Abstract
Ischemic stroke is a major cause of mortality and morbidity globally. Among the ischemic stroke subtypes, cardioembolic stroke is with poor functional outcome (Modified Rankin score ≥ 2). Early diagnosis of cardioembolic stroke will prove beneficial. This study examined the microRNAs targeting cluster of differentiation 46 (CD46), a potential biomarker for cardioembolic stroke. CD46 mRNA level was shown to be differentially expressed (p < 0.001) between cardioembolic stroke (median = 1.32) and non-cardioembolic stroke subtypes (large artery stroke median = 5.05; small vessel stroke median = 6.45). Bioinformatic search showed that miR-19a, -20a, -185 and -374b were found to target CD46 mRNA and further verified by luciferase reporter assay. The levels of miRNAs targeting CD46 were significantly reduced (p < 0.05) in non-cardioembolic stroke patients (large artery stroke median: miR-19a = 0.63, miR-20a = 0.42, miR-185 = 0.32, miR-374b = 0.27; small artery stroke median: miR-19a = 0.07, miR-20a = 0.06, miR-185 = 0.07, miR-374b = 0.05) as compared to cardioembolic stroke patients (median: miR-19a = 2.69, miR-20a = 1.36, miR-185 = 1.05, miR-374b = 1.23). ROC curve showed that the miRNAs could distinguish cardioembolic stroke from non-cardioembolic stroke with better AUC value as compared to CD46. Endogenous expression of CD46 in Human Umbilical Vein Endothelial Cells (HUVECs) were found to be regulated by miR-19a and miR-20a. Thus implicating that miR-19a and -20a may play a role in pathogenesis of cardioembolic stroke, possibly via the endothelial cells.
Introduction
Ischemic stroke is one of the leading causes of death and disabilities worldwide. It accounts for 80% of all reported stroke cases [1–3]. According to Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification [4], among the different ischemic stroke subtypes, cardioembolic stroke, a type of ischemic stroke characterized by the blood clot originating from a cardioembolic source, is associated with poor functional outcome (based on Modified Rankin score) in patients [5]. However, the current challenge for cardioembolic stroke diagnosis is the lack of an accurate diagnosis to segregate cardioembolic stroke cases from large artery stroke cases (which is another common ischemic stroke subtype). Hence, identification of specific biomarkers for cardioembolic stroke will prove beneficial.
Previous reports had highlighted a group of mRNAs which can potentially classify ischemic stroke patients into their various stroke subtypes [6], [7]. Among these mRNAs, there was an inhibitor of complement cascade, cluster of differentiation 46 (CD46) [8], which could distinguish cardioembolic stroke from non-cardioembolic stroke subtype [6], [7]. Components of complement cascade that had been found to be increased in ischemic stroke patients, have also been observed to be associated with stroke outcome [9–11]. This denotes that complement cascade is deregulated during ischemic stroke and hence can be a potential biomarker for ischemic stroke. Thus, it would be beneficial to investigate how the differential expression of CD46 contributes to the pathogenesis of the different stroke subtypes.
As the expression pattern of microRNAs (miRNAs) present in blood changes in response to different disease states, we studied them to obtain information on the molecular mechanism of cardioembolic stroke pathogenesis involving CD46. miRNAs are a class of short (18–22 nucleotides), endogenously expressed, non-coding RNAs that act as riboregulators of gene expression [12], [13]. Typically, miRNAs down-regulate gene expression of target mRNA through the process of RNA interference (RNAi) [13]. A single miRNA can target multiple genes and it can also have an overall effect on certain pathways. Hence, looking into miRNAs targeting CD46 can potentially uncover the differential pathway regulation between cardioembolic and non-cardioembolic stroke. Thus, this study was aimed to identify the miRNAs targeting CD46 as they may serve as potential biomarkers and to uncover the mechanism behind differential expression of CD46 between cardioembolic and non-cardioembolic stroke.
Materials and methods
Patient recruitment
This study was approved by the Medical Ethics Committee of the University Malaya Medical Centre (UMMC), Kuala Lumpur, Malaysia (Ref. No: 607.20) and the Institutional Review Board (IRB) of the National University of Singapore (NUS; NUS-IRB Ref Code: 08–381; Approval: NUS-676), and was carried out in accordance with the Declaration of Helsinki (2008). Written informed consent was obtained from patients and was carried out with the approval of the ethics committee and IRB. The patients were recruited from University Malaya Medical Centre (UMMC), Kuala Lumpur, Malaysia. The patients were admitted into UMMC via the Neurology service and Accident and Emergency (A&E) department. Ischemic stroke in patients were diagnosed through either magnetic resonance imaging (MRI) or computed tomography (CT) scan. Blood samples were collected from patients within 24 hours following admission. Peripheral blood was collected by venepuncture into BD Vacutainer with no additive (BD, Franklin Lakes, NJ, US; Cat# 366703) and was immediately aliquoted into Eppendorf tubes containing RNA later (Ambion, Life Technology, Carlsbad, CA, US) according to manufacturer’s protocol. These samples were stored at -80°C until required.
39 patients and 18 healthy controls were recruited for this study. Patients were classified according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification [4] (Table 1). The outcomes of the patients were assessed using the modified Rankin scale (mRS) [5] and dichotomized into good (mRS < 2) and poor outcome (mRS ≥ 2; Table 1).
Table 1. Patients’ demography.
| Control (n = 18) | CE stroke (n = 13) | Non-CE stroke (n = 26) | p-value (CE vs Non-CE) | |
|---|---|---|---|---|
| Age (Mean ± SD) | 42.87 ± 12.42 | 67.50 ± 16.13 | 62.84 ± 11.61 | 0.19 |
| Female, n (%) | 6(33.33%) | 5 (41.67%) | 10 (38.46%) | 0.27 |
| Dyslipidaemia, n (%) | NA | 9 (75.00%) | 19 (73.08%) | 0.30 |
| Diabetes mellitus, n (%) | NA | 5 (41.67%) | 14 (53.85%) | 0.21 |
| Hypertension, n (%) | NA | 5 (41.67%) | 20 (76.92%) | 0.03 |
| Ischemic heart disease, n (%) | NA | 4 (33.33%) | 6 (23.08%) | 0.24 |
| Atrial fibrillation, n (%) | NA | 10 (83.33%) | 0 (0.00%) | <0.001 |
| Previous stroke, n (%) | NA | 3 (25.00%) | 3 (11.54%) | 0.20 |
| Smoking, n (%) | NA | 0 (0.00%) | 5 (19.23%) | 0.13 |
| Alcohol, n (%) | NA | 0 (0.00%) | 1 (3.84%) | 0.68 |
| Modified Rankin Scale < 2, n (%) | NA | 1 (8.33%) | 6 (23.07%) | 0.21 |
The patients were classified according to cardioembolic stroke and non-cardioembolic stroke (large artery stroke and small vessel stroke). The various parameters were tested using Mann-Whitney U test. CE: cardioembolic. Non-CE: Non-cardioembolic.
Transfection of miRNAs in HUVECs
Human umbilical vein endothelial cells (HUVEC, CRL-1730; ATCC, US) were cultured using Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, US) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, US) and 1% penicillin-streptomycin (Gibco, Carlsbad, CA, US) and maintained at 37°C in 5% carbon dioxide. HUVECs were seeded at density of 3 X 104 cells in each well of 24 well plates. Human anti-miRNAs and miRNA mimics were transfected at final concentration of 30 nM in 50 μl of Opti-MEM (Gibco, Carlsbad, CA, US) complexed with 1 μl of NeoFx in 50 μl of Opti-MEM (Gibco, Carlsbad, CA, US) per well. Cells were cultured at 37°C in 5% carbon dioxide for 48 hours.
RNA isolation
Total RNAs (+miRNAs) were isolated from blood using Ambion Ribopure blood extraction kit (Ambion, Life Technology, Carlsbad, CA, US) according to manufacturer’s protocol. Total RNAs (including miRNAs) were isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, US) according to manufacturer’s protocol. The concentration and integrity of total RNAs were determined by the Nano-Drop ND-1000 Spectrophotometry (NanoDrop Tech, Wilmington, DE, US) and denaturing agarose gel electrophoresis.
Cloning of CD46 3’UTR fragment
Gene specific primers were used to amplify the region of CD46 3’UTR containing the seed regions of the miRNAs (forward primer, 5’- GACAGCCATAACAGGAGTGC– 3’; reverse primer, 5’–ACTTCCAGAGAAAACTGGTC– 3’). The PCR product was cloned into pMIR-REPORT Luciferase vector (Promega, Madison, WI, US) at the SpeI and HindIII multi-cloning sites.
Luciferase assay
Luciferase assay was performed according to Sepramaniam et al [14]. HeLa cells (CCL-2; ATCC, USA) were cultured using Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, US) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, US) and 1% penicillin-streptomycin (Gibco, Carlsbad, CA, US) and maintained at 37°C in 5% carbon dioxide. Cells were transfected with 50nM anti-miRNAs or miRNA mimics for 3 hours followed by 100 ng/well pMIR-REPORT Luciferase vector for 3 hours as previously shown by Sepramaniam et al [14] and Kaur et al [15]. The cells were lysed 48 h later for measurement of luciferase activity. Dual luciferase assay (Promega, Madison, WI, US) was used to quantitate the effects of anti- or pre-miRNA interaction with CD46 3’UTR. The assay was performed according to the manufacturer’s protocol. In all experiments, transfection efficiencies were normalized to those of cells co-transfected with the Renilla luciferase expressing vector (pRL-CMV; Promega, Madison, WI, US) at 10 ng/well.
Reverse transcription and real-time qualitative polymerase chain reaction (qPCR)
Reverse transcription followed by real-time qualitative PCR were carried out according to Kaur et al. [15] For gene quantification, reverse transcription was performed using TaqMan Reverse Transcription kit (Applied Biosystems, Foster City, CA, US) according to manufacturer’s protocol. Quantification of CD46 mRNA was performed using SYBR Green assay (Applied Biosystems, Foster City, CA, US) and GAPDH was used as an endogenous control [15] for normalization (CD46 forward primer, 5’-GGTGTTGCTGCTGTACTCCTTCT-3’; CD46 reverse primer, 5’-CCAATGAGCTCCATAGCTTCAA-3’; GAPDH forward primer, 5’-AACAGCGACACCCATCCTC-3’; GAPDH reverse primer, 5’-CATACCAGGAAATGAGCTTGACAA-3’)
For microRNA quantification, reverse transcription was performed using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, US) with microRNA-specific stem-loop primers. The microRNA expression was normalized to endogenous control GAPDH [15]. Subsequently, miRNA quantitation was performed using TaqMan chemistry. All reactions were conducted on Applied Biosystems 7900HT Fast Real-time PCR system (Applied Biosystems, Foster City, CA, US).
Protein gel and Western blot analysis
Protein was obtained from the phenol phase of the RNA extraction reaction (Invitrogen, Carlsbad, CA, US) and quantified using Bio-Rad protein assay (Bio-Rad, Hercules, CA, US). 40 μg of total protein was resolved using Bolt 4–12% Bis-Tris plus gel (Novex, Life Technologies, Grand Island, NY, US) and transferred using the iBlot gel transfer system (Novex, Life Technologies, Grand Island, NY, US). The membranes were probed using the iBind Flex Western Device (Thermo Fisher Scientific, Waltham, MA, US) according to manufacturer’s protocol, using primary antibodies for CD46 (1:1000; Abcam, Cambridge, UK; Cat# ab789), primary antibodies for β-Actin (1:5000; Bio-Rad, Hercules, CA, USA; Cat# MCA5775GA) and secondary antibodies (1:5000; horseradish peroxidase-conjugated goat anti-mouse; Bio-Rad, Hercules, CA, USA; Cat# 1721011). The proteins on the membranes were visualized via enhanced chemiluminescence (SuperSignal West; Thermo Fisher Scientific, Waltham, MA, USA) and exposed to hyperfilm (Amersham hyperfilm, GE healthcare lifescience, Little Chalfont, UK). The labelling intensities of the bands were quantitated using ImageJ software (National Institutes of Health, Bethesda, MD, US) [16].
Immunocytochemistry
Immunocytochemistry was performed on HUVECs treated with anti-miRNAs and miRNA mimics. The cells were fixed with 4% formaldehyde in phosphate-buffered saline for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 30 min, and blocked with 5% FBS in PBST for 30 min. CD46 was probed using primary antibodies for CD46 (1:100; Abcam, Cambridge, UK; Cat# ab789) and Texas Red conjugated secondary antibodies (1:200; Abcam, Cambridge, UK; Cat# ab7066). 0.1 μg/mL Hoechst 33342 (Biotium, Foster City, CA, USA) was used for visualising the nucleus. The images were viewed and analysed using LSM710 confocal imaging software (Carl Zeiss AG, Oberkochen, Germany).
In silico prediction
Bioinformatics prediction of miRNAs targeting CD46 was performed using available databases online: Targetscan version 6.2 (www.targetscan.org) [17–20], miRanda August 2010 release (www.microRNA.org) [21–24], microcosm version 5 (www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) [24–26], DIANA microT version 3 (http://diana.cslab.ece.ntua.gr/microT/) [27], [28], miRDB version 4 (mirdb.org/) [29], [30], miRWalk March 2011 update (www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) [31], PITA 2007 release (genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) [32], RepTar version 1.2 (reptar.ekmd.huji.ac.il/) [33] and StarBase version 2 (starbase.sysu.edu.cn/) [34], [35]. The binding energy of between the 3’UTR and the miRNA was determined using available software RNA22 version 2 (https://cm.jefferson.edu/rna22/Interactive/) [36].
In silico analysis
Bioinformatics prediction of miRNAs targeting CD46 was performed using available databases online. Pathway analysis was conducted using DIANA miRPath version 2 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index) [37]. Gene ontology (GO) analysis was performed using GeneCodis version 3 (http://genecodis.cnb.csic.es/) [38–40], using the list of predicted genes from miRPath.
Statistical and data analysis
Statistical significance was set at p-value <0.05. All analyses were performed using Microsoft Excel 2010 and SPSS version 16. MiRNA expression levels were expressed as fold change relative to the respective control samples. Hierarchical clustering was plotted using TIGR Multiple Experiment Viewer (TMeV; http://www.tm4.org/mev/) [41]
Results
CD46 mRNA expression in various stroke subtypes
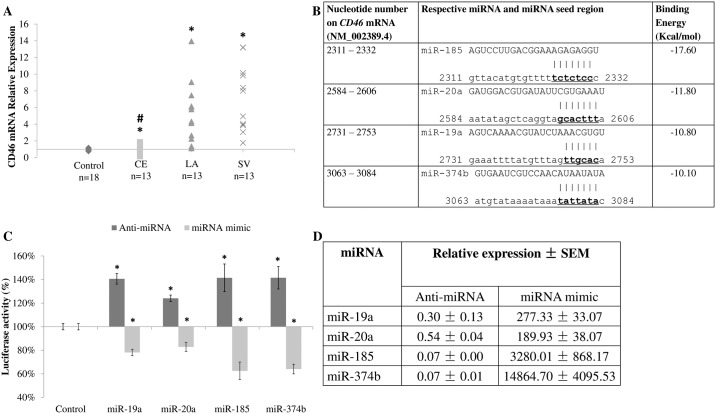
CD46 mRNA measurements were quantitated in ischemic stroke patients (Patients’ demography in Table 1). CD46 mRNA levels were found to be significantly higher (p-value < 0.001) in large artery (median = 5.05; interquartile range = 2.54–6.46) and small vessel stroke patients (median = 6.45; interquartile range = 3.87–9.90) as compared with cardioembolic stroke patients (median = 1.32; interquartile range = 0.86–1.58; Fig 1A). This observation is consistent with previous findings that CD46 mRNA was differentially expressed between cardioembolic stroke and non-cardioembolic stroke (large artery and small vessel stroke) [6], [7]. Thus, it is important to identify the miRNAs that target CD46 mRNA in order to demonstrate the potential of using miRNAs as biomarkers to distinguish cardioembolic stroke from non-cardioembolic stroke.
Fig 1. CD46 mRNA level in cardioembolic and non-cardioembolic (large artery and small vessel stroke) patients and miRNAs targeting CD46.
A. CD46 mRNA level in patients. Data expressed as relative expression with respect to control samples, with the number of patients and controls indicated. CE represents cardioembolic stroke. LA represents large artery stroke. SV represents small vessel stroke. Data in A are shown as relative expression. B. miRNA binding sites and binding energies. Nucleotides in bold denoted the seed region. C. Luciferase assay. D. miRNA levels in transfected HeLa cells. Data in C and D are shown as mean ± SD. All experiments were performed in n = 3. * denotes significant difference (p < 0.05) against control (A and C) using Mann-Whitney U test. # denotes significance between cardioembolic and non cardioembolic stroke (A) using Mann-Whitney U test.
Identification of miRNAs targeting CD46 mRNA
Bioinformatics analysis was performed to identify miRNAs targeting CD46 mRNA. A total of 9 miRNA prediction databases that were available online were used [17–36]. A total of 857 miRNAs (at least in one of the database) have been found to target CD46 3’UTR. This was narrowed down to 157 miRNAs by comparison with our group’s previously published data on miRNA profiles from ischemic stroke patients [42]. Among them, 12 miRNAs (miR-17, -18a, -19a, -19b, -20a, -20b, -106b, -181a, -181b, -185, -374b and -421) were predicted by 5 or more databases to target CD46 mRNA. Of these, miR-19a, -20a, -185 and -374b were finally selected as they had the lowest binding energies to the CD46 3’UTR (Fig 1B).
miR-19a, -20a, -185 and -374b targets CD46 mRNA
In order to validate that miR-19a, -20a, -185 and -374b target CD46 mRNA, luciferase reporter assay was performed. The 3’ untranslated region (3’UTR) of CD46 was sub-cloned into the pMIR-REPORT™ plasmid and used for co-transfection with respective anti-miRNA or miRNA mimics in HeLa cells. The success of transfection was verified by measuring the level of respective miRNAs in the transfected cells (Fig 1C). Cells transfected with anti-miRNAs exhibited an increase in relative luciferase activity, while cells transfected with miRNA mimics showed a reduction in relative luciferase activity (Fig 1D). The results demonstrated that CD46 mRNA is a bona fide target of miR-19a, -20a, -185 and -374b.
Differential miRNA expression between cardioembolic stroke and non-cardioembolic stroke subtypes
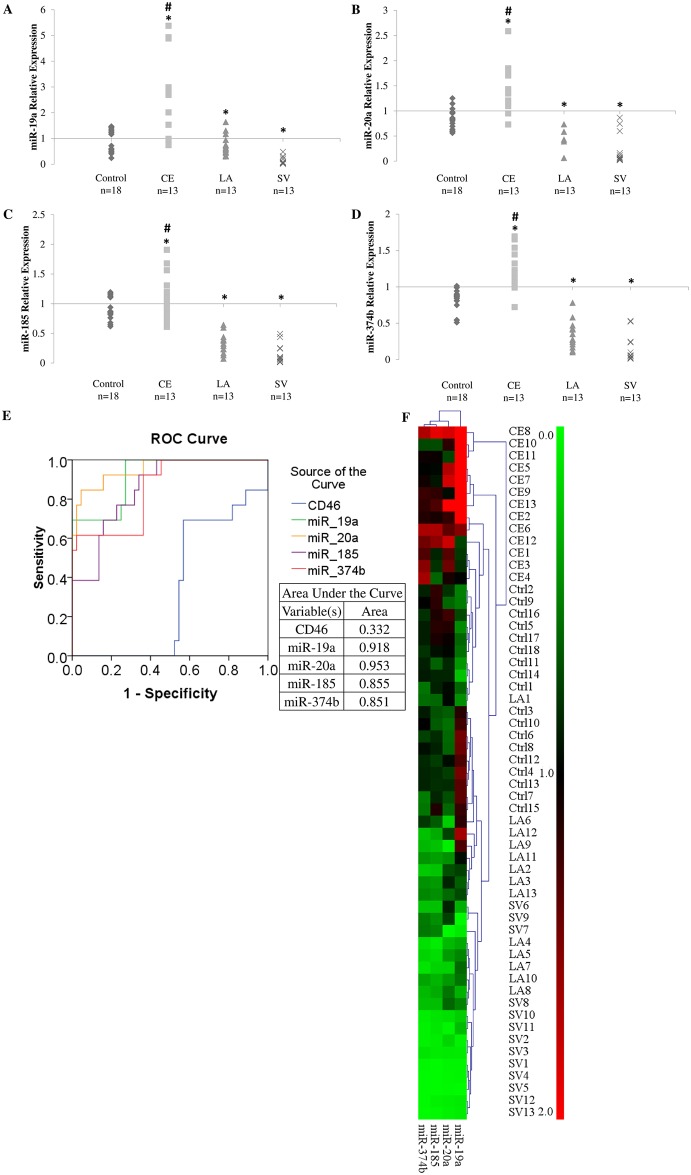
Our investigation showed that the expression of miR-19a, -20a, -185 and -374b were significantly reduced in large-artery stroke (Median: miR-19a = 0.63, miR-20a = 0.42, miR-185 = 0.32, miR-374b = 0.27; Interquartile range: miR-19a = 0.47–0.94, miR-20a = 0.26–0.68, miR-185 = 0.23–0.39, miR-374b = 0.20–0.42; Fig 2A–2D) and small vessel patients (Median: miR-19a = 0.07, miR-20a = 0.06, miR-185 = 0.07, miR-374b = 0.05; Interquartile range: miR-19a = 0.03–0.19, miR-20a = 0.03–0.16, miR-185 = 0.02–0.24, miR-374b = 0.01–0.23; Fig 2A–2D) when compared with the cardioembolic stroke patients (Median: miR-19a = 2.69, miR-20a = 1.36, miR-185 = 1.05, miR-374b = 1.23; Interquartile range: miR-19a = 0.98–2.98, miR-20a = 1.17–1.80, miR-185 = 0.85–1.30, miR-374b = 1.06–1.53; Fig 2A–2D). This is as expected based on our observation that the expression of CD46 was higher in the non-cardioembolic stroke patients, and thus strongly supports our earlier findings that these miRNAs target CD46 mRNA.
Fig 2. miRNAs level in cardioembolic and non-cardioembolic (large artery and small vessel stroke) patients.
A. miR-19a. B. miR-20a. C. miR-185. D. miR-374b. * denotes tested to be significantly different (p < 0.05) from control (A—D) using Mann-Whitney U test. # denotes tested to be statistically significant between cardioembolic and non cardioembolic stroke. E. Receiver operating characteristic (ROC) curve for CD46, miR-19a, -20a, -185 and -374b. F. Hierarchical clustering of control and patients according to miRNA levels. CE represents cardioembolic stroke. LA represents large artery stroke. SV represents small vessel stroke. Ctrl represents control. Data are shown as relative expression. Red represents up-regulation. Green represents down-regulation.
Furthermore, using the expression of CD46 and the expression of miRNAs targeting CD46 (miR-19a, -20a, -185 and -374b), a receiver operating characteristic (ROC) curve was obtained (Fig 2E). The plot shows that the miRNAs were more effective in distinguishing cardioembolic stroke patients from non-cardioembolic stroke patients and healthy controls (Fig 2E). This was denoted by the larger area under curve (AUC) value for the miRNAs than CD46 (AUC value: CD46 = 0.332, miR-19a = 0.918, miR-20a = 0.953, miR-185 = 0.855, miR-374b = 0.851). Using TMeV [41], the miRNA expression can be used to plot for hierarchical clustering, which showed segregation of cardioembolic stroke samples from controls and noncardioembolic stroke samples (Fig 2F). Hence, these miRNAs may prove to be useful biomarkers for cardioembolic stroke diagnosis.
In silico analysis of pathophysiological mechanism of miRNAs targeting CD46
In order to determine the possible mechanism behind the dysregulation of CD46 and its respective miRNAs, in silico analysis was performed to decipher the potential pathways and processes that were affected during the dysregulation. Pathway analysis was performed using DIANA miRPath version 2 [37]. The results showed that among the top 10 dysregulated pathways (based on enrichment score), there were 3 pathways (Wnt signalling pathway, TGF-beta signalling pathway and Gap junction) associated with endothelial cells (Table 2) [43–45]. Further investigation was conducted by focussing at biological processes and gene ontology (GO). GO analysis was performed on GeneCodis version 3 (Table 2) [38–40]. There were multiple biological processes associated with endothelial cells from the GO analysis, pointing to endothelial cells as the possible site of action for the dysregulated gene and miRNAs. Hence, further verification was performed with human umbilical vein endothelial cells (HUVECs) as a model.
Table 2. Table of pathways and Gene Ontology (GO) biological processes associated with miR-19a, -20a, -185 and -374b.
| KEGG Pathway | Enrichment Score | GO Biological Process | Enrichment Score |
|---|---|---|---|
| Axon guidance | 24.08 | cell adhesion | 22.90 |
| Wnt signaling pathway | 23.66 | blood coagulation | 14.72 |
| Pathways in cancer | 23.50 | angiogenesis | 13.83 |
| p53 signaling pathway | 21.19 | canonical Wnt receptor signaling pathway | 8.99 |
| Ubiquitin mediated proteolysis | 20.44 | platelet activation | 7.46 |
| Transcriptional misregulation in cancer | 20.10 | wound healing | 7.40 |
| TGF-beta signaling pathway | 20.08 | positive regulation of angiogenesis | 5.74 |
| Endocytosis | 19.75 | Wnt receptor signaling pathway, calcium modulating pathway | 5.57 |
| Melanogenesis | 18.83 | vascular endothelial growth factor receptor signaling pathway | 5.44 |
| Gap junction | 17.13 | negative regulation of canonical Wnt receptor signaling pathway | 5.03 |
| GnRH signaling pathway | 16.65 | Wnt receptor signaling pathway | 4.55 |
| Prostate cancer | 15.97 | Notch signaling pathway | 4.51 |
| MAPK signaling pathway | 15.18 | cell-matrix adhesion | 4.35 |
| HTLV-I infection | 15.18 | negative regulation of cell adhesion | 3.95 |
| Salivary secretion | 13.52 | transforming growth factor beta receptor signaling pathway | 3.95 |
| Calcium signaling pathway | 13.35 | regulation of cell adhesion | 3.65 |
| ABC transporters | 13.09 | positive regulation of endothelial cell migration | 3.60 |
| Melanoma | 12.52 | positive regulation of vasoconstriction | 3.60 |
| Regulation of actin cytoskeleton | 12.17 | positive regulation of endothelial cell proliferation | 3.37 |
| ErbB signaling pathway | 12.17 | blood vessel development | 3.17 |
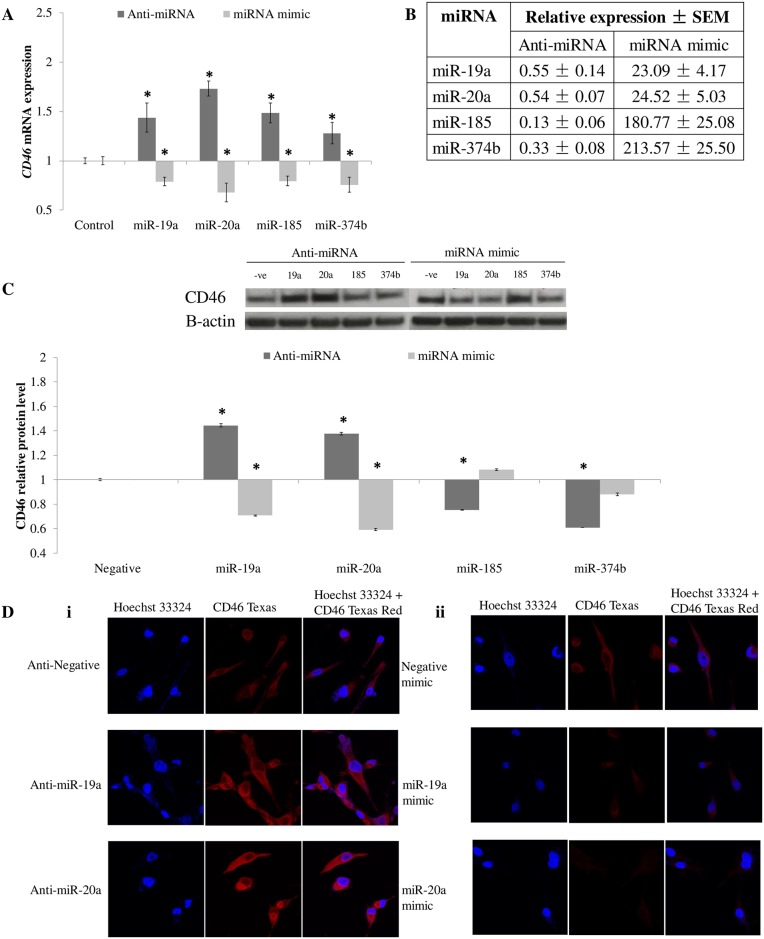
miR-19a and -20a regulate CD46 expression in HUVECs
In order to show miRNAs target CD46 mRNA in endothelial cells, HUVECs were transfected with either anti-miRNA or miRNA mimics. CD46 mRNA and protein were quantitated in these cells to determine the effect of miRNAs on CD46 endogenously (Fig 3). The success of transfection was also verified by measuring the level of respective miRNAs in the transfected cells (Fig 3B). As expected, the transfection of anti-miRNA caused an increase in CD46 mRNA level (anti-miR-19a = 1.43 ± 0.14, anti-miR-20a = 1.73 ± 0.07, anti-miR-185 = 1.48 ± 0.09, anti-miR-374b = 1.27 ± 0.10) while the introduction of miRNA mimic reduced the level of CD46 mRNA (miR-19a mimic = 0.79 ± 0.04, miR-20a mimic = 0.67 ± 0.09, miR-185 mimic = 0.79 ± 0.04, miR-374b mimic = 0.75 ± 0.07; Fig 3A).
Fig 3. Anti-miRNA and miRNA mimic transfection in HUVECs.
A. CD46 mRNA expression in transfected HUVECs. B. miRNA levels in transfected HUVECs. C. Western blot analysis and respective quatification of CD46 protein in transfected HUVECs.–ve: Negative; 19a: miR-19a; 20a: miR-20a; 185: miR-185; 374b: miR-374b. D. Immunocytochemistry of transfected HUVECs. All experiments were performed in n = 3. * denoted tested to be significantly different (p < 0.05) from control (A and C) using Mann-Whitney U test. Data in A and C were shown as mean ± SD, n = 3.
However, at the protein level, only miR-19a and -20a were shown to bring about an effect on CD46 protein level, which was quantified based on intensity of bands in Western blotting and measured using ImageJ software [16] The anti-miRNA transfected HUVECs had a significant increase in CD46 protein expression (anti-miR-19a = 1.44 ± 0.01, anti-miR-20a = 1.37 ± 0.00, anti-miR-185 = 0.75 ± 0.00, anti-miR-374b = 0.60 ± 0.00; Fig 3C; S1 Fig) while miRNA mimic transfection brought about a decrease in CD46 protein level (miR-19a mimic = 0.70 ± 0.00, miR-20a mimic = 0.59 ± 0.01, miR-185 mimic = 1.08 ± 0.00, miR-374b mimic = 0.87 ± 0.01; Fig 3C; S1 Fig). This was further verified through immunocytochemistry (Fig 3D). HUVECs transfected with anti-miRNAs (miR-19a and miR-20a) showed an increase in CD46 protein, denoted by the increase in red fluorescence as compared with control (Fig 3Di), whilst HUVECs transfected with miRNA mimics showed reduced red fluorescence (Fig 3Dii), signifying decrease in CD46 protein. These results demonstrated the relevance of miR-19a and -20a targeting CD46 in endothelial cells.
Discussion
Cardioembolic stroke is a subtype of ischemic stroke which is well-known for its high mortality and morbidity rate. Therefore, it is imperative to identify more accurate markers for cardioembolic stroke as it will have implications on the timely selection of appropriate therapy.
Jickling et al [6], [7] demonstrated that CD46 mRNA was among a list of 40 mRNA as potential markers for distinguishing ischemic stroke subtypes. Among these 40 mRNAs, CD46 seems to have a direct involvement in the pathophysiology of different ischemic stroke subtypes. This is not surprising as CD46 is part of the complement cascade which regulates inflammation, a crucial pathological process in the pathophysiology of ischemic stroke. In this study, we have shown that CD46 mRNA level to be significantly up-regulated in non-cardioembolic stroke while in cardioembolic stroke the CD46 mRNA level appears to be similar to that of the healthy controls. This observation is in line with the current literature which suggests that atherosclerosis, the underlying pathology of large artery and small vessel stroke, is largely driven by the process of inflammation [46].
Furthermore, we have identified endothelial cells as a possible location. It is noteworthy to mention that endothelial cell dysfunction is an important hallmark in atherosclerosis and it was previously reported that CD46 protein level was found to be up-regulated in atherosclerotic plaque [47], [48]. Hence, the up-regulation of CD46 mRNA found in patients with large artery and small vessel stroke is entirely in agreement with current evidence, signifying the crucial role that CD46 plays in the pathogenesis of large artery and small vessel stroke.
In addition, by using in silico prediction and biochemical methodology we have identified miR-19a, -20a, -185 and -374b to be targeting CD46. We have utilized luciferase assay, a direct method that is used to prove the binding between miRNA:mRNA pair in various complementary binding studies [49], [50]. However, when CD46 protein expression were measured, only miR-19a and miR-20a proved to be useful to significantly modulate CD46 in HUVECs. miR-19a has been reported to protect endothelial cells from lipopolysaccharide-induced apoptosis, which is a characteristic of atherosclerosis [51]. The down-regulation of miR-19a detected in large artery and small vessel stroke implies the increase in lipopolysaccharide-induced apoptosis leading to ischemic stroke onset. This demonstrates the potential mechanism of miR-19a and -20a in endothelial cells during atherosclerosis and even possibly contributing to large artery and small vessel stroke.
During the pathway analysis, complement cascade was highlighted to be regulated by miR-19a, -20a, -185 and -374b. The complement cascade also interacts with the coagulation cascade. Coagulation cascade plays an important role in the formation of blood clot, the underlying cause for most ischemic stroke cases. There is a subtle difference in the induction of coagulation cascade between cardioembolic stroke and non-cardioembolic stroke (large artery and small vessel strokes), which is emphasized in the different treatments between cardioembolic stroke and non-cardioembolic stroke [52], [53]. Non-cardioembolic strokes are treated with anti-platelet drugs as the process of blood clotting is most likely induced via the tissue factor pathway due to tissue damage, such as rupture of the atherosclerotic plaque [52]. On the other hand, cardioembolic stroke is treated with anti-coagulants, suggesting that the blood coagulation follows the intrinsic pathway associated with pooling of blood, which corresponds to the pathogenesis of blood clot in cardioembolic stroke [53]. It is noteworthy to mention that miR-19a was reported to possibly modulate tissue factor pathway inhibitor (TFPI) which could signify that the up-regulation of miR-19a in cardioembolic stroke led to a pro-coagulation state in cardioembolic stroke patients [54]. Also, miR-20a was reported in a panel of miRNAs that could distinguish etiology of various ischemic stroke subtypes [37]. This lends further support to the relevance of this miRNAs in the pathogenesis of cardioembolic stroke.
Interestingly, results from the GO analysis showed that there were biological processes involved in coagulation cascade (blood coagulation and platelet activation). This seems to suggest that the deregulation of the complement cascade, involving CD46 and miRNAs targeting CD46 (miR-19a, -20a, -185 and -374b) may be potential markers for the differential regulation of coagulation cascade between cardioembolic stroke and non-cardioembolic stroke. A preliminary in silico prediction was performed to identify potential mRNA targets of miR-19a, -20a, -185 and -374b within the complement and coagulation cascade. It is noteworthy to mention that these miRNAs target several other genes in the pathways (miR-19a: 9 targets; miR-20a: 2 targets; miR-185: 6 targets; miR-374b: 5 targets; Table 3). Hence, a further investigation into miR-19a, -20a, -185 and -374b modulation of complement and coagulation cascade in blood cells may provide further understanding of the molecular mechanism behind the difference in complement and coagulation cascade activities between cardioembolic stroke and non-cardioembolic stroke.
Table 3. Predicted targets of miR-19a, -20a, -185 and -374b in complement and coagulation cascade.
| hsa-miRNA | Predicted targets |
|---|---|
| miR-19a | AT3, CD46, CD55, F5, F13, PAI-1, TF, TFPI and PLAU |
| miR-20a | CD46 and PAI-1 |
| miR-185 | CD46, F7, F8, F11, F13 and PLAT |
| miR-374b | CD46, CD55, F5, PAI-2 and TFPI |
AT3: Anti-thrombin III. CD46: Cluster of differentiation 46. CD55: Cluster of differentiation 55. F5: Factor V. F7: Factor VII. F8: Factor VIII. F11: Factor XI. F13: Factor XIII. PAI-1: Plasminogen activator inhibitor-1. PAI-2: Plasminogen activator inhibitor-2. PLAT: tissue plasminogen activator. PLAU: urokinase-type plasminogen activator. TF: Tissue factor. TFPI: Tissue factor pathway inhibitor.
In the design of this study, GAPDH was used as an endogenous control for normalization of miRNA expression. The level of GAPDH was found to be consistent throughout all the experimental conditions. Despite the length of GAPDH mRNA being longer than the length of miRNA, numerous studies had used GAPDH as endogenous control for normalizing the level of miRNAs [55–59], which vindicates its role as an endogenous control for miRNA level normalization.
In brief, we have verified that CD46 mRNA is differentially expressed between cardioembolic stroke and non-cardioembolic stroke subtypes, where it was up-regulated in non-cardioembolic stroke. Furthermore, miR-19, -20a, -185 and -374b have been demonstrated to target CD46 mRNA and these miRNAs also showed correspondingly inversed expression when compared to the expression of CD46 mRNA where the expression of the miRNAs were down-regulated in non-cardioembolic stroke. In addition, endothelial cells in non-cardioembolic stroke were likely to be affected by this dysregulation. Among the miRNAs, miR-19a and -20a are the most likely candidates to be involved in up-regulating CD46 through their down-regulation in endothelial cells. Based on the ROC plot, miRNAs were more accurate in the diagnosis of cardioembolic stroke patients where miR-19a and -20a have been found to have stronger AUC values (miR-19a = 0.918, miR-20a = 0.953). Nonetheless, more data are needed to identify the threshold range of CD46 mRNA, miR-19, -20a, -185 and -374b for accurate diagnosis in stroke patients.
Supporting information
A. CD46. B. Beta-actin. 1. Anti-miR-374b. 2. Anti-miR-185. 3. Anti-miR-20a. 4. Anti-miR-19a. 5. Anti-Negative. 6. miR-374b mimic. 7. miR-185 mimic. 8. miR-20a mimic. 9. miR-19a mimic. 10. Negative mimic.
(TIFF)
Acknowledgments
This work was supported by the research grants from National Research Foundation (NRF): R-184-002-165-281, National Medical Research Council (NMRC/EDG): R-183-000-230-275, and the National Medical Research Council (NMRC/IRG: R-183-000-290-213) Singapore; and the University of Malaya, Kuala Lumpur, Malaysia (UM/HIR: E0067 & E0036).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the research grants from National Research Foundation (NRF): R-184-002-165-281, National Medical Research Council (NMRC/EDG): R-183-000-230-275, and the National Medical Research Council (NMRC/IRG): R-183-000-290-213, Singapore; and the University of Malaya (UM/HIR): E0067 & E0036. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Truelsen T, Begg S, Mathers C. The global burden of cerebrovascular disease. Global Burden of Diseases 2006 update. World Health Organization. 2006. http://www.who.int/healthinfo/statistics/bod_cerebrovasculardiseasestroke.pdf
- 2.Burke TA, Venketasubramanian RN. The epidemiology of stroke in the East Asian region: a literature-based review. Int J Stroke. 2006; 1(4): 208–15. 10.1111/j.1747-4949.2006.00060.x [DOI] [PubMed] [Google Scholar]
- 3.Sudlow CL, Warlow CP. Comparable studies of the incidence of stroke and its pathological types: results from an international collaboration. International Stroke Incidence Collaboration. Stroke. 1997; 28(3): 491–9. [DOI] [PubMed] [Google Scholar]
- 4.Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993; 24(1): 35–41. [DOI] [PubMed] [Google Scholar]
- 5.Farrell B, Godwin J, Richards S, Warlow C. The United Kingdom transient ischaemic attack (UK-TIA) aspirin trial: final results. J Neurol Neurosurg Psychiatry. 1991; 54(12): 1044–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jickling GC, Xu H, Stamova B, Ander BP, Zhan X, Tian Y, et al. Signatures of cardioembolic and large-vessel ischemic stroke. Ann Neurol. 2010; 68(5): 681–92. 10.1002/ana.22187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jickling GC, Stamova B, Ander BP, Zhan X, Liu D, Sison SM, et al. Prediction of cardioembolic, arterial, and lacunar causes of cryptogenic stroke by gene expression and infarct location. Stroke. 2012; 43(8): 2036–41. 10.1161/STROKEAHA.111.648725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liszewski MK, Farries TC, Lublin DM, Rooney IA, Atkinson JP. Control of the complement system. Adv Immunol. 1996; 61: 201–83. [DOI] [PubMed] [Google Scholar]
- 9.Song FY, Wu MH, Zhu LH, Zhang ZQ, Qi QD, Lou CL. Elevated Serum Mannose-Binding Lectin Levels Are Associated with Poor Outcome After Acute Ischemic Stroke in Patients with Type 2 Diabetes. Mol Neurobiol. 2015; 52(3): 1330–40. 10.1007/s12035-014-8941-0 [DOI] [PubMed] [Google Scholar]
- 10.Wang ZY, Sun ZR, Zhang LM. The relationship between serum mannose-binding lectin levels and acute ischemic stroke risk. Neurochem Res. 2014; 39(2): 248–53. 10.1007/s11064-013-1214-x [DOI] [PubMed] [Google Scholar]
- 11.Széplaki G, Szegedi R, Hirschberg K, Gombos T, Varga L, Karádi I, et al. Strong complement activation after acute ischemic stroke is associated with unfavorable outcomes. Atherosclerosis. 2009; 204(1): 315–20. 10.1016/j.atherosclerosis.2008.07.044 [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75(5): 843–54. [DOI] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391(6669): 806–11. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 14.Sepramaniam S, Armugam A, Lim KY, Karolina DS, Swaminathan P, Tan JR, Jeyaseelan K. MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J Biol Chem. 2010; 285(38): 29223–30. 10.1074/jbc.M110.144576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur P, Tan JR, Karolina DS, Sepramaniam S, Armugam A, Wong PT, Jeyaseelan K. A long non-coding RNA, BC048612 and a microRNA, miR-203 coordinate the gene expression of neuronal growth regulator 1 (NEGR1) adhesion protein. Biochim Biophys Acta. 2016; 1863(4): 533–43. 10.1016/j.bbamcr.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 16.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7): 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120(1): 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19(1): 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007; 27(1): 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011; 18(10): 1139–46. 10.1038/nsmb.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008; 36(Database issue): D149–53. 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010; 11(8): R90 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004; 2(11): e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003; 5(1): R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006; 34(Database issue): D140–4. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008; 36(Database issue): D154–8. 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009; 10: 295 10.1186/1471-2105-10-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009; 37(Web Server issue): W273–6. 10.1093/nar/gkp292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008; 24(3): 325–32. 10.1093/bioinformatics/btm595 [DOI] [PubMed] [Google Scholar]
- 30.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008; 14(6): 1012–7. 10.1261/rna.965408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011; 44(5): 839–47. 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 32.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007; 39(10): 1278–84. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- 33.Elefant N, Berger A, Shein H, Hofree M, Margalit H, Altuvia Y. RepTar: a database of predicted cellular targets of host and viral miRNAs. Nucleic Acids Res. 2011; 39(Database issue): D188–94. 10.1093/nar/gkq1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011; 39(Database issue): D202–9. 10.1093/nar/gkq1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014; 42(Database issue): D92–7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006; 126(6): 1203–17. 10.1016/j.cell.2006.07.031 [DOI] [PubMed] [Google Scholar]
- 37.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012; 40(Web Server issue): W498–504. 10.1093/nar/gks494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007; 8(1): R3 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009; 37(Web Server issue): W317–22. 10.1093/nar/gkp416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012; 40(Web Server issue): W478–83. 10.1093/nar/gks402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003; 34(2): 374–8. [DOI] [PubMed] [Google Scholar]
- 42.Sepramaniam S, Tan JR, Tan KS, DeSilva DA, Tavintharan S, Woon FP, et al. Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci. 2014; 15(1): 1418–32. 10.3390/ijms15011418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dejana E. The role of wnt signaling in physiological and pathological angiogenesis. Circ Res. 2010; 107(8): 943–52. 10.1161/CIRCRESAHA.110.223750 [DOI] [PubMed] [Google Scholar]
- 44.Walshe TE, Saint-Geniez M, Maharaj AS, Sekiyama E, Maldonado AE, D'Amore PA. TGF-beta is required for vascular barrier function, endothelial survival and homeostasis of the adult microvasculature. PLoS One. 2009; 4(4): e5149 10.1371/journal.pone.0005149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallez Y, Huber P. Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim Biophys Acta. 2008; 1778(3): 794–809. 10.1016/j.bbamem.2007.09.003 [DOI] [PubMed] [Google Scholar]
- 46.Hansson GK, Robertson AK, Söderberg-Nauclér C. Inflammation and atherosclerosis. Annu Rev Pathol. 2006; 1: 297–329. 10.1146/annurev.pathol.1.110304.100100 [DOI] [PubMed] [Google Scholar]
- 47.Seifert PS, Hansson GK. Complement receptors and regulatory proteins in human atherosclerotic lesions. Arteriosclerosis. 1989; 9(6): 802–11. [DOI] [PubMed] [Google Scholar]
- 48.Li SH, Szmitko PE, Weisel RD, Wang CH, Fedak PW, Li RK, et al. C-reactive protein upregulates complement-inhibitory factors in endothelial cells. Circulation. 2004; 109(7): 833–6. 10.1161/01.CIR.0000117087.27524.0E [DOI] [PubMed] [Google Scholar]
- 49.Liu CZ, Liu W, Zheng Y, Su JM, Li JJ, Yu L, He XD, Chen SS. PTEN and PDCD4 are bona fide targets of microRNA-21 in human cholangiocarcinoma. Chin Med Sci J. 2012; 27(2): 65–72. [PubMed] [Google Scholar]
- 50.Bohmer M, Sharbati J, Zur Bruegge J, Einspanier R, Sharbati S. Structural analysis of microRNA-target interaction by sequential seed mutagenesis and stem-loop 3' RACE. PLoS One. 2013; 8(11): e81427 10.1371/journal.pone.0081427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang WL, Zhang YF, Xia QQ, Zhu J, Yu X, Fan T, et al. MicroRNA-19a regulates lipopolysaccharide-induced endothelial cell apoptosis through modulation of apoptosis signal-regulating kinase 1 expression. BMC Mol Biol. 2015; 16: 11 10.1186/s12867-015-0034-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Lin WH, Zhao YD, Chen XY, Leung TW, Chen C,et al. The effectiveness of dual antiplatelet treatment in acute ischemic stroke patients with intracranial arterial stenosis: a subgroup analysis of CLAIR study. Int J Stroke. 2013; 8(8): 663–8. 10.1111/j.1747-4949.2012.00828.x [DOI] [PubMed] [Google Scholar]
- 53.Hallevi H, Albright KC, Martin-Schild S, Barreto AD, Savitz SI, Escobar MA, et al. Anticoagulation after cardioembolic stroke: to bridge or not to bridge? Arch Neurol. 2008; 65(9): 1169–73. 10.1001/archneur.65.9.noc70105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jickling GC, Ander BP, Zhan X, Noblett D, Stamova B, Liu D. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS One. 2014; 9(6): e99283 10.1371/journal.pone.0099283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Martino I, Visone R, Fedele M, Petrocca F, Palmieri D, Martinez Hoyos J, Forzati F, Croce CM, Fusco A. Regulation of microRNA expression by HMGA1 proteins. Oncogene. 2009; 28(11):1432–42. 10.1038/onc.2008.495 [DOI] [PubMed] [Google Scholar]
- 56.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008; 105(26): 9093–8. 10.1073/pnas.0803072105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008; 105(5): 1608–13. 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DH, Saetrom P, Snøve O Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008; 105(42): 16230–5. 10.1073/pnas.0808830105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baglìo SR, Devescovi V, Granchi D, Baldini N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013; 527(1): 321–31. 10.1016/j.gene.2013.06.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. CD46. B. Beta-actin. 1. Anti-miR-374b. 2. Anti-miR-185. 3. Anti-miR-20a. 4. Anti-miR-19a. 5. Anti-Negative. 6. miR-374b mimic. 7. miR-185 mimic. 8. miR-20a mimic. 9. miR-19a mimic. 10. Negative mimic.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.