Abstract
Photobleaching of fluorescent proteins (FPs) is a major limitation to their use in advanced microscopy, and improving photostability remains highly challenging due to limited understanding of its molecular mechanism. Here we discovered a new mechanism to increase FP photostability. Cysteine oxidation, implicated in only photobleaching before, was found to drastically enhance FP photostability to the contrary. We generated a far-red FP mStable by introducing a cysteine proximal to the chromophore. Upon illumination, this cysteine was oxidized to sulfinic and sulfonic acids, enabling mStable more photostable than its ancestor mKate2 by 12-fold and surpassing other far-red FPs. mStable outperformed in laser scanning confocal imaging and super-resolution structured illumination microscopy. Moreover, photosensitization to oxidize a cysteine similarly introduced in another FP mPlum also increased its photostability by 23-fold. This post-folding cysteine sulfoxidation cannot be simply substituted by the isosteric aspartic acid, representing a unique mechanism valuable for engineering better photostability into FPs.
Fluorescent proteins (FPs) have enabled novel approaches for studying biology and spurred advanced microscopies such as single molecule and super-resolution imaging1,2. All FPs are subjected to irreversible photobleaching, and therefore photostability is a critical property of consideration when choosing a FP for optical microscopy, which limits the duration of imaging, the number of images that can be acquired, and the resolution of single molecule-based technologies3. In contrast to substantial progress made in altering the wavelength and brightness of FPs4, improving their photostability has limited success for few FPs through directed evolution5–9. The general challenge lies in the presence of multiple possible pathways to deactivate the fluorescence of the chromophore, and molecular mechanisms to prevent chromophore photodestruction remain largely unknown7,10–13. Experimental and theoretical studies have concluded that O2 plays an important role in photobleaching of FPs14–18. In particular, oxidation of cysteine and methionine side chains proximal to the chromophore by singlet oxygen has been shown to lock the chromophore in nonfluorescent state13. Here we report mStable, a far-red FP that is over 12-fold more photostable than its ancestor and surpasses existing far-red FPs. We discovered a novel mechanism of enhancing photostability: oxidation of a cysteine residue close to the chromophore, instead of photobleaching, markedly increases the photostability of mStable.
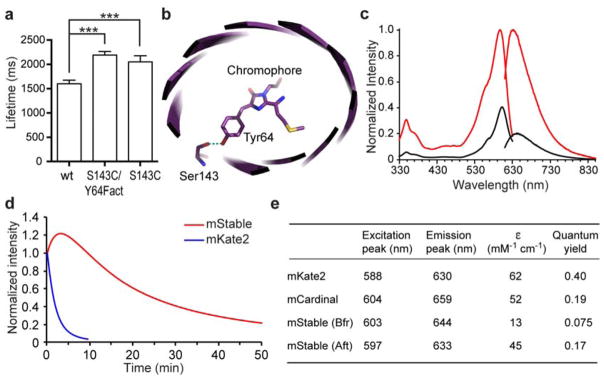
In a previous effort to increase the photon output of FPs19, we found that an mKate2 mutant (S143C/Y64Fact) showed an increased lifetime on the single molecule level than the wild-type (wt) mKate2 (Figure 1a). Side chain of Ser143 hydrogen bonds with the hydroxybenzylidene phenolate group of the chromophore to stabilize the cis fluorescent conformation of mKate2 (Figure 1b)20; that mutation of this Ser to a weaker hydrogen-bonding Cys increased the lifetime is counterintuitive. This observation promoted us to generate and characterize the mKate2 S143C mutant, which also showed increased lifetime in single molecule analysis (Figure 1a). Compared to the wt mKate2, the S143C mutation red shifted the excitation and emission peak to 603 nm and 644 nm, respectively (Figure 1c). When photobleaching was measured on protein micro-droplets using the standard wide-field continuous illumination by an arc-lamp3, mKate2 showed a t1/2 of 1.4 min (Figure 1d). In contrast, t1/2 of the S143C mutant was extended to 16.7 min, indicating a dramatic 12-fold increase of photostability. Gel filtration verified that the S143C mutant remained monomeric (Supplementary Figure S1). We thus named this S143C mutant as mStable for its increased photostability.
Figure 1. mStable, a far-red fluorescent protein showing unusual photostability.
(a) Single molecule measurements show that the lifetimes (mean ± s.e.) of an mKate2 mutant S143C/Y64Fact (2190 ± 73 ms, n = 364) and mutant S143C (2052 ± 125 ms, n = 37) were longer than that of the wild type mKate2 (1600 ± 74 ms, n = 193). *** P < 0.0001, unpaired t test. Fact, unnatural amino acid p-acetyl-L-phenylalanine. (b) Crystal structure of mKate (PDB ID 3BXB)20 showing Ser143 hydrogen bonding with the hydroxybenzylidene phenolate group of the chromophore. mKate2 is derived from mKate with mutations of V45A, S158A and K231R 26. (c) Fluorescence excitation and emission spectra of mStable before (black line) and after (red line) photoactivation. Emission spectra were taken at the excitation wavelength 600 nm, and emission was monitored at 640 nm for excitation spectra. (d) Photobleaching kinetics of purified mStable and mKate2 in aqueous microdroplets under arc-lamp illumination with 580/20 nm excitation filter. mKate2 t1/2 = 1.4 min; mStable t1/2 = 16.7 min. Note in this work, the t1/2s for mStable were all measured from the peak of photoactivation to 50% of the peak intensity. Each curve is the mean of three independent experiments. (e) Characteristics of mStable in comparison with mKate2 and mCardinal. Bfr and Aft indicate before and after photoactivation, respectively. ε, extinction coefficient.
Photobleaching measurements of mStable indicate there was a photoactivation step upon initial illumination (Figure 1d). We measured the fluorescence spectra, extinction coefficient and quantum yield of mStable before and after photoactivation, and compared them with those of the parent mKate2 and mCardinal21, another far-red FP with improved photostability (Figure 1c, 1e, Supplementary Figure S2). After photoactivation, the extinction coefficient and quantum yield of mStable both increased. The brightness of mStable after photoactivation is 31% of mKate2 and 77% of mCardinal, but is higher than many other far-red FPs including TagRFP657, E2-Crimson, eqFP670, IFP1.4 and iRFP21.
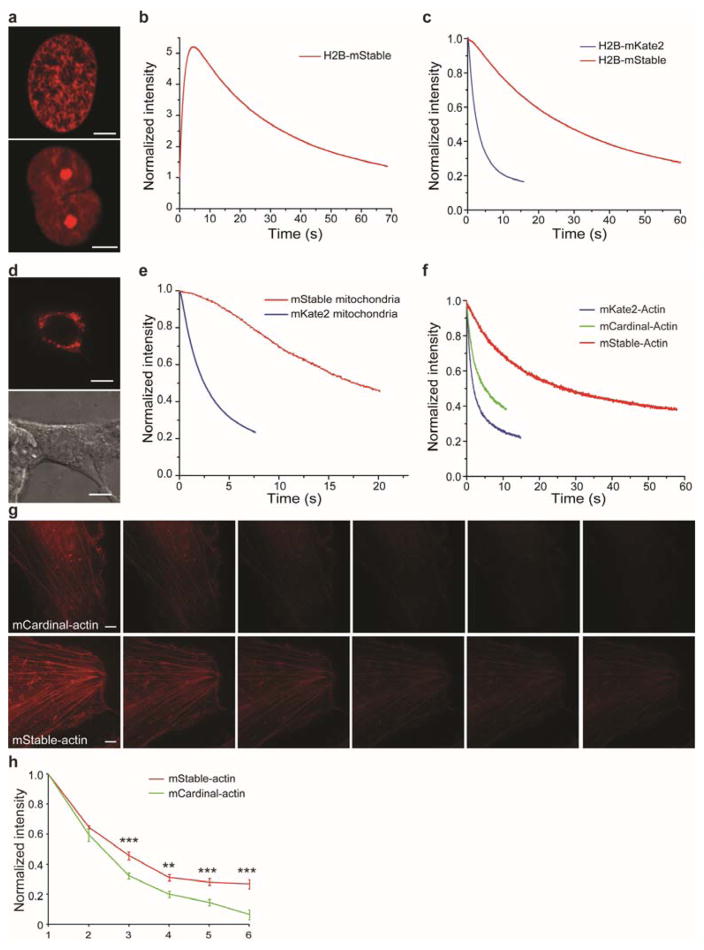
We next evaluated the photostability of mStable inside live mammalian cells. mStable was fused to the C-terminus of human histone H2B, and the resultant H2B-mStable was expressed in HEK293T cells. Live-cell imaging confirmed that mStable does not interfere with localization of histone (Figure 2a). Imaging and photobleaching was performed using laser-scanning confocal microscopy on H2B-mStable expressing cells. Photoactivation of mStable was also observed in live cells (Figure 2b) as seen in protein micro-droplets. For photobleaching, the same-emission-intensity method was used, in which different fluorescent proteins were excited with light of adjusted intensities to achieve the same emission intensity at the outset for a fair comparison3,7,21. For mStable, the same emission intensity was matched at the peak of photoactivation. In comparison to the similarly fused and expressed H2B-mKate2, the t1/2 increased from 2.6 s for mKate2 to 27.1 s for mStable (Figure 2c). mStable was also expressed in HEK293T cells at the mitochondrion by using a mitochondrial localization signal (Figure 2d), and laser-scanning confocal imaging measured t1/2 of 17.5 s for mStable and 2.4 s for mKate2 (Figure 2e). We also fused mStable and mKate2 to the N-terminus of human actin. Transient expression of these constructs in HeLa cells showed that mStable does not interrupt actin expression and localization (Figure 2g). Laser-scanning confocal imaging of these cells measured t1/2 of 2.1 s for mKate2-actin, while 25.6 s was obtained for mStable-actin (Figure 2f). These results indicate that mStable is 7–12 fold more photostable than its ancestor mKate2 for mammalian cell imaging.
Figure 2. mStable is more photostable than mKate2 and mCardinal for imaging in live mammalian cells.
(a) Confocal fluorescence images of mStable fused to human histone H2B expressed in HEK293T cells. Cells at two different phases are shown. Scale bar: 5 μm. (b) Imaging of mStable fused to human histone H2B expressed in HEK293T cells showed photoactivation followed by photobleaching (see also Figure S3). (c) Photobleaching kinetics of mStable and mKate2 fused to human histone H2B expressed in HEK293T cells. Each curve is the mean of three independent experiments. H2B-mKate2 t1/2 = 2.6 s; H2B-mStable t1/2 = 27.1 s. (d) Confocal fluorescence image (top) and differential interference contrast image (bottom) of HEK293T cells expressing mStable at mitochondria. Scale bar: 10 μm. (e) Photobleaching kinetics of mStable and mKate2 targeted to mitochondria. Each curve is the mean of three independent experiments. mKate2 t1/2 = 2.4 s; mStable t1/2 = 17.5 s. (f) Photobleaching kinetics of mStable, mKate2 and mCardinal fused to human actin expressed in HeLa cells. Each curve is the mean of three independent experiments. mKate2-actin t1/2 = 2.1 s; mCardinal t1/2 = 4.7 s; mStable t1/2 = 25.6 s. (g) Structured-illumination microscopy (SIM) images of mCardinal and mStable fused to actin in HeLa cells. A time series of SIM images were taken using the same settings for imaging both samples, and each SIM image was reconstructed from 100 raw images. Scale bar: 5 μm. (h) Photobleaching of mCardinal and mStable in the time series of SIM images. The mean intensity of each SIM image was normalized to that of the respective first SIM image. ** P < 0.01; *** P < 0.001, unpaired t test.
mCardinal is a recently developed far-red FP with overall photostability superior to other far-red FPs21. To compare mStable with mCardinal, we fused them to the N-terminus of human actin and expressed the proteins in HeLa cells. Photobleaching using laser-scanning microscopy measured t1/2 of 4.7 s for mCardinal-actin (Figure 2f), which is indeed better than mKate2-actin (2.1 s). In contrast, mStable-actin yielded 25.6 s for t1/2, 5.4-fold more stable than mCardinal-actin.
To further evaluate whether the high photostability of mStable can be beneficial for challenging imaging conditions, we performed super-resolution structured illumination microscopy (SIM) on mStable-labeled actin in live mammalian cells. SIM illuminates the sample in a series of orientations by laser light passing through a movable optical grating, creating Moiré fringes through which super-resolution information can be captured to reconstruct images with resolution beyond the diffraction limit22,23. This advanced wide-field imaging technology thus demands high photostability of the fluorophore to acquire a large number of images for reconstruction. mStable-actin and mCardinal-actin were respectively expressed in HeLa cells, and the cells were continuously imaged over time using SIM under the same conditions and settings. The samples were illuminated from 5 rotations with 5 phases for each rotation, and 4 images were averaged into one. A total of 100 raw images were thus acquired to reconstruct each SIM image. The resultant SIM images showed actin bundles in high clarity for both mStaple-actin and mCardinal-actin in the initial images (Figure 2g). As SIM imaging was repeated, the fluorescent intensity of mCardinal-actin was getting too weak to clearly discern the actin structure at repeat three, while the mStable-actin bundle remained clear in repeat six (Figure 2g). Normalized intensity decrease of SIM images over time for these two samples (Figure 2h) indicates that mStable lasted significantly longer than mCardinal. These results demonstrate that mStable fulfills the stringent SIM imaging requirements in live mammalian cells and performs better than mCardinal.
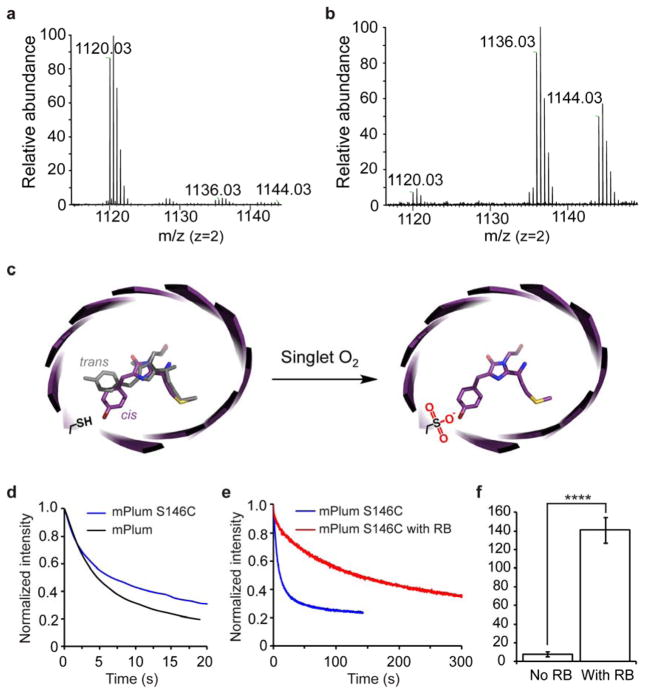
What is the molecular mechanism underlying the unusual photostability of mStable? The only difference between mStable and the parental mKate2 is the S143C mutation. Cys is structurally similar to Ser, but Cys, in contrast to Ser, has the tendency to be oxidized. However, oxidation of Cys side chain has been reported to be involved in photobleaching FPs only but never in enhancing their photostabilities13. We noticed there was a photoactivation step when photobleaching mStable with either continuous arc-lamp (Figure 1d) or intermittent laser-scanning illumination (Supplementary Figure S3), suggesting a light-induced modification of the chromophore or its nearby residues. We therefore characterized Cys143 before and after photoactivation of mStable using high-resolution Fourier transform ion trap mass spectrometry (FTMS). Before photoactivation, Cys143 of mStable stayed as the cysteine thiol (-SH) form (Figure 3a, Supplementary Figure S4–5). In contrast, after photoactivation, the side chain of Cys143 mainly presented as the di-oxidized sulfinic acid (63%) and tri-oxidized sulfonic acid (37%) forms (Figure 3b, Supplementary Figure S4–5). FTMS showed there was no change to the chromophore of mStable after photoactivation. In addition, mKate2, which has Ser143, showed no photoactivation and poorer photostability under the same illumination conditions used for mStable (Figure 1d, 2c, 2e, 2f). Moreover, the presence of sodium azide, a specific singlet oxygen quencher, significantly decreased the photostability of mStable by 21% (Supplementary Figure S6). Based on these results, we think that upon illumination the chromophore of mStable generates singlet oxygen, which oxidizes the proximal Cys143 thiol to sulfinic and sulfonic acids. The resultant sulfinic and sulfonic acids stabilize the chromophore in the cis fluorescent conformation to enhance the photostability (Figure 3c).
Figure 3. Cysteine oxidation enhances photostability.
(a) High resolution FTMS analysis of mStable purified and kept in dark. The observed peak with monoisotopic masses of 1120.03 Da corresponds to the Cys143-containing peptide TLGWEACTETLYPADGG LEGR in which the Cys is in the thiol form (expected [M+H]2+ = 1120.0271 Da). (b) High resolution FTMS analysis of the photoactivated mStable. Monoisotopic masses were observed corresponding to the Cys143-containing peptide in which Cys143 was di-oxidized (expected [M+H]2+ = 1136.0232 Da, measured 1136.02 Da) or tri-oxidized (expected [M+H]2+ = 1144.0211 Da, measured 1144.02 Da). Notably, intensity of the thiol cysteine peak (expected [M+H]2+ = 1120.0271 Da, measured 1120.03 Da) decreased concomitantly in the photoactivated mStable. Tandem MS/MS analyses of these peptides confirming the Cys143 oxidation states are shown in Supplementary Figure S4–5. (c) Proposed mechanism to explain the unusual photostability of mStable. (d) Photobleaching kinetics under laser illumination shows that introduction of Cys into mPlum at the corresponding position (S146C) increased photostability by 20%. wt mPlum t1/2 = 5 s; mPlum(S146C) t1/2 = 6 s. (e) Photobleaching kinetics of mPlum(S146C) under laser illumination with and without Rose Bengal (RB) photosensitization. After RB-mediated oxidation, mPlum(S146C) was separated using size chromatography for photobleaching measurement. Each curve is the mean of four independent experiments. (f) Photostability of mPlum(S146C) increased 23-fold after oxidation via RB sensitization. No RB t1/2 = 6 s; with RB t1/2 = 140 s. **** P < 0.0001, unpaired t test.
Cys143 oxidation represents a unique mechanism to enhance the photostability of mStable. In mKate, Ser143 is known to hydrogen bond with the hydroxybenzylidene phenolate group of the chromophore to facilitate the chromophore switching to the cis conformation, the fluorescent state20. In mStable before photoactivation, mutation of Ser143 to Cys abolishes this hydrogen bonding ability, so that the chromophore can populate in either the cis (fluorescent state, purple in Figure 3c) or the trans (nonfluorescent state, grey in Figure 3c) conformation. After mStable is photoactivated, the sulfinic or sulfonic acid formed at Cys143 site may become a stronger hydrogen bonding partner than Ser and Cys in stabilizing the cis chromophore. In addition, Cys oxidation extends the side chain length at site 143, and this extension may prevent the chromophore from switching back to the nonfluorescent trans conformation via steric hindrance, effect of which is impossible through the shorter Ser or Cys 20. In an attempt to mimic sulfinic acid isosterically while retaining its hydrogen-bonding ability, we mutated Cys143 to Asp in mStable. Interestingly, the mStable(C143D) mutant was nonfluorescent when expressed in mammalian cells (Supplementary Figure S7). Circular dichroism analyses of mStable(C143D) purified from E. coli showed that the red chromophore did not form and the β-barrel structure was disrupted (Supplementary Figure S8), suggesting that Asp143 interferes with protein folding and chromophore formation. This observation highlights the uniqueness of sulfinic/sulfonic acid generated via post-folding modification of Cys143 in that it evades structural perturbation that could be caused by the isosteric Asp residue directly incorporated through protein translation. Further directed evolution of the mStable (C143D) mutant may generate variants that are capable of folding/maturing and retain the high photostability of mStable.
Would a Cys positioned at a corresponding position in a different far-red FP have similar photostabilizing effect? We mutated Ser146 into Cys in mPlum24, and found that the mPlum(S146C) mutant showed 20% increase in t1/2 over the wt mPlum (Figure 3d). This extent of increase was less than that measured for mStable. We reasoned that mPlum might not efficiently oxidize Cys146 via singlet oxygen generated by its chromophore. We thus incubated mPlum(S146C) with Rose Bengal, a small molecule photosensitizer for generating singlet oxygen. After Rose Bengal photosensitization, oxidation of Cys146 side chain to sulfinic acid and sulfonic acid was confirmed by MS/MS (Supplementary Figure S10), and the extent of Cys146 oxidation was increased from 36% to 91% (Supplementary Figure S9). Consistently, the t1/2 of mPlum(S146C) increased from 6 s to 140 s upon Rose Bengal photosensitization (Figure 3e, 3f). This 23-fold increase in photostability of mPlum(S146C) (but not of wt mPlum) suggests that Cys oxidation, not Cys alone, is capable of and necessary for enhancing photostability, further supporting our proposed mechanism.
In summary, we report here a far-red FP mStable with photostability surpassing existing far-red FPs. Cysteine oxidation, a damaging modification reported in the past, transforms into a beneficial mechanism for enhancing the photostability of far-red FPs. It also represents a unique protein modification in that it occurs after protein folding and at a site (buried within the β-barrel) not readily accessible by modifying enzymes. Highly photostable FPs should be valuable for advanced microscopic applications. Cys oxidation may lead to new avenues (such as Cys scanning) for addressing the challenge of engineering better photostability into various FPs.
METHODS
Mass spectrometry
mStable samples kept in dark or treated with light activation were digested by trypsin (200 ng, Sigma, Proteomics grade) overnight at 37 °C in 50 mM ammonium bicarbonate buffer. The resulting peptides were analyzed by high resolution Fourier transform ion trap MS on a Thermo LTQ (linear trap quadrupole)-Orbitrap XL mass spectrometer (ThermoFisher) as previously described25. mPlum samples untreated or treated with rose bengal photosensitization were dissolved in 8 M Urea (pH 8.5), reduced with 2 mM DTT for 20 min, and alkylated with 10 mM iodoacetamide for 15 min. Sample was diluted to 2 M urea with 100 mM Tris (pH 8.5), and digested with trypsin overnight at 37 °C. Digestion was terminated with formic acid at final concentration 5%. Digested peptides were analyzed with 40 min HPLC gradient and flow rate at 300 nL min−1. Mass spec data was collected in data-dependent mode on Orbitrap Elite (ThermoFisher). Full scans (400–1600 m/z) were performed in Orbitrap. MS2 scans were triggered for top 10 peaks in LTQ and dynamic exclusion time of 18 seconds was used. Raw data was searched against mStable or mPlum and regular contaminant protein sequences. Precursor ion tolerance and fragment ion tolerance were set to 4.5 ppm and 0.5 Da, separately. Two miss cleavage sites were allowed, and oxidation of methionine was used as variable modification. Identification results were filtered with threshold (protein False Discovery Rate less than 1%).
Photobleaching measurements
Arc-lamp photobleaching of purified protein in aqueous microdroplets in PBS (pH = 7.4) were performed exactly as previously described3. Laser photobleaching was performed on a Zeiss light scanning microscope (LSM 700). Live-cell photobleaching was performed on live HeLa or HEK293T cells. Bleaching of purified proteins was performed on protein samples deposited onto a nickel-nitrilotriacetic acid coated glass slide. Bleaching was achieved in conjunction with the acquisition of a series of images over time. Details are provided in Supporting Information.
Supplementary Material
Acknowledgments
Lei Wang wishes to dedicate this paper to the memory of his mentor Dr. Roger Y. Tsien, who passed away on August 24, 2016. We thank Paul A. Steinbach for help with arc-lamp photobleaching in aqueous microdroplets, Wolfgang Fischer for help with mass spectrometry and Michael Z. Lin for providing plasmids pmCardinal-Actin-C-18 and pmTagRFP-T-H2B-6. L.W. acknowledges support from US National Institutes of Health (1R01GM118384-01).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supporting methods for molecular cloning and mutagenesis, protein expression and purification, gel filtration chromatography, cell culture, absorption and fluorescence spectroscopy, extinction coefficient and quantum yield measurement, single molecule measurement, photobleaching measurements, structured-illumination microscopy, and CD spectra measurement. Supplementary table 1 and figures S1–S10. This material is available free of charge via the Internet.
References
- 1.Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 4.Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. J Cell Sci. 2011;124:157–160. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mena MA, Treynor TP, Mayo SL, Daugherty PS. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat Biotechnol. 2006;24:1569–1571. doi: 10.1038/nbt1264. [DOI] [PubMed] [Google Scholar]
- 6.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredj A, Pasquier H, Demachy I, Jonasson G, Levy B, Derrien V, Bousmah Y, Manoussaris G, Wien F, Ridard J, Erard M, Merola F. The single T65S mutation generates brighter cyan fluorescent proteins with increased photostability and pH insensitivity. PLoS One. 2012;7:e49149. doi: 10.1371/journal.pone.0049149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAnaney TB, Zeng W, Doe CF, Bhanji N, Wakelin S, Pearson DS, Abbyad P, Shi X, Boxer SG, Bagshaw CR. Protonation, photobleaching, and photoactivation of yellow fluorescent protein (YFP 10C): a unifying mechanism. Biochemistry. 2005;44:5510–5524. doi: 10.1021/bi047581f. [DOI] [PubMed] [Google Scholar]
- 11.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 12.Dean KM, Lubbeck JL, Binder JK, Schwall LR, Jimenez R, Palmer AE. Analysis of red-fluorescent proteins provides insight into dark-state conversion and photodegradation. Biophys J. 2011;101:961–969. doi: 10.1016/j.bpj.2011.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan C, Adam V, Byrdin M, Ridard J, Kieffer-Jaquinod S, Morlot C, Arcizet D, Demachy I, Bourgeois D. Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein. J Am Chem Soc. 2013;135:15841–15850. doi: 10.1021/ja406860e. [DOI] [PubMed] [Google Scholar]
- 14.Greenbaum L, Rothmann C, Lavie R, Malik Z. Green fluorescent protein photobleaching: a model for protein damage by endogenous and exogenous singlet oxygen. Biol Chem. 2000;381:1251–1258. doi: 10.1515/BC.2000.153. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez-Banzo A, Nonell S, Hofkens J, Flors C. Singlet oxygen photosensitization by EGFP and its chromophore HBDI. Biophys J. 2008;94:168–172. doi: 10.1529/biophysj.107.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regmi CK, Bhandari YR, Gerstman BS, Chapagain PP. Exploring the diffusion of molecular oxygen in the red fluorescent protein mCherry using explicit oxygen molecular dynamics simulations. J Phys Chem B. 2013;117:2247–2253. doi: 10.1021/jp308366y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigorenko BL, Nemukhin AV, Polyakov IV, Khrenova MG, Krylov AI. A Light-Induced Reaction with Oxygen Leads to Chromophore Decomposition and Irreversible Photobleaching in GFP-Type Proteins. J Phys Chem B. 2015;119:5444–5452. doi: 10.1021/acs.jpcb.5b02271. [DOI] [PubMed] [Google Scholar]
- 18.Roy A, Carpentier P, Bourgeois D, Field M. Diffusion pathways of oxygen species in the phototoxic fluorescent protein KillerRed. Photochem Photobiol Sci. 2010;9:1342–1350. doi: 10.1039/c0pp00141d. [DOI] [PubMed] [Google Scholar]
- 19.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10:885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu J, Haynes RD, Corbel SY, Li P, Gonzalez-Gonzalez E, Burg JS, Ataie NJ, Lam AJ, Cranfill PJ, Baird MA, Davidson MW, Ng HL, Garcia KC, Contag CH, Shen K, Blau HM, Lin MZ. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 23.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takimoto JK, Dellas N, Noel JP, Wang L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol. 2011;6:733–743. doi: 10.1021/cb200057a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.