Abstract
Aim:
In the current study, we analysised only the articles that investigate serum proteome profile of cirrhosis patients or HCC patients versus healthy controls.
Background:
Increased understanding of cancer biology has enabled identification of molecular events that lead to the discovery of numerous potential biomarkers in diseases. Protein-protein interaction networks is one of aspect that could elevate the understanding level of molecular events and protein connections that lead to the identification of genes and proteins associated with diseases.
Methods:
Gene expression data, including 63 gene or protein names for hepatocellular carcinoma and 29 gene or protein names for cirrhosis, were extracted from a number of previous investigations. The networks of related differentially expressed genes were explored using Cytoscape and the PPI analysis methods such as MCODE and ClueGO. Centrality and cluster screening identified hub genes, including APOE, TTR, CLU, and APOA1 in cirrhosis.
Results:
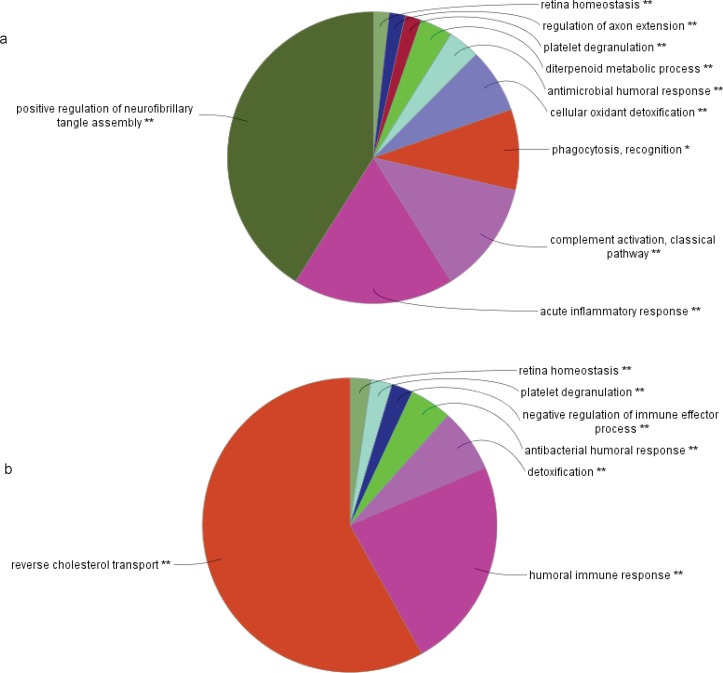
CLU and APOE belong to the regulation of positive regulation of neurofibrillary tangle assembly. HP and APOE involved in cellular oxidant detoxification. C4B and C4BP belong to the complement activation, classical pathway and acute inflammation response pathway. Also, it was reported TTR, TFRC, VWF, CLU, A2M, APOA1, CKAP5, ZNF648, CASP8, and HSP27 as hubs in HCC. In HCC, these include A2M that are corresponding to platelet degranulation, humoral immune response, and negative regulation of immune effector process. CLU belong to the reverse cholesterol transport, platelet degranulation and human immune response. APOA1 corresponds to the reverse cholesterol transport, platelet degranulation and humoral immune response, as well as negative regulation of immune effector process pathway.
Conclusion:
In conclusion, this study suggests that there is a common molecular relationship between cirrhosis and hepatocellular cancer that may help with identification of target molecules for early treatment that is essential in cancer therapy.
Key Words: Cirrhosis, Hepatocellular carcinoma, Protein-Protein Interaction Network, Gene ontology
Introduction
Hepatocellular carcinoma (HCC) is the one of the most common malignancy in the world (1). It usually occurs following previous liver disease, such as chronic hepatitis B or C and liver cirrhosis (LC) (2). Most HCCs develop following by chronic liver diseases (3). Multiple factors such as genetic and epigenetic changes have been reported in HCC patients (4). cDNAmicroarray studies were designed to identify abnormally expressed gene sets for HCC (5-7). On the other hand, some studies have been surveyed proteomic profile of HCC patients and have been introduced some new biomarkers (8). Many polymorphisms alter biologic pathways of carcinogenesis, including inflammation (IL1B, TNFA and TGF) (9, 10); oxidative stress (SOD2, MPO) (11, 12); DNA repair (MTHFR, XRCC3) (13, 14) and cell cycle (MDM2) (15) and TP53 (16). At least, in half of HCC patients the P53 cell cycle pathway alters with frequent TP53 mutations (12%- 48%) (17-19). Apart from genetic aberrations, alterations in protein expression have been reported such as Alpha- Fetoprotein (AFP), Des-Gamma-Carboxy (Abnormal), Prothrombin (DCP), Transforming Growth Factor-Beta (TGF-Beta), Serum Alpha-1-Fucosidase, Human Carbonyl Reductase 2, Tumor-Specific Growth Factor (TSGF), and Epidermal Growth Factor Receptor Family (EGFR) (20). At present, diagnostic methods included alpha-fetoprotein (AFP) and magnetic resonance imaging (MRI). Currently, the accepted biomarker for diagnosis of HCC is AFP; But the sensitivity and specificity of this agent are not satisfactory (21, 22). On the other way, most patients often have advanced in stage of disease at the time of diagnosis because the lack of special sign (23). Hence, there is an urgent demand to find specific biomarkers of HCC, which can be used specifically and sensitively in diagnosis as well as in prognosis and therapeutic evaluation. Liver cirrhosis (LC) and its associated complications are introduced as essential factors in morbidity and mortality worldwide (24, 25). The diagnostic methods of cirrhosis are based on the combined results of clinical and imaging examinations (24). Clinical symptoms and laboratory data of liver diseases frequently overlap; thus interpretation a differential diagnosis is difficult (26). Liver biopsy, is performed in patients with ambiguous diagnostic results. But this way is an invasive method which imposes pain to patients. For this, a non-invasive method for early diagnosis of hepatic fibrosis is needed. Some genetic liver disease that predispose to early cirrhosis with related mutations have been reported. Cystic fibrosis by altering activity of CFTR, Wilson disease by increased levels the ATP7B, hereditary hemochromatosis by iron-induced lipid peroxidation causes hepatocellular injury, Glycogen storage disease type IV by The altered stored glycogen impairs the osmotic pressure within the hepatocyte and Cholesteryl ester storage disease by Accumulation of cholesteryl are the known ones (26). Since the biological heterogeneity of liver diseases, it is difficult to distinguish HCC and other liver diseases such as cirrhosis or fatty liver, simply by the clinical symptoms or pathophysiological characteristics (27). Ideally, for timely treatment, the biomarker(s) that could recognize the fibrosis in the early stages of hepatic disorder to prevent the progression of cirrhosis to HCC is required (28). Interaction networks might give information of the functions of newly discovered proteins (29-31). The protein network analysis provides a scientific model that improves understanding of the mechanisms underlying human diseases (32-38). The centralized applications of PPI networks to disease addition to the identification of genes and proteins associated with diseases turn around on the study of network properties to find their relation to disease states. Classification of network- based disease and the identification of disease-related sub networks are another applications of the PPI network (39). Our goal is to present the PPI of cirrhosis and HCC patients versus healthy control group and comparison with each other to find common and sensitive points to introduce possible biomarker (s) in these diseases development and subsequently drug target discovering. This investigation was designed based on proteomic studies in cirrhosis and HCC patients published since 1997.
Material and Methods
Data Collection
In this study, the inclusion criteria were the proteomic studies on the human species involved in the comparison between the serum or plasma of patients (cirrhosis or hepatocellular carcinoma) and healthy control. Exclusion criteria were the studies on non-human sample and the studies on samples of tissue or cell lines, saliva, CSF, and urine. It has also been eliminated the papers which were compared serum or plasma of patients with the groups except healthy control. There was no limitation in methods in proteomic study. A number of 73 papers about HCC and cirrhosis proteomic profiling were reviewed. We manually evaluated the publications and it was selected the articles in line with the above conditions. Duplicated proteins and genes were eliminated. Finely, 29 genes for cirrhosis and 63 genes for HCC were extracted. Genes and proteins were presented in tables 1 and 2. Uniprot accession number of selected genes were retrieved from web site (uniprot.org).
Table 1.
A number of genes in cirrhosis derived from articles include of serum proteomic profile
| Protein name(references) | Uniprot code | Gene name | Protein name(references) | Uniprot code | Gene name |
|---|---|---|---|---|---|
| ApolipoproteinA-1(47) | P02647 | APOA1 | apolipoprotein E(46) | P02649 | APOE |
| Haptoglobin(48) | P00738 | HP | C4b-binding protein (46)alpha chain | P04003 | C4BPA |
| Alpha1-Antitrypsin(28) | P01009 | SERPINA1 | Glutathione peroxidase 3(46) | P22352 | GPX3 |
| Ceruloplasmin(49) | P00450 | CP | Beta-2-glycoprotein 1(28) | P02749 | APOH |
| Transthyretin(50) | P02766 | TTR | complement component 4B(46) | P0C0L5 | C4B |
| glycoprotein 1(46, 50) | P02750 | LRG1 | Zinc-2-glycoprotein(28) | P25311 | AZGP1 |
| Complement factor H-related protein 1(28) | Q03591 | CFHR1 | Immunoglobulin gamma-2-chainc(50) | P01859 | IGHG2 |
| Apolipoprotein L1(51) | O14791 | ApoL1 | complement factor B(46) | P00751 | CFB |
| Transgelin (28) | Q01995 | TAGLN | complement component 4A(46) | P0C0L4 | C4A |
| Paraoxonase/ arylesterase 1(46) | P27169 | PON1 | amyloid P component(46) | P02743 | APCS |
| alpha-1-microglobulin/ bikunin (46) | P02760 | AMBP | Inter-alpha-trypsin inhibitor heavy chain H4(28) | Q14624 | ITIH4 |
| haptoglobin-related protein(46) | P00739 | LRG | Complement C3(28) | P01024 | C3 |
| Hemopexin(50) | P02790 | HPR | 1 antichymotrypsin(46) α | P01011 | SERPINA3 |
| α-1 acid glycoprotein(50) | P02763 | ORM1 | CD5-like antigen(50) | O43866 | CD5L |
| clusterin(50) | P10909 | CLU |
Table 2.
A number of genes in hepatocellular carcinoma derived from articles include of serum proteomic profiling
| Protein name(references) | Gene name | Uniprot code | Protein name(references) | Gene name | Uniprot code |
|---|---|---|---|---|---|
| Annexin A6(53) | ANXA6 | P08133 | Mannose receptor, C type 1-like 1(52) | MRC1L1 | B9EJA8 |
| Complement component 9 (53) | C9 | P02748 | Vascular cell adhesion protein 1(52) | VCAM1 | E9PDD2 |
| Ceruloplasmin(53) | CP | P00450 | Fibrinogen β chain, isoform CRA_e(52) | FGB | D3DP13 |
| serum amyloid A4(53) | SAA4 | P35542 | Proteinase 3(52) | PRTN3 | D6CHE9 |
| serum amyloid A2(53) | SAA2 | P0DJI9 | Hemoglobin delta-beta fusion protein(52) | HBD/HBB | Q5XTR9 |
| Transthyretin(52, 54) | TTR | P02766 | Polymeric immunoglobulin | PIGR | P01833 |
| Clusterin(54) | CLU | P10909 | receptor(52) Fos-related antigen 2(52) |
FOSL2 | C9JCN8 |
| haptoglobin α2 chain(54) | HP | P00738 | Cyclin-dependent kinase-like 1(52) | CDKL1 | Q00532 |
| heat-shock protein 27(55) α-fetoprotein(52) | Hsp27 | P04792 | cytoskeleton-associated protein5(52) | CKAP5 | Q14008 |
| Apolipoprotein A-I (46) | AFP APOA1 | P02771 P02647 | Transferrin receptor protein 1 (52) Transmembrane protein 200C(52) | TFRC TMEM200C | P02786 A6NKL6 |
Protein-Protein Interaction Analysis
PPI network is the basic skeleton for proteins to determine their functions in the system biology (40). Specialization the interactions of proteins in a given proteome could reveal the biochemistry of the cell (41). It also helps in the identification of drug targets by introducing hubs (42). The PPI network was visualized using the Cytoscape 3.2.1 software. MINT, Reactome-FLs, databases were used for this topology visualization. We used Molecular Complex Detection (MCODE) to analyze the characteristics of the networks. The MCODE clusters was based on the topology to find the densely connected region. Gene ontology categories were analyzed to identify the function of each highly connected region that was generated by the MCODE. These include Kappa statistic ≥ 0.5, enrichment and Bonferroni step down method for probability value correction (43). The degree of functional enrichment for a given cluster was quantitatively assessed (P-value) using the ClueGO tool (44). ClueGO integrates gene ontology (GO) terms and creates a functionally organized GO/pathway term network. It can analyze genes and comprehensively visualizes functionally grouped terms (45).
Results
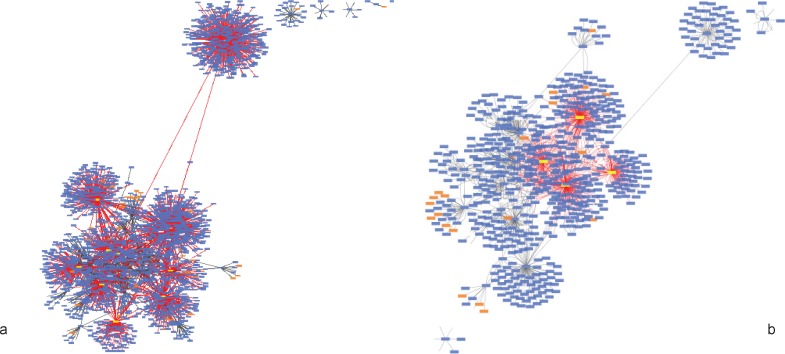
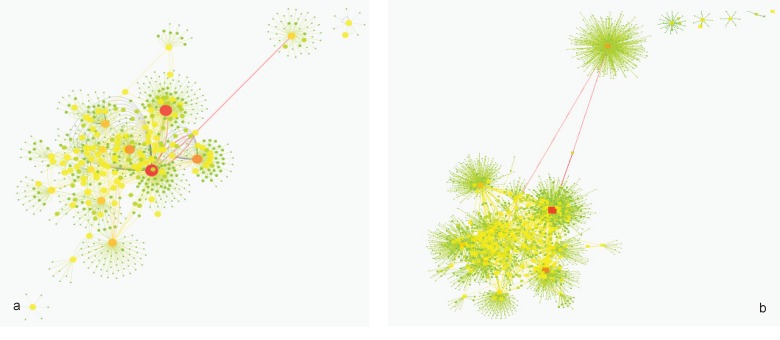
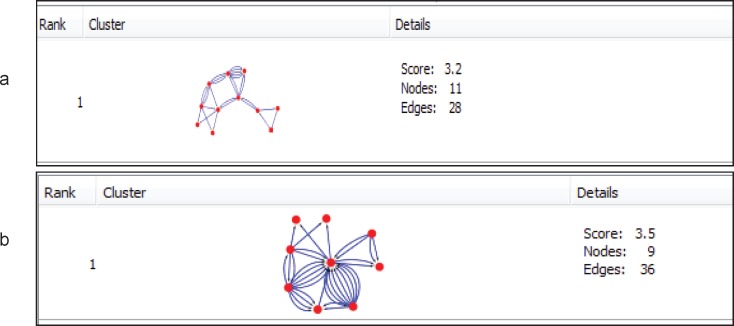
Twenty-nine (29) genes in cirrhosis and sixty-three (63) genes in HCC with differential gene expression were distinguished via literature survey. Genes and proteins were presented in tables 1 and 2 for cirrhosis liver and HCC respectively. Hub is a node with a number of links that greatly exceeds the average (57). Cytoscape analysis revealed a great number of close interconnections that can be seen in Figure 1 (a for cirrhosis liver and b related to HCC). Based on degree values, about 15% of the initial proteins are selected as hub proteins. APOE, TTR, CLU and APOA1 as related hub proteins of cirrhosis and TTR, CLU, APOA1, TFRC, VWF, CKAP5, A2M, ZNF648, mCASP8, HSP27 as hub proteins of HCC are highlighted in figure 1 and tabulated in table 3. In figure 2 is represented based on the node size and color changes of the nodes. As the circles get bigger and their color change from green to red, their value of the degree and the betweenness centrality increase. The related subnetworks were introduced by MCODE (figure 4). Among the genes that involved in cirrhosis, APOE determined as seed while genes of HCC have no seed. ClueGo is also a Cytoscape software for gene ontology and pathway enrichment analysis. The analyzed results of MCODE by ClueGo for biological process is shown in figure 4.
Figure 1.
This figure shows a PPI network, which consists of 651 nodes and 1305 edges for cirrhosis (a) and 2024 nodes and 3817edges for HCC (b). The highlighted nodes APOE, TTR, CLU related to cirrhosis and TTR CLU, APOA1, TFRC, VWF, CKAP 5, A2M, ZNF648, CASP8, HSP27 for hepatocellular carcinoma
Table 3.
A number of genes with significant centrality value derived from figure 1, a for cirrhosis liver and b is related to HCC, based on two fundamental centrality properties analysis (Degree and Betweeness centrality
| Unipr otcode | Gene name | Degree | Betweenness centrally | ||
|---|---|---|---|---|---|
| a | P02647 | APOA1 | 238 | 0.43889 | |
| P10909 | CLU | 216 | 0.319182 | ||
| P02766 | TTR | 136 | 0.142145 | ||
| P02649 | APOE | 131 | 0.165061 | ||
| b | Uniprotcode | Gene name | Degree | Betweenness centrally | |
| P04792 | HSP27 | 846 | 0.444179 | ||
| Q14790 | CASP8 | 544 | 0.162997 | ||
| Q5T619 | ZNF648 | 400 | 0.361564 | ||
| Q14008 | CKAP5 | 271 | 0.19395 | ||
| P02647 | APOA1 | 238 | 0.159222 | ||
| P01023 | A2M | 230 | 0.131263 | ||
| P10909 | CLU | 216 | 0.101279 | ||
| P02786 | TFRC | 146 | 0.076278 | ||
| P04275 | VWF | 146 | 0.093813 | ||
| P02766 | TTR | 136 | 0.040481 | ||
Figure 2.
Mapping degree and betweenness parameters of candidate protein interaction network to node size and color; (a) cirrhosis patients and (b) HCC patients. More details are accessible in the text
Figure 4.
Functional distribution of biological process of modules of (a) cirrhosis and (b) HCC (b) (P<0.05). These include Kappa statistic ≥ 0.5, enrichment and Bonferroni step down method for probability value correction. The stars show the pathways with P-value <0.05. The pathways with two stars have more significant score rather than one star
Discussion
The importance of human molecular interaction networks no summarized only in reveals protein function with their inter- relationships, even its clear view of fundamental human biology as well as disease progression, diagnosis, and treatment (58).
The protein Networks analysis provides a model that elevates systems-level understanding of the mechanisms of diseases (59, 60) to analyze therapeutic drugs and their targets (61-63) and discovering the novel network-based biomarkers (64). In this study, protein network of HCC and cirrhosis patient has investigated. This analysis can lead to figure out a better understanding of the etiology of both liver disease and the specified pattern of gene expression in cirrhosis and HCC. On the other hand, therapeutic targets and diagnostic biomarkers can be accelerated by targeting the specific hub genes. Previous studies introduced cirrhosis as one of the reasons in HCC development (65) then finding molecular common points to be expected between two diseases. The importance of PPI analysis has been reported in cancer related genes (34, 36). Hub genes have virtual conception to study due to their centrality role in a PPI network (66). As represented in figure 1, protein interaction networks of HCC and cirrhosis are made up of numerous nodes that provide hub selection. APOE, TTR, CLU and APOA1 as hub protein are introduced for cirrhosis and TTR, CLU, APOA1, TFRC, VWF, CKAP5, A2M, ZNF648, CASP8 and HSP27 (HSPB1) are the related hub proteins to HCC. The all introduced hub proteins (except TTR for HCC) are bottlenecks (cut off 0.05 is used for betweenness centrality). APOA1 and CLU are the two hub-bottlenecks common between the two diseases. As it is presented in the table 3, APOA1 and CLU are characterized by the most values of degree and betweenness centralities for cirrhosis. ApoA1 is the main protein component of high density lipoprotein in plasma, which is involved in the formation of most plasma cholesterol esters (67) .This protein potently suppresses tumor growth and metastasis in multiple animal tumor models (68) . The validated changes of expression of APOA1 accompanied by a few proteins have the potential for development into high-performance tests used in the diagnosis and or monitoring of HCC and LC patients (46). CLU is a Golgi molecular chaperone involved in BAX- antiapoptotic processes, activation of the phosphatidylinositol 3-kinase/ protein kinase B pathway, promotion of angiogenesis, mediation of the nuclear factor kappa B (NF-κB) pathway and modulation of extra-cellular signal-regulated kinase (ERK) signaling. A number of biological processes, including programmed cell death (Down regulation allows for p53activation and cell death), lipid transport, membrane recycling and cell adhesion (69, 70). Serum clusterin was introduced as more specific and sensitive biomarker than AFP in distinction of HBV-cirrhosis with HCC base on HBV-cirrhosis (71). It also has been shown that clusterin may be a useful marker in the evaluation of prognosis of patients with alcoholic cirrhosis and severity of liver disease (72). CLU involved in BAX- antiapoptotic processes and it’s down regulation allows p53 activation and cell death (73). MCODE clustering algorithm (center based) demonstrates the possible presence of similar functional protein in the two PPI networks. As it is shown in figure 3 there are 2 clusters for the two PPI networks that are approximately similar. The PPI network in HCC has no seed, but in cirrhosis C4BPA is introduced as seed protein. More information about the roles of the 2 clusters in the biological processes is presented in figure 4. ClueGo provided functional annotation (BP) of the studied modules. Biological process analysis revealed some similarities between the two diseases. The significant role of the immune system and a few common pathways show the closeness of the two diseases. Some of the important proteins are involved in the terms. The highlighted roles of APOA1 in the revers cholesterol transport pathway and CLU in positive regulation of neurofibrillary tangle assembly pathway in the two major pathways of studied diseases are identified. CLU is related to reverse cholesterol transport, platelet degranulation and human immune response pathways. However, APOA1 is involved in reverse cholesterol transport, platelet degranulation and humoral immune response negative regulation of immune effector process pathways.
Figure 3.
MCODE algorithm analysis demonstrates clusters based on the number of interconnections in the large network of protein-protein interactions for (a) cirrhosis disease and (b) HCC disease
Protein-protein interaction analysis and pathway assessment showed a closed molecular relationship between cirrhosis and HCC. The finding pointed to the significant role of APOA1 and CLU as the common two biomarker in development of cirrhosis and HCC diseases.
Acknowledgment
This research has been derived from the PhD. thesis of Akram Safaei.
References
- 1.Bosch FX, Ribes J, Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271–85. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 2.Davis G, Dempster J, Meler J, Orr D, Walberg M, Brown B, Berger B, et al. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent) 2008;21:266–80. doi: 10.1080/08998280.2008.11928410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meer S, Robert A, Siersema PD, Erpecum K. Urveillance for hepatocellular carcinoma in chronic liver disease: Evidence and controversies. World J Gastroenterol. 2013;19:6744–56. doi: 10.3748/wjg.v19.i40.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nature Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 5.Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–37. [PubMed] [Google Scholar]
- 6.Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U S A. 2001;98:15089–94. doi: 10.1073/pnas.241522398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MW, Yue ZN, Geiss GK, Sadovnikova NY, Carter VS, Boix L, et al. Identification of novel tumor markers in hepatitis C virus- associated hepatocellular carcinoma. Cancer Res. 2003;63:859–64. [PubMed] [Google Scholar]
- 8.Megger DA, Naboulsi W, Meyer HE, Sitek B. Proteome analyses of hepatocellular carcinoma. J Clin Transl Hepatol. 2014;2:23–30. doi: 10.14218/JCTH.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarhuni A, Guyot E, Rufat P, Sutton A, Bourcier V, Grando V, et al. Impact of cytokine gene variants on the prediction and prognosis of hepatocellular carcinoma in patients with cirrhosis. J Hepatol. 2014;61:342–50. doi: 10.1016/j.jhep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Kato N, Hoshida Y, Yoshida H, Taniguchi H, Goto T, et al. Interleukin-1β gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology. 2003;37:65–71. doi: 10.1053/jhep.2003.50017. [DOI] [PubMed] [Google Scholar]
- 11.Nahon P, Sutton A, Rufat P, Ziol M, Akouche H, Laguillier C, et al. Myeloperoxidase and superoxide dismutase 2 polymorphisms comodulate the risk of hepatocellular carcinoma and death in alcoholic cirrhosis. Hepatology. 2009;50:1484–93. doi: 10.1002/hep.23187. [DOI] [PubMed] [Google Scholar]
- 12.Nahon P, Sutton A, Rufat P, Charnaux N, Mansouri A, Moreau R, et al. A variant in myeloperoxidase promoter hastens the emergence of hepatocellular carcinoma in patients with HCV- related cirrhosis. J Hepatol. 2012;56:426–32. doi: 10.1016/j.jhep.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Yuan JM, Lu SC, Van Den Berg D, Govindarajan S, Zhang ZQ, Mato JM, et al. Genetic polymorphisms in the methylenetetrahydrofolate reductase and thymidylate synthase genes and risk of hepatocellular carcinoma. Hepatology. 2007;46:749–58. doi: 10.1002/hep.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Long X, Ma Y, Qu DY, Liu YG, Huang ZQ, Huang YZ, et al. The polymorphism of XRCC3 codon 241 and AFB1-related hepatocellular carcinoma in Guangxi population, China. Ann Epidemio. 2008;18:572–8. doi: 10.1016/j.annepidem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Dharel N, Kato N, Muroyama R, Moriyama M, Shao R-X, Kawabe T, et al. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–71. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 16.Yoon YJ, Chang HY, Ahn SH, Kim JK, Park YK, Kang DR, et al. MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Carcinogenesis. 2008;29:1192–6. doi: 10.1093/carcin/bgn090. [DOI] [PubMed] [Google Scholar]
- 17.Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–31. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 18.Woo HG, Wang XW, Budhu A, Kim YH, Kwon SM, Tang ZY, et al. Association of TP53 mutation with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroent erology. 2011;140:1063–70. doi: 10.1053/j.gastro.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behne T, Copur MS. Biomarkers for hepatocellular carcinoma. Int J Hepatol. 2012;2012:859076. doi: 10.1155/2012/859076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J proteome Res. 2011;10:1828–36. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- 22.Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World Gastroenterol. 2010;16:418–24. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Serag HB, Mason AC, Key C. Trends in survival of patients with hepatocellular carcinoma between 1977 and 1996 in the United States. Hepatology. 2001;33:62–5. doi: 10.1053/jhep.2001.21041. [DOI] [PubMed] [Google Scholar]
- 24.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–51. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Liu J, Lin C, Wang H, Jiang Y, Wang J, et al. Peroxiredoxin 2: a potential biomarker for early diagnosis of hepatitis B virus related liver fibrosis identified by proteomic analysis of the plasma. BMC Gastroenterol. 2010;10:115. doi: 10.1186/1471-230X-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scorza M, Elce A, Zarrilli F, Liguori R, Amato F, Castaldo G. Genetic diseases that predispose to early liver cirrhosis. Int J Hepatol. 2014;2014:713754. doi: 10.1155/2014/713754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072–7. doi: 10.1002/hep.26130. [DOI] [PubMed] [Google Scholar]
- 28.Gangadharan B, Antrobus R, Dwek RA, Zitzmann N. Novel serum biomarker candidates for liver fibrosis in hepatitis C patients. Clin Chem. 2007;53:1792–9. doi: 10.1373/clinchem.2007.089144. [DOI] [PubMed] [Google Scholar]
- 29.Re M, Mesiti M, Valentini G. A fast ranking algorithm for predicting gene functions in biomolecular networks. IEEE/ACM Trans Comput Biol Bioinform. 2012;9:1812–8. doi: 10.1109/TCBB.2012.114. [DOI] [PubMed] [Google Scholar]
- 30.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. Gene MANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesiti M, Re M, Valentini G. Think globally and solve locally: secondary memory-based network learning for automated multi- species function prediction. Giga Science. 2014;3:5–10. doi: 10.1186/2047-217X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safaei A, Tavirani MR, Oskouei AA, Azodi MZ, Mohebbi SR, Nikzamir AR. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol Hepatol Bed Bench. 2016;9:114–23. [PMC free article] [PubMed] [Google Scholar]
- 33.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7:17–31. [PMC free article] [PubMed] [Google Scholar]
- 34.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Tavirani MR, Seyyedi SS. Protein-Protein Interaction Network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8:215–22. [PMC free article] [PubMed] [Google Scholar]
- 35.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi- Nejad A, Rostami-Nejad M, Rahmatirad S. Protein Clustering and Interactome Analysis in Parkinson and Alzheimer’s Diseases. Arch Iran Med. 2016;19:101–9. [PubMed] [Google Scholar]
- 36.Zali H, Tavirani MR. Meningioma protein-protein interaction network. Arch Iran Med. 2014;17:262–72. [PubMed] [Google Scholar]
- 37.Zamanian Azodi M, Peyvandi H, Rostami-Nejad M, Safaei A, Rostami K, Vafaee R, et al. Proteinprotein interaction network of celiac disease. Gastroenterol Hepatol Bed Bench. 2016;9:268–77. [PMC free article] [PubMed] [Google Scholar]
- 38.Safaei A, Tavirani MR, Oskouei AA, Azodi MZ, Mohebbi SR. Evaluation of protein clustering of pancreatic cancer. Arvand J Health Med Sci. 2016;1:68–77. [Google Scholar]
- 39.Chautard E, Thierry-Mieg N, Ricard-Blum S. Interaction networks: from protein functions to drug discovery A review. Pathologie Biologie. 2009;57:324–33. doi: 10.1016/j.patbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009;5:e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein- protein interaction detection: methods and analysis. Int J Proteomics. 2014;2014:147648. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedamallu CS, Posfai J. Open source tool for prediction of genome wide protein-protein interaction network based on ortholog information. Source Code Biol Med. 2010;5:8. doi: 10.1186/1751-0473-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivera CG, Vakil R, Bader JS. NeMo: Network Module identification in Cytoscape. BMC Bioinformatics. 2010;18:1471–2105. doi: 10.1186/1471-2105-11-S1-S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–3. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fye HK, Wright-Drakesmith C, Kramer HB, Camey S, da Costa AN, Jeng A, et al. Protein profiling in hepatocellular carcinoma by label-free quantitative proteomics in two west African populations. PLoS One. 2013;8:e68381. doi: 10.1371/journal.pone.0068381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bedossa P, Poynard T, Abella A, Aubert A, Pignon JP, Naveau S, et al. Apolipoprotein A1 is a serum and tissue marker of liver fibrosis in alcoholic patients. Alcohol Clin Exp Res. 1989;13:829–33. doi: 10.1111/j.1530-0277.1989.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 48.Poon T, Chan H, Leung H, Lo A, Lau R, Hui A, et al. Liver cirrhosis–specific glycoforms of serum proteins in chronic hepatitis B infection: identification by lectin affinity chromatography and quantitative proteomic profiling. Hong Kong Med J. 2015;21:22–6. [PubMed] [Google Scholar]
- 49.Montaser-Kouhsari L. Plasma myeloperoxidase activity and apolipoprotein A-1 expression in chronic hepatitis B patients. Arch Iran Med. 2011;14:254–8. [PubMed] [Google Scholar]
- 50.Sarvari J, Mojtahedi Z, Taghavi SAR, Kuramitsu Y, Shahrabadi MS, Ghaderi A, et al. Differentially expressed proteins in chronic active hepatitis, cirrhosis, and HCC related to HCV infection in comparison with HBV infection: a proteomics study. Hepat Mon. 2013;13:e8351. doi: 10.5812/hepatmon.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gangadharan B, Antrobus R, Dwek RA, Zitzmann N. Novel serum biomarker candidates for liver fibrosis in hepatitis C patients. Clin Chem. 2007;53:1792–9. doi: 10.1373/clinchem.2007.089144. [DOI] [PubMed] [Google Scholar]
- 52.Gao HJ, Chen YJ, Zuo D, Xiao MM, Li Y, Guo H, et al. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol Med. 2015;12:246–54. doi: 10.7497/j.issn.2095-3941.2015.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang MH, Tyan YC, Jong SB, Huang YF, Liao PC, Wang MC. Identification of human hepatocellular carcinoma-related proteins by proteomic approaches. Anal Bioanal Chem. 2007;388:637–43. doi: 10.1007/s00216-007-1263-6. [DOI] [PubMed] [Google Scholar]
- 54.Sarvari J, Mojtahedi Z, Kuramitsu Y, Fattahi MR, Ghaderi A, Nakamura K, et al. Comparative proteomics of sera from HCC patients with different origins. Hepat Mon. 2014;14:e13103. doi: 10.5812/hepatmon.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng JT, Liu YK, Song HY, Dai Z, Qin LX, Almofti MR, et al. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581–8. doi: 10.1002/pmic.200401309. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Wang X, Li S, Hu H, Zhang D, Hu P, et al. The role of von Willebrand factor as a biomarker of tumor development in hepatitis B virus-associated human hepatocellular carcinoma: a quantitative proteomic based study. J Proteomics. 2014;106:99–112. doi: 10.1016/j.jprot.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Agarwal S, Deane CM, Porter MA, Jones NS. Revisiting date and party hubs: novel approaches to role assignment in protein interaction networks. PLoS Comput Biol. 2010;6:e1000817. doi: 10.1371/journal.pcbi.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nature Genet. 2009;41:415–23. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yıldırım MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug— target network. Nature Biotech. 2007;25:1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 62.Winter C, Kristiansen G, Kersting S, Roy J, Aust D, Knösel T, et al. Google goes cancer: improving outcome prediction for cancer patients by network-based ranking of marker genes. PLoS Comput Biol. 2012;8:e1002511. doi: 10.1371/journal.pcbi.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Re M, Valentini G. Network-based drug ranking and repositioning with respect to DrugBank therapeutic categories. IEEE/ACM Trans Comput Biol Bioinform. 2013;10:1359–71. doi: 10.1109/TCBB.2013.62. [DOI] [PubMed] [Google Scholar]
- 65.Simonetti RG, Cammà C, Fiorello F, Politi F, D’Amico G, Pagliaro L. Hepatocellular carcinoma. Dig Dis Sci. 1991;36:962–72. doi: 10.1007/BF01297149. [DOI] [PubMed] [Google Scholar]
- 66.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 67.Gray J, Chattopadhyay D, Beale GS, Patman GL, Miele L, King BP, et al. A proteomic strategy to identify novel serum biomarkers for liver cirrhosis and hepatocellular cancer in individuals with fatty liver disease. BMC Cancer. 2009;9:271. doi: 10.1186/1471-2407-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zamanian-Daryoush M, Lindner D, Tallant TC, Wang Z, Buffa J, Klipfell E, et al. The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol Chem. 2013;288:21237–52. doi: 10.1074/jbc.M113.468967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sansanwal P, Li L, Sarwal MM. Inhibition of intracellular clusterin attenuates cell death in nephropathic cystinosis. J Am Soc Nephrol. 2015;26:612–25. doi: 10.1681/ASN.2013060577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin CC, Tsai P, Sun HY, Hsu MC, Lee JC, Wu IC, et al. Apolipoprotein J, a glucose-upregulated molecular chaperone, stabilizes core and NS5A to promote infectious hepatitis C virus virion production. J Hepatol. 2014;61:984–93. doi: 10.1016/j.jhep.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Liu YH, Mai SJ, He LJ, Liao YJ, Deng HX, et al. Evaluation of serum clusterin as a surveillance tool for human hepatocellular carcinoma with hepatitis B virus related cirrhosis. J Gastroenterol Hepatol. 2010;25:1123–8. doi: 10.1111/j.1440-1746.2009.06205.x. [DOI] [PubMed] [Google Scholar]
- 72.Hogåsen K, Homann C, Mollnes TE, Graudal N, Hogåsen AKM, Hasselqvist P, et al. Serum clusterin and vitronectin in alcoholic cirrhosis. Liver. 1996;16:140–6. doi: 10.1111/j.1600-0676.1996.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 73.Norouzinia M, Asadzadeh H, Shalmani HM, Al Dulaimi D, Zali MR. Clinical and histological indicators of proximal and distal gastric cancer in eight provinces of Iran. Asian Pac J Cancer Prev. 2012;13:5677–9. doi: 10.7314/apjcp.2012.13.11.5677. [DOI] [PubMed] [Google Scholar]