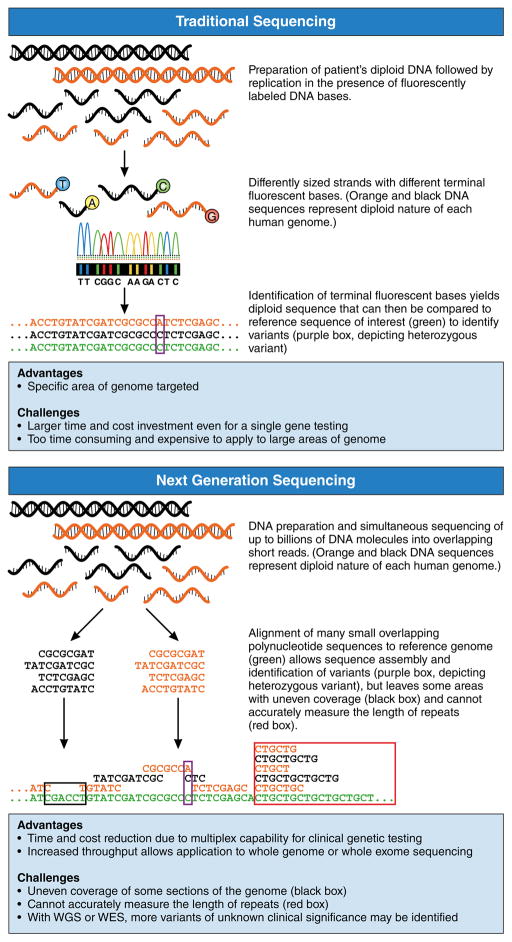
The initial sequencing of the human genome took nearly a decade to complete. Today, the same can be accomplished in hours using next generation sequencing (NGS). Prior to the invention of NGS, genetic testing was accomplished by traditional Sanger sequencing. In cardiovascular medicine, this approach typically involved the sequencing of a region of interest in a single affected patient’s DNA and then comparison of that sequence to the human genome reference sequence. Variants found were then compared to a panel of sequences from unaffected people, often anonymous blood donors. This strategy has been the mainstay of genetic testing to assess small numbers of disease-related genes across cardiovascular and non-cardiovascular diseases. However, this approach is limited in its scalability.
NGS builds upon these basic principles, extending them to allow billions of DNA molecules to be sequenced simultaneously. This multiplex technology yields billions of small polynucleotide reads, called “short reads.” Mapping and assembly of these short reads to a reference genome then allows discovery of genetic variants at scale, (Figure 1) increasing the speed and reducing the cost of sequencing by orders of magnitude.1 Thus, NGS can easily be applied to panels of genes, every gene, or the whole genome in clinical settings. This scientific leap in sequencing technology has occurred over just 15 years and has brought the genetic code to the forefront of diagnosis and therapy, making it increasingly important for clinicians to be aware of its utility, as well as its limitations, in practice.
Figure 1. Comparison of Traditional Sequencing and Next Generation Sequencing (NGS).
In traditional sequencing, DNA is replicated in the presence of fluorescently labeled bases, yielding differently sized strands with different terminal bases. These terminal bases are identified in order to assemble the entire sequence and compare it to the reference genome. In NGS, DNA is simultaneously sequenced into billions of overlapping short reads that are then assembled by alignment to the reference genome. Here, each diploid genome is represented by black and orange copies. Green sequence represents reference genome. Purple boxes illustrate heterozygous variants, the black box illustrates the problem of uneven coverage and the red box indicates the inability of short reads to accurately sequence large areas of repeated sequence.
NGS in Clinical Cardiovascular Medicine
Within cardiovascular medicine, NGS is used in the diagnosis of Mendelian disease, i.e., disease caused by a single variant in a single gene. Cardiovascular Mendelian diseases span multiple syndromes such as long QT syndrome, familial hypercholesterolemia, hypertrophic and familial dilated cardiomyopathies, and Marfan syndrome. The yield of genetic testing with NGS-based platforms for identifying causative variants is variable across these diseases, ranging from up to 80% in long QT syndrome to as low as 20–40% in dilated cardiomyopathy.2 Characterizing the underlying genetic cause of these cases reassures the patient, directs family screening and fertility planning, and can often actually guide therapy. For example, identifying the causal gene in long QT syndrome allows precision targeting of therapy in subsets of disease caused by potassium versus sodium channel dysfunction. Further, clarifying that left ventricular hypertrophy is due to variants in transthyretin (TTR) or Fabry’s disease can allow targeted therapies including RNA silencing, isoform stabilizers and enzyme replacement. Several commercially available disease-specific genetic testing panels exist, and vary in the number of genes tested and reported diagnostic yield.2 In cases for which these panels are unrevealing, broader sequencing strategies such as exome or genome sequencing may be helpful.
In the case of unrevealing genetic panel testing, WGS or WES can yield important information about potentially pathogenic variants. NGS offers significant technological advances over traditional sequencing for this purpose. Specifically, it makes broad sequencing for the identification of previously unreported pathogenic variants in multiple family members feasible. This was previously accomplished with genome-wide scans followed by mapping of identified variation to nearby potentially pathogenic genes, however, using NGS for this purpose facilitates the location of potentially pathogenic variation directly to the gene of interest. This is particularly powerful in patients with a clear family history and living affected family members because segregation analysis of candidate variants can be performed to clarify their role. If the variant segregates with the disease in question, (meaning that all disease-affected individuals carry the variant of interest), then this lends significant support to this variant’s pathogenicity. However, if any disease-affected individual does not carry the suspect variant, then the variant is not likely pathogenic for the syndrome of interest. In addition to multiple reports of clinical integration of genomic data, reports regarding the use of whole genome and exome sequencing in patients with diagnostic odysseys report solve rates of at least 25%.3 Also, emerging rapid turnaround diagnosis of inherited disease and mosaicism can contribute positively to improved patient care.4
Challenges of NGS in the Clinical Setting
The speed and cost effectiveness of NGS is balanced by challenges in appropriately characterizing certain areas of the genome and certain types of variation. Capture approaches for selective sequencing and sequencing chemistry itself both contribute to uneven coverage. For example, areas of the genome with high guanine and cytosine base content (“GC content”) may be more difficult to expose to replication reactions because of the high energy bonds made between the two strands of DNA at these bases. Additionally, short reads cannot accurately characterize regions of repeated sequence that are longer than the short reads themselves. For example, in myotonic dystrophy-related cardiomyopathy, disruption of the causative gene occurs due to an abnormally high number of repeats of a section of its sequence. Because a single short read is not long enough to encompass all repeats, alignment of different lengths of repeated sequence cannot accurately determine the total number of repeats (Figure 1). Finally, our algorithms for detecting certain forms of variation such as small insertions and deletions remain sub-optimal. Overall, an estimated 20–25% of exonic bases thought to be involved in human disease lie in regions of the genome with low NGS coverage or accuracy, and tools to improve the speed and accuracy of analysis and sequencing are in constant development.4
An additional inevitable consequence of broader sequencing by NGS is the challenge of assigning pathogenicity to newly identified variants. Humans differ at millions of genetic positions, some commonly found in the population, some rarely found, and some unique to one individual. Yet, only a very small number of these identified variants are truly causative of disease. Thankfully, the increasing availability of sequencing data from tens of thousands of unaffected people helps narrow the list of rare variants to those that are most suspicious. However, even with these recent advances, there will remain some variants for which it is impossible to determine potential causality. Such variants are classified as “variants of uncertain significance”, a term commonly abbreviated to “VUS.” Such variants, in genes known to cause disease, will be reported by genetic tests – a small number in multi-gene panels and many more in the setting of WGS and WES. Self-evidently, a VUS in a gene known to cause disease can create significant clinical equipoise regarding its use in predictive testing and diagnosis.
Thus, categorizing each variant with respect to the evidence for its pathogenicity is extremely important, as false positives and negatives are of high consequence to patient care. Variant review includes a thorough investigation of 1) previous reports of disease associated with that particular variant, 2) segregation with disease within families, 3) its rarity in the healthy population, and 4) experimental evidence or computational prediction that variation in the gene of interest can cause disease. Because of the subjectivity of this process, moderate disagreement with respect to variant classification exists between commercial providers, national databases and even expert reviewers at the same center.5 These findings, combined with variation in diagnostic yield between syndromes, make genetic counselors fundamental to the clinical care of families with inherited disease. Their expertise is crucial in variant review and treating the psychosocial challenges of receiving and living with a diagnosis of genetic disease.
The Future of NGS in Clinical Practice
The successful application of clinical genomics will require the expansion and sharing of current genetic and clinical databases as well as the development of a more precise disease taxonomy. Such precision medicine is becoming increasingly possible with advanced mobile technology, integration of the genome into the medical record and high throughput sequencing platforms for gene expression.4 Thus, the human genome is a powerful clinical tool already being employed in cardiovascular medicine, and its potential applications in precision diagnostics and therapeutics are on the horizon.
Footnotes
Conflict of Interest Disclosures: E.A.A. is a co-founder of, and an advisor at, Personalis Inc.
Citations
- 1.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturm AC, Hershberger RE. Genetic testing in cardiovascular medicine: current landscape and future horizons. Curr Opin Cardiol. 2013;28:317–325. doi: 10.1097/HCO.0b013e32835fb728. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17:507–522. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 5.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. The Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]