Abstract
We describe here the main features of ‘Lachnoclostridium urinimassiliense’ strain Marseille-P2804T (= CSUR P2804) and ‘Lachnoclostridium phocaeense’ strain Marseille-P3177T (= CSUR P3177) that were isolated from urine samples after kidney transplantation in two women.
Keywords: Culturomics, kidney transplantation, Lachnoclostridium phocaeense, Lachnoclostridium urinimassiliense, taxonomy
As a part of the diversity's exploration of the urinary microbiota by culturomics [1], [2], we investigated urine samples of adult kidney transplant recipients. The patients provided signed informed consent, and the agreement of the local ethics committee of the IFR48 (Marseille, France) was obtained under number 09-022.
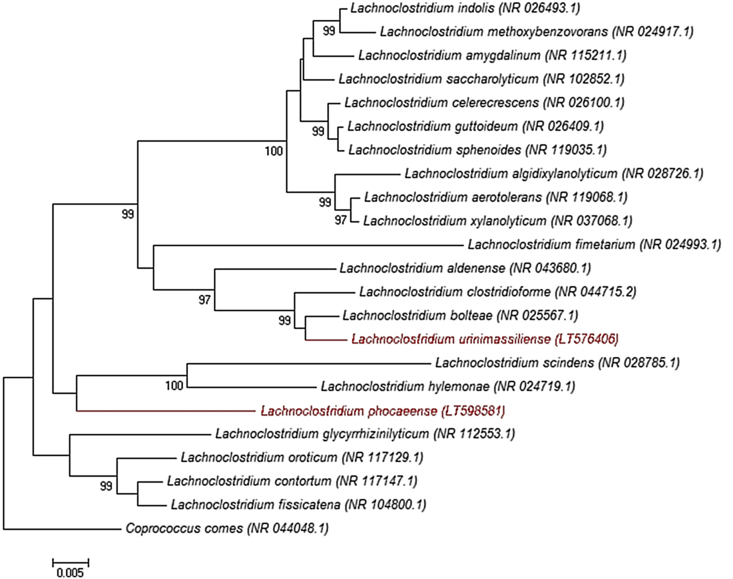
The bacterial strains Marseille-P2804 and Marseille-P3177 were isolated from urine samples after kidney transplantation in two women (37 and 51 years old respectively). We initially preincubated the urine samples at 37°C for 96 hours in an anaerobic blood culture bottle (BACTEC Lytic/10 Anaerobic/F Culture Vials; Becton-Dickinson, Pont de Claix, France) enriched with 5% sterilized rumen by microfiltration (0.2 μm pore diameter). Then the preincubated samples were seeded on 5% sheep's blood–enriched Columbia agar medium (bioMérieux, Marcy l'Etoile, France). A pure culture of the strain Marseille-P2804 was obtained after 3 days of incubation at 37°C under anaerobic atmosphere (AnaeroGen; Oxoid, Dardilly, France), while the strain Marseille-P3177 was obtained after 5 days of incubation under the same temperature and atmospheric conditions. Agar grown colonies were circular, translucent whiteish and glistening with a diameter between 0.7 and 1 mm. Strain Marseille-P2804 cells were Gram-positive bacilli, nonmotile and pointed rod shaped, ranging in diameter from 600 to 800 nm. Strain Marseille-P3177 cells were motile Gram-positive bacilli, ranging in diameter from 250 to 350 nm. Both strains Marseille-P2804 and Marseille-P3177 do not exhibit oxidase or catalase activities. Strains were not identified by systematic matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [3]. Thus, the complete 16S rRNA gene was sequenced using fD1-rP2 primers as previously described [4] and a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). The strain Marseille-P2804 exhibited a sequence similarity of 98.4% with Lachnoclostridium bolteae strain WAL 16351T (GenBank accession no. NR025567) [5], while the strain Marseille-P3177 exhibited a sequence similarity of 94.6% with Lachnoclostridium contortum strain ATCC 25540 (GenBank accession no. NR_117147) [6] (Fig. 1).
Fig. 1.
Phylogenetic tree showing position of Lachnoclostridium urinimassiliense strain Marseille-P2804T and Lachnoclostridium phocaeense strain Marseille-P3177T relative to other phylogenetically close neighbours. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 90% were retained. Coprococcus comes was used as outgroup. Scale bar indicates 0.5% nucleotide sequence divergence.
Consequently, according to the 16S rRNA gene sequence similarity to delineate a new species of prokaryotes [7], [8], we propose to classify the strains Marseille-P2804 and Marseille-P3177 as the representative strains of new members of the recently described genus Lachnoclostridium [9] within the family Lachnospiraceae in the phylum Firmicutes and the creation of two new species: ‘Lachnoclostridium urinimassiliense’ sp. nov. (u.ri.ni.mas.sil.ien'se, from u.ri.ni L. neut. adj., ‘urine,’ and mas.si.li.en'se L. neut. adj. massiliense, ‘pertaining to Massilia,’ the Latin name of Marseille, France, where the organism was first isolated) and ‘Lachnoclostridium phocaeense’ sp. nov. (pho.cae.en'se, L. fem. adj. phocaeense, referring to Phocaea, the Latin name of the Greek people who founded Marseille, France, where the organism was first isolated) and considering the strains Marseille-P2804T and Marseille-P3177T as the type strains of ‘Lachnoclostridium urinimassiliense’ sp. nov. and ‘Lachnoclostridium phocaeense’ sp. nov. respectively.
MALDI-TOF MS spectrum
MALDI-TOF MS spectra of ‘Lachnoclostridium urinimassiliense’ strain Marseille-P2804T and ‘Lachnoclostridium phocaeense’ strain Marseille-P3177T are available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under accession number LT576406 for Marseille-P2804T and LT598581 for Marseille-P3177T.
Deposit in a culture collection
Strain Marseille-P2804T and Marseille-P3177T were deposited in the Collection de Souches de l'Unité des Rickettsies (CSUR, WDCM 875) under numbers P2804 and P3177 respectively.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of interest
None declared.
References
- 1.Hilt E.E., McKinley K., Pearce M.M., Rosenfeld A.B., Zilliox M.J., Mueller E.R. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. J Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y., Liu C., Molitoris D.F., Tomzynski T.J., Lawson P.A., Collins M.D. Clostridium bolteae sp. nov., isolated from human sources. Syst Appl Microbiol. 2003;26:84–89. doi: 10.1078/072320203322337353. [DOI] [PubMed] [Google Scholar]
- 6.Holdeman L.V., Cato E.P., Moore W.E.C. Eubacterium contortum (Prévot) comb. nov.: emendation of description and designation of the type strain. Int J Syst Bacteriol. 1971;21:304–306. [Google Scholar]
- 7.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152. [Google Scholar]
- 8.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 9.Yutin N., Galperin M.Y. A genomic update on clostridial phylogeny: gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]