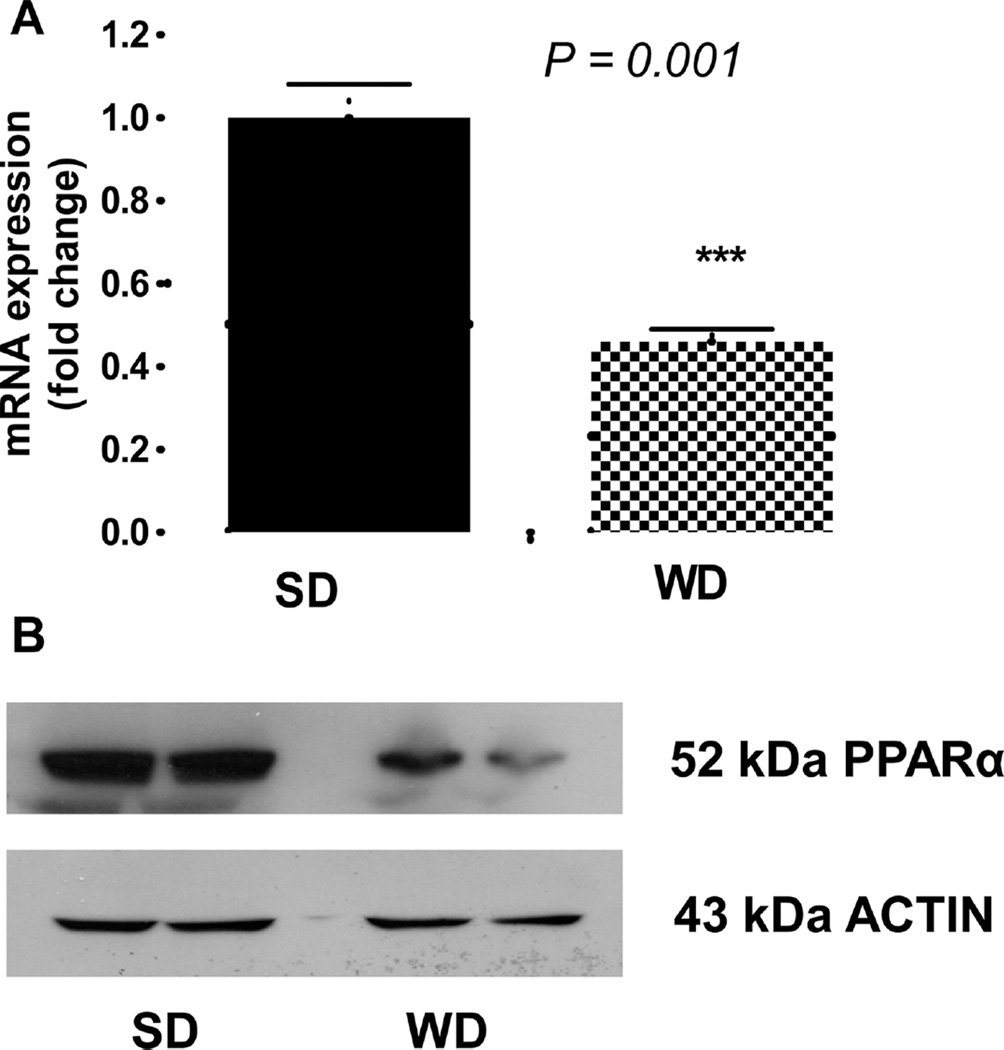
The upstream pathway analysis of plasma proteome dynamics revealed that many of the analyzed proteins are connected to PPARα, a ligand-activated transcriptional factor that regulates hepatic lipid metabolism and APR (Figure 7). Although a major physiological role of PPARα is to control fatty acid oxidation in response to fasting and high-fat feeding, PPARα is also an important modulator of lipid homeostasis through regulation of genes involved in lipoprotein metabolism. As an anti-inflammatory protein, PPARα also regulates the cytokine signaling pathway that is critical in hepatic APR reactions.47 Activation of PPARα impairs cytokine signaling pathways, reduces triglyceride-rich particle production, and promotes HDL metabolism. In contrast, suppression of PPARα expression and activity has been associated with lipotoxicity and atherosclerosis. To test whether the WD-induced alteration in plasma proteome dynamics is due to suppression of PPARα, we quantified the hepatic protein abundance by Western blot and gene expression with rtPCR. WD significantly reduced both gene (Figure 8A) and protein expression of PPARα (Figure 8B).
Figure 7.
Effect of WD on dynamic plasma proteome networks. Unsupervised upstream pathway analysis of proteins with measured turnover rate results from the 2H2O metabolic-labeling experiment revealed that observed expression patterns were significantly associated with the suppression of PPARα. The outer circle of proteins was experimentally measured, where a green color represents a decreased k value and red represents an increased k value in WD compared to that in SD. Note that a gray color indicates no change. The activities of proteins (k-values) at the center were predicted to be activated by the upstream analysis as indicated with red color.
Figure 8.
Effect of WD on hepatic expression of the PPARα gene and protein. After 12 weeks of a diet experiment, mice were euthanized, and liver samples were analyzed. (A) RT-PCR of hepatic PPARα mRNA (means ± SD; *, P < 0.05; n = 6 per group). (B) Western blot of hepatic PPARα from WD and SD-fed LDLR−/− mice with actin used as a loading control.
Pro-Inflammatory Index of apoB-Depleted Plasma
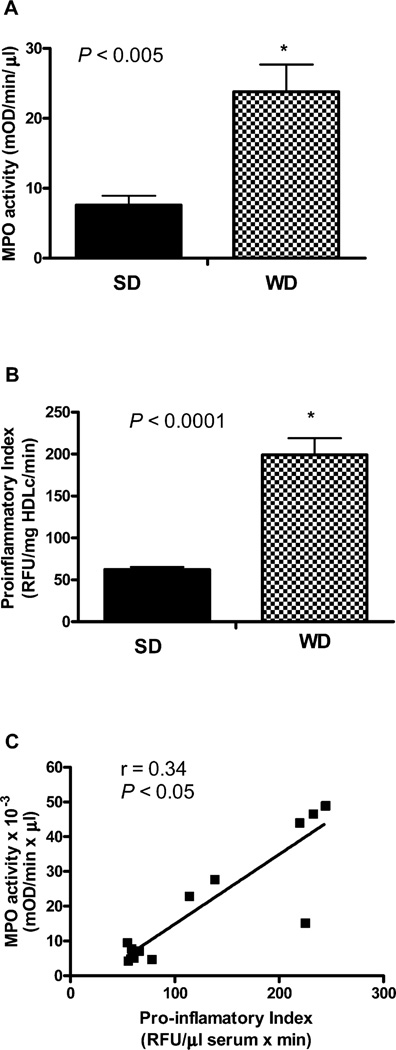
Because our flux studies revealed that a WD resulted in the APR-induced proinflammatory alterations of plasma proteome dynamics, we analyzed the proinflammatory index of apoB-depleted plasma. This assay determines the capacity of plasma to inhibit or aggravate Cu2+-induced total oxidation. In addition, we measured the activity of MPO, a neutrophil-derived heme protein that is mechanistically linked to hepatic oxidative stress and inflammation in NAFLD. As a catalyst of post-translational oxidative modification of plasma proteins, MPO is also involved in atherosclerosis.10 The WD resulted in a 4-fold increase in plasma MPO activity (Figure 9A). Consistent with these results and proinflammatory alterations of plasma proteome dynamics, we also found that WD leads to a more than 2-fold increase in the proinflammatory property of plasma (P < 0.0005) (Figure 9B). Interestingly, the WD-induced increase in plasma MPO activity was positively correlated with the proinflammatory index of apoB-depleted plasma (r = 0.3, P < 0.05) (Figure 9C).
Figure 9.
Effect of WD on proinflammatory properties of apoB-depleted plasma in LDLR−/− mice. After 12 weeks of a diet experiment, mice were euthanized and plasma samples were analyzed (n = 6/group). WD results in increased plasma MPO activity (A) associated with the proinflammatory index of HDL (B). Plasma MPO activity was significantly correlated with proinflammatory index of apoB-depleted plasma (r = 0.34, P < 0.05) (C).
DISCUSSION
The main goal of this study was to identify the temporal signatures of plasma proteins that would aid in our understanding of the pathophysiological mechanisms involved in NAFLD and associated atherosclerosis. For this purpose, we coupled traditional measurements of static proteomics with 2H2O-based metabolic labeling to assess alterations in proteome dynamics. Our results show that WD-induced alterations in plasma protein turnover lead to proinflammatory remodeling of apoB-depleted plasma proteome composition. These changes are consistent with the suppression of PPARα, a ligand-activated transcription factor involved in APR, HDL metabolism, and the development of both NAFLD and atherosclerosis. Thus, this study, for the first time, determined WD-induced alterations in plasma proteome dynamics and thereby identified the liver-secreted protein mediators that mechanistically link NAFLD and atherosclerosis.
Plasma protein abundance ranges from 5 pg/mL (inter-leukin-6) to 50 mg/mL (albumin), and accurate quantification of low-abundance proteins requires depletion of high-abundance proteins.30 Despite their interference with the analysis of the minor plasma peptides, the high-abundance proteins represent many physiologically important proteins secreted from the liver. Quantitative analysis of many of these high-abundance proteins is diagnostically useful for assessing the liver function, immune status, disorders of coagulation or fibrinolysis, and other diseases. ApoB-depletion used in this study allowed identification and quantification of more than a hundred proteins, including the kinetics of 57 distinct proteins associated with HDL.
On the basis of static proteomics, it was recently shown that plasma proteome composition is altered in coronary artery disease and several other diseases associated with chronic inflammation, including chronic kidney disease, rheumatoid arthritis, type 2 diabetes, psoriasis, aging, and NAFLD.6,7,17,48–50 Although static quantitation of proteins as biomarkers is important, these results do not explain the mechanisms of these altered levels. Changes in protein abundance are the net result of an imbalance between protein synthesis and degradation. To study the role of a diet-induced hepatic acute phase reaction on plasma proteome dynamics, we coupled label-free quantification with a 2H2O-based metabolic-labeling approach to assess both the fractional turnover and RPR of individual plasma proteins with different functions. Our steady-state flux analysis relies on the assumption that concentrations of analyzed proteins are stable during kinetic measurements. To ensure steady-state metabolic conditions, the 2H2O-based turnover study was performed in adult mice after 12 weeks on either a SD or a WD. The stable levels of analyzed proteins during the one-week labeling experiment demonstrate a steady-state condition (Table S4). The seven-day labeling experiment with the frequent blood sampling during the early hours of 2H2O exposure allows one to capture the turnover rates of plasma proteins with a wide range of half-lives that span from 3.0 h (haptoglobin, WD group) to 516.0 h (hemoglobin, SD group).
Liver-secreted proteins are involved in lipid metabolism, APR, innate immunity, and regulation of endothelial health. Our label-free quantification studies demonstrated that WD leads to proatherogenic and proinflammatory remodeling of the plasma proteome in a mouse model of NAFLD, a disease that is associated with hepatic and systematic inflammation and atherosclerosis. Our metabolic labeling-based kinetic study revealed that these changes were directly related to altered dynamics of HDL proteins. Notably, dyslipidemia-induced hepatic oxidative stress and inflammation in a mouse model of NAFLD caused increased fractional turnover rate (k) and relative production rate (RPR) of multiple proinflammatory proteins. Furthermore, our kinetic study discovered several dynamic signatures of a diet-induced NAFLD that were not identified by simple quantification of protein expression, demonstrating that protein turnover measurements can provide useful information on protein homeostasis that cannot be observed by conventional protein expression measurements.
Consistent with the role of inflammation in the pathogenesis of both NAFLD and atherosclerosis, our results demonstrated that one of the most prominent WD-induced changes in proteome dynamics was related to the activation of the complement pathway involved in innate and adaptive immunity. In addition to lectin and classical pathways, the complement system is initiated by the activation of the alternative pathway. The alternative pathway begins by binding of factor B (FB) to complement 3b (C3b) generated from spontaneous hydrolysis of complement 3 (C3). Properdin stabilizes the complex C3bBF by delaying its decay, whereas factors I and H act together to limit the formation of this complex. Increased flux of FB observed in our study suggests activation of the alternative complement pathway, which is in agreement with reduced C3 expression due to its increased utilization in this pathway. This was also in agreement with the increased levels of properdin, a positive regulator of complement activation that amplifies the alternative pathway. Our metabolic labeling study demonstrated that changes in properdin expression were related to its reduced turnover.
The alternative pathway along with the classic and lecithin pathways converges into the terminal pathway, which is activated upon the generation of C5 convertase enzyme. C5b produced from C5 convertase cleavage serves as an anchor for the assembly of terminal complement complex C5–9 by stepwise binding to C6, C7, C8 and C9. Increased fluxes of C6 and C8 observed in our study indicate that WD-induced NAFLD is also associated with activation of the terminal complement pathway. Consistent with the role of the complement system in NAFLD and atherosclerosis,46,51 these results suggest that increased flux of complement proteins in plasma may contribute to atherosclerosis in NAFLD.
Our pathway analysis based on the proteome dynamics data suggests that PPARα is the upstream transcriptional regulator of altered plasma protein flux that results from consumption of a WD. As a ligand-activated nuclear receptor, PPARα induces gene transcription after heterodimerization with retinoic X receptor (RXR). PPARα regulates hepatic APR reaction, including activation of the complement system and cytokine-signaling pathway through its effects on NF-κB activation.52–54 On the other hand, inflammation suppresses PPARα expression,47 stimulates the complement pathway, and impairs reverse cholesterol transport function of HDL with the simultaneous impairment of apoAI and PON1 expression.12,55 Consistent with this notion and a known role of PPARα as a modulator of the inflammatory APR in the liver,56 we found that the WD resulted in a reduced expression of both PPARα gene and protein in the liver. On the basis of these results, we propose that the WD-induced production of hepatic cytokines suppresses hepatic PPARα, leading to activation of NF-κB that transcriptionally upregulates complement pathways and down-regulates apoAI and PON1 production. Thus, in addition to its direct anti-inflammatory action in the vascular wall, PPARα regulates hepatic CVD factors through its control of hepatic synthesis of circulatory proteins involved in atherosclerosis.
PPARα expression is regulated by nutritional status and hormonal signals (e.g., leptin, growth factor, insulin). Fasting increases and glucose decreases PPARα expression, leading to diminished fatty acid oxidation, triglyceride accumulation, and lipotoxicity.47,52 In addition, regulation of PPARα activity occurs by phosphorylation,57 post-translational ubiquitination,58 SUMOylation,59 and acetylation,47 which can regulate PPARα expression through increased proteosomal degradation. Thus, we speculate that WD-induced hyperglycemia and hyperlipidemia result in transcriptional suppression and/or post-translational acetylation of PPARα, which leads to a reduction in its expression through increased degradation. Further studies are warranted to assess the effect of WD on post-translational modification and stability of PPARα.
The alterations observed in the kinetics of plasma proteins, including those involved in complement pathway, lipid metabolism, cholesterol transport, oxidative stress, and inflammation are in agreement with the results of the previous study on the effect of a WD on hepatic gene expression in LDLR−/− mice.46 Similar to this study, our proteome dynamics results demonstrated that a WD leads to increased production of apoAIV but decreased production of ITIH2, proteins involved in dyslipidemia and proteolysis inhibition, respectively. Likewise, our proteome dynamics study confirmed the activation of an alternative complement pathway reported in this study.46 Thus, the plasma proteome flux analysis may provide a new strategy for the measurement of hepatic temporal gene expression.
The results of this study also have relevance for the NAFLD and its associated CVD. As a hepatic manifestation of metabolic syndrome, NAFLD is associated with dyslipidemia, inflammation, and atherosclerosis. It is also known that endothelial function is impaired in patients with NASH.3 Hepatic inflammation plays a key role in disease progression from simple steatosis to NASH, and it is an independent predictor of endothelial dysfunction in NASH.2 Inflammation is also involved in atherosclerosis. In future studies, it would be important to determine whether proinflammatory remodeling of plasma proteome dynamics in patients with NASH predisposes them to atherosclerosis.
In summary, our study demonstrated that WD-induced NAFLD is associated with proinflammatory alterations of the plasma proteome. Our proteome dynamics approach provides evidence that production rates of proinflammatory proteins are increased and antioxidant proteins are reduced in response to WD-induced APR. Our pathway analysis revealed that the changes observed in the plasma proteome were related to transcriptional regulation of hepatic proinflammatory genes. The ability to combine studies of proteome dynamics with functional pathway analysis may have broad applications for other disease states and suggests potential targets for therapeutic modulation.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health Grant RO1GM112044 and the American Heart Association Grants 13IRG14700011 and 15GRNT25500004. Mass spectrometry instrument used in this study was purchased using NIH grant 1s10RR031537-01. We thank Dr. Min Li (CCF) for technical assistance.
ABBREVIATIONS
- GC–MS
gas chromatography–mass spectrometry
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- APR
acute phase response
- FCR
fractional catabolic rate
- FSR
fractional synthesis rate
- RPR
relative production rate
- SD
standard diet
- WD
western diet
- TBW
total body water
- LDLR−/−
LDL receptor-deficient
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- CVD
cardiovascular disease
- HDL
high-density lipoprotein
- PON1
paraoxonase 1
- MPO
myeloperoxidase
- SC
spectral counts
- NSAF
normalized spectral abundance factor
- MDS
multidimensional scaling
- IPA
ingenuity pathway analysis
- GO
Gene Ontology
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Label-free quantification methods, proteomics results, relative quantification of protein expression on a WD, IPA analysis, and complement system pathway schematic (PDF)
- Spectral counts, relative protein expression data for SD vs WD and in LDLR−/− mice, and metabolic conditioning upon 2H2O labeling (XLSX)
The authors declare the following competing financial interest(s): SF Previs is currently employed by Merck. The other authors have no conflict of interest to disclose.
REFERENCES
- 1.Targher G, Bertolini L, Padovani R, Rodella S, Zoppini G, Zenari L, Cigolini M, Falezza G, Arcaro G. Relations between carotid artery wall thickness and liver histology in subjects with nonalcoholic fatty liver disease. Diabetes Care. 2006;29(6):1325–1330. doi: 10.2337/dc06-0135. [DOI] [PubMed] [Google Scholar]
- 2.Edmison J, McCullough AJ. Pathogenesis of non-alcoholic steatohepatitis: human data. Clinics in liver disease. 2007;11(1):75–104. doi: 10.1016/j.cld.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Targher G. Associations between liver histology and early carotid atherosclerosis in subjects with nonalcoholic fatty liver disease. Hepatology. 2005;42(4):974–975. doi: 10.1002/hep.20894. discussion 975. [DOI] [PubMed] [Google Scholar]
- 4.Larter CZ, Yeh MM, Haigh WG, Van Rooyen DM, Brooling J, Heydet D, Nolan CJ, Teoh NC, Farrell GC. Dietary modification dampens liver inflammation and fibrosis in obesity-related fatty liver disease. Obesity. 2013;21(6):1189–1199. doi: 10.1002/oby.20123. [DOI] [PubMed] [Google Scholar]
- 5.Navab M, Berliner JA, Subbanagounder G, Hama S, Lusis AJ, Castellani LW, Reddy S, Shih D, Shi W, Watson AD, Van Lenten BJ, Vora D, Fogelman AM. HDL and the inflammatory response induced by LDL-derived oxidized phospholipids. Arterioscler., Thromb., Vasc. Biol. 2001;21(4):481–488. doi: 10.1161/01.atv.21.4.481. [DOI] [PubMed] [Google Scholar]
- 6.Riwanto M, Rohrer L, Roschitzki B, Besler C, Mocharla P, Mueller M, Perisa D, Heinrich K, Altwegg L, von Eckardstein A, Luscher TF, Landmesser U. Altered activation of endothelial anti-and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation. 2013;127(8):891–904. doi: 10.1161/CIRCULATIONAHA.112.108753. [DOI] [PubMed] [Google Scholar]
- 7.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 2007;117(3):746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidsson P, Hulthe J, Fagerberg B, Camejo G. Proteomics of apolipoproteins and associated proteins from plasma high-density lipoproteins. Arterioscler., Thromb., Vasc. Biol. 2010;30(2):156–163. doi: 10.1161/ATVBAHA.108.179317. [DOI] [PubMed] [Google Scholar]
- 9.Toth PP, Barter PJ, Rosenson RS, Boden WE, Chapman MJ, Cuchel M, D’Agostino RB, Davidson MH, Davidson WS, Heinecke JW, Karas RH, Kontush A, Krauss RM, Miller M, Rader DJ. High-density lipoproteins: a consensus statement from the National Lipid Association. J. Clin. Lipidol. 2013;7(5):484–525. doi: 10.1016/j.jacl.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Smith JD. Myeloperoxidase, inflammation, and dysfunctional high-density lipoprotein. J. Clin. Lipidol. 2010;4(5):382–388. doi: 10.1016/j.jacl.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Wu Z, Riwanto M, Gao S, Levison BS, Gu X, Fu X, Wagner MA, Besler C, Gerstenecker G, Zhang R, Li XM, DiDonato AJ, Gogonea V, Tang WH, Smith JD, Plow EF, Fox PL, Shih DM, Lusis AJ, Fisher EA, DiDonato JA, Landmesser U, Hazen SL. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J. Clin. Invest. 2013;123(9):3815–3828. doi: 10.1172/JCI67478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khovidhunkit W, Memon RA, Feingold KR, Grunfeld C. Infection and inflammation-induced proatherogenic changes of lipoproteins. J. Infect. Dis. 2000;181(Suppl 3):S462–S472. doi: 10.1086/315611. [DOI] [PubMed] [Google Scholar]
- 13.Yu C, Xu C, Xu L, Yu J, Miao M, Li Y. Serum proteomic analysis revealed diagnostic value of hemoglobin for nonalcoholic fatty liver disease. J. Hepatol. 2012;56(1):241–247. doi: 10.1016/j.jhep.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui MS, Fuchs M, Idowu MO, Luketic VA, Boyett S, Sargeant C, Stravitz RT, Puri P, Matherly S, Sterling RK, Contos M, Sanyal AJ. Severity of nonalcoholic fatty liver disease and progression to cirrhosis are associated with atherogenic lipoprotein profile. Clin. Gastroenterol. Hepatol. 2015;13(5):1000–1008. e3. doi: 10.1016/j.cgh.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan LR, Wang DX, Liu H, Zhang XX, Zhao H, Hua L, Xu P, Li YS. A pro-atherogenic HDL profile in coronary heart disease patients: an iTRAQ labelling-based proteomic approach. PLoS One. 2014;9(5):e98368. doi: 10.1371/journal.pone.0098368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mange A, Goux A, Badiou S, Patrier L, Canaud B, Maudelonde T, Cristol JP, Solassol J. HDL proteome in hemodialysis patients: a quantitative nanoflow liquid chromatography-tandem mass spectrometry approach. PLoS One. 2012;7(3):e34107. doi: 10.1371/journal.pone.0034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holzer M, Trieb M, Konya V, Wadsack C, Heinemann A, Marsche G. Aging affects high-density lipoprotein composition and function. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2013;1831(9):1442–1448. doi: 10.1016/j.bbalip.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronsein GE, Pamir N, von Haller PD, Kim DS, Oda MN, Jarvik GP, Vaisar T, Heinecke JW. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J. Proteomics. 2015;113:388–399. doi: 10.1016/j.jprot.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A, Ong JP, Goodman Z, Chandhoke V. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology. 2005;42(3):665–674. doi: 10.1002/hep.20838. [DOI] [PubMed] [Google Scholar]
- 20.Bell LN, Theodorakis JL, Vuppalanchi R, Saxena R, Bemis KG, Wang M, Chalasani N. Serum proteomics and biomarker discovery across the spectrum of nonalcoholic fatty liver disease. Hepatology. 2010;51(1):111–120. doi: 10.1002/hep.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al Rifai M, Silverman MG, Nasir K, Budoff MJ, Blankstein R, Szklo M, Katz R, Blumenthal RS, Blaha MJ. The association of nonalcoholic fatty liver disease, obesity, and metabolic syndrome, with systemic inflammation and subclinical atherosclerosis: the Multi-Ethnic Study of Atherosclerosis (MESA) Atherosclerosis. 2015;239(2):629–633. doi: 10.1016/j.atherosclerosis.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rachdaoui N, Austin L, Kramer E, Previs MJ, Anderson VE, Kasumov T, Previs SF. Measuring proteome dynamics in vivo: as easy as adding water? Mol. Cell. Proteomics. 2009;8(12):2653–2663. doi: 10.1074/mcp.M900026-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasumov T, Ilchenko S, Li L, Rachdaoui N, Sadygov RG, Willard B, McCullough AJ, Previs S. Measuring protein synthesis using metabolic (2)H labeling, high-resolution mass spectrometry, and an algorithm. Anal. Biochem. 2011;412(1):47–55. doi: 10.1016/j.ab.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Willard B, Rachdaoui N, Kirwan JP, Sadygov RG, Stanley WC, Previs S, McCullough AJ, Kasumov T. Plasma proteome dynamics: analysis of lipoproteins and acute phase response proteins with 2H2O metabolic labeling. Mol. Cell. Proteomics. 2012;11(7):M111. doi: 10.1074/mcp.M111.014209. 014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasumov T, Dabkowski ER, Shekar KC, Li L, Ribeiro RF, Jr, Walsh K, Previs SF, Sadygov RG, Willard B, Stanley WC. Assessment of cardiac proteome dynamics with heavy water: slower protein synthesis rates in interfibrillar than subsarcolemmal mitochondria. Am. J. Physiol Heart Circ Physiol. 2013;304(9):H1201–H1214. doi: 10.1152/ajpheart.00933.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shekar KC, Li L, Dabkowski ER, Xu W, Ribeiro RF, Jr, Hecker PA, Recchia FA, Sadygov RG, Willard B, Kasumov T, Stanley WC. Cardiac mitochondrial proteome dynamics with heavy water reveals stable rate of mitochondrial protein synthesis in heart failure despite decline in mitochondrial oxidative capacity. J. Mol. Cell. Cardiol. 2014;75:88–97. doi: 10.1016/j.yjmcc.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasumov T, Li L, Li M, Gulshan K, Kirwan JP, Liu X, Previs S, Willard B, Smith JD, McCullough A. Ceramide as a mediator of non-alcoholic Fatty liver disease and associated atherosclerosis. PLoS One. 2015;10(5):e0126910. doi: 10.1371/journal.pone.0126910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasumov T, Willard B, Li L, Li M, Conger H, Buffa JA, Previs S, McCullough A, Hazen SL, Smith JD. 2H2O–based high-density lipoprotein turnover method for the assessment of dynamic high-density lipoprotein function in mice. Arterioscler., Thromb., Vasc. Biol. 2013;33(8):1994–2003. doi: 10.1161/ATVBAHA.113.301700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah V, Herath K, Previs SF, Hubbard BK, Roddy TP. Headspace analyses of acetone: a rapid method for measuring the 2H–labeling of water. Anal. Biochem. 2010;404(2):235–237. doi: 10.1016/j.ab.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Lassman ME, McLaughlin TM, Somers EP, Stefanni AC, Chen Z, Murphy BA, Bierilo KK, Flattery AM, Wong KK, Castro-Perez JM, Hubbard BK, Roddy TP. A rapid method for cross-species quantitation of apolipoproteins A1, B48 and B100 in plasma by ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2012;26(2):101–108. doi: 10.1002/rcm.5296. [DOI] [PubMed] [Google Scholar]
- 32.Walsh MT, Atkinson D. Solubilization of low-density lipoprotein with sodium deoxycholate and recombination of apoprotein B with dimyristoylphosphatidylcholine. Biochemistry. 1983;22(13):3170–3178. doi: 10.1021/bi00282a021. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat. Genet. 1993;3(3):266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 35.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 36.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82(6):2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 39.Navab M, Hama SY, Hough GP, Subbanagounder G, Reddy ST, Fogelman AM. A cell-free assay for detecting HDL that is dysfunctional in preventing the formation of or inactivating oxidized phospholipids. J. Lipid Res. 2001;42(8):1308–1317. [PubMed] [Google Scholar]
- 40.Ji X, Xu H, Zhang H, Hillery CA, Gao HQ, Pritchard KA., Jr Anion exchange HPLC isolation of high-density lipoprotein (HDL) and on-line estimation of proinflammatory HDL. PLoS One. 2014;9(3):e91089. doi: 10.1371/journal.pone.0091089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rensen SS, Slaats Y, Nijhuis J, Jans A, Bieghs V, Driessen A, Malle E, Greve JW, Buurman WA. Increased hepatic myeloperoxidase activity in obese subjects with nonalcoholic steatohepatitis. Am. J. Pathol. 2009;175(4):1473–1482. doi: 10.2353/ajpath.2009.080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bio-informatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 43.Shah AS, Tan L, Long JL, Davidson WS. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 2013;54(10):2575–2585. doi: 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan D, Navab M, Bruce C, Fogelman AM, Jiang XC. PLTP deficiency improves the anti-inflammatory properties of HDL and reduces the ability of LDL to induce monocyte chemotactic activity. J. Lipid Res. 2004;45(10):1852–1858. doi: 10.1194/jlr.M400053-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Moerland M, Samyn H, van Gent T, Jauhiainen M, Metso J, van Haperen R, Grosveld F, van Tol A, de Crom R. Atherogenic, enlarged, and dysfunctional HDL in human PLTP/ apoA-I double transgenic mice. J. Lipid Res. 2007;48(12):2622–2631. doi: 10.1194/jlr.M700020-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Recinos A, 3rd, Carr BK, Bartos DB, Boldogh I, Carmical JR, Belalcazar LM, Brasier AR. Liver gene expression associated with diet and lesion development in atherosclerosis-prone mice: induction of components of alternative complement pathway. Physiol. Genomics. 2004;19(1):131–142. doi: 10.1152/physiolgenomics.00146.2003. [DOI] [PubMed] [Google Scholar]
- 47.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 2006;116(3):571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alwaili K, Bailey D, Awan Z, Bailey SD, Ruel I, Hafiane A, Krimbou L, Laboissiere S, Genest J. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2012;1821(3):405–415. doi: 10.1016/j.bbalip.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Tolle M, Huang T, Schuchardt M, Jankowski V, Prufer N, Jankowski J, Tietge UJ, Zidek W, van der Giet M. High-density lipoprotein loses its anti-inflammatory capacity by accumulation of pro-inflammatory-serum amyloid A. Cardiovasc. Res. 2012;94(1):154–162. doi: 10.1093/cvr/cvs089. [DOI] [PubMed] [Google Scholar]
- 50.Vivekanandan-Giri A, Slocum JL, Byun J, Tang C, Sands RL, Gillespie BW, Heinecke JW, Saran R, Kaplan MJ, Pennathur S. High density lipoprotein is targeted for oxidation by myeloperoxidase in rheumatoid arthritis. Ann. Rheum. Dis. 2013;72(10):1725–1731. doi: 10.1136/annrheumdis-2012-202033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rensen SS, Slaats Y, Driessen A, Peutz-Kootstra CJ, Nijhuis J, Steffensen R, Greve JW, Buurman WA. Activation of the complement system in human nonalcoholic fatty liver disease. Hepatology. 2009;50(6):1809–1817. doi: 10.1002/hep.23228. [DOI] [PubMed] [Google Scholar]
- 52.Zambon A, Gervois P, Pauletto P, Fruchart JC, Staels B. Modulation of hepatic inflammatory risk markers of cardiovascular diseases by PPAR-alpha activators: clinical and experimental evidence. Arterioscler., Thromb., Vasc. Biol. 2006;26(5):977–986. doi: 10.1161/01.ATV.0000204327.96431.9a. [DOI] [PubMed] [Google Scholar]
- 53.Zandbergen F, Plutzky J. PPARalpha in atherosclerosis and inflammation. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2007;1771(8):972–982. doi: 10.1016/j.bbalip.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mogilenko DA, Kudriavtsev IV, Shavva VS, Dizhe EB, Vilenskaya EG, Efremov AM, Perevozchikov AP, Orlov SV. Peroxisome proliferator-activated receptor alpha positively regulates complement C3 expression but inhibits tumor necrosis factor alpha-mediated activation of C3 gene in mammalian hepatic-derived cells. J. Biol. Chem. 2013;288(3):1726–1738. doi: 10.1074/jbc.M112.437525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGillicuddy FC, de la Llera Moya M, Hinkle CC, Joshi MR, Chiquoine EH, Billheimer JT, Rothblat GH, Reilly MP. Inflammation impairs reverse cholesterol transport in vivo. Circulation. 2009;119(8):1135–1145. doi: 10.1161/CIRCULATIONAHA.108.810721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gervois P, Kleemann R, Pilon A, Percevault F, Koenig W, Staels B, Kooistra T. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-alpha activator fenofibrate. J. Biol. Chem. 2004;279(16):16154–16160. doi: 10.1074/jbc.M400346200. [DOI] [PubMed] [Google Scholar]
- 57.Diradourian C, Girard J, Pegorier JP. Phosphorylation of PPARs: from molecular characterization to physiological relevance. Biochimie. 2005;87(1):33–38. doi: 10.1016/j.biochi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Blanquart C, Barbier O, Fruchart JC, Staels B, Glineur C. Peroxisome proliferator-activated receptor alpha (PPARalpha) turnover by the ubiquitin-proteasome system controls the ligand-induced expression level of its target genes. J. Biol. Chem. 2002;277(40):37254–37259. doi: 10.1074/jbc.M110598200. [DOI] [PubMed] [Google Scholar]
- 59.Pourcet B, Pineda-Torra I, Derudas B, Staels B, Glineur C. SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR. J. Biol. Chem. 2010;285(9):5983–5992. doi: 10.1074/jbc.M109.078311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.