Abstract
During metastatic progression, circulating cancer cells become lodged within the microvasculature of end-organs, where a majority die from mechanical deformation. Though this phenomenon was first described over a half-century ago, the mechanisms enabling certain cells to survive this metastasis-suppressive barrier remain unknown. By applying whole-transcriptome RNA-sequencing (RNA-seq) technology to isogenic cancer cells of differing metastatic capacity, we identified a mutation encoding a truncated form of the pannexin-1 (PANX1) channel, PANX1 1–89, as recurrently enriched in highly metastatic breast cancer cells. PANX1 1–89 functions to permit metastatic cell survival during traumatic deformation in the microvasculature by augmenting ATP release from mechanosensitive PANX1 channels activated by membrane stretch. PANX1-mediated ATP release acts as an autocrine suppressor of deformation-induced apoptosis via P2y-purinergic receptors. Finally, small-molecule therapeutic inhibition of PANX1 channels is found to reduce the efficiency of breast cancer metastasis. These data suggest a molecular basis for metastatic cell survival upon microvasculature-induced biomechanical trauma.
Introduction
Primary tumour cells entering the blood stream are rapidly transported away from their site of origin and disseminated throughout the body. These circulating cancer cells eventually land in the microvascular beds of secondary end-organs, where they are deformed within capillaries of smaller diameter and lower compliance1. This mechanical stress is responsible for the loss of up to greater than 90% of cancer cells entering the small vessels2–6. Despite this barrier to metastatic progression, subsets of cancer cells are able to endure such mechanical stress, thereby maintaining an opportunity to infiltrate the parenchyma of organs and ultimately form lethal metastatic colonies. While recent work has revealed genes and biological processes regulating steps of metastatic progression7–12, the molecular mechanisms that enable select cancer cells to survive microvascular deformation are not understood. By analyzing recurrent alterations in the mutational spectra of in vivo-selected metastatic human breast cancer cells, we have identified extracellular ATP-release via pannexin-1 (PANX1) channels as a pro-metastatic signal that permits the survival of cancer cells while they are deformed in the vasculature of metastatic organs.
Results
Identification of mutations enriched in highly metastatic breast cancer cells
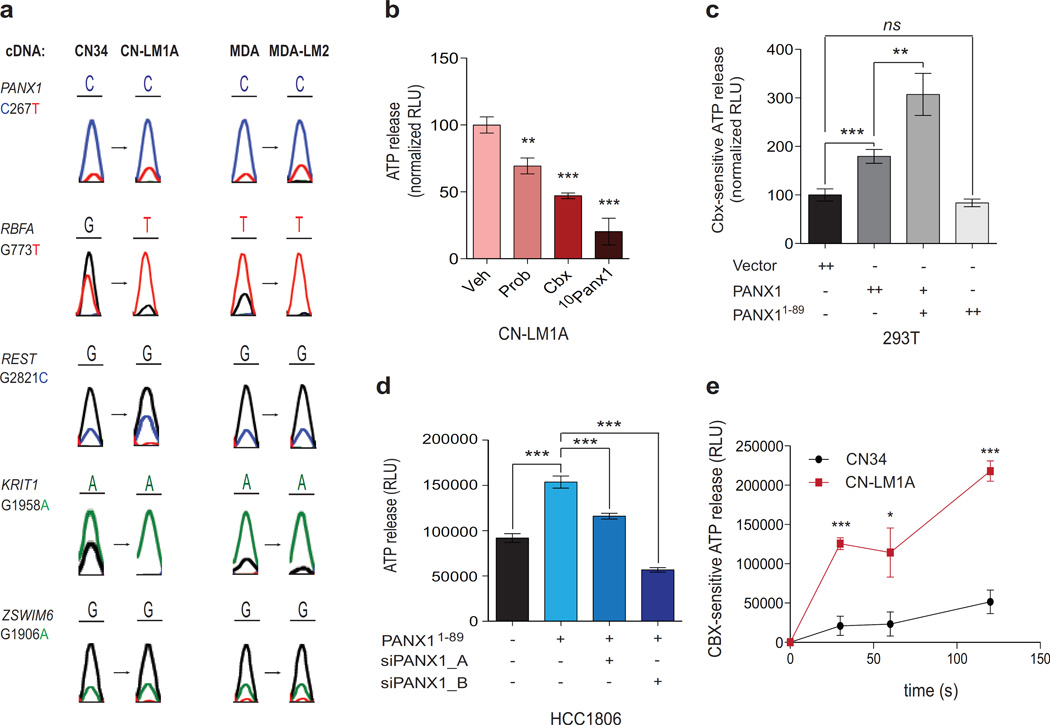
Despite great advancements in tumour sequencing technology, it remains to be determined whether mutations exist that function specifically to drive metastasis13. This is likely a consequence of the expansive genetic heterogeneity present in bulk tumour tissue. For instance, low frequency (<10%) sub-clonal mutations comprise the majority of all mutations present in breast tumours14,15. To circumvent this problem, we performed a systematic screen for functional non-synonomous single nucleotide variants (nSNVs) with allelic frequencies that were recurrently enriched through in vivo selection for metastatic sub-clones. We reasoned that the discovery and functional characterization of such mutations might reveal molecular signaling pathways not yet known to play a role in metastasis biology. To this end, we performed whole-transcriptomic RNA-sequencing (RNA-seq) of in vivo-selected16,17 highly metastatic human breast cancer cell sub-lines, CN-LM1A and MDA-LM2, as well as the CN34 and MDA-MB-231 parental lines from which they were derived (Supplementary Figure 1a and Supplementary Table 1). We postulated that the nSNVs most likely to confer a metastatic advantage should be expressed in the transcriptomes of both CN-LM1A and MDA-LM2 sub-lines at allelic frequencies significantly higher than those quantified in their less metastatic parental populations CN34 and MDA-MB-231, respectively. Our discovery framework (see Methods) implicated four missense variants and a single nonsense variant as potential promoters of metastasis (Fig. 1a and Supplementary Figure 1b). The genes bearing the missense nSNVs include the mitochondrial ribosome-binding factor RBFA, the transcription factor REST, the integrin-β1 regulator KRIT1 and the zinc-finger-containing gene ZSWIM6. The nonsense variant, PANX1 C268T, was significantly enriched in the mRNA transcripts of the pannexin-1 ATP channel of both sub-lines. Sanger sequencing of cDNA independently confirmed the enrichment of these mutant alleles in metastatic breast cancer cells (Fig. 1a). These mutations implicate five genes as potential regulators of breast cancer metastasis.
Figure 1. PANX1 1–89 augments PANX1-mediated ATP release in metastatic breast cancer cells.
a, Sanger sequencing traces from the cDNA of CN34, CN-LM1A, MDA-MB-231 and MDA-LM2 cells at the nSNV alleles predicted to result in non-neutral substitutions by PolyPhen-2. b, Quantification of PANX1-mediated ATP release from CN-LM1A cells pretreated for 10 min with PBS, 2mM PB, 500 µM CBX, or 100 µM 10Panx1 peptide; n = 4. c, Quantification of PANX1-mediated ATP release from HEK293T cells transfected with 8 µg control vector (n = 8), 8 µg wild-type PANX1 (n = 8), 5 µg wild-type PANX1 and 3 µg PANX1 1–89 (n = 7), or 8 µg PANX1 1–89 (n = 3), and pretreated for 10 min with 500 µM carbenoxolone (CBX) or an equivalent volume of PBS. d, Quantification of ATP release from PANX1 1–89-expressing HCC1806 breast cancer cells transfected with short interfering RNAs (siRNAs) against full-length endogenous PANX1 or control siRNA; n = 12. e, Time-course measurements of CBX-sensitive ATP release from CN34 parental cells and the CN-LM1A metastatic derivative sub-line pretreated with CBX (500 µM) or PBS for 10 min; n = 4. Error bars, s.e.m., ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Experimental results presented are representative and were independently replicated at least two times with two independent cell lines.
PANX11–89 enhances pannexin-1 channel activity
The therapeutic targeting of proteins expressed on the surface of cancer cells by antibodies such as anti-HER2 (Herceptin) or anti-CD20 (Rituximab) have had major impacts on the survival of patients with breast cancer and lymphoma, respectively. Because the PANX1 mutation alters a cell-surface channel protein, we reasoned that, if functional, it too might offer potential for therapeutic targeting. Allele-specific RNA-seq (Supplementary Figure 1c) and Sanger sequencing of genomic DNA (Supplementary Figure 1d, e) validated the transcriptomic and genomic enrichment of the PANX1 C268T allele in the highly metastatic derivative sub-lines, respectively. PANX1 encodes the monomeric subunit of a heximeric plasma membrane channel that, when activated, mediates the release of ATP from cells into the extracellular space18–27—a well-established autocrine/paracrine intravascular signaling mechanism28,29. The PANX1 C268T nonsense mutation substitutes a premature termination codon for the glutamine codon at position 90 of the 426 amino acid PANX1 protein. Prior to testing this mutation in PANX1 channel activity assays, we wanted to know whether the metastatic human breast cancer cells in which PANX1 C268T was identified express functional PANX1 channels. Indeed, treatment of the CN-LM1A and MDA-LM2 metastatic sub-lines with three established PANX1 inhibitors—probenecid (Prob)30,31, carbenoxolone (CBX)22,25,30,32,33, or the more potent mimetic peptide 10Panx1 (Supplementary Figure 2a)22,26,30—significantly reduced extracellular ATP release (Fig. 1b and Supplementary Figure 2b, c), suggesting that highly metastatic breast cancer cells mediate substantial ATP release through PANX1 channels. To determine whether PANX11–89 alters ATP release via PANX1 channels, we measured extracellular ATP release from cells expressing wild-type PANX1 alone, wild-type PANX1 with PANX11–89, or PANX11–89 alone. PANX1-mediated ATP release was quantified by measuring the reduction in ATP release in the presence of CBX22,25,30,32,33. When co-expressed with full-length PANX1, PANX11–89 significantly enhanced the release of ATP through PANX1 channels (Fig. 1c and Supplementary Figure 2d). However, ATP release was not enhanced when PANX11–89 was expressed alone in PANX1-deficient human cells (Fig. 1c) or in PANX1-null mouse embryonic fibroblasts (MEFs) (Supplementary Figure 2e), suggesting that mutant PANX11–89 requires the presence of wild-type full-length PANX1 for its effect and does not homo-oligomerize to form functional ATP release channels. The expression of PANX11–89 was also sufficient to significantly augment PANX1-mediated ATP release in HCC1806, BT549 and MDA-MB-468—three distinct human breast cancer cell lines harboring wild-type endogenous PANX1 (Fig. 1d and Supplementary Figure 2f, g). This effect was significantly reduced upon knockdown of endogenous wild-type PANX1 (Fig. 1d and Supplementary Figure 2h), further suggesting that PANX11–89 mediates the enhancement of extracellular ATP release from breast cancer cells specifically through full-length PANX1 channels. Consistent with these findings, highly metastatic CN-LM1A and MDA-LM2 sub-lines secreted nearly 5-fold more PANX1-mediated ATP than their less metastatic parental cell populations without expressing higher levels of the PANX1 gene (Fig. 1e and Supplementary Figure 2i, j).
PANX1 channel activity promotes metastatic cell survival in the vasculature
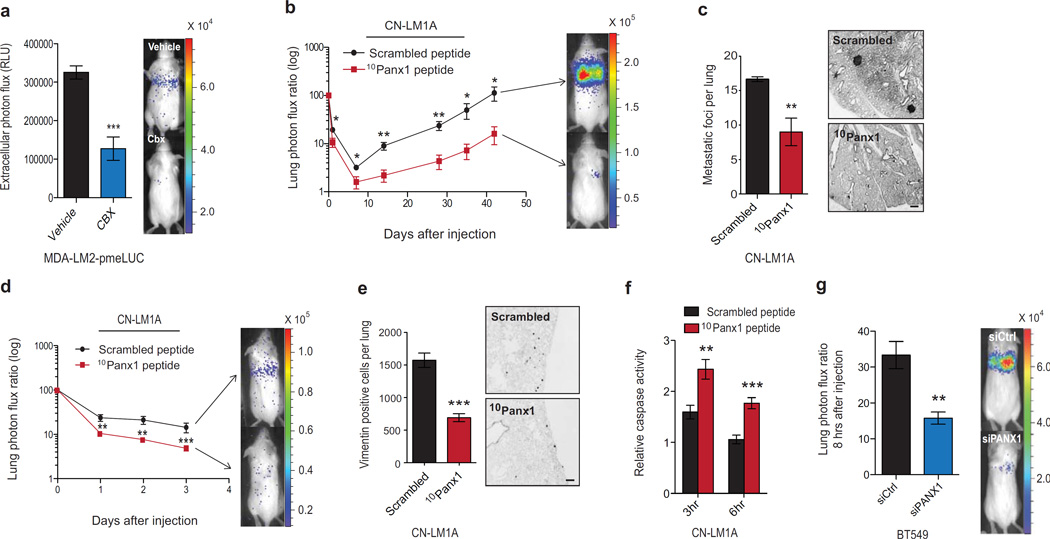
Given these findings, we went on to study whether activated PANX1 channels play a role in cancer metastasis. We first asked whether PANX1 channels are activated in vivo. To address this, we injected breast cancer cells expressing plasma membrane-anchored extracellular luciferase into the venous circulation of mice, and quantified the extracellular ATP released from cancer cells in vivo. Five minutes after venous injection, we observed significant extracellular ATP bioluminescent signal detected in the lungs of mice, which was attenuated upon pharmacological pre-treatment of cells with the PANX1 inhibitor CBX (Fig. 2a). To determine whether the observed early activation of PANX1 channels is necessary for efficient metastasis, we acutely blocked PANX1 channels by pre-incubating CN-LM1A and MDA-LM2 cells with 10Panx1 or its corresponding scrambled peptide and assessed the cells’ metastatic activity through tail-vein lung colonization assays. This acute (30 min) inhibition of PANX1 significantly inhibited metastasis as evidenced by reductions in metastatic signal at early time-points that persisted at 6-week endpoints (Fig. 2b, c and Supplementary Figure 2k, l), suggesting that PANX1 channel activity increases the metastatic capacity of human breast cancer cells.
Figure 2. Breast cancer cell PANX1 activity within the pulmonary vasculature promotes lung metastasis.
a, Quantitative bioluminescence imaging of extracellular ATP release by cancer cells in the lung vasculature 5 min after tail-vein injection of 1 × 105 MDA-LM2 cells expressing plasma membrane-anchored extracellular luciferase (MDA-LM2-pmeLUC). MDA-LM2-pmeLUC cells were pretreated for 10 min with either CBX (500 µM; n = 5) or PBS (n = 7) prior to injection into FVB/NJ mice. b, Quantitative bioluminescence imaging of lung metastasis after the injection of 1 × 105 highly metastatic CN-LM1A breast cancer cells pretreated with 100 µM 10Panx1 (n = 6) or scrambled peptide (n = 7), into NOD scid (NS) mice. c, Day 42 quantification of metastatic foci from H&E-stained lungs (left) and representative images from vimentin-stained lungs (right) of mice injected with CN-LM1A cells pretreated with 10Panx1 or scrambled peptide; n = 5. Scale bar, 0.5 mm. d, Daily quantitative imaging plot of lung bioluminescence subsequent to the injection of 1 × 105 metastatic CN-LM1A breast cancer cells pretreated (30 min) with 100 µM 10Panx1 or scrambled peptides, into NS mice; n = 7. e, Lungs from mice were extracted at day 3, sectioned and stained for vimentin and the numbers of vimentin-positive cancer cells were quantified; n = 5. Scale bar, 0.25 mm. f, In vivo quantification of luciferase-based caspase-3/7 activity at 3 and 6 hrs after tail-vein injection of 1 × 105 CN-LM1A breast cancer cells, pretreated with 100 µM 10Panx1 or scrambled peptide, into NS mice; n = 5. g, Quantitative bioluminescence imaging of cancer cells in the lungs 8-hours after the injection of 2 × 105 BT549 cells transfected with short interfering RNAs (siRNAs) against PANX1 or control siRNA; n = 5. Error bars, s.e.m., *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Experiments b-f are representative and were replicated at least two times in independent cell lines. Bioluminescent and histological images are representative of the median.
To confirm that PANX1 inhibition blunts early metastatic dissemination, we performed tail-vein injections of CN-LM1A and MDA-LM2 metastatic cells pretreated with 10Panx1 and quantified lung bioluminescence daily, over three days (Fig. 2d, e and Supplementary Figure 3a). Consistent with a role for activated PANX1 channels in metastatic dissemination, inhibition of PANX1 led to a significant decrease in lung bioluminescence as early as day one (Fig. 2d and Supplementary Figure 3a). Histological examination of lungs from these mice also revealed a reduction in the number of disseminated breast cancer cells detected histologically (Fig. 2e and Supplementary Figure 3b). The acute impact of PANX1 channel inhibition on cancer-cell lung bioluminescence suggested a block in metastatic progression in the vasculature. In support of this, PANX1 blockade did not inhibit proliferation, invasion, transendothelial migration or anchorage-independent cell survival capacity (Supplementary Figure 3c–g). We therefore hypothesized that PANX1 inhibition might increase cell death within the microvasculature. To test this, we performed tail-vein injections of CN-LM1A and MDA-LM2 metastatic cells pre-incubated with 10Panx1 or scrambled peptides and quantified in vivo caspase activity using a luciferase-based reporter. In support of enhanced intravascular cell death, in vivo caspase activity was significantly augmented upon PANX1 inhibition at early time points (3 and 6 hrs) post-injection (Fig. 2f and Supplementary Figure 3h)—well before monocyte recruitment (24 hrs)34 or cancer cell extravasation (48 hrs)34,35 occur. Consistent with these findings, acute siRNA-mediated knockdown of PANX1 channels in the independent metastatic breast cancer cell line BT549 significantly decreased cancer cell lung bioluminescence at 8-hours post venous injection (Fig. 2g), further supporting the role of PANX1 channels in promoting microvascular survival during metastatic dissemination.
ATP release via mechanosensitive PANX1 channels promotes metastatic cell survival during intravascular deformation via purinergic signaling
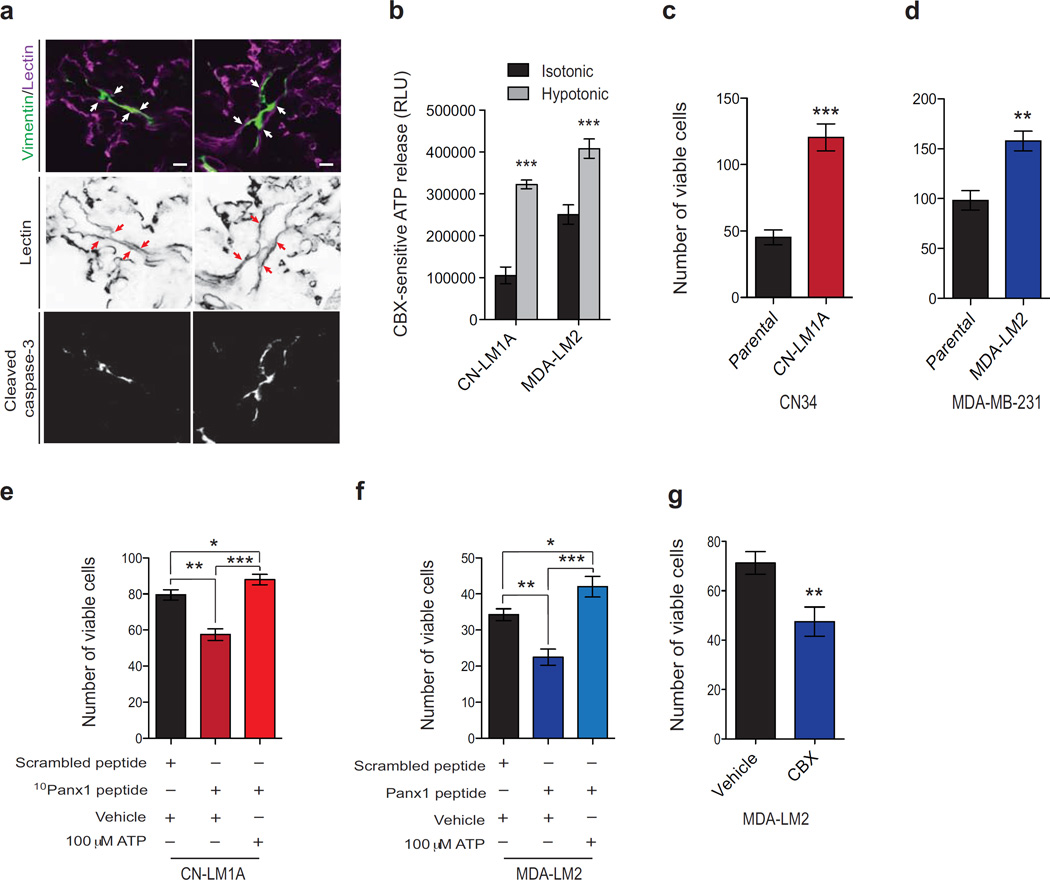
Confocal microscopy revealed that, at the time when PANX1 activity was found to promote survival (3 hrs post-injection), cancer cells were confined within the pulmonary vasculature and many had become morphologically elongated in small blood vessels (Fig. 3a). Consistent with quantitative in vivo caspase measurements, a number of these intravascular cancer cells displayed activated caspase immunoreactivity (Fig. 3a). Of the many mechanisms responsible for the inefficiency of metastasis, intravascular death is responsible for the loss of up to greater than 90% of the cancer cells entering the microvasculature of secondary organs2–6. Considering that caspase-positive breast cancer cells were substantially elongated in the small lung vessels soon after injection (Fig. 3a), and that PANX1 is a mechanosensitive channel that opens during plasma membrane stretch18–21, we wondered whether PANX1 activation during cancer cell deformation in the vasculature might underlie its observed role in early cancer-cell survival. Hypotonic cell swelling—an established perturbation that imparts plasma membrane stretch in a well-controlled manner6,19–21,36—significantly increased the amplitude of PANX1-mediated ATP release in CN-LM1A and MDA-LM2 breast cancer cells (Fig. 3b). Accordingly, we found CN-LM1A and MDA-LM2 cells to be significantly more resistant to lethal hypotonic stretch relative to their parental population (Fig. 3c, d). We then tested the role of PANX1 activity in promoting survival during cell deformation by hypotonically stretching CN-LM1A and MDA-LM2 cells in the presence of the 10Panx1 inhibitor or scrambled control peptides. The number of viable cancer cells remaining after incubation in hypotonic solution was significantly reduced when cells were subjected to PANX1 channel inhibition (Fig. 3e, f). Moreover, PANX1 inhibition also reduced cell survival after centrifugal stretch (Fig. 3g and Supplementary Figure 4a). Importantly, cell viability was fully rescued in the PANX1-inhibited cells through addition of extracellular ATP (Fig. 3e, f). These data suggest that ATP signaling at the cell surface mediates the observed survival phenotype.
Figure 3. Mechanosensitive PANX1 channels release ATP to increase cancer cell survival during intravascular membrane stretch.
a, Representative images of mouse lungs stained for cancer cells and blood vessels (green and magenta, respectively, top panel), blood vessels (black, middle panel) or cleaved caspase-3 (white, bottom panel) 3 hrs after tail-vein injection of 1 × 105 CN-LM1A cells. Arrows indicate endothelium. Scale bar, 10 µm. b, Quantification of PANX1-mediated CBX-sensitive ATP release from CN-LM1A and MDA-LM2 cells during 5 min exposure to isotonic (100% PBS) or hypotonic (70% PBS) solution; n = 4 (CN-LM1A isotonic), n = 3 (CN-LM1A hypotonic), n = 4 (MDA-LM2 isotonic), n = 4 (MDA-LM2). c, Quantification of viable, trypan blue-negative, CN34 and CN-LM1A cells after 1 hr extreme hypotonic (12.5% PBS) stretch; n = 4. d, Quantification of viable, trypan blue-negative CN34, CN-LM1A, MDA-MB-231 and MDA-LM2 cells after 1 hr extreme hypotonic (12.5% PBS) stretch; n = 4. e, Quantification of viable, trypan blue-negative, CN-LM1A cells after 1 hr incubation in extremely hypotonic (12.5% PBS) solution in the presence of scrambled peptide (100 µM), 10Panx1 peptide (100 µM) or 10Panx1 peptide (100 µM) and 100 µM ATP; n = 4. f, Quantification of viable, trypan blue-negative, MDA-LM2 cells after 1 hr incubation in extremely hypotonic (12.5% PBS) solution in the presence of scrambled peptide (100 µM), 10Panx1 peptide (100 µM) or 10Panx1 peptide (100 µM) and 100 µM ATP; n = 4. g, Quantification of viable, trypan blue-negative MDA-LM2 cells with 30 min Boyden chamber centrifugation (3,800 rpm) after cells were pre-treated for 10 min with 500 µM CBX or PBS; n = 4. Error bars, s.e.m., *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Experiments b–g are representative and were replicated at least two times with two independent cell lines.
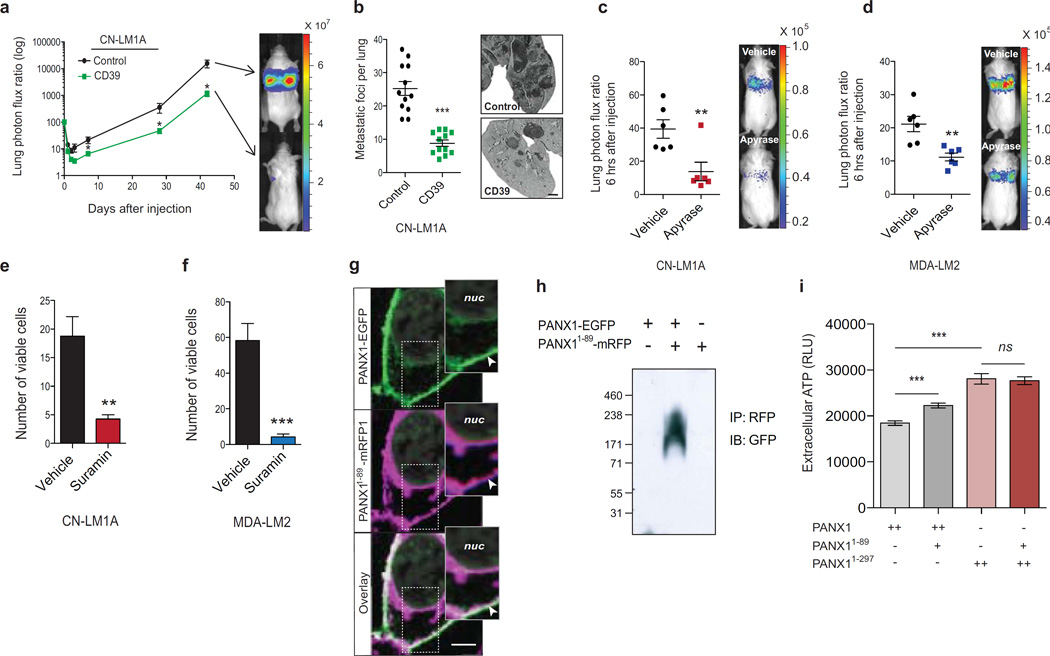
Because intravascular purinergic signaling at the plasma membrane could affect multiple cellular processes28, we asked whether ATP release through PANX1 channels could be responsible for its metastasis-enhancing effects. To address this, we expressed CD39, a plasma membrane-anchored extracellular ATP hydrolase, in CN-LM1A cells and tested their ability to metastasize. CD39 expression, which substantially depleted extracellular ATP at the plasma membrane (Supplementary Figure 4b), significantly reduced metastatic lung signals at early (Supplementary Fig. 4c) and late (Fig. 4a) time-points, and substantially reduced the number of metastatic foci in the lungs (Fig. 4b). Consistent with ATP being necessary for cancer cell survival in the vasculature, CN-LM1A and MDA-LM2 lung bioluminescence was significantly reduced 6 hrs after intravenous co-injection of cells with apyrase, a potent extracellular ATP hydrolase22,37 (Fig. 4c, d). Additionally, the viability of CN-LM1A and MDA-LM2 cells was dramatically reduced upon their exposure to hypotonic solution supplemented with suramin (Fig. 4e, f)—a broad-spectrum antagonist of ATP-binding P2y-purinergic receptors (P2yRs)38, suggesting that ATP release stimulated by plasma membrane stretch activates P2yRs whose signaling protects cancer cells from lethal mechanical injury. These data reveal a role for extracellular ATP release through mechanosensitive PANX1 channels as a cancer-cell-autonomous survival signal during critical deformation of the plasma membrane.
Figure 4. Extracellular ATP enhances metastatic survival via breast cancer cell-autonomous purinergic signaling.
a, Quantitative bioluminescence imaging of lung metastasis after tail-vein injection of 1 × 106 metastatic CN-LM1A cells, expressing CD39 or control vector, into NS mice; n = 12. b, Lungs from day 42 were extracted, H&E stained, and the numbers of metastatic foci were quantified; n = 12. Scale bar, 1 mm. c, Quantitative imaging of lung bioluminescence at 6 hrs post tail-vein injection of 1 × 105 CN-LM1A breast cancer cells pretreated (30 min) and co-injected with apyrase (2U/ml) into FVB/NJ mice; n = 6. d, Quantitative imaging of lung bioluminescence at 6 hrs post tail-vein injection of 4 × 104 MDA-LM2 breast cancer cells pretreated (30 min) and co-injected with apyrase (2U/ml) into FVB/NJ mice; n = 6. e, Quantification of viable, trypan blue-negative, CN-LM1A cells after 15 min incubation in extremely hypotonic (12.5% PBS) solution in the presence of suramin (50 µM) or water vehicle; n = 4. f, Quantification of viable, trypan blue-negative MDA-LM2 cells after 15 min incubation in extremely hypotonic (12.5% PBS) solution in the presence of suramin (50 µM) or water vehicle; n = 4. g, Confocal microscopy images of HEK293T cells expressing PANX1-EGFP (green) and PANX1 1–89-mRFP (magenta). Co-localization at the plasma membrane is shown by channel overlay (white). Nuclear (nuc) DAPI stain is indicated. Arrowheads indicate PANX1-EGFP and PANX1 1–89-mRFP1 colocalization at the plasma membrane. Scale bar, 5 µm. h, Co-immunoprecipitation of PANX1 1–89-RFP from protein-crosslinked (2mM DSP) HEK293T cells expressing PANX1-EGFP, PANX1-EGFP and PANX1 1–89-mRFP, or PANX1 1–89-mRFP. Anti-GFP antibody was used to detect the wild-type PANX1 in complex with mutant PANX1 1–89. Molecular weights are indicated. i, Quantification of ATP release from HEK293T cells transfected with 5 µg wild-type PANX1 (n = 8), 5 µg wild-type PANX1 and 2.5 µg PANX1 1–89 (n = 9), 5 µg C-terminus-deleted PANX11–297 (n = 11) or 5 µg PANX11–297 and 2.5 µg PANX1 1–89 (n = 12). Error bars, s.e.m., ns, nonsignificant, *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Experiments c–f are representative and were replicated at least two times with independent cell lines. Bioluminescent and histological images are representative of the median.
Enhanced PANX1-mediated ATP release is sufficient to increase metastatic efficiency
To assess PANX1 activity in gain-of-function experiments, we utilized the PANX1-activating mutation PANX11–89, because metastatic breast cancer cells did not tolerate the supraphysiological overexpression of wild-type PANX1 channels, presumably secondary to critical losses of ATP. We first wanted to further characterize the molecular association between PANX11–89 and wild-type PANX1 channels. Since PANX1 homo-oligomerizes to form functional channels39 and metastatic cells express both wild-type and mutant PANX1, we hypothesized that PANX11–89 interacts with full-length PANX1. In support of this, we detected prominent co-localization of full-length PANX1 and truncated PANX11–89 at the plasma membrane (Fig. 4g). To directly test whether PANX11–89 interacts with full-length PANX1, we performed reciprocal in vitro co-immunoprecipitation experiments that revealed PANX11–89 to associate with full-length PANX1 (Supplementary Figure 4d, e). To confirm this interaction, we tested whether PANX11–89 complexes with full-length PANX1 in live cells by performing crosslinking of cellular proteins in culture. Co-immunoprecipitation of PANX11–89 from protein-crosslinked cells revealed the mutant protein to be in close association with wild-type PANX1 in vivo (Fig. 4h and Supplementary Figure 4f, g). We also found that the increase in ATP-release mediated by PANX11–89 was independent of caspase signaling, a known activator of PANX1 channel activity in pre-apoptotic cells22,40, as mutation of the caspase cleavage site in PANX1 did not impair its ATP release enhancement (Supplementary Figure 4h). Interestingly, PANX11–89 lost the ability to augment ATP release when co-expressed with PANX11–297, a mutant lacking the intracellular C-terminus domain that has been shown to interact with the pore of the channel to impede ATP release22,40 (Fig. 4i), suggesting that this domain may be required for channel activation by PANX11–89. These findings reveal PANX11–89 to be an activator of PANX1 channels in breast cancer and implicate the N-terminal domain of PANX1 as a positive regulator of channel activation in metastatic breast cancer cells.
We then tested whether PANX11–89 is sufficient to promote metastasis in cancer cells expressing wild-type PANX1 channels. Indeed, expressing PANX11–89 in MDA-MB-468 and HCC1806 cells, which do not express mutant PANX1 endogenously, led to significantly increased metastatic dissemination in orthotopic (Fig. 5a, b) and tail-vein (Fig. 5c, d) metastasis assays. These experiments also revealed the non-organ-specific pro-metastatic effect of PANX11–89, as cells expressing PANX11–89 displayed an enhanced capacity to spread systemically (Fig. 5b, c) and to colonize distant organs such as the lung, liver and bone (Fig. 5d). We next sought to determine the impact of PANX11–89 on intravascular metastatic cell survival. Expression of PANX11–89 in BT549 and MDA-MB-468 cells was sufficient to significantly increase cancer-cell lung bioluminescence 18 hrs after injection (Fig. 5e and Supplementary Figure 5a) and decrease in vivo cell death (activated-caspase signal) at 3- and 6-hours after injection (Fig. 5f and Supplementary Figure 5b). PANX11–89 expression also significantly enhanced the survival of BT549 and MDA-MB-468 cells during hypotonic stretch (Fig. 5g and Supplementary Figure 5c). Moreover, the addition of apyrase to PANX11–89-expressing cells in hypotonic solution completely abrogated the survival advantage afforded by PANX11–89 expression (Fig. 5g and Supplementary Figure 5c), suggesting that extracellular ATP is responsible for the enhanced viability resulting from mutant PANX1 expression. These data reveal that PANX11–89-induced PANX1 channel activity enhances the efficiency of metastasis by promoting metastatic breast cancer cell survival during physical deformation.
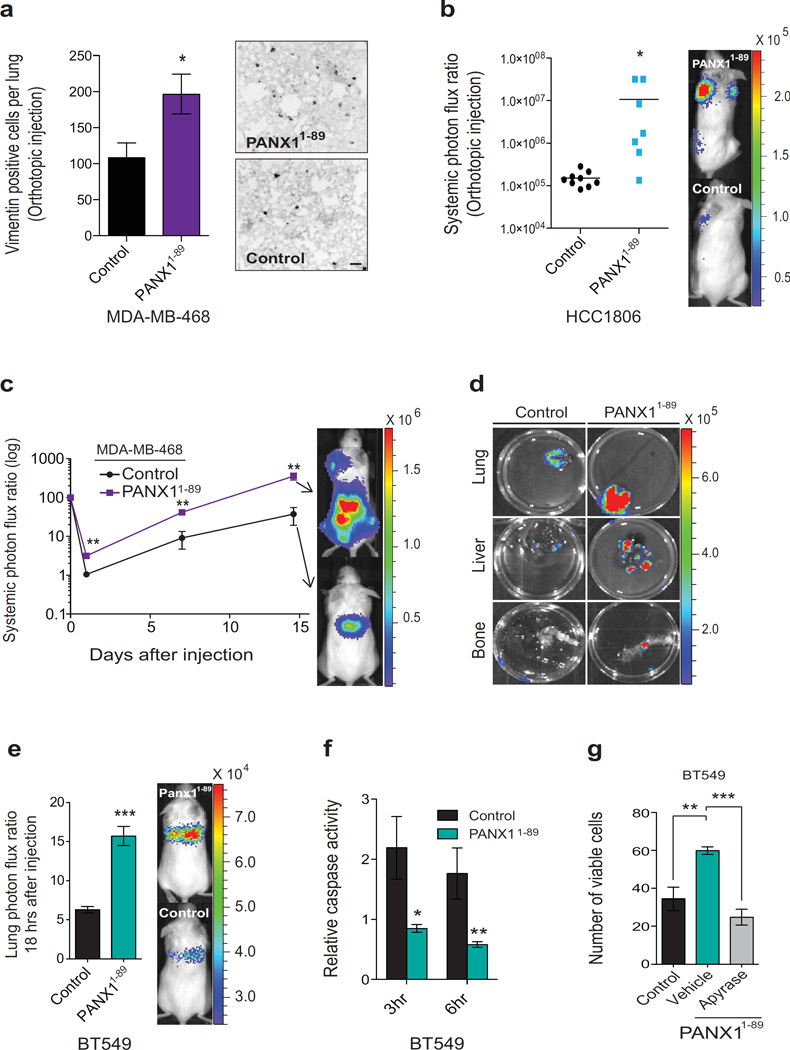
Figure 5. Mutational augmentation of PANX1 channel activity enhances the metastatic efficiency of breast cancer cells.
a, The numbers of vimentin positive breast cancer cells in the lung were counted one week after the extraction of size-matched mammary fat pad primary tumours generated by the orthotopic injection of 2.5× 105 MDA-MB-468 cells expressing PANX1 1–89 or control vector into NOD scid gamma (NSG) mice; n = 4. Scale bar, 100 µm. b, Quantitative bioluminescence imaging of systemic metastasis one week after the extraction of size-matched mammary fat pad tumours generated by the orthotopic injection of 5 × 105 HCC1806 breast cancer cells expressing PANX1 1–89 (n = 7) or control vector (n = 9) into NSG mice. c, Quantitative bioluminescence imaging of systemic metastasis after tail-vein injection of 1 × 106 MDA-MB-468 breast cancer cells, expressing PANX1 1–89 or control vector, into NSG mice; n = 4. d, Ex vivo bioluminescence imaging of metastatic target organs (lung, liver and bone) 14 days after tain-vein injection of MDA-MB-468 cells. e, Quantitative imaging of lung bioluminescence 18 hrs post tail-vein injection of 1 × 106 BT549 cells, expressing PANX1 1–89 or a control vector, into NSG mice; n = 6. f, In vivo quantification of luciferase-based caspase-3/7 activity at 3 and 6 hrs post tail-vein injection of 1 × 106 BT549 cells, expressing either PANX1 1–89 or a control vector, into NS mice; n = 4 (3hr control), n = 5 (3hr PANX1 1–89), n = 4 (6hr control), n = 5 (6hr PANX1 1–89). g, Quantification of viable, trypan blue-negative BT549 cells expressing PANX1 1–89 or a control vector after 1 hr extreme hypotonic (12.5% PBS) stretch in the presence of succinate buffer or apyrase (2U/ml); n = 3. Error bars, s.e.m., *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Orthotopic experiments replicated in two independent triple-negative cell human breast cancer cell lines. Experiments e–g are representative and were replicated at least two times in at least two independent cell lines. Bioluminescent and histological images are representative of the median.
Pharmacological inhibition of PANX1 channels reduces breast cancer metastasis
Finally, we investigated whether pharmacological inhibition of activated PANX1 channels on metastatic cells could suppress metastatic progression. CN-LM1A cells pretreated with CBX, a PANX1 channel inhibitor approved for the treatment of gastroesophogeal reflux disease (GERD) in the UK, displayed dramatically reduced lung-metastatic activity (Supplementary Figure 5d, e). To better assess the clinical potential of this therapy, we treated mice with two distinct CBX regimens prior to introducing metastatic breast cancer cells into the circulation (Fig. 6a). We found that seven days of CBX therapy significantly inhibited the ability of highly metastatic MDA-LM2 cells to colonize the lungs (Fig. 6b, c). Importantly, this treatment was well tolerated and did not impact body weight (Supplementary Figure 5f). Strikingly, reducing this treatment to two doses of CBX within 24 hrs of cancer cell injection significantly reduced the number of highly metastatic CN-LM1A cells in the lungs at as early as 24 hrs (Fig. 6d, e) as well as lung metastatic colonization at four weeks (Fig. 6f, g and Supplementary Figure 5g). These data suggest that pharmacological PANX1 inhibition could have the potential to blunt the development of new distant metastases in patients with metastatic primary tumors.
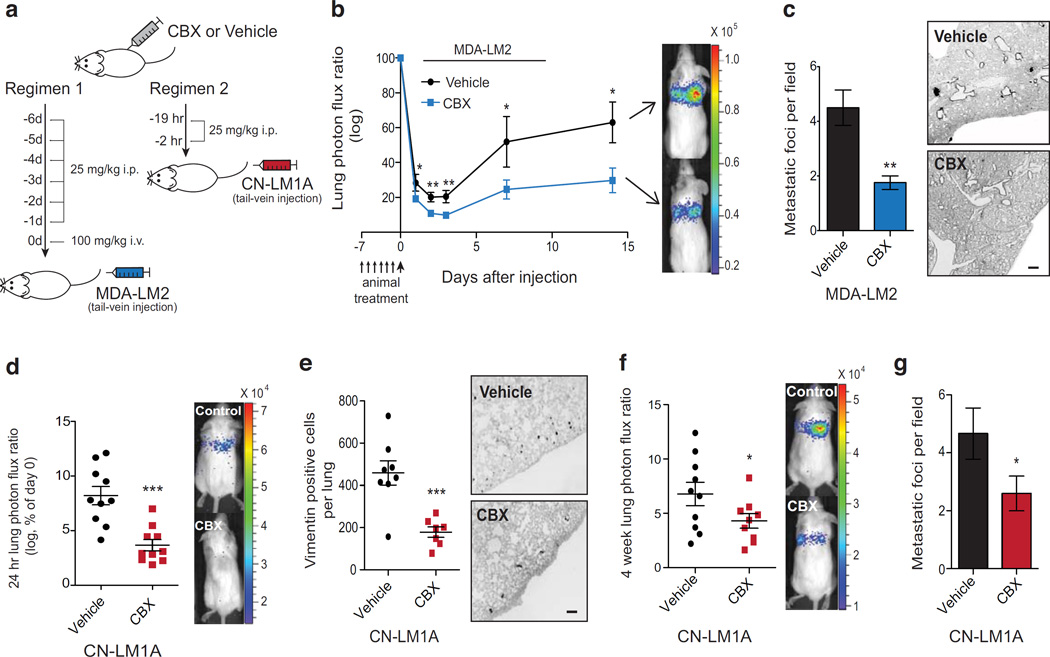
Figure 6. Pharmacological PANX1 channel blockade reduces breast cancer metastasis to the lungs.
a, Schematic depicting the two in vivo CBX therapy regimens tested. b, Quantitative bioluminescence imaging of lung metastasis after tail-vein injection of 5 × 104 MDA-LM2 breast cancer cells into NS mice pretreated daily with 25 mg/kg i.p. CBX or an equivalent volume of PBS for six days and with 100 mg/kg i.v. CBX or an equivalent volume of PBS 30 min prior to cancer cell injection; n = 8 (vehicle), n = 9 (CBX). c, Quantification of the number of metastatic foci at 2-weeks from H&E-stained lungs (left) and representative images from vimentin-stained lungs (right); n = 4. Scale bar, 0.5 mm. d, Quantitative bioluminescence imaging lungs 24 hrs after tail-vein injection of 1 × 105 CN-LM1A breast cancer cells into NS mice pretreated with 25 mg/kg i.p. CBX or an equivalent volume of PBS at 19 and 2 hrs prior to cancer cell injection; n = 10. e, Lungs were extracted at 24 hrs, sectioned and stained for vimentin and the number of vimentin-positive cancer cells were quantified; n = 8 (vehicle), n = 7 (CBX). Scale bar, 0.25 mm. f, Quantitative bioluminescence imaging of breast cancer cells in the lungs at 4 weeks after tail-vein injection of 1 × 105 CN-LM1A breast cancer cells into NS mice pretreated with 25 mg/kg i.p. CBX (n = 9) or an equivalent volume of PBS (n = 10) at 19 and 2 hrs prior to cancer cell injection. g, Quantification of the number of metastatic foci at 4-weeks from H&E-stained lungs; n = 10 (vehicle), n = 9 (CBX). Error bars, s.e.m., *, P < 0.05; **, P < 0.01; ***, P < 0.001 by a one-tailed Student’s t-test. n represents biological replicates. Therapeutic PANX1 inhibition experiments were performed using two independent triple negative breast cancer cell lines. Bioluminescent and histological images are representative of the median.
Discussion
By applying next-generation sequencing analyses to in vivo-selected highly metastatic sub-populations, we have identified a mutation in a channel that promotes metastatic progression. This study illustrates how the identification of a sub-clonal mutation that is recurrently enriched in the transcriptomes of highly metastatic human cancer cells can serve as a molecular clue for uncovering biological mechanisms that regulate a complex step of cancer metastasis. Our identification of a PANX1 channel-activating mutation, PANX11–89, led to our finding that PANX1 channel activity can promote the metastasis of certain breast cancer cells by improving their ability to withstand traumatic deformation in the microvasculature of target organs. Though the extent to which PANX1 activity plays a role in human cancer metastasis remains unknown, we propose a working-model based our findings whereby augmented ATP release through mechanosensitive PANX1 channels results in the cell-autonomous enhancement of cancer cell survival during microvasculature-induced plasma membrane stretch—an important barrier to metastatic progression (Fig. 6). Our work suggests that ATP released via PANX1 channels mediates this effect through purinergic receptors on the cell surface, which have been implicated in survival signaling during deformation-induced injury37.
While we have identified ATP release and its action through purinergic P2Y receptor(s) as mediating the survival effects during breast cancer cells stretch, the identity of the specific purinergic receptor(s) remains unknown. Systematic investigation of P2Y receptors involved in PANX1-mediated ATP-dependent mechanical survival should reveal the P2Y receptor(s) specifically responsible for mediating the pro-survival effects. Moreover, future studies will uncover the signal transduction pathways and their link to pro-survival genes that mediate cell survival by this pathway.
We have found that the truncated PANX11–89 protein augments ATP release from cells. The ability of PANX11–89 to augment ATP release requires full-length wild-type PANX1 expression. The molecular/structural basis for the suggested interaction between PANX11–89 and full-length PANX1 remains to be characterized. Crystallographic studies of PANX11–89 in association with full-length PANX1 should be quite illuminating with respect to the mechanism of enhanced PANX1 channel opening. One potential mechanism by which PANX11–89 augments ATP release via PANX1 channels might be through the loss of the auto-inhibitory C-terminal domain that is lacking in the PANX11–89 mutation40. It is possible that other truncating mutations lacking the PANX1 C-terminal, as well as missense mutations interfering with the tail-to-pore interaction, could also lead to increased channel activity.
We have shown that enhanced ATP release through PANX1 can promote breast cancer cell survival in the context of mechanical deformation. It is also possible that ATP or breakdown products of ATP, such as ADP, AMP or adenosine, could also regulate metastatic progression at later time-points by modulating the microenvironment through effects on immune cell effectors.
PANX1 channel opening is herein identified as a potential druggable target whose therapeutic inhibition may reduce the development of clinical metastases. Patients at high risk for relapse that may particularly benefit from PANX1 blockade therapy include those undergoing diagnostic or therapeutic procedures that may cause cancer cells to enter the vasculature during mechanical tumour perturbation. Fine needle aspiration of human breast tumours has been shown to cause shedding of cancer cells into the bloodstream41, which are rapidly transported to distant sites. Likewise, experiments in mice have found that operative trauma during surgical dissection can lead to hematogenous dissemination and subsequent metastasis42,43. Therapeutic inhibition of PANX1 with CBX—a drug with proven safety in humans—has the potential to reduce the development of distant metastases in such cases. CBX could also represent a starting point for the development of more potent PANX1 inhibitors through medicinal chemistry. It will be important for future studies to examine whether augmented PANX1 activity, via activating mutations or increased gene expression, plays a broader role in cancer metastasis by additional tumour types.
Methods
Cell culture
BT549 and MDA-MB-468 cells were a generous gift from S. Chandarlapaty (Human Oncology & Pathogenesis Program, Memorial Sloan-Kettering Cancer Center). CN34, MDA-MB-231 cells and their respective sub-lines, CN-LM1A and MDA-LM2, were propagated as previously described16,44. HEK293T, PANX1-null MEFs, and MDA-MB-468 cells were cultured in DMEM-based media supplemented with 10% FBS, glutamine, pyruvate, penicillin, streptomycin and fungizone. BT549 and HCC1806 lines were cultured in RPMI-based media supplemented with 10% FBS, glutamine, pyruvate, penicillin, streptomycin and fungizone. Cells were periodically tested for Mycoplasma contamination. CN32, MDA-MB-231, CN-LM1A and MDA-LM2 cell lines were authenticated by RNA-seq profiling. All transfections were performed using Lipofectamine 2000 (Life Technologies).
RNA sequencing, read alignment, allele frequency quantification and SNV calling
Total RNA was extracted using the MiRvana kit (Ambion) and reverse transcribed using the cDNA First-Strand Synthesis kit (Life Technologies). For whole-transcriptome sequencing, cDNA libraries were generated using the mRNA Sequencing Sample Preparation Kit (Illumina, 2009) according to the manufacturer’s instructions and sequencing-by-synthesis was performed using the GAIIx sequencer (Illumina). For read alignment, the quality of the reads generated by high-throughput sequencing was first examined using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The reads were then trimmed and filtered by quality and aligned against the human genome (release hg18) using the TopHat aligner45, which builds exon models de novo from the RNA-seq data and aligns reads across splice junctions. PCR duplicates that might interfere with SNV calling were removed. A statistical framework was then implemented to identify SNVs from the read alignments based on the assumption that biological variations will be found in a significant number of reads, while non-biological variations will not. A Poisson-Binomial distribution46 was used to calculate the p-values of significance, taking into account the total number of reads at each genomic position, number of reads with mismatches, as well as the error rate at each sequencing cycle (e.g. typically the sequencing error is higher during later stages of sequencing). P-values were corrected for multiple hypotheses using the Benjamini-Hochberg method47 and the false discovery rate was controlled to 1%. SNVs identified in less than 10 unique reads were removed from further analysis. The SNVs were also filtered to remove known single nucleotide polymorphisms (SNPs) reported in dbSNP (v130) and annotated with our in-house programs (T.D.S. and O.E., in preparation). To minimize the false-positive rate and enable subsequent statistical analyses, we sequenced biological replicates of each cell line. Low-confidence mutations were excluded by removing known polymorphisms, low coverage (<10×) nSNVs and any nSNV not detected in both biological RNA-seq replicates of each cell line. To identify nSNVs significantly enriched during metastatic selection, we quantified the differences in nSNV allelic frequencies between the highly metastatic sub-lines and their respective poorly metastatic parental lines. Metastasis-enriched nSNVs were defined as those that displayed an increase in allelic frequency over the four rounds of RNA-seq and met the criteria for significance (p < 0.05, q ≤ 0.25) by a one-tailed Student’s t-test (Supplementary Fig. 1A). Two additional computational algorithms (SNVmix48 and VarScan49) independently validated these methods. SNV allelic ratios were calculated by dividing the number of unique reads containing a given variant by the total number of unique reads at each position50. Amino acid substitutions resulting from the nSNVs were ranked by the likelihood of being non-neutral using the computational tool PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)51.
Allele-specific RNA-seq
Two-step PCR reactions of the PANX1 C268T allele in biological triplicates of each cell line were performed using the barcoded primers listed in Supplementary Table 3. PCR amplicons were then sequenced using the HiSeq2000 (Illumina) platform. Position specific quantification of each allele was performed and used to calculate the PANX1 C268T allelic frequency of each cell line replicate.
Sanger sequencing
Sanger sequencing was performed by GENEWIZ. Sequencing primers are listed in Supplementary Table 3.
Animal studies
All animal work was conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee at The Rockefeller University. Seven- to eight-week-old age-matched female NOD/SCID mice were used for breast cancer tail-vein lung colonization and systemic metastatic colonization assays17,44. PANX1-null mice (Shestopalov lab, University of Miami) were bred for the generation PANX1-null MEFs at embryonic day 14 (E14). For the in vitro PANX1 inhibition metastasis assays, 1 × 105 LM1A or 4 × 104 LM2 cells in 100 µl were incubated for 30 min with either 100 µM 10Panx1 (Tocris), 100 µM scrambled peptide (Tocris), 500 µM CBX (Sigma-Aldrich) or an equivalent volume of PBS vehicle and injected intravenously into the lateral tail-vein. For the in vivo therapeutic PANX1 inhibition metastasis assays, mice were weighed, treated with CBX or an equivalent volume of PBS by intraperitoneal injections at the indicated times and doses, and then tail-vein injected with 1 × 105 LM1A or 4 × 104 LM2 cells. For the PANX1 extracellular ATP release assays, 1 × 105 MDA-LM2 cells expressing plasma membrane-anchored luciferase52 (LM2-pmeLUC) were preincubated with CBX (500 µM) or an equivalent volume of PBS vehicle and injected into seven-week-old age-matched female FVB/NJ mice. For orthotopic metastasis assays, two primary tumours per NSG mouse were generated through bilateral injections of 2.5 × 105 MDA-MB-468 or 5.0 × 105 HCC1806 breast cancer cells into the lower mammary fat pads. The resulting tumours were measured every three days, size-matched, extracted at 100 mm3 and metastatic cell dissemination and colonization was noninvasively assayed through bioluminescence imaging as previously described17. For the long-term metastasis assays involving extracellular ATP depletion, 1 × 106 CN-LM1A cells, expressing the plasma membrane-anchored extracellular ATP hydrolase CD39 or control vector, were injected into the lateral tail-vein of NS mice. For the acute extracellular ATP depletion assays, cells were preincubated with apyrase (2U/ml; NEB) or an equivalent volume of succinate buffer control and injected into seven-week-old age-matched female FVB/NJ mice. For the truncated PANX1 in vivo metastasis assays, 5 × 105 cellsper 100 µl PBS were introduced intravenously through tail-vein (MDA-MB-468 and BT549). Caspse activity was measured in vivo through retro-orbital injections of 0.75 mg of VivoGlo™ Caspase 3/7 Substrate (Z-DEVD-Aminoluciferine Sodium Salt) (Promega)53 per mouse.
Generation of PANX1-null MEFs
MEFs from E14 PANX1-null embryos were generated as previously described54.
Generation of retrovirus and stable overexpression cells
Generation of retroviral-overexpressing cells was performed as previously described17,44,55. Primers used to generate overexpression constructs are listed in Supplementary Table 3. C-terminus fluorescent protein-tagged PANX1 and PANX11–89 were cloned into the respective pcDNA3-EGFP and pcDNA3-mRFP1 vectors with a GRPLE linker. Plasma-membrane anchored extracellular luciferase was cloned as previously described52. CD39 (clone 5762493, Open Biosystems) was stably expressed using the lentiviral pLenti vector system. For bioluminescent tracking of MDA-MB-468, BT549 and HCC1806 cancer lines, cells were labeled with a triple-fusion protein reporter construct as previously described17.
siRNA-mediated full-length-PANX1-specific messenger RNA (mRNA) knockdown
siRNAs were transfected into HCC1806 cells using Lipofectamine reagent according to the manufacturer’s protocol. Forty-eight hours after transfection, the cellular RNA was extracted and full-length PANX1 mRNA levels were quantified as described previously17. At ninety-six hours after transfection, the cells were subjected to extracellular ATP release assays as described below. siRNA specifically designed to target endogenous full-length PANX1 mRNA without interfering with PANX11–89 overexpression was purchased from Life Technologies (ID: 134470, siRNA_A) and Thermo Scientific (custom siRNA sequence: 5’-AGGAAGCACCUGAGAGUAUU-3’, siRNA_B). BLOCK-iT control siRNAs were obtained from Invitrogen.
Analysis of mRNA expression
Expression of mRNA was quantified as described previously17. Primers can be found in Supplementary Table 3.
Immunofluorecence and confocal microscopy
Cells expressing fluorescently tagged proteins were fixed in 4% paraformaldehyde, stained with DAPI (Roche), mounted using ProLong Gold Antifade reagent (Life Technologies), and imaged using the Leica TCS SP5 II system.
Immunoprecipitation and immunoblotting
Cellular lysates were prepared by lysing cells (10–40 million) overnight in ice-cold RIPA buffer containing protease and phosphatase inhibitors (Roche). The next day, cellular debris was removed by centrifugation (12,000 rpm) for 20 min at 4 °C. A 50 µL slurry of Anti-FLAG M2 Magnetic Beads (Sigma-Aldrich) was added to the supernatant and rocked for 4 hrs at 4 °C. The beads were then washed three times with ice-cold lysis buffer. The immunoprecipitated proteins were eluted by denaturation in Laemmli buffer at 95 °C for 5 min, separated using SDS–PAGE, transferred to a PVDF membrane (Pierce), blocked and probed using a primary antibody to the N-terminal of PANX1 (1:1,000; ZMD.695 from Life Technologies), which has not been validated specifically for the detection of PANX11–89. Bound antibodies were chemi-luminescently detected using horseradish peroxidase–conjugated secondary antibodies (1:10,000), ECL Western Blotting Substrate (Pierce) and the SRX-101A (Konica Minolta) developer, according to the manufacturer’s instructions.
Protein crosslinking immunoprecipitation and immunoblotting
Dithiobis[succinimidyl propionate] (DSP) was dissolved in anhydrous DMSO, diluted in PBS to formulate crosslinking solutions at the concentrations indicated, and used to crosslink cells in culture according to the manufacturer’s instructions (Pierce). Crosslinked cells were then washed twice in PBS and lysed using ice-cold RIPA buffer supplemented with protease and phosphatase inhibitors (Roche). Lysates were collected by cell scraping, sonicated and rotated at 4 °C for 4–12 hrs. Cellular debris was removed by centrifugation (12,000 rpm) for 20 min at 4 °C. Protein concentrations were measured using the BCA protein assay (Pierce). For immunoprecipitation of crosslinked proteins, a 50 µL slurry of Anti-RFP Magnetic Beads (MBL International) was added to 200 µg protein lysates diluted in 750 µL Co-IP lysis buffer (Thermo Scientific) and rocked for 4 hrs at 4 °C. The beads were then washed three times in Co-IP lysis buffer, resuspended in Non-Reducing Lane Marker Sample Buffer (Thermo Scientific), boiled at 95 °C for 5 min, and separated using SDS-PAGE on a Novex 3–8% Tris-Acetate gel (Life Technologies). For crosslinked PANX1 complex immunoblotting, primary antibodies to the N-terminal of PANX1 (1:1000; ZMD.695 from Life Technologies), GFP (1:1000, ab290 from Abcam) and RFP (1:1000; biotin conjugated ab34771 from Abcam) were used. Bound antibodies were chemi-luminescently detected using horseradish peroxidase–conjugated secondary antibodies (1:10,000), ECL Western Blotting Substrate (Pierce) and the SRX-101A (Konica Minolta) developer, according to the manufacturer’s instructions.
ATP release assays
Cells were seeded in quadruplicate at 100,000–200,000 cells per well in 24-well plates and grown overnight. Each well was then washed with 200 µl PBS. For PANX1 inhibition, cells were incubated at room temperature for 10 min in PBS supplemented with one of the following reagents: CBX (500 µM), PB (2 mM; Life Technologies), 10Panx1 (100 µM) or an equivalent dose of the appropriate vehicle control (100% PBS or scrambled peptide). The wash or pretreatment solution was then aspirated, replaced with 200 µM fresh 100% PBS or 70% PBS (hypotonic stretch assays) for the indicated times, harvested and transferred to microcentrifuge tubes, and then spun at 1,000 rpm for 2 min at room temperature. Supernatants were transferred to 96-well plates and ATP was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions. For hypotonic stretch assays, the numbers of viable cells remaining post-stretch were counted for each treatment condition to rule out the contribution of cell lysis to extracellular ATP levels.
Histology and Immunofluorescence
For histological quantification of single breast cancer cells, mice lungs were extracted at one and three days post cancer cell injection. By administering solutions through the heart and trachea, lungs were perfused and fixed with PBS and 4% paraformaldehyde, respectively. Slices were mounted using ProLong Gold Antifade Reagent (Life Technologies). For histological quantification of metastatic foci, mice lungs were extracted at the indicated time-points, perfused and fixed as above, and sent out for sectioning and H&E staining (Histoserv). Quantification of cells and metastatic foci from histological step-sections was performed in a blinded manner. Immunohistochemical staining for human vimentin (1:20, clone V9, Vector Laboratories) was performed on paraffin-embedded sections and visualized with Vectastain ABC kit (PK-6012) and DAB chromagen (SK-4105, Vector Laboratories). Vimentin-positive macroscopic nodules were counted for experimental analysis.
For endothelial labeling, 100 mg of biotinlyated lectin dissolved in PBS was injected retro-orbitally and allowed to circulate for 5 min prior to fixation. Mice were subsequently sacrificed, and their lungs were extracted without perfusion and fixed in 4% paraformaldehyde overnight. For immunofluorescence, frozen lung sections (10 microns thick) were stained with antibodies against human vimentin (1:40, clone V9, Vector Laboratories) and cleaved caspase-3 (1:2000, clone 5A1E, Cell Signaling Technology). Primary antibodies were visualized with Alexa Fluor Dye-conjugated secondary antibodies (1:200, Life Technologies.) Stained sections were mounted using ProLong Gold Antifade Reagent (Life Technologies). Images were obtained using Zeiss scanning laser confocal microscope (LSM 510).
Hypotonic deformation cell viability assays
Cells were seeded in quadruplicate at 100,000 cells per well in 24-well plates and grown overnight. When comparing wild-type and mutant PANX1-expressing cells, replicate plates of cells were counted at the time of the assay for normalization. Each well was then washed once with 200 µl 100% PBS. For PANX1 inhibition, cells were incubated with 100 µM 10Panx1 or scrambled peptides for 10 min in 100% PBS. The wash or pretreatment solution was then aspirated, and the cells were stretched in 200 µl hypotonic solution (12.5% PBS) supplemented with the indicated reagents or vehicle controls for the times indicated. For ATP rescue experiments, 100 µM ATP (NEB) was added to the 10Panx1 hypotonic solution, and an equivalent volume of water was added to the control hypotonic solution. For pan-P2yR inhibition experiments, suramin (50 µM; Sigma-Aldrich) or water vehicle control was added to the pretreatment solution. For ATP depletion experiments, apyrase (2 U/ml) or an equivalent volume of succinate buffer control was added to the hypotonic solution. After deformation, the cells were gently washed twice with 100% PBS, trypsinized, stained with trypan blue (Sigma-Aldrich) and the remaining viable cells were quantified.
Centrifugal deformation cell viability assays
Cells were pretreated with CBX (500 µM) or PBS for 10 min in suspension, then washed and seeded in quadruplicate at 100,000 cells per Boyden chamber in fresh DMEM-based media. The chambers were placed in 24-well plates and centrifuged for 30 min at 3,800 rpm. Viable cells were stained with trypan blue (Sigma-Aldrich) and quantified.
Cancer cell proliferation
For molecular PANX1 inhibition assays, 5 × 103 cancer cells overexpressing either the autoinhibitory C-terminal domain or control vector were seeded in quadruplicate in a 96 well plate in a 100 µl volume of DMEM-based media containing 10% FBS. At days 0, 3, and 5, cells were trypsinized and viable cells were counted using a hemocytometer. Proliferation counts were normalized to day 0. For peptide inhibition proliferation assays, cancer cells were pretreated with 100 µM 10Panx1 peptide or vehicle for 15 min and seeded into 24 well plates (25,000 cells/well) under the same conditions as pretreatment. After 24 hrs, cancer cells were trypsinized and viable cells were counted using a hemocytometer.
Invasion assays
Invasion assays were performed as previously described 44. Briefly, serum starved sub-lines were pretreated for 15 min with 100 µM of 10Panx1 peptide or scrambled peptide in 0.2% FBS DMEM-based media, seeded onto Trans-well invasion chambers (BD Biosciences) under the same treatment conditions and incubated for 18–20 hrs. Cells that had invaded were counted in five fields per insert and then quantified using ImageJ.
Transendothelial migration assays
Transendothelial migration assays were performed as previously described 44. Briefly, serum starved Cell Tracker Green CMFDA-labeled (Life Technologies) MDA-LM2 cells were pretreated for 15 min with 100 µM of 10Panx1 peptide or scrambled peptide in 0.2% FBS DMEM-based media, seeded on a monolayer of HUVEC cells under the same treatment conditions (50,000 cells/well) and incubated for 18 hrs. Migrated cells were counted in five fields per insert and then quantified using ImageJ.
Anchorage-independent survival
Cancer cells (5,000 cells/well) were seeded in quadruplicate on 96-well Ultra-Low Attachment Surface plates (Corning) in the presence of 100 µM of 10Panx1 peptide or scrambled peptide in 10% FBS DMEM-based media and incubated for 36 hours. Cell suspensions were then transferred to microcentrifuge tubes, isolated by centrifugation (1,000 rpm) for 5 min, trypsinized and counted using a hemocytometer.
Statistics and Representative Images
No statistical methods were used to predetermine sample size. The experiments were not randomized and, other than histological nodule and single cell quantification, the investigators were not blinded to allocation during experiments and outcome assessment. For all animal studies, discrepancies between n for each data set are the result of death, illness or technical challenges unrelated to the experimental conditions being compared. No randomization or blinding was used for the animal studies. For in vitro experiments, differences in n are due to technical issues during experimental setup or implementation and are unrelated to the experimental outcomes. Exclusion criteria for all studies were pre-established. The statistical tests used to compare experimental samples and cohorts are well established and previously described. One-tailed Student’s t-test is the most common statistical analysis implemented in this study. To minimize unrecognized assumptions during data analysis, we often used independent statistical methods for individual experiments. All experimental results presented with representative images, as defined in the figure legends, have been replicated at least two times. Bioluminescent and histological images are representative of the median.
Supplementary Material
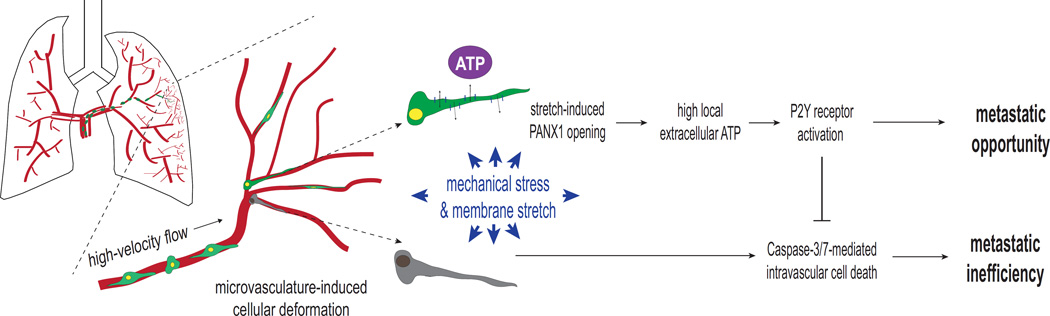
Figure 7. Proposed working-model of PANX1-mediated ATP release as a regulator of intravascular metastatic cell survival.
Schematic showing stretch-induced ATP release through mechanosensitive PANX1 channels activating a cancer cell-autonomous purinergic signaling pathway that inhibits cell death during mechanical stress in the microvasculature of target organs. Those cancer cells enabled to secrete the levels of extracellular ATP necessary to permit intravascular survival via autocrine purinergic signaling pathways, or the adjacent cells lodged within the same tumor cell cluster, are afforded an opportunity to undergo the subsequent steps of the metastatic cascade—extravasation, colonization, re-initiation and proliferation.
Acknowledgments
We thank C. Alarcon, N. Pencheva, A. Nguyen, J. Ross and Saeed Tavazoie for comments on previous versions of this manuscript. We thank C. David Allis, B. Chait, T. Kapoor and V. Ruta for their insightful comments on this work. We thank C. Zhao of the Rockefeller Genomics Resource Center and J. Xiang of the Weill Cornell Medical College Genomics Resources Core Facility for assistance with next-generation RNA-sequencing. We thank S. Chandarlapaty at MSKCC for providing the BT549, MDA-MB-468 and HCC1806 breast cancer cell lines. We thank V. Shestopalov for providing the PANX1-null mice. J. Massague generously provided metastatic sub-lines. P.W.F. is a biomedical fellow in the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program supported by NIH MSTP grant GM07739 and an Anderson Cancer Center Fellow at Rockefeller University. N.H is an Anderson Cancer Center Fellow at Rockefeller University. S.F.T. is a Department of Defense (DOD) Era of Hope Scholar, a DOD Scholar-Innovator Award Recipient and a Rita Allen Foundation Scholar.
Footnotes
Accession Numbers
RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE45162.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
S.F.T. conceived the project and supervised all research. P.W.F. and S.F.T. wrote the manuscript. P.W.F. and S.F.T. designed the experiments. P.W.F., S.Z., N.H., and C.M. performed the experiments. T.D.S., H.G., P.W.F. and O.E. designed and performed the computational analysis.
The authors declare no competing financial interests.
References and Notes
- 1.ZEIDMAN, I. The fate of circulating tumors cells. I. Passage of cells through capillaries. Cancer Res. 1961;21:38–39. [PubMed] [Google Scholar]
- 2.Weiss L, Nannmark U, Johansson BR, Bagge U. Lethal deformation of cancer cells in the microcirculation: a potential rate regulator of hematogenous metastasis. Int J Cancer. 1992;50:103–107. doi: 10.1002/ijc.2910500121. [DOI] [PubMed] [Google Scholar]
- 3.Wong CW, et al. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61:333–338. [PubMed] [Google Scholar]
- 4.Kienast Y, et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 5.Weiss L. Biomechanical interactions of cancer cells with the microvasculature during hematogenous metastasis. Cancer Metastasis Rev. 1992;11:227–235. doi: 10.1007/BF01307179. [DOI] [PubMed] [Google Scholar]
- 6.Weiss L, Harlos JP, Elkin G. Mechanism of mechanical trauma to Ehrlich ascites tumor cells in vitro and its relationship to rapid intravascular death during metastasis. IntJ Cancer. 1989;44:143–148. doi: 10.1002/ijc.2910440125. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korpal M, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min J, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–68. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Locovei S, Bao L, Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proc Natl Acad Sci USA. 2006;103:7655–7659. doi: 10.1073/pnas.0601037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ransford GA, et al. Pannexin 1 contributes to ATP release in airway epithelia. Am J Respir Cell Mol Biol. 2009;41:525–534. doi: 10.1165/rcmb.2008-0367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seminario-Vidal L, et al. Rho signaling regulates pannexin 1-mediated ATP release from airway epithelia. J Biol Chem. 2011;286:26277–26286. doi: 10.1074/jbc.M111.260562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chekeni FB, et al. Pannexin 1 channels mediate find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467:863–867. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandilos JK, Bayliss DA. Physiological mechanisms for the modulation of pannexin 1 channel activity. J Physiol. 2012;590:6257–6266. doi: 10.1113/jphysiol.2012.240911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacVicar BA, Thompson RJ. Non-junction functions of pannexin-1 channels. Trends Neurosci. 2010;33:93–102. doi: 10.1016/j.tins.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Thompson RJ, et al. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science. 2008;322:1555–1559. doi: 10.1126/science.1165209. [DOI] [PubMed] [Google Scholar]
- 26.Thompson RJ, Zhou N, MacVicar BA. Ischemia opens neuronal gap junction hemichannels. Science. 2006;312:924–927. doi: 10.1126/science.1126241. [DOI] [PubMed] [Google Scholar]
- 27.Schenk U, et al. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci Signal. 2008;1:ra6. doi: 10.1126/scisignal.1160583. [DOI] [PubMed] [Google Scholar]
- 28.Corriden R, Insel PA. Basal release of ATP: an autocrine-paracrine mechanism for cell regulation. Sci Signal. 2010;3:re1. doi: 10.1126/scisignal.3104re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher D, Strilic B, Sivaraj KK, Wettschureck N, Offermanns S. Platelet-Derived Nucleotides Promote Tumor-Cell Transendothelial Migration and Metastasis via P2Y2 Receptor. Cancer Cell. 2013;24:130–137. doi: 10.1016/j.ccr.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Gulbransen BD, et al. Activation of neuronal P2×7 receptor-pannexin-1 mediates death of enteric neurons during colitis. Nat Med. 2012;18:600–604. doi: 10.1038/nm.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silverman W, Locovei S, Dahl G. Probenecid, a gout remedy, inhibits pannexin 1 channels. Am J Physiol Cell Physiol. 2008;295:C761–C767. doi: 10.1152/ajpcell.00227.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruzzone R, Barbe MT, Jakob NJ, Monyer H. Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J Neurochem. 2005;92:1033–1043. doi: 10.1111/j.1471-4159.2004.02947.x. [DOI] [PubMed] [Google Scholar]
- 33.Ma W, Hui H, Pelegrin P, Surprenant A. Pharmacological characterization of pannexin-1 currents expressed in mammalian cells. J Pharmacol Exp Ther. 2009;328:409–418. doi: 10.1124/jpet.108.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian B, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 36.Li A, et al. Mechanisms of ATP release, the enabling step in purinergic dynamics. Cell Physiol Biochem. 2011;28:1135–1144. doi: 10.1159/000335865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belete HA, Hubmayr RD, Wang S, Singh RD. The role of purinergic signaling on deformation induced injury and repair responses of alveolar epithelial cells. PLoS One. 2011;6:e27469. doi: 10.1371/journal.pone.0027469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, Piskuric NA, Vollmer C, Nurse CA. P2Y2 receptor activation opens pannexin-1 channels in rat carotid body type II cells: potential role in amplifying the neurotransmitter ATP . J Physiol. 2012;590:4335–4350. doi: 10.1113/jphysiol.2012.236265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambrosi C, et al. Pannexin1 and Pannexin2 channels show quaternary similarities to connexons and different oligomerization numbers from each other. J Biol Chem. 2010;285:24420–24431. doi: 10.1074/jbc.M110.115444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandilos JK, et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J Biol Chem. 2012;287:11303–11311. doi: 10.1074/jbc.M111.323378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu XC, Chow LW. Fine needle aspiration may shed breast cells into peripheral blood as determined by RT-PCR. Oncology. 2000;59:217–222. doi: 10.1159/000012164. [DOI] [PubMed] [Google Scholar]
- 42.Murthy SM, et al. The influence of surgical trauma on experimental metastasis. Cancer. 1989;64:2035–2044. doi: 10.1002/1097-0142(19891115)64:10<2035::aid-cncr2820641012>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 43.Demicheli R, Retsky MW, Hrushesky WJ, Baum M, Gukas ID. The effects of surgery on tumor growth: a century of investigations. Ann Oncol. 2008;19:1821–1828. doi: 10.1093/annonc/mdn386. [DOI] [PubMed] [Google Scholar]
- 44.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 45.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Liu JS. Statistical Applications of the Poisson-Binomial and Conditional Bernouilli Distributions. Statistica Sinica. 1997;7:875–892. [Google Scholar]
- 47.Benjamini YHT. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 48.Goya R, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–736. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pellegatti P, Falzoni S, Pinton P, Rizzuto R, Di Virgilio F. A novel recombinant plasma membrane-targeted luciferase reveals a new pathway for ATP secretion. Mol Biol Cell. 2005;16:3659–3665. doi: 10.1091/mbc.E05-03-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biserni A, Martorana F, Roncoroni C, Klaubert D, Maggi A, Ciana P. Identification of Apoptotic Cells in Reporter Mice Using Modified Luciferin. TOP (TRANSGENIC OPERATIVE PRODUCTS) SRL. 2010 [Google Scholar]
- 54.Xu J. Preparation, Culture, and Immortalization of Mouse Embryonic Fibroblasts. Current Protocols in Molecular Biology 28.1. 2005:8. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 55.Pencheva N, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.