Abstract
Lung cancer is a leading cause of cancer death, and late diagnosis is one of the most important reasons for the high mortality rate. microRNAs (miRNAs) are key players in gene regulation and therefore in tumorigenesis. As far as lung carcinogenesis is concerned, miRNAs open novel fields in biomarker research, in diagnosis, and in therapy. In this review we focus on miR-21 in lung cancer and especially on how miR-21 is involved 1) as a biomarker in response or resistance to therapy or 2) as a therapeutic target.
Keywords: lung cancer, miR-21, therapeutic target, NSCLC, response to therapy
Introduction
Lung cancer remains the number one cause of cancer-related deaths in both men and women, accounting for more than 1.4 million deaths per year.1 It is classified into two main histological subtypes, non-small-cell lung cancer (NSCLC), which accounts for 85% of cases, and small-cell lung cancer (SCLC).2 Although significant improvements have been made toward the early detection of the disease and early diagnosis and treatment, the 5-year overall survival (OS) rate for patients with NSCLC has not improved markedly.3 High mortality is mainly due to late disease presentation at an advanced stage, due to tumor heterogeneity, limited understanding of lung cancer biology, and largely, due to the development of resistance to therapies. Thus, the identification of novel treatment strategies is critical and essential for lung cancer management. Moreover, there is an urgent need to find accurate biomarkers which could detect lung cancer with high sensitivity and specificity in the early state of disease.
microRNAs (miRNAs) are endogenous small noncoding RNAs (17–22 nucleotides), which seem to play a crucial role in tumorigenesis by regulating gene expression at the posttranscriptional level.4,5 The first miRNA was discovered in 1993 by Lee et al,6 and the term microRNA was first presented in 2001.7 Nowadays, more than 2,000 miRNAs have been identified in the human genome, and the vast majority of them are deregulated in cancer cells.4,8 The fact that each miRNA has the ability to target multiple genes within a pathway9 makes miRNAs one of the most abundant classes of regulatory genes in humans, regulating up to 30% of human protein coding genes.10,11 Due to their important role in many cancer types, miRNAs comprise some of the most attractive targets for therapeutic intervention in cancer today.
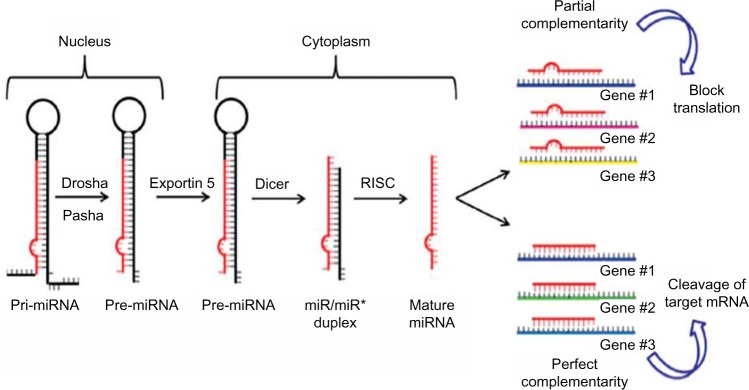
The biogenesis of miRNAs is a multistep process beginning in the nucleus where miRNAs are initially transcribed by RNA polymerase II into a precursor primary miRNA.12 Then the primary miRNA is processed in the nucleus by the RNase III enzyme Drosha and the protein Pasha/DGCR8 into pre-miRNAs (approximately 70-nucleotide).13 Subsequently, this pre-miRNA is transported out of the nucleus into the cytoplasm via Exportin-5,14 where Dicer1 cleaves the hairpin loop of pre-miRNA.15 The functional strand of the mature miRNA is then loaded together with Argonaute (Ago2) proteins16 into the RNA-induced silencing complex (RISC)17 and finally, the mature single-stranded miRNA is loaded into the RISC, which mediates the degradation or translation inhibition of target mRNA by binding to its seed sequence in the target mRNA’s 3′-UTR (Figure 1).18,19 miRNAs can be secreted extracellularly bound to lipoproteins20 or secreted in cell-derived extracellular vesicles such as exosomes as a method of cell-to-cell communication.21 Recently, Melo et al22 showed that breast cancer-associated exosomes contain miRNAs associated with the RISC-loading complex and that these exosome miRNAs display cell-independent capacity to process pre-miRNAs into mature miRNAs.
Figure 1.
Biogenesis of miRNAs.
Note: *Indicates that this is the precursor form of miRNA.
Abbreviations: miRNAs, microRNA; RISC, RNA-induced silencing complex.
miRNAs may act not only within cells, but also at other sites within the body via their circulation in peripheral blood and other body fluids.23 Several studies have shown that serum and plasma miRNAs (called circulating miRNAs) present great promise as novel noninvasive biomarkers for the early diagnosis of various cancers due to their ease of access and long-term stability.24–26 However, as the accurate quantification of circulating miRNAs was more challenging than expected recently, Ferracin et al27 used droplet digital PCR technology in order to assess the absolute levels of nine of the most abundant miRNAs (miR-320a, miR-21-5p, miR-378a-3p, miR-181a-5p, miR-3156-5p, miR-2110, miR-125a-5p, miR-425-5p, miR-766-3p) in serum and plasma of breast, colorectal, lung, and melanoma cancer patients and in healthy controls. According to the final data of this study, there is a clear indication that circulating miRNAs are present in the circulation at levels that can change according to the cancer status and that only one miRNA, namely miR-21-5p, was consistently increased in the plasma of all cancer patients.27
In this review we summarize recent advances in the identification and characterization of miRNAs in lung cancer and especially the role of miR-21 in lung cancer and how miR-21 may be used to facilitate patient diagnosis, prognosis, and response or resistance to therapy.
miRNAs and biology of lung cancer
A growing number of studies suggest an important involvement of miRNAs in various steps of lung carcinogenesis. Several miRNAs are involved in the regulation of cell cycle and lung cancer growth, including in apoptosis, angiogenesis, invasion, and metastasis by targeting different or similar genes.28–31
Apoptosis
The expression of B-cell lymphoma 2 (BCL-2) family gene, one of the central regulators of apoptosis, is regulated by numerous miRNAs. It has been reported that downregulation of miR-7 and miR-335 mediated induction of apoptosis in A549 cells via targeting BCL-2 which subsequently led to an increase in the activities of apoptotic proteins caspase-3 and caspase-7 in cells with apoptotic nuclei.32,33 Moreover, very recently Huang et al34 showed that miR-181 was down-regulated both in NSCLC tissues and cell lines and that this reduction was associated with increased BCL-2 levels. They suggested that miR-181 exerts its proapoptotic function mainly through targeting BCL-2 expression.
Angiogenesis
Phosphatase and tensin homolog (PTEN), a well-known regulator of Akt pathway, has been reported as a target gene of miR-494.35 In this way, miR-494 has a proangiogenic activity through activation of Akt/eNOS pathway. In contrast, miR-200 inhibits angiogenesis through direct and indirect mechanisms especially by targeting interleukin-8 (IL8) and CXCL1 secreted by the tumor endothelial and cancer cells.36
Invasion and metastasis
miR-21 posttranscriptionally downregulates the expression of the tumor suppressor PTEN and subsequently, stimulates growth and invasion in NSCLC.37 Moreover, inhibition of miR-21 expression reduces proliferation, migration, and invasion of A549 cells by upregulating the expression of the programmed cell death protein 4 (PDCD4).38 Li et al39 showed that the expression of miR-183/96/182 cluster is strongly correlated with miR-200 and epithelial–mesenchymal transition in NSCLC as both of these miRNA families target FOXF2, which can potently regulate epithelial–mesenchymal transition, invasion, and metastasis in lung cancers by transcriptional repression of E-cadherin and miR-200.
miR-21
miR-21 is one of the most commonly observed aberrant miRNAs in human cancers and is one of the first miRNAs to be described as an oncomir. A large-scale miRNA analysis on 540 samples in six different types of solid tumors has shown that miR-21 was the only miRNA upregulated in all cancer types.40 Several studies have shown that miR-21 is upregulated in breast,41,42 ovarian,43 colorectal,44 prostate,45,46 pancreatic,47 thyroid cancer,48 as well as in glioma.49,50 Many studies have also clearly shown that miR-21 is deregulated in lung cancer.51,52
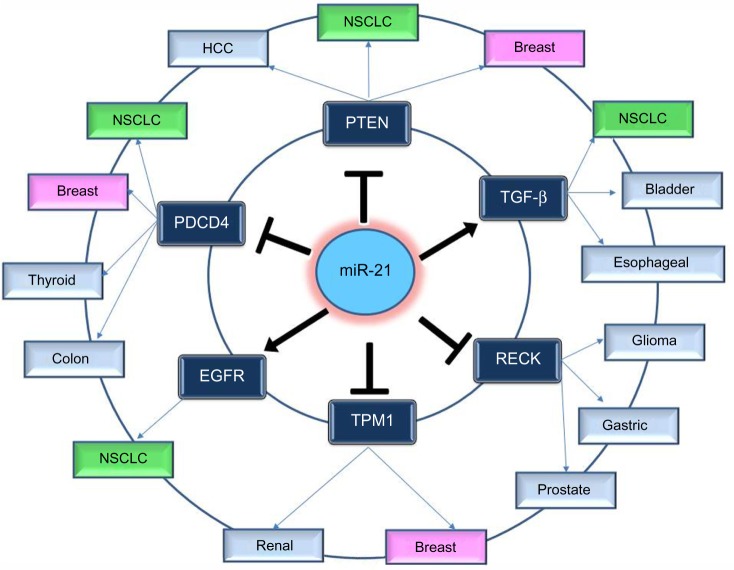
In order to understand the way that miR-21 expression is involved in so many different types of cancer, many functional studies have been performed. These studies predicted and validated a number of target genes of miR-21; most of them are tumor suppressors. The most notable and experimentally validated direct targets of miR-21 are discussed in the following sections and Figure 2.
Figure 2.
Notable and experimentally validated direct targets of miR-21.
Abbreviations: NSCLC, non-small-cell lung cancer; HCC, hepatocellular carcinoma; PTEN, Phosphatase and tensin homolog; PDCD4, programmed cell death protein 4; TPM1, Tropomyosin 1; EGFR, epidermal growth factor receptor; TGF-β, transforming growth factor-b.
PTEN
Inhibition of miR-21 in cultured hepatocellular carcinoma cells increased the expression of PTEN tumor suppressor, and decreased tumor cell proliferation, migration, and invasion.53 The effect of miR-21 on PTEN expression was also assessed in NSCLC cell lines and in NSCLC tumor samples; it was shown once more that miR-21 represses PTEN and stimulates growth and invasion in NSCLC.37,54,55 High miR-21/low PTEN expression levels indicated a poor tyrosine kinase inhibitor (TKI) clinical response and shorter OS in NSCLC patients and moreover the in vitro assays have showed that miR-21 was upregulated concomitantly to downregulation of PTEN in PC-9 gefitinib resistant cells in comparison with PC-9 cells.56 Overexpression of PTEN gene could also mimic the functions of miR-21 inhibitor in NSCLC cells, while siRNA-mediated PTEN knockdown could reverse the effects on NSCLC cells induced by overexpression of miR-21.54
PDCD4
Knockdown of miR-21 in colon cancer cells led to reduced extravasation and distal metastasis because of the rise of the protein expression levels of PDCD4.44 miR-21 negatively regulates PDCD4 at the posttranscriptional level in colon cancer,44 breast cancer,57 and medullary thyroid carcinoma.58 Moreover, inhibition of miR-21 expression reduces proliferation, migration, and invasion of A549 NSCLC cells by upregulating the expression of PDCD4.38,59
Tropomyosin 1
This gene is a member of the tropomyosin (Tm) family, which plays an important role in cancer cell transformation60 and is a direct target of miR-21. Tropomyosin 1 (TPM1) carries a putative miR-21 binding site, which is responsible for regulation by miR-21 at the translational level.61 Inhibition of miR-21 leads to an increase in TPM1 protein expression levels in breast cancer, but inhibition of TPM1 through miR-21 overexpression results in progressive cell migration and invasion.61 Moreover, the role of miR-21 as regulator in TPM1 was also demonstrated in renal cell carcinoma.62
Transforming growth factor-b
Overexpression of transforming growth factor-b (TGF-β) has been described in several tumor tissues, and it was demonstrated that it is associated with tumor progression and metastasis.63,64 Researches have been performed to study the correlated expression of TGF-β and miR-21 in bladder tumors,65 esophageal cancer,66 and lung cancer.67 In lung cancer, TGF-β1, a central pathological mediator of fibrotic diseases, enhanced miR-21 expression in primary pulmonary fibroblasts, and increasing miR-21 levels promoted the pro-fibrogenic activity of TGF-β1 in fibroblasts.68
Epidermal growth factor receptor pathway
A significant correlation between epidermal growth factor receptor (EGFR) and miR-21 levels in lung carcinoma cell lines and the suppression of miR-21 by an EGFR-TKI suggest that the EGFR signaling is a pathway positively regulating miR-21 expression.51 The association of miR-21 expression with the acquired resistance to EGFR-TKIs in NSCLC cell lines, animal models, and advanced NSCLC patients was further studied by Li et al,69 who found that miR-21 could induce EGFR-TKI resistance through inhibiting PTEN and PDCD4 expression and activating the PI3K/Akt signal pathway. In addition to the miRNA microarray data showing higher level of miR-21 in EGFR mutation cases, the in vitro analyses using NSCLC cell lines also showed that the activated EGFR signaling upregulates miR-21 expression. A statistically significant positive correlation was observed between miR-21 expression level and p-EGFR level in NSCLC cell.51,70
RECK
miR-21 regulation of RECK expression was detected in several types of cancer such as in glioma71 and in prostate cancer.72 RECK decreases the amount of active MMP-2 and MMP-9 and inhibits metastatic activity in vitro and in vivo through modulation of these MMPs, which are known to be involved in cancer progression.73
miR-21 as a prognostic and diagnostic biomarker in lung cancer
Yanaihara et al74 were the first group that analyzed miRNA microarray data of lung cancer specimens from NSCLC adenocarcinoma patients and indicated that high miR-155 expression was a significantly unfavorable prognostic factor. On the basis of the microarray results, they further selected miR-17, miR-21, and miR-155 to validate in three additional cohorts of NSCLC adenocarcinoma patients and finally suggested that elevated miR-21 expression is associated with worse prognosis in NSCLC patients.75 Additionally, Lu et al76 studied in a cohort of 357 stage I NSCLC patients the expression levels of several miRNAs and found the prognostic value of two miRNA signature. The prognostic value of mature miR-21 in NSCLC was first shown in 2008; in this study it was shown that miR-21 was upregulated in fresh frozen NSCLC tissues in respect to corresponding adjacent noncancerous tissues and that this overexpression correlated with OS.52 Two meta-analyses summarized the data from different studies and concluded that overexpression of miR-21 was an independent prognostic factor for NSCLC.77,78 Moreover, in a meta-analysis study from a total of 141 prognostic miRNAs, it was identified that four miRNAs (miR-155, -21, -34, and let-7) appeared more frequently, and miR-21 and miR-155, especially, were found to be independent prognostic markers associated with unfavorable survival and recurrence in lung cancer.79
Multiple studies have confirmed since then that high expression of miR-21 in plasma and tissues is correlated with worse prognosis in NSCLC patients and could be an independent prognostic marker.30,80–82 However, there are few studies focusing on the potential predictive value of miRNAs for early-stage NSCLC. A study performed by Tang et al80 concluded that overexpression of circulating miR-21 and miR-155 in plasma and lower expression of circulating miR-145 could distinguish lung cancer patients from healthy smokers with 69.4% sensitivity and 78.3% specificity and that this pattern is associated with poor survival. Liu et al81 showed that serum miR-21 may be a potential novel biomarker for the diagnosis of NSCLC as miR-21 has been found to be significantly elevated in the serum of NSCLC patients with lower survival. Studies examining postoperative lung cancer patients also found that miR-21 serum levels significantly decreased in response to successful surgeries, with higher miR-21 expression corresponding to shorter survival time and disease recurrence.82 With regard to the plasma, Geng et al83 showed that five plasma miRNAs (miR-20a, miR-145, miR-21, miR-223, and miR-221) could be used as promising biomarkers in early screening of NSCLC based on the results from plasma samples of 126 early-stage NSCLC patients, 42 noncancerous pulmonary disease patients, and 60 healthy controls.
miR-21 as a predictive biomarker in lung cancer
One of the most important issues facing current cancer research is resistance to treatments such as chemotherapy, radiotherapy, and targeted therapies (Table 1). Recent studies are showing that miRNAs can be potentially used as predictive biomarkers in various types of cancer. Blower et al84 first reported that variable miR-21 levels can change sensitivity to the cytotoxic action of drugs in opposite directions suggesting the existence of different mechanisms determining toxicity in cancer cell lines. Several studies have shown that miRNAs can control response to therapy in tumors.85,86 Correlation of miR-21 expression and response to therapy has been studied in many types of cancer. More specifically in: 1) breast cancer where miR-21 overexpression mediates resistance to trastuzumab,87 2) leukemia where miR-21 expression is upregulated in the daunorubicin-resistant leukemia cell line,88 and 3) gastric cancer where miR-21 overexpression induces resistance to cisplatin.89
Table 1.
miR-21 controls key biological processes driving to tumorigenesis and therapeutic response in lung cancer
| Target | Biological process | Resistance/sensitivity | Reference |
|---|---|---|---|
| EGFR | Angiogenesis, survival, and proliferation | Gefitinib, TKI | 92,96 |
| BCL-2 | Apoptosis | Platinum | 90 |
| PTEN | Proliferation | Gefitinib, cisplatin | 56,91 |
| Caspase-8, TRAF-7 | Inhibition of NF-κB activation | TRAIL | 94 |
Abbreviations: TKI, tyrosine kinase inhibitor; PTEN, Phosphatase and tensin homolog; EGFR, epidermal growth factor receptor; BCL-2, B-cell lymphoma 2.
Regarding miRNA expression at the primary tumor, it was reported that NSCLC patients with lower expression of miR-21 seem to have substantial benefits from an adjuvant platinum-based chemotherapy as compared with patients with higher miR-21 expression tumors.90 Additionally, it was found that miR-21 induced the resistance of the NSCLC cell line to cisplatin via targeting the 3′-UTR of PTEN to decrease its expression posttranscriptionally.91 Using miR-21 mimics and its inhibitor, Yang et al91 found that exogenous miR-21 promoted cell survival when exposed to cisplatin treatment, whereas miR-21 inhibition led to the opposite function.
It is now well established that EGFR TKIs, such as gefitinib and erlotinib, constitute the best therapeutic option in first, second, and maintenance setting for NSCLC patients carrying EGFR mutations.92 Unfortunately, half of the patients who initially respond to EGFR-TKI acquire resistance to EGFR-TKI within the 1st year, and in 50% of these cases, a secondary EGFR-T790M gatekeeper mutation has been identified.92,93 miR-21 overexpression was found to reduce sensitivity to gefitinib in PC9 cells by downregulating PTEN and activating Akt and ERK pathways, which enhanced invasive ability and induced apoptosis.56 Furthermore, in the same study, Shen et al56 investigated whether alteration of miR-21/PTEN expression correlates with TKI sensitivity using 47 NSCLC tumor samples from TKI-treated patients. As expected, patients with progressive disease had significantly higher miR-21 levels and lower PTEN expression than patients having partial response and stable disease. These findings are all in agreement and suggest involvement of high miR-21/low PTEN expression in modulation of TKI sensitivity in NSCLC.56
In lung cancer, very recently, Jeon et al94 have shown that overexpression of miR-21, miR-30, and miR-100 could modulate the response to tumor necrosis factor-related apoptosis-inducing ligand by targeting fundamental tumor suppressor genes such as caspase-8, caspase-3, TRAF-7, and FoxO3a and oncogenes such as PI3K and cyclins. They report that in the resistant cells, downmodulation of caspase-8 and TRAF-7 by miR-21 and miR-100 activates NF-κB, which in turn, by a positive feedback loop, induces the transcriptional activation of miR-21 and miR-100.94
Circulating miR-21 in serum was recently shown to be an independent prognostic indicator for NSCLC patients who developed resistance to EGFR-TKI since it was found to be significantly increased in disease progression; high levels of circulating miR-21 is a poor prognostic factor for advanced NSCLC patients treated with EGFR-TKIs.69 Plasma miR-21 was identified as an early detection and chemosensitivity biomarker in patients with advanced NSCLC.95 In this study, Wei et al95 demonstrated that plasma levels of miR-21 were found to be significantly higher in the stable and progressive disease group than in the partial chemotherapeutic response group (cisplatin or carboplatin based), whereas no statistical significance was found between the partial chemotherapeutic response group and the control group. miR-21 overexpression in plasma of NSCLC patients was found to be an independent predictor of response to gefitinib; patients with higher expression of miR-21 in plasma had a significant improvement in OS following an adjuvant therapy with gefitinib compared to those with low expression.96 Plasma miR-21 expression is associated with resistance to platinum-based chemotherapy in NSCLC; downregulation of miR-21 by a locked nucleic acid (LNA) anti-miR-21 in vitro and in vivo enhanced the sensitivity of A549 cells to cis-diamminedichloride platinum II (DDP) through apoptotic signaling.97
Regarding resistance to radiotherapy, Liu et al98 reported that upregulation of miR-21 could inhibit the sensitivity of NSCLC cells to irradiation, while downregulation of miR-21 could significantly enhance the sensitivity of NSCLC cells to irradiation. The potential molecular mechanism of this resistance was further studied by Ma et al,99 suggesting that the effects of miR-21 overexpression on A549 cell apoptosis enhanced by irradiation depend on the activation of PI3K/Akt signaling pathway.
It is important to point out that according to recent findings, exosomes also seem to get involved in the resistance to therapy in lung cancer. As Xiao et al100 showed, exposure of A549 cells to DDP could cause cells to release more exosomes than in normal conditions, and the interaction of these exosomes with other A549 cells could increase the resistance of these A549 cells to DDP. When exposed to DDP, A549 cells were stimulated and the expression level of miR-21 increased significantly both in exosomes and in cancer cells.
miR-21 as a therapeutic target in lung cancer
Restoring or downregulating the expression levels of specific miRNAs could be a promising approach to treat cancer; there are two ways to develop miRNA-based therapeutics: 1) miRNA mimics known as “miRNA replacement therapy”, which are used to restore a loss of function of tumor suppressor genes and 2) miRNA antagonists (antagomirs), which are synthetic RNAs complementary to a specific targeted miRNA that block its function and allow in this way its target mRNA to be translated.101 This approach has a very limited use for clinical applications till now.
The first miRNA replacement therapy, MRX34, is a liposome formulate miR-34 mimic that is intravenously injected.102 This potential drug has entered human clinical trials for patients with unresectable primary liver cancer or solid cancers with liver involvement (http://www.clinicaltrials.gov, NCT01829971). Besides, Santaris Pharma is conducting an early clinical trial, which is currently ongoing, using LNA-anti-miR-122 (PSC-3649) in healthy human subjects. The results of these above-cited studies will provide valuable information on the pharmacokinetic and safety profile of these potential drugs. Finally, inhibition of miR-21 was chosen as one of the promising therapeutic strategies for treating hepatocellular carcinoma, and a miR-21 inhibitor (RG-012) is currently being developed by Regulus Therapeutics.
Numerous studies in lung cancer cells lines use an LNA-based miR-21 (anti-miR-21) in order to knock down miR-21 expression. Anti-miR-21 significantly inhibited growth and induced death in A549 cells through apoptotic signaling51,97 and protected against metastasis of human lung cancer cells through inhibition of NADPH oxidase.103 In the in vivo study, a xenograft tumor assay was designed to manipulate the effect of anti-miR-21, meanwhile, tumor growth curve was recorded and immunohistochemistry of the xenograft tumor was performed.73 As expected, miR-21 inhibitor suppresses NSCLC growth in the xenograft model. Moreover, Xu et al97 transfected A549 cells with LNA anti-miR-21 or scramble control into female nude mice followed by treatment with DDP or phosphate buffer saline and noticed that the size of the implanted tumors (A549 transfected with LNA anti-miR-21) treated with DDP were reduced in comparison with the controls, this enhanced the evidence that downregulation of miR-21 by LNA anti-miR-21 increased the in vivo chemosensitivity. As experiments with the xenograft carcinoma model indicate that one transient transfection with anti-miR-21 is sufficient to cause substantial inhibition of tumor growth, this raises the possibility that anti-miR-21 may have potential therapeutic value.
Future perspectives
miRNA-based therapeutics could be a remarkably smart way for targeting therapy in cancer, since one miRNA in charge of a specific function or mechanism can simultaneously target several downstream genes. miR-21 overexpression is involved in many oncogenic pathways of lung cancer and other malignancies. The development of a targeted therapeutic that aims to silence miR-21 seems to be very promising. However, there are some limitations such as how to reach a sufficient dose within the cells in order to achieve efficient miRNA inhibition and which is the right dose of miR-21. However, most studies so far have been performed in vitro, in vivo studies based on systemic or localized target delivery of anti-miR-21 are still required to be carried out. As miR-21 is overexpressed in most human tumors, therapeutic delivery of anti-miR-21 may still be beneficial for a large number of cancers for which no cure is available. However, there is no doubt that significant work remains to be done, and it is necessary to wait for the pharmacokinetic results of the clinical trials in order to evaluate the safety and the efficiency of these molecules.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 8.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky N. MicroRNA target predictions in animals. Nat Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 16.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. RNA. 2009;15:814–822. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 20.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids – the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15:827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Ferracin M, Lupini L, Salamon I, et al. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget. 2015;6:14545–14555. doi: 10.18632/oncotarget.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 29.White NM, Fatoohi E, Metias M, Jung K, Stephan C, Yousef GM. Metastamirs: a stepping stone towards improved cancer management. Nat Rev Clin Oncol. 2011;8:75–84. doi: 10.1038/nrclinonc.2010.173. [DOI] [PubMed] [Google Scholar]
- 30.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong S, Zheng Y, Jiang P, Liu R, Liu X, Chu Y. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci. 2011;7:805–814. doi: 10.7150/ijbs.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Li M, Zhang R, et al. Effect of miR-335 upregulation on the apoptosis and invasion of lung cancer cell A549 and H1299. Tumour Biol. 2013;34:3101–3109. doi: 10.1007/s13277-013-0878-9. [DOI] [PubMed] [Google Scholar]
- 34.Huang P, Ye B, Yang Y, Shi J, Zhao H. MicroRNA-181 functions as a tumor suppressor in non-small cell lung cancer (NSCLC) by targeting Bcl-2. Tumour Biol. 2015;36:3381–3387. doi: 10.1007/s13277-014-2972-z. [DOI] [PubMed] [Google Scholar]
- 35.Mao G, Liu Y, Fang X, et al. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis. 2015;18(3):373–382. doi: 10.1007/s10456-015-9474-5. [DOI] [PubMed] [Google Scholar]
- 36.Pecot CV, Rupaimoole R, Yang D, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Meng H, Peng Q, et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015;22:23–29. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Tan Q, Yan M, et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 42.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 44.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 45.Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folini M, Gandellini P, Longoni N, et al. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sondermann A, Andreghetto FM, Moulatlet AC, et al. miR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32(6):521–530. doi: 10.1007/s10585-015-9724-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Zhang J, Jia Q, et al. Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep. 2010;24:195–201. doi: 10.3892/or_00000846. [DOI] [PubMed] [Google Scholar]
- 50.Quintavalle C, Donnarumma E, Iaboni M, et al. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. 2013;32:4001–4008. doi: 10.1038/onc.2012.410. [DOI] [PubMed] [Google Scholar]
- 51.Seike M, Goto A, Okano T, et al. miR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 53.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372(1–2):35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W, Bai W, Zhang W. miR-21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non-small cell lung cancer A549 cells. Clin Transl Oncol. 2014;16(8):708–713. doi: 10.1007/s12094-013-1135-9. [DOI] [PubMed] [Google Scholar]
- 56.Shen H, Zhu F, Liu J, et al. Alteration in miR-21/PTEN expression modulates gefitinib resistance in non-small cell lung cancer. PLoS One. 2014;9:e103305. doi: 10.1371/journal.pone.0103305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 58.Pennelli G, Galuppini F, Barollo S, et al. The PDCD4/miR-21 pathway in medullary thyroid carcinoma. Hum Pathol. 2015;46:50–57. doi: 10.1016/j.humpath.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helfman DM, Flynn P, Khan P, Saeed A. Tropomyosin as a regulator of cancer cell transformation. Adv Exp Med Biol. 2008;644:124–131. doi: 10.1007/978-0-387-85766-4_10. [DOI] [PubMed] [Google Scholar]
- 61.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 62.Lv L, Huang F, Mao H, et al. MicroRNA-21 is overexpressed in renal cell carcinoma. Int J Biol Markers. 2013;28:201–207. doi: 10.5301/JBM.2013.10831. [DOI] [PubMed] [Google Scholar]
- 63.Gorsch SM, Memoli VA, Stukel TA, et al. Immunohistochemical staining for TGF-β1 associates with disease progression in human breast cancer. Cancer Res. 1992;52:6949–6952. [PubMed] [Google Scholar]
- 64.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis – correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 65.Monfared H, Ziaee SA, Hashemitabar M, et al. Co-regulated expression of TGF-β Variants and miR-21 in bladder cancer. Urol J. 2013;10:981–987. [PubMed] [Google Scholar]
- 66.Zhang Y, Pan T, Zhong X, Cheng C. Nicotine upregulates microRNA-21 and promotes TGF-β-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014;35:7063–7072. doi: 10.1007/s13277-014-1968-z. [DOI] [PubMed] [Google Scholar]
- 67.Yao Q, Cao S, Li C, Mengesha A, Kong B, Wei M. Micro-RNA-21 regulates TGF-β-induced myofibroblast differentiation by targeting PDCD4 in tumor-stroma interaction. Int J Cancer. 2011;128:1783–1792. doi: 10.1002/ijc.25506. [DOI] [PubMed] [Google Scholar]
- 68.Liu G, Friggeri A, Yang Y, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Ren S, Li X, et al. miR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer. 2014;83:146–153. doi: 10.1016/j.lungcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Ma X, Kumar M, Choudhury SN, et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:10144–10249. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haffner MC, Mosbruger T, Esopi DM, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexius-Lindgren M, Andersson E, Lindstedt I, Engström W. The RECK gene and biological malignancy – its significance in angiogenesis and inhibition of matrix metalloproteinases. Anticancer Res. 2014;34:3867–3873. [PubMed] [Google Scholar]
- 74.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 75.Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu Y, Govindan R, Wang L, et al. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33:1046–1054. doi: 10.1093/carcin/bgs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu W, Xu B. MicroRNA-21 identified as predictor of cancer outcome: a meta-analysis. PLoS One. 2014;9:e103373. doi: 10.1371/journal.pone.0103373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma XL, Liu L, Liu XX, et al. Prognostic role of microRNA-21 in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2012;13:2329–2334. doi: 10.7314/apjcp.2012.13.5.2329. [DOI] [PubMed] [Google Scholar]
- 79.Yang M, Shen H, Qiu C, et al. High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer. 2013;49:604–615. doi: 10.1016/j.ejca.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 80.Tang D, Shen Y, Wang M, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 81.Liu XG, Zhu WY, Huang YY, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 82.Lee JH, Voortman J, Dingemans AM, et al. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One. 2011;6:e21300. doi: 10.1371/journal.pone.0021300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 85.Donahue TR, Nguyen AH, Moughan J, et al. Stromal microRNA-21 levels predict response to 5-fluorouracil in patients with pancreatic cancer. J Surg Oncol. 2014;110:952–959. doi: 10.1002/jso.23750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oue N, Anami K, Schetter AJ, et al. High miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. 2014;134:1926–1934. doi: 10.1002/ijc.28522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gong C, Yao Y, Wang Y, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bai H, Xu R, Cao Z, Wei D, Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011;585:402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 89.Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–168. doi: 10.1016/j.tox.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 90.Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y. miRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. 2012;13:330–340. doi: 10.4161/cbt.19073. [DOI] [PubMed] [Google Scholar]
- 91.Yang Z, Fang S, Di Y, Ying W, Tan Y, Gu W. Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PLoS One. 2015;10:e0121547. doi: 10.1371/journal.pone.0121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 93.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 94.Jeon YJ, Middleton J, Kim T, et al. A set of NF-κB-regulated microRNAs induces acquired TRAIL resistance in Lung cancer. Proc Natl Acad Sci U S A. 2015;112(26):E3355–E3364. doi: 10.1073/pnas.1504630112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei J, Gao W, Zhu CJ, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30:407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen Y, Tang D, Yao R, et al. MicroRNA expression profiles associated with survival, disease progression, and response to gefitinib in completely resected non-small-cell lung cancer with EGFR mutation. Med Oncol. 2013;30:750. doi: 10.1007/s12032-013-0750-1. [DOI] [PubMed] [Google Scholar]
- 97.Xu L, Huang Y, Chen D, et al. Downregulation of miR-21 increases cisplatin sensitivity of non-small cell lung cancer. Cancer Genet. 2014;207:214–220. doi: 10.1016/j.cancergen.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 98.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 99.Ma Y, Xia H, Liu Y, Li M. Silencing miR-21 sensitizes non-small cell lung cancer A549 cells to ionizing radiation through inhibition of PI3K/Akt. Biomed Res Int. 2014;2014:617868. doi: 10.1155/2014/617868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao X, Yu S, Li S, et al. Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs with “antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 102.Mirna Therapeutics Press release: Mirna therapeutics is first to advance MicroRNA into the clinic for cancer. [Accessed September 7, 2015]. Available from: http://www.mirnarx.com.
- 103.Yan S, Liu G, Pei C, et al. Inhibition of NADPH oxidase protects against metastasis of human lung cancer by decreasing microRNA-21. Anticancer Drugs. 2015;26:388–398. doi: 10.1097/CAD.0000000000000198. [DOI] [PubMed] [Google Scholar]