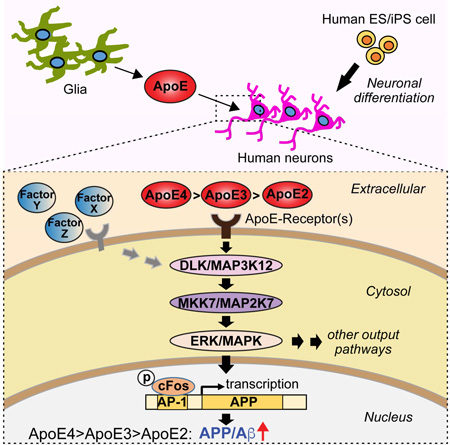
SUMMARY
Human ApoE apolipoprotein is primarily expressed in three isoforms (ApoE2, ApoE3, and ApoE4) that differ only by two residues. ApoE4 constitutes the most important genetic risk factor for Alzheimer’s disease (AD), ApoE3 is neutral, and ApoE2 is protective. How ApoE isoforms influence AD pathogenesis, however, remains unclear. Using ES cell-derived human neurons, we show that ApoE secreted by glia stimulates neuronal Aβ-production with an ApoE4>ApoE3>ApoE2 potency rank order. We demonstrate that ApoE-binding to ApoE-receptors activates dual leucine-zipper kinase (DLK), a MAP-kinase kinase kinase that then activates MKK7 and ERK1/2 MAP-kinases. Activated ERK1/2 induces cFos phosphorylation, stimulating the transcription factor AP-1, which in turn enhances transcription of amyloid-β precursor protein (APP) and thereby increases amyloid-β levels. This molecular mechanism also regulated APP transcription in mice in vivo. Our data describe a novel signal transduction pathway in neurons whereby ApoE activates a non-canonical MAP-kinase cascade that enhances APP transcription and amyloid-β synthesis.
Keywords: Apolipoprotein E (ApoE), Alzheimer’s disease, MAP-kinase signaling, amyloid precursor protein (APP), beta amyloid (Aβ), cFos, CRISPR, CRISPRi, dual leucine-zipper kinase (DLK), transcription
Graphical Abstract
INTRODUCTION
Apolipoprotein E (ApoE), a major component of LDL and VLDL lipoproteins circulating in blood, mediates uptake of lipoproteins into cells by binding to ApoE receptors, principally the LDL-receptor (Goldstein and Brown, 2015). ApoE is also highly expressed in brain, primarily in astrocytes, but its function in brain remains unclear (Holtzman et al., 2012; Wang and Eckel, 2014). In addition to LDL-receptors, several related ApoE-receptors are expressed throughout the body. Many of these other ApoE-receptors bind additional ligands and perform central functions during development. For example, the LDL-receptor-related proteins VLDLR and ApoER2 bind reelin and regulate cortical development (Trommsdorff et al., 1999), LDL-receptor-related proteins 5 and 6 (Lrp5 and Lrp6) are co-receptors for wnts (Tamai et al., 2000), and LDL-receptor-related protein Lrp2 is an auxiliary receptor for sonic hedgehog (Christ et al., 2012). Moreover, ApoE exhibits signaling properties in dissociated cultures of rodent neurons, and activates MAP-kinases (Ohkubo et al., 2001; Qiu et al., 2004). Several MAP-kinases are prominently expressed in brain, including the MAP-kinase kinase kinase DLK (for ‘dual leucine-zipper kinase’), which has been implicated both in the degeneration and regeneration of neurons (Tedeschi and Bradke, 2013; Lu et al., 2014). In C. elegans and Drosophila, regeneration of injured neurons requires DLK activation (Hammarlund et al., 2009; Yan et al., 2009; Shin et al., 2012). In rodents, conversely, DLK mediates neuronal cell death during development and contributes to excitotoxic neurodegeneration (Chen et al., 2012). However, the precise relation of ApoE-receptor binding to MAP-kinase activation has not been explored.
Human ApoE is expressed in three genetic isoforms, ApoE2, ApoE3, and ApoE4, that encode proteins which differ only in two residues. Strikingly, the ApoE4 allele is the most important genetic risk factor for Alzheimer’s disease (AD), whereas the ApoE2 allele is protective (Strittmatter et al., 1993). How ApoE isoforms predispose to AD, or protect against it, is incompletely understood (Holtzman et al., 2012; Kanekiyo et al., 2014). A central feature of AD pathology is formation of amyloid-β (Aβ) oligomers and plaques (Pimplikar et al., 2010; De Strooper and Karran, 2016; Goedert, 2015). Although the precise role of Aβ in AD remains debated, Aβ clearly constitutes a central component of AD pathogenesis that correlates with neuronal cell death and that may perform a causative role in AD pathogenesis (Goedert, 2015; De Strooper and Karran, 2016). Both in AD patients and in cognitively apparently normal people, ApoE4 is associated with increased Aβ-accumulation in brain (Schmechel et al., 1993; Fouquet et al., 2014). ApoE binds Aβ in its delipidated form, but this binding is probably not responsible for the effect of ApoE4 on Aβ-accumulation because ApoE does not bind Aβ as a native lipoprotein (Verghese et al., 2013). ApoE4 enhances Aβ-oligomerization, suggesting that ApoE4 could predispose to AD by increasing toxic Aβ oligomers (Hashimoto et al., 2012). Mice express only a single ApoE allele; exchanging endogenous ApoE in mice with human ApoE2, ApoE3, and ApoE4 produces complex effects that include changes in Aβ aggregation and clearance (Castellano et al., 2011; Fryer et al., 2005).
Collectively, these observations suggest that ApoE may normally function in brain as a lipid-transport and/or signaling molecule, and that ApoE4 may predispose to AD because it affects the production, clearance, and/or toxicity of Aβ (Holtzman et al., 2012). However, these observations are based on experiments that analyzed glial cells and neurons together – either in vitro or in vivo – and focused on rodents. Here, we took a different approach, and examined the effects of recombinant ApoE2, ApoE3, and ApoE4 on human neurons that were cultured without glia to exclude the confounding influence of secreted factors produced by glia, including glial ApoE. We show that ApoE is indeed a signaling molecule, demonstrate that ApoE activates an unusual MAP-kinase signaling cascade, document that this cascade stimulates cFos phosphorylation and APP-gene transcription, and show that this stimulation leads to increased APP and Aβ synthesis. Most importantly, we show that the intrinsic efficacy in activating this pathway differs dramatically between ApoE2, ApoE3, and ApoE4, mirroring their relative effect on AD pathogenesis. Thus, our data uncover an ApoE-dependent signaling pathway that may perform a general role in brain as well as contribute a significant component to AD pathogenesis.
RESULTS
ApoE proteins stimulate neuronal Aβ-production with an ApoE4>ApoE3>ApoE2 potency rank order
We trans-differentiated human ES cells into human excitatory neurons using transient expression of Ngn2 (Fig. 1A, 1B, S1A; Zhang et al., 2013). In previous studies (e.g., Pak et al., 2015; Patzke et al., 2016), we co-cultured human neurons with mouse glia because glia secretes multiple factors that support survival and synaptogenesis (Ullian et al., 2001). However, here we aimed to develop an approach that would allow examining the effects of glial factors on neurons, making it necessary to culture human neurons without glia. Towards this goal, we compared human neurons cultured on glia with neurons cultured on mouse embryonic fibroblasts (MEFs), or on matrigel alone (Fig. 1B, 1C). We found that under all conditions, human neurons appeared to develop normally with no major change in dendritic arborization, but neurons plated on matrigel alone did not survive as well as neurons co-cultured with glia or MEFs (Fig. 1C). Therefore, we utilized human neurons cultured on MEFs as a standard approach.
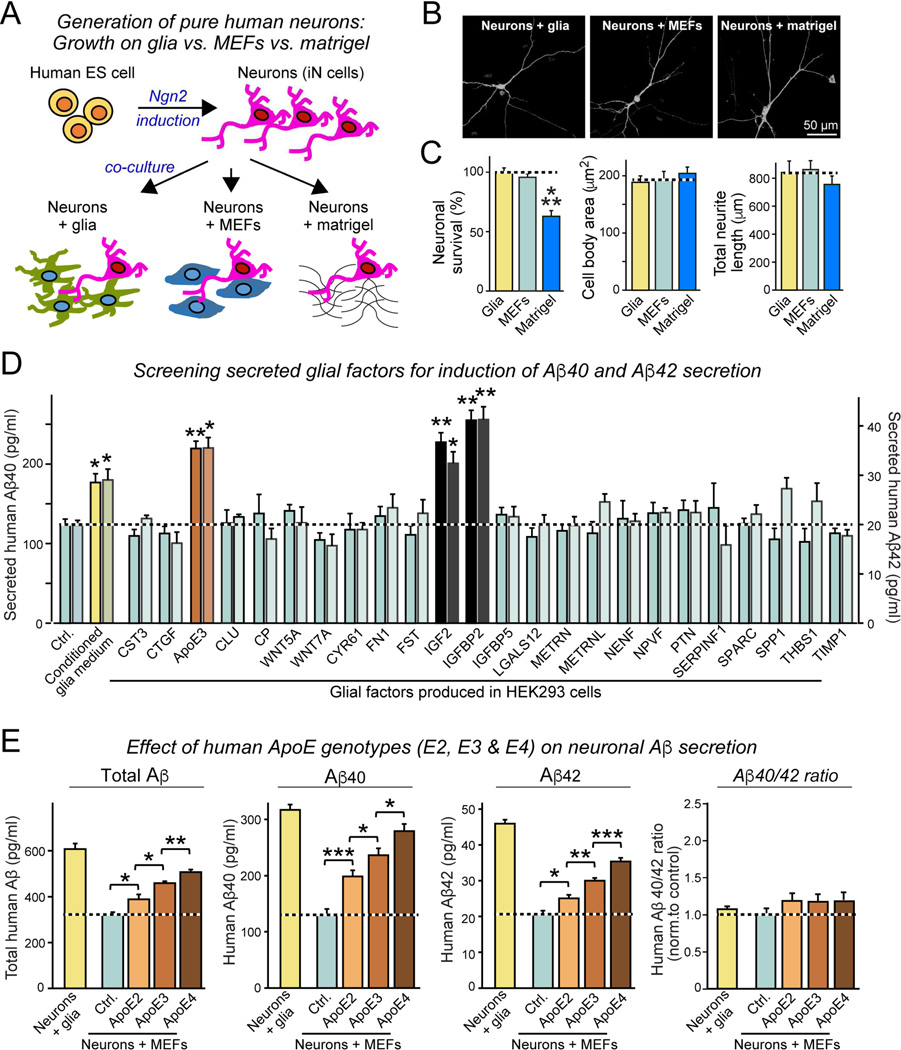
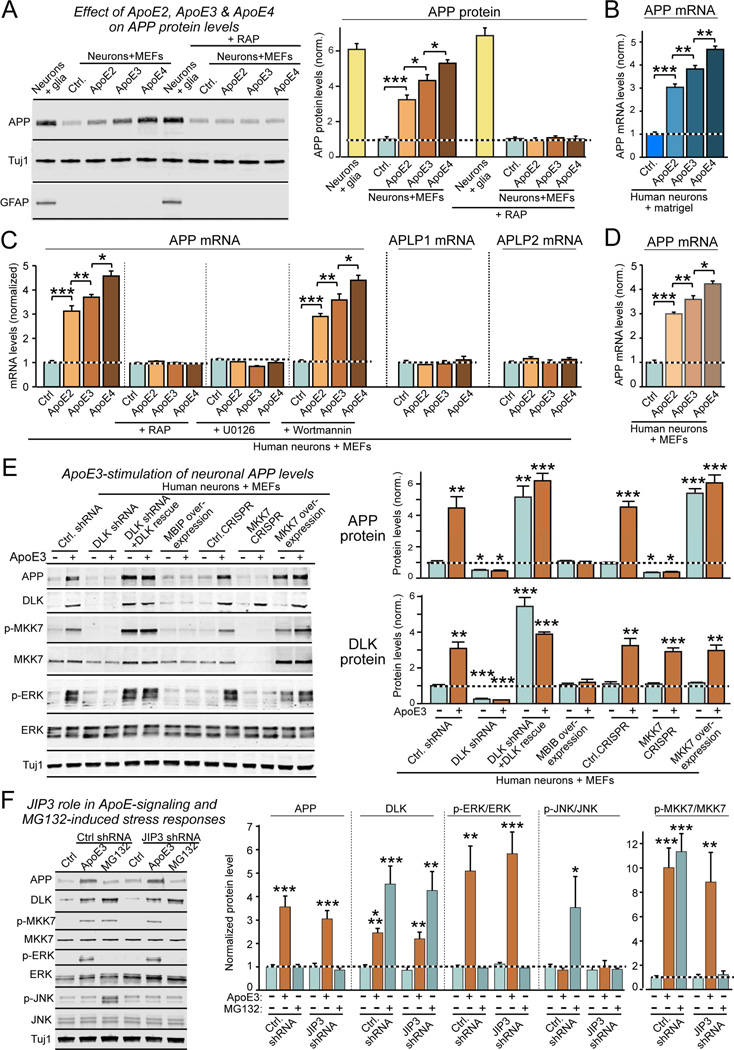
Figure 1. ApoE stimulates Aβ-production with an ApoE4>ApoE3>ApoE2 potency rank order in human neurons cultured in the absence of glia or serum.
(A) Experimental design. Human neurons (iN cells) were generated from H1 ES cells by forced expression of neurogenin-2 (Ngn2), and cultured either on mouse glia (green) which secrete copious amounts of ApoE, on murine embryonic fibroblasts (MEFs) which secrete no ApoE (blue), or on matrigel (black lines).
(B) Representative images of human neurons at day 10 after induction (D10) cultured on glia, MEFs, or matrigel, and sparsely labeled by EGFP transfection.
(C) Survival, soma size, and neurite length of human neurons cultured on glia, MEFs, or matrigel.
(D) Screening 24 secreted proteins that are abundantly produced by cultured mouse glia reveals three factors (ApoE, IGF2, and IGFBP2) that induce Aβ40 and Aβ42 secretion from human neurons cultured on MEFs. Various factors were produced as human proteins in HEK293 cells (names reflect gene symbols), and added to human neurons on MEFs at D10. Media from treated neurons were analyzed by ELISA at D12.
(E) ApoE2, ApoE3, and ApoE4 (all at 10 µg/ml; from D10-12) differentially stimulate Aβ40 and Aβ42 secretion by human neurons cultured on MEFs with an ApoE4>ApoE3>ApoE2 potency rank order, thereby partially rescuing the decrease in Aβ40 and Aβ42 secretion when neurons are co-cultured with MEFs instead of glia. Summary graphs show total concentrations of Aβ (left), Aβ40 (left center), and Aβ42 (right middle) in the medium, and of the Aβ42/Aβ40 ratio (right) measured by ELISA at D12. For cellular Aβ, Aβ40, and Aβ42 levels and for similar results for neurons generated from iPS cells, see Fig. S1.
Data are means ± SEM (n≥3 independent experiments); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons. For additional data and controls, see Fig. S1
As expected, neurons grown in the absence of glia exhibited decreased synaptogenesis. Interestingly, however, neurons co-cultured with glia secreted 2–3 fold more Aβ40 and Aβ42 than neurons co-cultured with MEFs, suggesting that a secreted glial signaling factor stimulates neuronal Aβ-synthesis. Indeed, adding conditioned medium from glial cultures to human neurons significantly stimulated Aβ40 and Aβ42 production (Fig. 1D). We thus asked which glial factor(s) might be responsible. Based on RNAseq data from glia, we selected 24 abundant secreted glial proteins that we produced in transfected HEK293 cells. Adding these factors to neurons cultured on MEFs revealed that three proteins, ApoE, Igf2, and IgfBP2, significantly increased Aβ-production (Fig. 1D). Since ApoE is an abundant secreted glial proteins that is critically important for AD pathogenesis (Holtzman et al., 2012) and is nearly undetectable in neurons or MEFs (Fig. S1B), the induction of Aβ-secretion by ApoE was intriguing, prompting us to focus on studying its mechanism of action.
We first asked whether the three different human genetic ApoE variants that are differentially linked to AD –ApoE2, ApoE3, and ApoE4– differentially stimulate Aβ-synthesis in human neurons. When we incubated neurons cultured on MEFs with recombinant human ApoE2, ApoE3, or ApoE4 (all at 10 µg/ml for 48 h), we found that all three ApoE isoforms robustly enhanced Aβ-synthesis, but with strikingly different efficacy (ApoE4>ApoE3>ApoE2). This differential efficacy was observed for total Aβ, Aβ40, or Aβ42, and for secreted as well as cellular Aβ (Fig. 1E, S1C, S1D), and was unrelated to α-or β-secretase expression (Fig. S1E). Moreover, the differential efficacy of ApoE2, ApoE3, or ApoE4 was independently reproduced with human neurons generated from different iPS cell lines (Fig. S1F, S1G). When we added ApoE to neurons co-cultured with glia, however, we observed no additional increase in Aβ-production, presumably because ApoE and other glial factors already maximally stimulate Aβ synthesis (Fig. 1D, S1H). Control experiments confirmed that our Aβ measurements monitored exclusively human neuronal Aβ synthesis, and were not contaminated by Aβ from other sources (Fig. S1I, S1J). Thus, ApoE secreted by cultured glia massively stimulates Aβ synthesis in human neurons, and ApoE2, ApoE3, and ApoE4 exhibit differential potency in stimulating Aβ-synthesis with a relative effect size that parallels the effect size of these risk factors on AD pathogenesis (Strittmatter et al., 1993).
ApoE2, ApoE3, and ApoE4 differentially activate ERK1/2 phosphorylation by binding to surface ApoE-receptors
How does ApoE stimulate Aβ synthesis? In rodent neurons, ApoE activates multiple signaling pathways, including MAP-kinases (Ohkubo et al., 2001; Qiu et al., 2004). Confirming these studies, we found that ApoE increased ERK1/2 phosphorylation in human neurons 2- to 5-fold (Fig. 2A). Strikingly, ApoE2, ApoE3, and ApoE4 again exhibited the same differential efficacy in this stimulation as for Aβ synthesis (ApoE4>ApoE3>ApoE2).
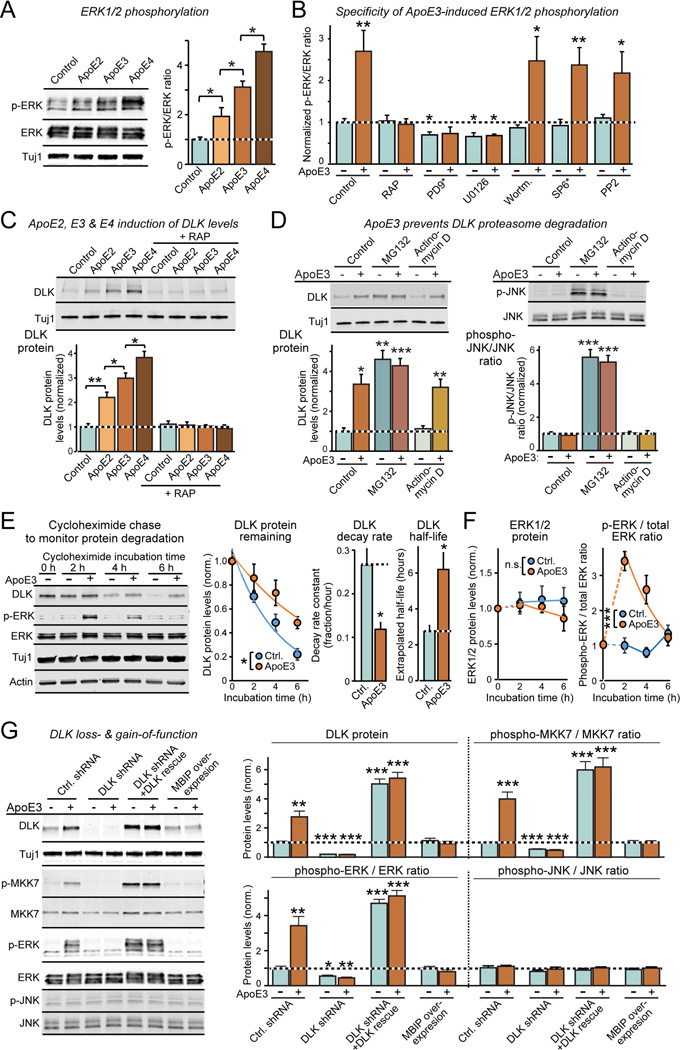
Figure 2. ApoE stimulates a non-canonical DLK→MKK7→ERK1/2 MAP-kinase signalling cascade in human neurons.
(A) ApoE (10 µg/ml, applied at D10 for 2 h) activates ERK1/2 phosphorylation in human neurons cultured on MEFs with an ApoE4>ApoE3>ApoE2 potency rank order (left, representative immunoblots; right, summary graphs).
(B) ApoE3-induced ERK1/2-phosphorylation is abolished by the ApoE-receptor blocker RAP and by inhibitors of MEKs (PD9* [PD98059] and U0126; both 50 µM), but not by inhibitors of JNK (SP6* [SP600125]; 25 µM), PI3K kinase (Wortmannin; 0.1 µM), or Src kinase (PP2; 10 µM). Drugs were applied 30 min before a 2 h ApoE3 (10 µg/ml) incubation at D10. For additional data, see Fig. S2.
(C) ApoE2, ApoE3, and ApoE4 cause a rapid, 2–3 fold increase in DLK in human neurons cultured on MEFs with an ApoE4>ApoE3>ApoE2 potency rank order; recombinant RAP that blocks ApoE-receptor binding also blocks ApoE-induced increases in DLK (left, representative immunoblots; right, summary graphs from neurons at D10 treated for 2 h with 10 µg/ml ApoE).
(D) Proteasome inhibitor MG132 (10 µM, applied for 2 h at D10) increases DLK in human neurons cultured on MEFs similar to ApoE3 (10 µg/ml), thereby occluding the effect of ApoE3 (left); in addition, MG132 stimulates JNK phosphorylation (right). The transcription inhibitor actinomycin D (1 µg/ml) has no effect on the ApoE-induction of DLK (left) or JNK phosphorylation (right).
(E) ApoE3 significantly slows the rapid turnover of DLK protein (measured by immunoblotting after addition of the protein synthesis inhibitor cycloheximide (0.1 g/l); left, representative blot; center, summary plots of the fraction of DLK remaining as a function of time after cycloheximide addition; right, summary graphs of the DLK decay rates and calculated half-lives as a function of ApoE3).
(F) ApoE3 has no effect on ERK1/2 protein levels that are stable, but ApoE3-stimulated ERK1/2 phosphorylation decays in parallel with DLK protein levels after protein synthesis inhibition (left, summary plots of the fraction of ERK1/2 remaining as a function of time after cycloheximide addition; right, summary plot of the phospho-ERK/total ERK ratio).
(G) ApoE3 strongly stimulates MKK7 and ERK1/2 phosphorylation but not JNK phosphorylation in human neurons cultured on MEFs (at D10); DLK knockdown with an shRNA or DLK inhibition by MBIP overexpression block ApoE3-induced MKK7 and ERK1/2 phosphorylation, whereas DLK overexpression constitutively activates MKK7 and ERK1/2 phosphorylation (left, representative immunoblots; right, summary graphs).
Data are means ± SEM (n≥3 independent experiments); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post-hoc test in pairwise comparisons in A and C and comparisons to control in B and D, and with two-way ANOVA in E and F.
Using ApoE3 as a standard ApoE isoform, we probed the mechanism of ApoE-induced ERK1/2 activation. RAP, a protein that prevents binding of ApoE to surface receptors (Herz et al., 1991), blocked ApoE-triggered phosphorylation of ERK1/2 (Fig. 2B, S2A, S2B), suggesting that ApoE acts by binding to ApoE-receptors. The MAP-kinase kinase inhibitors U0126 and PD98059 (which primarily but not exclusively target MEK1/2) also blocked ApoE-stimulated ERK1/2 phosphorylation. In contrast, inhibitors of the PI3-kinase Akt (Wortmannin), cJun N-terminal kinase (JNK; SP600125), and src kinase (PP2) had no effect (Fig. 2B, S2A, S2B). Therefore, ApoE activates a MAP-kinase signaling pathway in human neurons by binding to ApoE receptors, and does so with a potency rank order of ApoE4>ApoE3>ApoE2 similar to Aβ production.
ApoE activates a non-canonical DLK→MKK7→ERK1/2 MAP-kinase cascade
Human neurons express high levels of the MAP-kinase kinase kinase DLK (Fig. S2C, S2D), which is an attractive candidate for neuronal signaling because it has been implicated in axonal regeneration, synaptogenesis, and neurodegeneration (reviewed in Tedeschi and Bradke, 2013; Lu et al., 2014). Indeed, we found that ApoE induced a 2- to 4-fold increase in DLK protein levels in human neurons co-cultured with MEFs; this induction again was blocked by RAP, which had no effect on baseline DLK levels (Fig. 2C). Again, the three human ApoE isoforms increased DLK levels with the same differential potency as for the stimulation of Aβ synthesis and ERK1/2 phosphorylation (ApoE4>ApoE3>ApoE2; Fig. 2C). Neurons expressed high levels of DLK mRNA, but newly synthesized DLK protein was rapidly degraded (Fig. 2E). Proteasome inhibition with MG132 dramatically increased DLK protein levels under control conditions, and occluded subsequent ApoE3-induced increases of DLK levels (Fig. 2D). ApoE3 had no effect on phosphorylation of JNK1/2/3, the most abundant MAP-kinase in neurons, although JNK1/2/3 phosphorylation was stimulated by MG132, which induces a cellular stress response that activates JNKs (Fig. 2D).
The fact that the proteasome inhibitor MG132 mimics and occludes the effect of ApoE on DLK levels suggests that ApoE increases DLK levels by decreasing rapid proteasomal degradation of DLK. To test this hypothesis, we blocked new protein synthesis in human neurons with cycloheximide in the presence and absence of ApoE3, and measured the decay rate of previously synthesized DLK and ERK1/2 proteins, as well as the degree of ERK1/2 phosphorylation (Fig. 2E, 2F). We found that DLK was indeed rapidly degraded in the absence of ApoE3, whereas ERK1/2 were relatively stable. ApoE3 significantly decreased the DLK decay rate, but had no effect on ERK1/2 protein levels (Fig. 2E, 2F). As above, ERK1/2 phosphorylation was strongly stimulated by ApoE3, and decayed in parallel with DLK levels after cycloheximide addition (Fig. 2F, S2E). Thus, ApoE activates DLK by decreasing its rapid proteasomal degradation.
Is the ApoE-induced DLK activation responsible for ApoE-stimulated ERK1/2 phosphorylation? To address this question, we inhibited DLK using shRNA-mediated knockdowns or overexpression of MBIP (for MUK/DLK binding inhibitory protein, an endogenous inhibitor of DLK; Fukuyama et al., 2000). In addition, we constitutively increased DLK activity by overexpression on the background of the DLK knockdown (Fig. 2G, S2F). We then measured the effect of these manipulations, which did not impair neuronal survival (Fig. S2G), on ApoE3-induced increases in DLK protein levels and in ERK1/2 phosphorylation, and additionally monitored phosphorylation of MKK7, a MAP-kinase kinase that is a major DLK target in neurons (Merritt et al., 1999).
We found that ApoE3 stimulated MKK7 phosphorylation as strongly as ERK1/2 phosphorylation (~4–5 fold; Fig. 2G). DLK inhibition by knockdown or MBIP overexpression blocked ApoE3-stimulation of MKK7 and ERK1/2 phosphorylation. Conversely, DLK overexpression induced constitutive ERK1/2 and MKK7 phosphorylation, independent of ApoE3 (Fig. 2G). None of these manipulations increased JNK1/2/3 phosphorylation, used as a negative control. Knockdown of LZK (a.k.a. MAP3K13), another abundant MAP-kinase kinase kinase in neurons, also had no effect on ApoE3-induced increases in DLK protein and ERK1/2 phosphorylation (Fig. S2H, S2I).
Collectively, these data suggest the hypothesis that ApoE activates DLK by increasing its levels; DLK then phosphorylates MKK7, which in turn phosphorylates ERK1/2 but not JNKs (Fig. 3A). Activated phospho-ERK1/2 may then increase Aβ production by enhancing the levels of amyloid-β precursor protein (APP; see below). Although attractive, this hypothesis differs from current concepts of MAP-kinase signaling pathways which suggest that MKK7 primarily phosphorylates JNKs instead of ERKs (Morrison, 2012). Thus, we directly tested whether MKK7 mediates ApoE3-induced ERK1/2 phosphorylation.
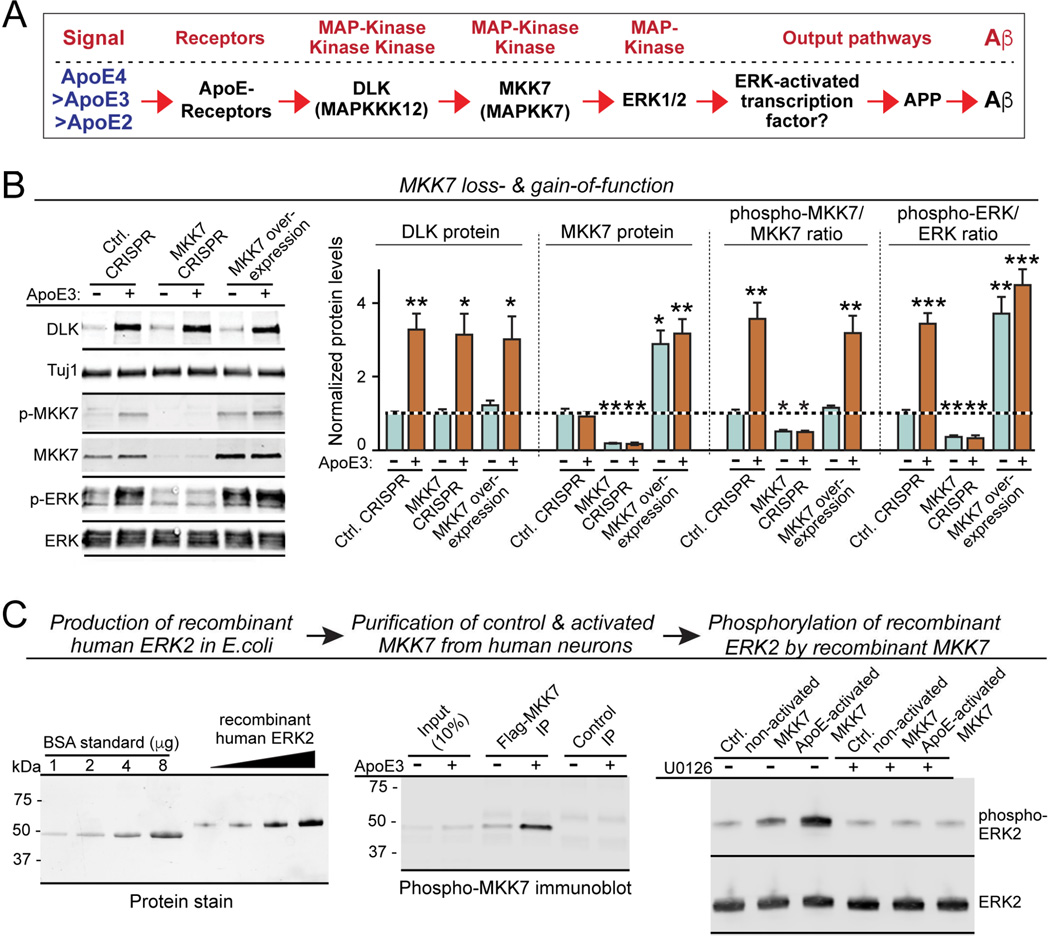
Figure 3. Validation of the non-canonical ApoE-stimulated DLK→MKK7→ERK1/2 MAP-kinase cascade that excludes JNK activation.
(A) Diagram of the proposed ApoE-induced non-canonical MAP-kinase signaling pathway.
(B) MKK7 inactivation by CRISPR blocks ApoE3 induction of ERK1/2 phosphorylation, while MKK7 overexpression constitutively activates ERK1/2 phosphorylation independent of ApoE3. Data are means ± SEM (n≥3 independent experiments); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post-hoc test, comparing all conditions to the control without ApoE treatment. For additional data and reagent validation, see Fig. S2.
(C) In vitro kinase assay with purified recombinant proteins demonstrates that ApoE3-activated MKK7 directly phosphorylates ERK2. Recombinant human ERK2 was produced in E. coli (left, stain-free SDS-polyacrylamide gel visualized by UV illumination), and naïve or ApoE-activated MKK7 was immunopurified from human neurons that overexpressed Flag-tagged MKK7 and were treated at D10 for 2 h with control or ApoE medium (center; phospho-MKK7 immunoblot). Recombinant ERK2 was then incubated for 30 min at 30 oC in the absence (test) and presence of the MEK inhibitor U0126 (50 µM; used as a further control) with Flag-beads containing immunoprecipitated control or ApoE-activated MKK7, or with control HA beads. Samples were analyzed by immunoblotting (right).
We inhibited MKK7 in human neurons by CRISPR or activated it by overexpression (Fig. 3B, S2J). Neither manipulation impaired the ApoE3-induced increase in DLK levels, but both dramatically altered ApoE3-dependent downstream ERK1/2 phosphorylation. MKK7 CRISPR suppressed baseline ERK1/2 phosphorylation and blocked ApoE3-induced ERK1/2 phosphorylation, whereas MKK7 overexpression constitutively enhanced ERK1/2 phosphorylation and rendered it ApoE3-independent (Fig. 3B). Thus, DLK-mediated MKK7 phosphorylation mediates ERK1/2 phosphorylation in human neurons.
To ensure that MKK7 actually directly phosphorylates ERK1/2, we produced recombinant ERK2 in bacteria, and purified naïve and phosphorylated MKK7 by immunoprecipitation from human neurons that overexpressed Flag-tagged MKK7 and that had been treated with control medium or ApoE3 (Fig. 3C). We then incubated recombinant ERK2 with immunoprecipitated naïve or ApoE3-activated MKK7 in the absence and presence of the MEK inhibitor U0126, and measured ERK2 phosphorylation. We observed that MKK7 directly phosphorylated ERK2; ERK2 phosphorylation by MKK7 was enhanced by prior ApoE3-dependent activation of MKK7, and blocked by U0126 (Fig. 3C). These data confirm that ApoE activates a non-canonical MAP-kinase signal transduction pathway consisting of a DLK→MKK7→ERK1/2 phosphorylation cascade (Fig. 3A).
ApoE-mediated MAP-kinase activation stimulates APP gene transcription
To test how ApoE-induced stimulation of ERK1/2 enhances Aβ-production, we monitored the effect of ApoE on the levels of APP as the precursor to Aβ (Fig. 3A). We found that addition of ApoE2, ApoE3, and ApoE4 to human neurons co-cultured with MEFs increased both APP mRNA and protein levels 3- to 5-fold, again with the same ApoE4>ApoE3>ApoE2 potency rank order observed above (Fig. 4A–4C). This result suggests that ApoE stimulates APP gene transcription. The same ApoE-dependent increases in APP mRNA levels were observed in human neurons plated on matrigel (Fig. 4B) and were found in independently derived neurons differentiated from iPS cell lines (Fig. S3A, S3B). The effects of ApoE on APP levels were concentration- and time-dependent, and were specific to neurons (Fig. S3C–S3E). Strikingly, ApoE only enhanced APP, but not APLP1 and APLP2 mRNA levels, despite the high homology between APP and APLP1 and APLP2 (Fig. 4C, S3A, S3B). In addition to ApoE, the other glial factors that increased Aβ-levels in our initial screen also acted by stimulating APP synthesis (Fig. S4A, S4B).
Figure 4. ApoE increases APP expression 3-4-fold with an ApoE4>ApoE3>ApoE2 potency rank order by activating the DLK→MKK7→ERK1/2 MAP-kinase pathway.
(A) Human neurons synthesize ~5-fold less APP when cultured on MEFs instead of glia; addition of ApoE2, ApoE3, or ApoE4 (each 10 µg/ml applied from D10-12) stimulates APP synthesis in human neurons on MEFs with a ApoE4>ApoE3>ApoE2 potency rank order, which is blocked by the ApoE-receptor antagonist RAP (left, representative immunoblots; right, summary graphs; see also Fig. S5).
(B) ApoE2, ApoE3, and ApoE4 increase APP mRNA levels 3–4 fold with a ApoE4>ApoE3>ApoE2 potency rank order in human neurons cultured on matrigel only.
(C) ApoE2, ApoE3, and ApoE4 increase only APP, but not APLP1 or APLP2 mRNA levels in human neurons on MEFs with an ApoE4>ApoE3>ApoE2 potency rank order; ApoE-induced APP mRNA increase is inhibited by RAP and the MAP-kinase inhibitor U0126, but not by the PI3K inhibitor Wortmannin.
(D) Recombinant cholesterol-free ApoE2, ApoE3, and ApoE4 produced in bacteria stimulate APP mRNA levels in human neurons on MEFs similar to recombinant ApoE2, ApoE3, and ApoE4 produced in HEK293 cells.
(E) Inhibition of DLK by shRNAs or MBIP blocks ApoE3-induced increases in APP protein levels, while DLK and MKK7 overexpression during rescue experiments constitutively increases APP protein levels independent of ApoE3. Note that DLK protein levels were not affected by MKK7 manipulations (left, representative blots; right, summary graphs).
(F) Knockdown of the JNK scaffold JIP3 has no effect on ApoE3-induced activation of the DLK→MKK7→ERK1/2 signal transduction cascade, but blocks induction of the JNK MAP-kinase cascade during the stress response to MG132. Human neurons on MEFs were infected with lentiviruses expressing a control shRNA or a JIP3 shRNA at D4, treated with control medium, ApoE3 (10 µg/ml), or MG132 (0.1 g/l) for 2 hours at D10, and analyzed by immunoblotting (left, representative blots; right, summary graphs).
Data are means ± SEM (n≥3 independent experiments for all bar graphs); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post-hoc test in pairwise comparison (A–D, F) or comparisons to controls (E). For further data and controls, see Fig. S4, S5.
The ApoE-induced increase in APP mRNA levels was blocked by RAP and by U0126 but not by Wortmannin, suggesting that it is dependent on ApoE-binding to surface receptors and on ApoE-induced activation of MAP-kinases (Fig. 4A, 4C). Previous studies suggested that ApoE-containing lipoproteins activate neurons by providing cholesterol (Mauch et al., 2001). Since ApoE protein produced in transfected HEK293 cells is secreted as a lipidated particle containing cholesterol, we tested cholesterol-free ApoE produced in E. coli, but observed the same 3- to 5-fold enhancement in APP mRNA levels with the same ApoE4>ApoE3>ApoE2 potency rank order as with HEK293-cell derived ApoE (Fig. 4D, S1C). Thus, ApoE acts by a cholesterol-independent, direct receptor action, as also suggested by its sensitivity to RAP. Moreover, we observed that inhibition of DLK by shRNAs or MBIP overexpression and inhibition of MKK7 by CRISPR both blocked the ApoE-induced increase in APP protein (Fig. 4E, S4D–S4F). DLK or MKK7 overexpression, conversely, constitutively increased APP mRNA and protein levels (Fig. 4E). Thus, activation of surface ApoE-receptors activates a non-canonical DLK→MKK7→ERK signaling pathway that increases APP mRNA and APP protein levels.
Similar to Aβ synthesis, ApoE3 had no effect on APP levels in human neurons co-cultured with glia (Fig. S1H, S5A–S5C), presumably because glial factors fully activate the non-canonical MAP-kinase pathway that stimulates APP transcription. Consistent with this hypothesis, DLK knockdowns or DLK inhibition by MBIP dramatically decreased APP and Aβ40 and Aβ42 levels in neurons co-cultured with glia, whereas DLK overexpression significantly increased these levels (Fig. S5A–S5C).
These data, viewed together, suggest that ApoE stimulates APP transcription via a non-canonical MAP-kinase pathway that involves ERKs but not JNKs. Consistent with this hypothesis, ApoE3 is taken up into the cell by a receptor-dependent pathway (Fig. S6A). The exclusion of JNKs in ApoE-dependent signaling is surprising because JNKs are canonical substrates for MKK7 (Morrison, 2012), JNKs have been identified as downstream targets for DLK in neurodegeneration (Chen et al., 2012), and APP directly binds to JNK scaffolding proteins (Scheinfeld et al., 2002). To further examine the role of JNK signaling in ApoE signaling, we tested the role of JIP3, the predominant JNK scaffold in neurons. JIP3 knockdown had no effect on the ApoE3-mediated increase in APP and DKL levels or MKK7 and ERK1/2 phosphorylation, but inhibited the MG132-mediated stimulation of MKK7 and JNK phosphorylation as expected (Fig. 4F, S4C). Thus, the JNK-pathway is not required for ApoE signaling.
AP-1 transcription factor activation mediates ApoE stimulation of APP transcription
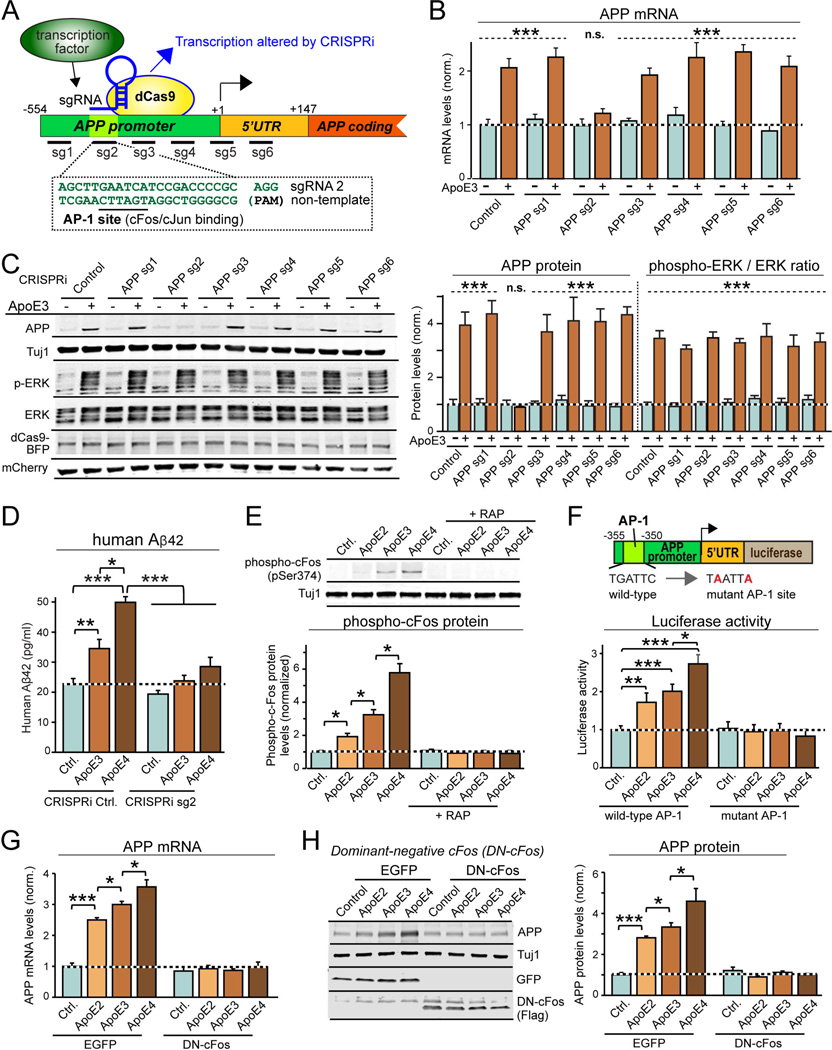
How does ApoE selectively increase APP, but not APLP1 and APLP2 transcription? To examine this question, we used CRISPR-interference (CRISPRi) mapping with guide RNAs that were targeted to APP gene promoter sequences (Fig. 5A; Larson et al., 2013). We found that of six guide RNAs tested, only one guide RNA (sg2) blocked ApoE3-induced increases in APP protein and mRNA levels (Fig. 5B, 5C, S6B). Importantly, CRISPRi with sg2 did not impair ApoE3-induced ERK1/2 phosphorylation, and had no effect on baseline APP mRNA and protein levels. Moreover, CRISPRi with sg2 abolished ApoE-induced stimulation of Aβ42 secretion from human neurons co-cultured with MEFs without altering baseline Aβ42 secretion (Fig. 5D), and decreased Aβ42 secretion from human neurons co-cultured with glia (Fig. S6C).
Figure 5. ApoE-mediated activation of cFos-containing transcription factor AP-1 stimulates APP-gene transcription.
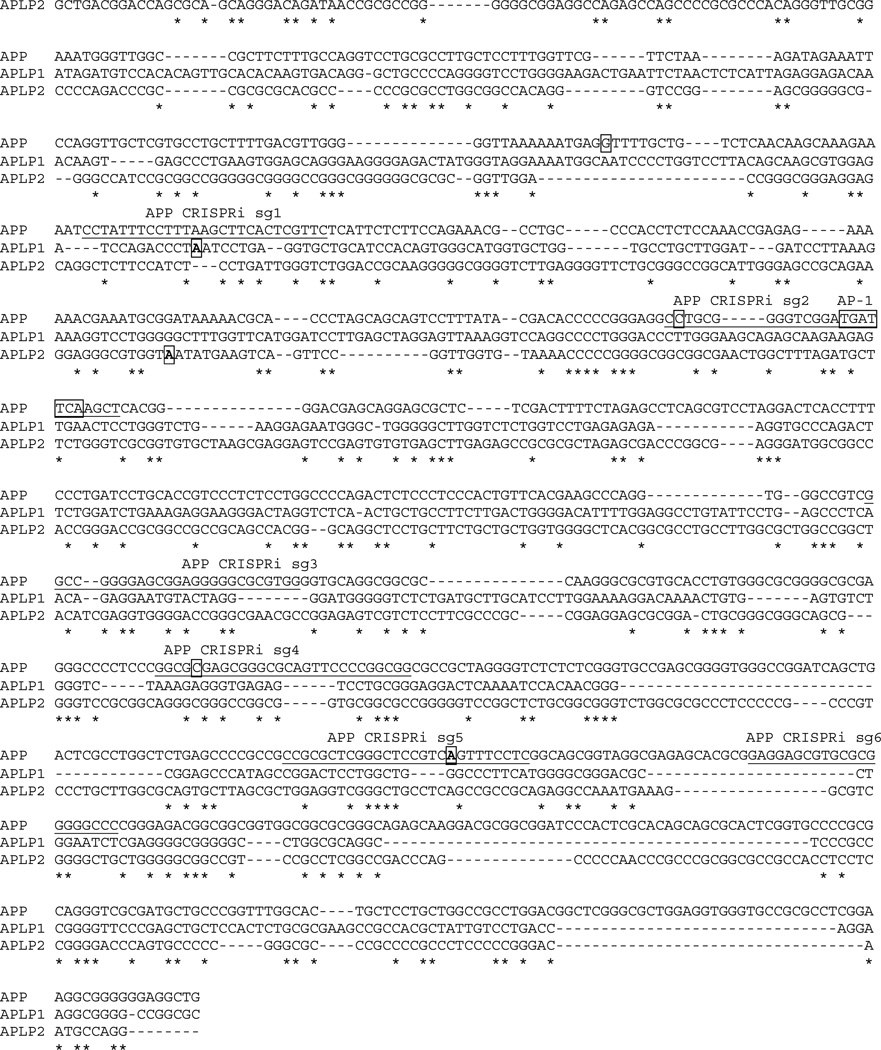
(A) CRISPRi strategy to identify APP promoter sequences required for ApoE-stimulation of APP-transcription using guide RNAs (sg1 to sg6) covering the proximal human APP-promoter (note that sg2 targets conserved AP-1 binding site).
(B) CRISPRi of AP-1 binding sequence in human APP-promoter blocks ApoE3-induced increase in APP mRNA levels. Human neurons on MEFs were infected at D4 with lentiviruses co-expressing BFP-tagged dCas9, various sgRNAs, and mCherry. Neurons were treated at D10 with ApoE3 (10 µg/ml), and APP mRNA levels were measured at D12.
(C) CRISPRi of AP-1 binding sequence in human APP-promoter blocks ApoE3-induced increase in APP protein but not in ERK1/2-phosphorylation. Experiments were performed as for B, except that neurons were analyzed by quantitative immunoblotting (left, representative blots; right, summary graph).
(D) CRISPRi of AP-1 binding sequence in human APP-promoter suppresses ApoE-induced, but not basal Aβ42 secretion from human neurons on MEFs. Experiments were performed as described for (B).
(E) ApoE activates cFos phosphorylation in human neurons co-cultured with MEFs with an ApoE4>ApoE3>ApoE2 rank potency order; cFos phosphorylation is blocked by the ApoE-receptor antagonist RAP (top, representative immunoblots; bottom, summary graphs).
(F) AP-1 binding site in human APP-promoter mediates ApoE stimulation of APP-gene transcription. Human neurons cultured on MEFs were infected at D4 with lentiviruses containing APP-promoter-driven firefly luciferase and constitutively expressed renilla luciferase (internal control), treated with ApoE (10 µg/ml) at D10, and analyzed at D12 (top, schematic of promoter reporter construct; bottom, summary graph of luciferase expression normalized to the renilla luciferase control).
(G & H) Dominant-negative cFos (DN-cFos) suppresses ApoE-induction of APP mRNA levels (G) and APP protein (H).
Data are means ± SEM (n≥3 independent experiments for all bar graphs); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001; n.s., not significant) was evaluated with one-way ANOVA with Tukey’s post-hoc test or Student’s t-test. In (B) and (C), the difference between –ApoE3 and +ApoE3 is significant for control, sg1 and 3–6 groups (p<0.001 as indicated), but not for sg2 (n.s.). For additional data, see Fig. S6.
The sg2-targeted sequence in the APP-gene promoter conforms remarkably well to the consensus sequence of dimeric AP-1 transcription factors containing cFos, cJun, and/or other partners, suggesting that ApoE-stimulated MAP-kinase signaling activates APP transcription by stimulating AP-1 (Fig. 5A). To test this hypothesis, we examined cFos phosphorylation (Monje et al., 2003). Strikingly, ApoE2, ApoE3 and ApoE4 stimulated cFos phosphorylation 2- to 6-fold in human neurons co-cultured with MEFs, and acted with the same potency rank order of ApoE4>ApoE3>ApoE2 as DLK protein levels, MKK7- and ERK1/2-phosphorylation, APP transcription, and Aβ-synthesis (Fig. 5E). Moreover, using an APP promoter reporter construct with wild-type or mutant AP-1 transcription factor binding sites (Wade et al., 1992), we confirmed that ApoE2, ApoE3 and ApoE4 enhanced transcription from the APP promoter in an AP-1-dependent manner, again with the same ApoE4>ApoE3>ApoE2 potency rank order (Fig. 5F). Furthermore, ApoE3 not only stimulated transcription of APP, but also that of cFos (Fig. S6D), which is a target of AP-1 in a positive feedback loop (Karin, 1995). Again, stimulation of APP and cFos transcription by ApoE3 was specific, as it was not observed with MG132-induced stress and was not inhibited by JIP3 knockdown (Fig. S6D). In addition, ApoE3 did not stimulate cJun transcription, which, however, was induced by MG132 in a JIP3-dependent manner.
Finally, to further probe the role of cFos in ApoE-stimulated APP transcription, we tested the effects of dominant-negative cFos (DN-cFos) overexpressed in human neurons (Olive et al., 1997). DN-cFos did not alter baseline APP mRNA and protein levels, but completely blocked increases in these levels induced by ApoE2, ApoE3, or ApoE4 (Fig. 5G, 5H). Thus, ApoE activates a DLK-dependent MAP-kinase signaling pathway that induces cFos phosphorylation, which stimulates AP-1 and enhances APP synthesis via a direct effect on the APP-gene promoter.
DLK-activated AP-1-dependent signaling regulates APP transcription in mouse neurons in culture and in vivo
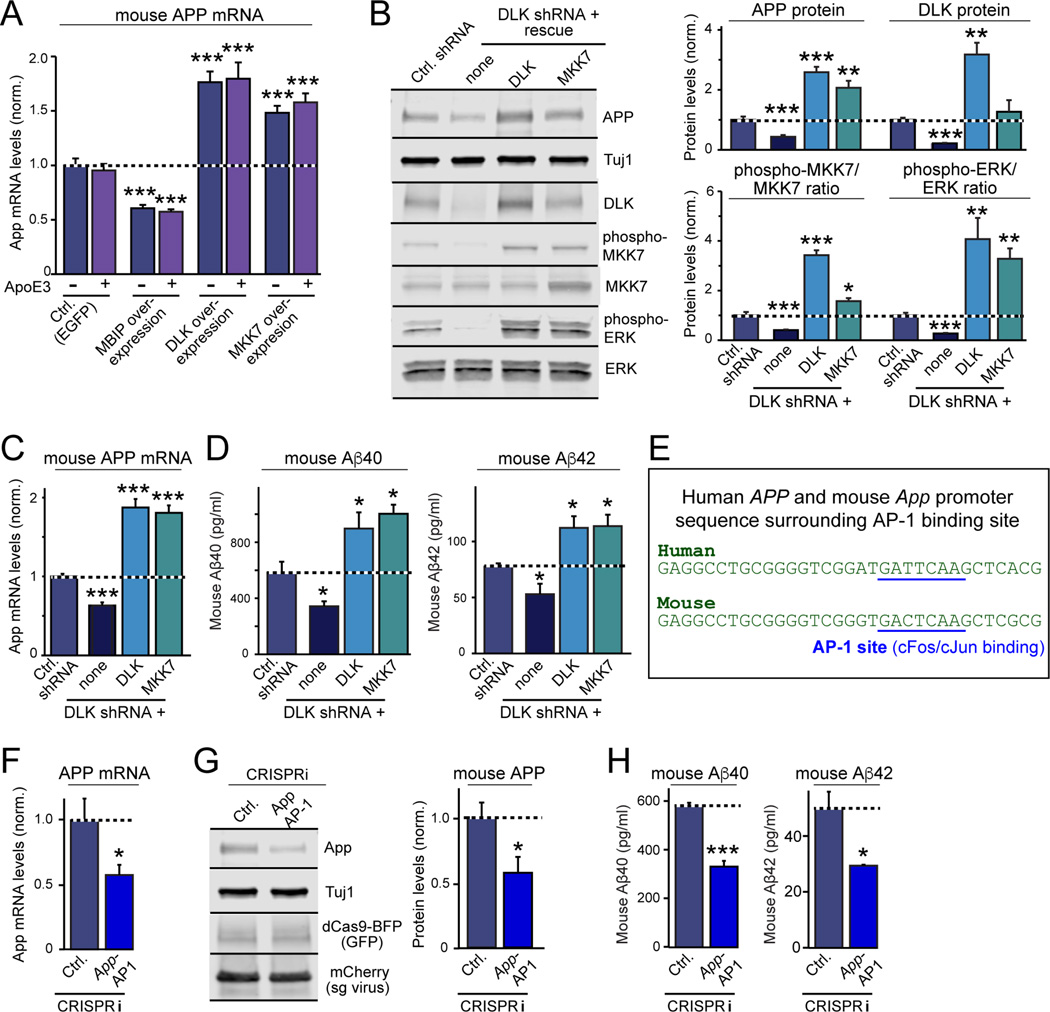
Do mechanisms similar to those we defined in relatively immature human neurons operate in more mature mouse neurons? To address this question, we examined dissociated cultures of neurons and glia from mouse cortex or hippocampus. ApoE had no effect on APP levels or Aβ-synthesis in these cultures, which is expected since ApoE and other factors are abundantly produced by their constituent glia (Fig. 6A, S7A, S7B). However, DLK knockdowns decreased the steady-state levels of MKK7 and ERK1/2 phosphorylation, APP mRNA and APP protein levels, and Aβ40 and Aβ42 secretion in these neuron/glia cultures, suggesting that the same DLK→MKK7→ERK1/2→APP signaling pathway controls App transcription in these neurons (Fig. 6B–D, S7C, S7D). Conversely, DLK and MKK7 overexpression in rescue experiments on the background of the DLK knockdown caused the opposite effect, with 2-to 4-fold increases in the steady-state levels of MKK7 and ERK1/2 phosphorylation, APP mRNA and protein levels, and Aβ40 and Aβ42 secretion (Fig. 6B–D, S7D). Thus, glial factors likely stimulate neuronal APP and Aβ synthesis by the same pathway in dissociated mouse neuron/glia cultures as in human neurons.
Figure 6. The DLK- and AP-1-dependent signaling pathway controlling App gene transcription and Aβ-synthesis is conserved in mouse neurons.
All experiments were carried out in dissociated mixed mouse neuron/glia cultures in which endogenous glia factors maximally stimulate ApoE-dependent signaling pathways.
(A) Exogenous ApoE3 has no effect on APP mRNA levels in neuron/glia cultures from mouse cortex, but inhibition of DLK by MBIP overexpression decreases APP mRNA levels, whereas DLK or MKK7 overexpression increase APP mRNA levels. Neuron/glia cultures were transduced with lentiviruses at DIV4, treated with ApoE3 (10 µg/ml) at DIV10, and analyzed by RT-PCR at DIV12.
(B) DLK knockdown decreases APP and DLK protein levels in neuron/glia cultures from mouse hippocampus, and additionally suppresses steady-state phosphorylation of ERK1/2 and MKK7, whereas rescue overexpression of either DLK or MKK7 increases APP protein levels and MKK7 and ERK1/2 phosphorylation.
(C & D) DLK knockdown decreases APP mRNA levels (C) and Aβ secretion (D) in neuron/glia cultures from mouse hippocampus, whereas rescue overexpression of either DLK or MKK7 increases APP mRNA levels (C) and Aβ40 and Aβ42 secretion (D).
(E) Alignment of the human APP- and murine App-promoter sequences containing the AP-1 binding site demonstrates a high degree of conservation.
(F & G) CRISPRi-mediated inhibition of AP-1 binding to the App-promoter in neuron/glia cultures from mouse hippocampus suppresses APP mRNA (F) and protein levels (G).
(H) CRISPRi-mediated inhibition of AP-1 binding to the App-promoter decreases Aβ40 and Aβ42 secretion in neuron/glia cultures from mouse hippocampus.
Data are means ± SEM (n≥3 independent experiments for all bar graphs); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and comparing to control with Tukey’s post-hoc multiple comparisons [(A) to (D)] and Student’s t test [(F) to (H)]. For additional data, see Fig. S7.
Next, we asked whether stimulation of App transcription in mouse neuron/glia cultures is also mediated by the AP-1 dependent activation of the mouse App promoter. Comparison of the human APP and mouse App promoter sequences revealed that the AP-1 binding sequence is conserved evolutionary (Fig. 6E). Using a guide RNA for the AP-1 binding sequence in the mouse App promoter, we found that CRISPRi significantly suppressed APP mRNA and protein levels and decreased Aβ40 and Aβ42 secretion in mouse neuron/glia cultures (Fig. 6F–6H, S7E–S7G). Thus, the same signaling pathway controls Aβ-production in human and mouse neurons.
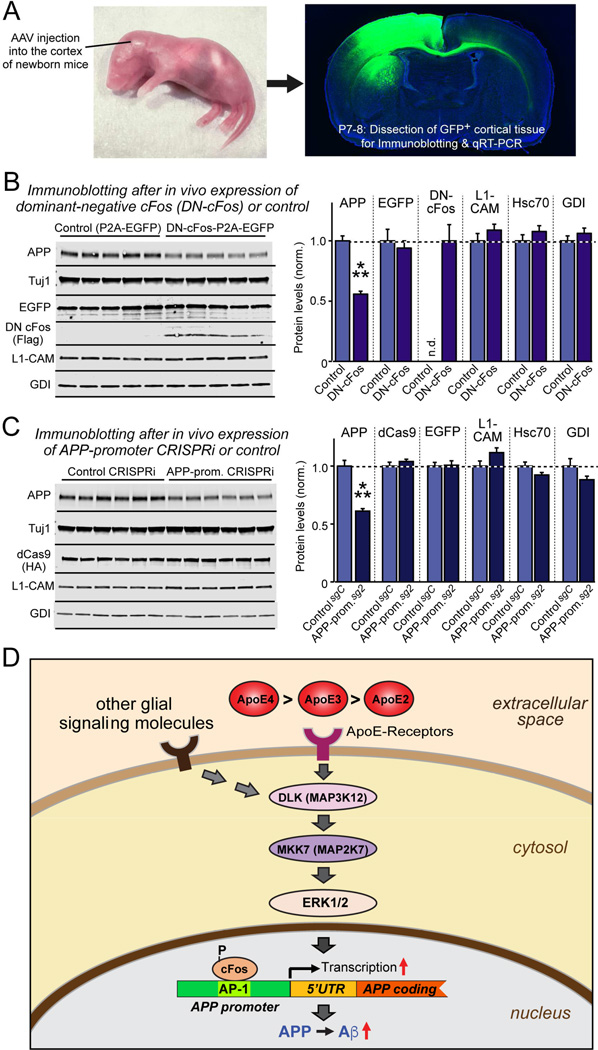
Finally, we tested whether the pathway that controls APP production via cFos activation in cultured neurons operates in vivo. Owing to the multitude of secreted glial factors that stimulate the pathway, we could not address this question by manipulating mouse ApoE as one of multiple signaling factors. Instead, we tested in mice whether the downstream, final regulatory event, the activation of App transcription via AP-1 binding to its promoter, operates in vivo. For this purpose, we stereotactically injected into the cortex of newborn mice AAVs that express a control protein or DN-cFos, the dominant-negative form of cFos, or AAVs that express dCAS9 with a control guide RNA or the guide RNA directed to the AP-1 binding sequence of the App promoter, thereby attempting to perform in vivo CRISPRi (Fig. 7A, S7H). We found that both manipulations significantly suppressed APP expression at the protein and mRNA levels (Fig. 7B, 7C, S7I, S7J). L1-CAM, Hsc70, and GDI as control proteins were not changed. Both manipulations only partially decreased APP levels, as would be expected for an impairment of only one element in the App promoter, and thus provided a specificity control. To the best of our knowledge, CRISPRi has not previously been attempted in vivo, and its applicability for promoter mapping in living mice enables attractive future studies that obviate the need for expensive transgenic experiments. Thus, the AP-1-dependent regulation of APP expression operates physiologically in vivo.
Figure 7. cFos-dependent signaling pathway regulates mouse App gene transcription in vivo.
(A) Experimental design. AAVs (encoding EGFP alone or EGFP and dominant-negative cFos (DN-cFos); and EGFP plus dCAS9 with either a control guide RNA or a guide RNA directed to the App promoter AP-1 binding site) were stereotactically injected into the cortex of anesthetized newborn mice (left), and cortex expressing EGFP was analyzed at P7-P8 (right).
(B & C) Suppression of cFos-signaling using DN-cFos (A; n = 5 mice for test and control) or CRISPRi of the AP-1 binding sequence of the App promoter (B; n = 6 mice for test and control) selectively decreases APP expression in vivo (for mRNA measurements, see Fig. S7). Data are means ± SEM (n.d., not detectable); statistical significance was evaluated with Student’s t-test (***, p<0.001).
(D) Schematic of the ApoE-signaling pathway that controls APP transcription and Aβ production via activation of the DLK MAP-kinase cascade. See text for details. ApoE is proposed to increase AD risk by causing an incremental chronic increase in APP abundance and Aβ secretion, with ApoE4 being more, and ApoE2 being less efficacious than ApoE3 in a parallel to their effects on AD risk.
DISCUSSION
In order to examine the biological functions of ApoE in brain and to explore the contribution of different ApoE isoforms to AD pathogenesis, we here have studied human neurons that were cultured in complete isolation from glial cells and in the absence of serum. This approach enabled us to monitor neuronal responses to different ApoE isoforms in the absence of confounding glial signals, such as endogenous ApoE that is abundantly secreted by cultured glia. Our results reveal that ApoE activates a non-canonical MAP-kinase signaling pathway by binding to cell-surface ApoE-receptors. This pathway consists of DLK as MAP-kinase kinase kinase that phosphorylates MKK7 as MAP-kinase kinase, which in turn phosphorylates ERK1/2 as MAP-kinases (Fig. 7D). Activated ERK1/2 then phosphorylates cFos, a subunit of AP-1 transcription factors; cFos phosphorylation stimulates AP-1-dependent APP gene transcription via an AP-1-binding site in the APP-gene promoter. The resulting increase in APP enhances Aβ levels. Importantly, we show that the three genetic isoforms of human ApoE (ApoE2, ApoE3, and ApoE4) exhibit differential potency in stimulating the DLK→MKK7→ERK1/2 MAP-kinase pathway, cFos phosphorylation, APP-gene transcription, and Aβ-synthesis. This differential potency may be relevant for AD since ApoE4 constitutes the most important genetic AD risk factor, whereas ApoE2 protects against AD (Strittmatter et al., 1993; Holtzman et al., 2012; Kanekiyo et al., 2014; Wang and Eckel, 2014). We posit that even though ApoE is only one of several signaling factors that drive APP and Aβ synthesis in neurons, the higher signaling efficacy of ApoE4 than ApoE3 in stimulating APP and Aβ synthesis may cause a cumulative effect over an individual’s lifetime, thus accounting for the increased Aβ-concentrations and AD incidence in individuals expressing ApoE4.
In our experiments, the culture of human neurons maintained independent of glia or serum allowed unprecedented control over exogenous signals acting on these neurons. Moreover, application of CRISPRi to map transcription-factor-binding sequences in a promoter in cultured cells and in vivo might prove generally useful for mapping promoter elements. Conceptually, the finding that ApoE activates a non-canonical MAP-kinase pathway involving ERK1/2 phosphorylation by MKK7 but not JNKs, and that ApoE acts here in a receptor-dependent, cholesterol-independent manner were surprising. The upstream mechanism involved – stabilizing DLK by preventing its rapid proteasomal degradation – was also unexpected, as was the downstream effector pathway whereby cFos-phosphorylation stimulates APP transcription via a specific AP-1 binding sequence that is evolutionarily conserved and that functions in vivo. Remarkably, ApoE upregulated only APP, but not its close homologs APLP1 and APLP2. Thus, our studies describe an unanticipated signal transduction pathway that links ApoE-signaling to Aβ-synthesis in human neurons.
Definition of the non-canonical MAP-kinase signal pathway mediated by DLK, MKK7, and ERK1/2 connects ApoE-dependent signaling to DLK, which performs important additional functions in synapse formation and neuronal regeneration (Yan et al., 2009; Hammarlund et al., 2009; Shin et al., 2012). However, different from other DLK activities that appear to involve MKK4 and JNKs (Tedeschi and Bradke, 2013), we observed that ApoE-induced stimulation of DLK activates MKK7 and ERK1/2, which then regulates gene transcription via activation of cFos, suggesting that DLK may be a key signaling platform whose precise functional readout depends on the location of that signal and the state of the neurons. Our in vivo experiments in mice show that the downstream signaling mechanisms that we defined normally regulate APP expression, documenting the physiological significance of this pathway. Although the upstream ApoE signal is redundant with other glial signals, redundancy of a function doesn’t mean it is unimportant. Moreover, chronic small changes in the efficacy of a function – such as ApoE2 vs. ApoE3 vs. ApoE4 signaling – could cumulatively cause major effects over the decades of the life of a person.
Our observation that ApoE2, ApoE3, and ApoE4 differentially activate APP transcription and Aβ-synthesis reveals a striking parallel to the contribution of these genetic ApoE variants to AD risk, suggesting that the differential activation of Aβ synthesis by ApoE2, ApoE3, and ApoE4 may contribute to their role in AD pathogenesis. However, we do not mean to imply that the differential efficacy of ApoE2, ApoE3, and ApoE4 in activating APP transcription and Aβ-synthesis is the only mechanism by which these genetic isoforms influence AD risk. In recent years, major progress was achieved in the understanding of APP cleavage by α-, β-, and γ-secretases, and plausible hypotheses describe how these proteases may contribute to AD pathogenesis and how rare missense mutations in APP and γ-secretase may cause familial AD (De Strooper and Karran, 2016; Goedert, 2015). How APP transcription is regulated, and what differentiates the APP gene from the closely related APLP1 and APLP2 genes that are not linked to AD, is much less clear. Genetic variations in the APP promoter correlate with AD risk (Lahiri et al., 2005; Brouwers et al., 2006), and APP gene duplications cause early-onset AD (Rovelet-Lecrux et al., 2006; Sleegers et al., 2006), showing that APP expression is indeed important for AD pathogenesis. The finding that ApoE increases APP transcription 4–6 fold in human neurons by activation of cFos-containing AP-1 transcription factors suggests that it may be possible to influence APP transcription pharmacologically as an approach to AD prevention or even therapy.
Our results raise important new questions, both about the mechanism and the conceptual implications of the signaling pathway that we describe. For example, which ApoE receptor mediates activation of DLK, and how is the ApoE signal transduced? ApoE receptors bind to adaptor proteins, such as Disabled-1 and X11/Mints, thereby mediating signal transduction (Gotthardt et al., 2000). Clearly, a similar pathway could operate in DLK activation. Another mechanistic question regards the differential efficacy of ApoE2, ApoE3, and ApoE4 in activating ApoE receptors – is this effect due to different binding affinities, or to different agonist potencies? This important issue can be addressed once the relevant ApoE receptor for signaling in neurons has been identified (Fig. 7D). Among broader questions, those regarding the general function of the ApoE-stimulated MAP-kinase signaling pathway may be paramount – what is its overall role, and which other biological processes are regulated by this pathway? Moreover, what is the biological function of APP, an issue that has been debated for decades, and why is expression of APP but not of APLP1 or APLP2 increased by ApoE signaling? These and other questions raised by the present studies provide fertile grounds for future advances in understanding not only brain signaling pathways, but also their contribution to AD pathogenesis.
STAR METHODS
KEY RESOURCES TABLE attached
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Thomas C. Südhof (tcs1@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human neurons induced from embryonic stem (ES) cells and iPS cells
H1 human ES cells were obtained from WiCell Research Resources (Wicell, WI), and iPS cell lines EB and SKC from the Stanford Stem Cell Core. Human neurons were generated from H1 and iPS cells essentially as described (Zhang et al., 2013) with modifications described in METHOD DETAILS.
Mice
CD1 wildtype mice Crl:CD1(ICR) purchased from Charles River Laboratories were used in vitro for dissociated cultures of hippocampal neurons, glia and murine embryonic fibroblasts (MEFs), and in vivo for AAV injections (see METHOD DETAILS). Both male and female animals were used, and randomly allocated to experimental groups. All animal usage followed the guidelines of and was overseen by the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University, the Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the Panel on Euthanasia of the American Veterinary Medical Association. All animals were housed in the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) accredited Research Animal Facility at Stanford Institutes of Medicine.
METHOD DETAILS
Culture of various principal types of cells
Human embryonic stem (ES) cells and induced pluripotent stem (iPS) cells. H1 human ES cells were obtained from WiCell Research Resources (Wicell, WI), maintained in feeder-free condition using mTeSR1 medium (Stem Cell Technologies), and used at intermediate (~50) passage numbers to generate human induced neuronal (iN) cells (Zhang et al., 2013). Cells were cultured as described (Zhang et al., 2013).
Mouse glia were cultured from the cortex of newborn CD1 mice. The cortex was dissected, digested with papain 10 U/ml for 20 minutes at 37°C, and harshly triturated. Cells were plated onto T75 flasks in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). After reaching 80–90% confluence, glial cultures were trypsinized and re-plated at lower density (1:3–4) in DMEM + 10% FBS. This re-plating procedure was repeated 2–3 times (typically 3–4 days apart) to remove mouse neurons before cultured glia were used for analysis or co-culture experiments with human neurons. No antibiotics or other drugs were used in glial cultures.
Murine embryonic fibroblasts (MEFs) were isolated from mouse embryos of CF-1 strain (Harlan Laboratories, Inc.) harvested at 12.5–13.5 postcoitum (p.c.). Briefly, embryos were dissected out of terminally anesthetized mice. The head and internal organs were removed, and the remaining carcasses were finely minced, trypsinized into single-cell suspensions, and plated onto T75 flasks. The cultured MEFs (P0) were frozen or expanded for up to 3 times (P3) before they were used for experiments.
Mouse hippocampal cortical neurons were cultured from newborn CD1 mice according to standard procedures (Zhang et al., 2013). In short, hippocampi or cortices were dissected, papain-digested, and triturated. The dissociated cortical neurons were plated on matrigel-coated glass coverslips, and maintained in Neurobasal-A (NBA) medium with B-27 supplement. No FBS was ever added into the mouse cortical cultures. The lentiviral infection if applicable was performed at DIV4, and ApoE treatment (10 µg/ml) was applied from DIV 7. Cultures were harvested on DIV12-14 for indicated biochemical assays.
Generation of neurons (iN cells) from human ES and iPSC cells
Human neurons (iN cells) were generated from H1 and iPS cells essentially as described (Zhang et al., 2013) with the following modifications (Fig. S1A): ES cells were detached with accutase and plated onto matrigel-coated 6-well plates (4×104 cells/well) on day −2. Lentiviruses expressing Ngn2 and rtTA were prepared as described below, and added to the ES cells in fresh mTeSR1 medium (Stem Cell Technologies) on day −1. Doxycycline (2 mg/l, to activate Ngn2 expression) was added on day 0 (D0) in DMEM-F12 medium with N2 supplement without morphogens. Puromycin (1 mg/l) was added on D1 in fresh DMEM-F12/N2 + doxycycline medium for selection up to 24 hours. On D2, differentiating neurons were detached with accutase and re-plated on cultured mouse glia, MEFs, or just matrigel-coated 24-well plates (200,000 cells/well), and maintained in NBA/B-27 medium with no doxycycline. Various lentiviral infection of neurons were performed on D4 as described below; ApoE incubations were initiated on D10 and lasted until neurons were analyzed for different parameters. For mRNA and protein measurements, the assays were performed on D12 after ApoE treatments of 2 days, unless otherwise specified in time course studies. For immunoblotting analysis of protein phosphorylation, the ApoE was administered for 2 hours, and neuronal cultures were harvested immediately.
Recombinant ApoE and RAP proteins, and glia-secreted factors
Recombinant ApoE2, ApoE3, and ApoE4 were produced both in HEK293 cells (FreeStyle 293-F cell, ThermoFisher) and bacteria (E. coli BL21 strain). Expression vectors for human ApoE2, ApoE3, and ApoE4 were constructed using an ApoE3 cDNA in pDONR221 vector (Gateway recombination system) that was obtained from Harvard Medical School (Harvard PlasmID Database, clone ID: HsCD00044600). Site-directed mutagenesis was used to generate ApoE2 (C526T) and ApoE4 (T388C) cDNAs. The ApoE2, ApoE3, and ApoE4 cDNAs were cloned into the lentiviral vector pLX304 (Addgene plasmid # 25890) using Gateway LR Clonase II (Invitrogen); the control plasmid was FUGW (Addgene plasmid # 14883), used to monitor transfection efficiency. 293-F cells were cultured in suspension in serum-free FreeStyle 293 Expression Medium (ThermoFisher, protein-free and chemically defined formulation) and transfected with control or ApoE expression plasmids using lipid-based FreeStyle MAX Reagent (ThermoFisher) following the manufacturer’s instructions. Supernatants from transfected 293-F cells were harvested 6 days after transfection and assessed by SDS-PAGE to determine the purity and yields of recombinant ApoE proteins (Fig. S1C). Stain-Free precast gels (Bio-Rad) were used for rapid fluorescent detection of proteins as an alternative to Coomassie staining. Upon UV illumination, the trihalo compounds contained in Stain-Free gels covalently bind to tryptophan residues of analyzed proteins and generate fluorescence. After centrifugation of ApoE2, ApoE3, and ApoE4 for 4 hours at 50,000g, more than 90% of the ApoE remained in the supernatant with apparently equal amounts of ApoE2, ApoE3, and ApoE4. The ApoE supernatants were diluted to proper concentration by fresh NBA/B27 (typically 50–80X) and added into cultured neurons for ApoE stimulation. The supernatant from control- (FUGW-) transfected 293-F cells was diluted the same way and added onto neurons as the control condition, in order to control the possible influences from other HEK293-secreted proteins. HEK293 ApoE proteins were used in all ApoE stimulation experiments unless otherwise indicated.
Bacterial ApoE proteins were expressed in E. coli BL21 strain (Invitrogen, Cat#C600003)as GST fusion proteins in modified pGEX-KG vectors harboring in multiple cloning site (BamHI-EcoRI) a cleavage site recognized by human rhinovirus 3C protease (PreScission Protease): LEVLFQ/GP (DNA sequence: ctggaagttctgttccaggggccc). The cDNAs encoding ApoE2, ApoE3, and ApoE4 (mature, 299 amino acids) obtained as described for the HEK293 cell expression experiments were cloned into BamHI and HindIII sites. Protein expression was induced in bacteria grown to OD 0.5 (measured at 600 nm) with Isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.05 mM) for 6 h at room temperature. Bacteria were then pelleted and resuspended in solubilization buffer (0.5 mg/ml lysozyme in PBS, 1 mM PMSF, 1 mM EDTA, and an EDTA-free protease inhibitor cocktail). Cell lysis was achieved by liquid nitrogen freezing/thawing and sonication. After centrifugation to remove insoluble components of cell lysates, proteins were affinity-purified with glutathione sepharose beads and collected by cleavage with rhinovirus 3C protease (PBS, 16 hours at 4 °C).
For production of recombinant RAP, pGEX-KG-RAP (generous gift of Dr. Joachim Herz, UT Southwestern Medical Center) was expressed in BL21 bacteria, and GST-RAP was purified as described above, except that thrombin cleavage (10 U per mg protein, overnight at 4°C) was used instead of rhinovirus 3C protease as a final step.
The 24 glia-secreted factors selected for screening experiments (Fig. 1D, S3D–3E) were produced from HEK293T cells transfected with plasmids encoding human proteins by a standard calcium phosphate protocol (Zhang et al., 2013). The culture medium was collected 72 hours after transfection, filtered with a 0.2 µm syringe filter to remove cell debris, diluted (1:5) with fresh growth medium Neurobasal-A/B27, and added into cultures of human neurons on MEFs at D10. At D12, the culture medium of neuronal cultures was harvested for Aβ ELISA (Fig. 1D), immunoblotting and qRT-PCR for APP and several control proteins (Fig. S3D–S3E). The human gene cDNA plasmids used for transfection were acquired by Gateway recombination cloning (Invitrogen). The destination vector is pLX304 (CMV promoter, Addgene plasmid # 25890), and the following clones were obtained from Harvard Medical School: APOE, HsCD00044600; CLU, HsCD00040309; CP, HsCD00348042; CST3, HsCD00043456; CTGF, HsCD00378376; CYR61, HsCD00040135; FN1, HsCD00296172; FST, HsCD00042431; IGF2, HsCD00366367; IGFBP2, HsCD00076522; IGFBP5, HsCD00368361; LGALS12, HsCD00369800; HsCD00323559; METRNL, HsCD00339900; NENF, HsCD00377723; NPVF, HsCD00295230; PTN, HsCD00082359; SERPINF1, HsCD00371068; SPARC, HsCD00042420; SPP1, HsCD00040432; THBS1, HsCD00437810; THBS2, HsCD00379263; TIMP1, HsCD00039987; WNT5A, HsCD00375640; WNT7A, HsCD00042006. The control plasmid is FUGW (EGFP driven by CMV promoter, Addgene plasmid # 14883)
Lentivirus and adeno-associated virus (AAV) production and administration
Lentiviruses were produced in HEK293T cells as described (Zhang et al., 2013). In short, HEK293T cells were transfected using Ca2+-phosphate in T75 flasks with a given lentiviral plasmid and the three helper plasmids pRSV-REV, pMDLg/pRRE, and VSV-G. The culture medium was harvested 40–44 hours after transfection, and virus particles were pelleted by ultra-centrifugation. The viral pellet was resuspended in MEM (100 µl per starting dish) and aliquots of the viral solution were snap-frozen in liquid nitrogen for storage. For viral transduction, 2–5 µl of lentiviral solutions were added to the culture medium of 6-well plate (for H1 ES cells) or a 24-well plate (for human neuronal cultures). Lentiviruses were produced from the following plasmids: i. Lentiviruses to trans-differentiate ES cells into neurons (TetO-Ngn2-P2A-puromycin and rtTA; Fig. S1A; Zhang et al., 2013); ii. Lentiviruses for overexpression of DLK for rescue (pLX304-DLK; clone ID: HsCD00413295), of MBIP for inhibiting DLK (pLX304-MBIP; HsCD00420627), and of MKK7 (pCW45-MKK7; HsCD00298961); for all lentiviral plasmids were obtained from Harvard Medical School; iii, Lentiviruses for overexpression of dominant-negative cFos (DN cFos, Olive et al., 1997)(cDNA from Addgene plasmid # 33353 and cloned into AgeI and EcoRI sites on lentiviral vector FUGW Addgene plasmid # 14883); and iv. Lentiviruses for shRNA-mediated DLK or LZK knockdown, for CRISPR-mediated inhibition of MKK7 expression, and for CRISPRi of the APP promoter as described in details below.
AAVs packaged with AAV-DJ capsids were used for high efficiency of in vivo neuronal infection. AAV vectors with pHelper and pRC-DJ were transfected into HEK293 cells, which were collected and lysed 72 hr later. The virus preparations were concentrated from cell lysates by fractioning with iodixanol gradient (40%) and filtering with 100,000 MWCO tube filter. The virus titer was measured by infecting HEK293 cells and adjusted to 1.0×107 infectious units/µl for brain injection. The AAV vectors used in DN-cFos experiment (Fig. 7B, S7H–S7J) express P2A-EGFP (control) or DN cFos-P2A-EGFP under the control of Synapsin-1 promoter. The AAV vectors used in CRIPSRi experiments (Fig. 7C, S7H–J) are described in the following section and also express EGFP as a marker for dissection of infected brain tissues for further assays.
The AAVs were injected into the prefrontal cortex of P0–P1 mouse pups. Neonatal mice were first cryo-anesthetized in an ice-cold chamber, and then secured in a prone position with a custom-made clay mold and adhesive bandage. A syringe pump injector equipped with pulled glass micropipette of beveled tip at ~50 µm diameter was used for injections. The insertion location of micropipette was guided by vascular anatomy, rostral to inferior cerebral vein and close to superior sagittal sinus. Each pup received one injection in only one hemisphere, with a total of 0.5 µl of AAV suspension at 2 different depths. The overall survival was nearly 100%. The injected pups were sacrificed at P7–8, and the brains were harvested for dissection of infected tissues marked by green fluorescence under blue-light illumination from a hand-hold LED lamp (Nightsea Inc., excitation 440–460 nm, see Fig. S7H) The dissected tissues were then lysed for immunoblotting and qPCR (Fig. 7A–7C, S7I–S7J).
Suppression of gene expression using RNAi, CRISPR/Cas9, or CRISPRi/dCas9
RNAi of DLK, LZK or JIP3. shRNAs to DLK (sequence: ACTCGTATTCCTTGTACATAG, TRC number: TRCN0000231658), to LZK (sequence: CCTGATGAGTTAGCTGATAAA, TRCN000019471) and to JIP3 (sequence: GTTCTCAGTGCGCGATGATTT, TRCN0000422388) and the control shRNA (sequence: TAAGGCTATGAAGAGATAC, SHC016) were purchased from Sigma-Aldrich in lentiviral vector pLKO.1-puro. The DLK shRNA targets the 3’UTR of DLK mRNA and does not affect expression of rescue DLK, while the control shRNA (non-target, Sigma-Aldrich SHC016) contains at least four mismatches to any human or mouse gene and was demonstrated by the manufacturer to target zero gene using microarray analyses.
MKK7 CRISPR. Lentiviral CRISPR/Cas9-mediated inhibition of human MKK7 expression was performed using a plasmid (lentiCRISPR v2; Addgene plasmid # 52961) that co-expresses mammalian codon-optimized Cas9 nuclease with a single guide RNA (sgRNA). The MKK7 sgRNA (sequence: GCTTCAGCTTTGCTTCCAGG) targets exon 1 with a cleavage site at amino acid 13 and was designed using web-based tools (Doench et al., 2014). The control sgRNA targets EGFP (EGFP sgRNA4; sequence: GGAGCGCACCATCTTCTTCA; Addgene plasmid # 51763) and was cloned into the same Cas9-expressing vector.
CRISPRi of the human APP promoter was performed essentially as described (Larson et al., 2013). Six sgRNAs were designed to probe the human APP promoter region from 554 base pairs 5’ to 147 base pairs 3’ of the major transcription start site +1 at nucleotide 9001 in GenBank D87675.1 (Fig. 5A). The sgRNA sequences are: sg1, ATTTCCTTTAAGCTTCACTCGTT; sg2, GCGGGGTCGGATGATTCAAGCT (AP-1 site); sg3, GCCGGGGAGCGGAGGGGGCGCG; sg4, GCGCGAGCGGGCGCAGTTCCCCGG; sg5, CGCTCGGGCTCCGTCAGTTTCCT; sg6, GAGGAGCGTGCGCGGGGGCCC. One sgRNA was also designed to target the well-conserved AP-1 site on mouse APP promoter, (sgApp AP-1, GGGTCGGGTGACTCAAGCTCGCG). For in vitro experiments (Fig. 7A–7C, S7H–S7J), the sgRNAs were cloned into a mCherry-co-expressed lentiviral plasmid (pgRNA-humanized; Addgene plasmid # 44248). A synthetic sgRNA targeting GFP was inserted into this pgRNA-humanized to construct the control plasmid (pU6-sgGFP-NT1; Addgene plasmid # 46914). The catalytically dead Cas9 (dCas), fused to blue fluorescent protein (BFP) and containing tandem nuclear localization sequences (2X NLS), was expressed via another lentiviral plasmid, pHR-SFFV-dCas9-BFP (Addgene plasmid # 46910). For in vivo experiments of AAV injection, the expression of sgApp AP-1 and a control sgRNA sequence targeting LacZ (sgLacZ, sequence: TGCGAATACGCCCACGCGAT) was driven by human U6 promoter on an AAV vector that expresses EGFP as a marker under the control of ubiquitin promoter. The dCas9, HA-tagged, was expressed via another AAV plasmid, AAV-CMV::NLS-dSaCas9(D10A,N580A)-NLS-3×HA (Addgene plasmid # 61594). The efficacy of the inhibition of gene expression by RNAi, CRISPR/Cas9 or CRISPRi/dCas9 was assessed by qRT-PCR and immunoblotting.
Immunofluorescence labeling experiments
Immunofluorescence staining experiments and image acquisition and analyses were performed essentially as described (Pak et al., 2015; Patzke et al., 2016). Briefly, cultured neurons were fixed in 4% paraformaldehyde, 4% sucrose in PBS, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 5% goat serum in PBS. Cells were incubated with primary antibodies diluted in blocking buffer overnight at 4°C, washed 3 times, a nd incubated with secondary antibodies in blocking buffer for 1 h at room temperature. Samples were then mounted on glass slides for confocal imaging. The primary antibodies were listed in Key Resources Table. The Alexa-488, Alexa-546-, and Alexa-633-conjugated secondary antibodies were used (Invitrogen). For analysis of neuronal cell body size and total neurite length (Fig. 1B–C), neurons were sparsely labeled with GFP by liposome transfection (Lipofectamin 2000, Life Technologies) with a FUGW construct (Addgene plasmid # 14883) at D9 and fixed at D10 to obtain fluorescent images of individual neurons. To compare the expression pattern of DLK and MKK7 between neurons and glia, co-cultures of human neurons and mouse glia were fixed and immunostained at D12 (Fig. S2B).
Immunofluorescence signals were visualized using a Nikon A1 confocal microscope with constant image settings. For image analyses, Z-stacked images were converted to maximal projection images and analyzed using NIS-Elements Confocal software (Nikon). Neurons were randomly chosen in confocal images.
For experiments investigating neuronal cell density (Fig. 1C, S3B), lentiviral transduction of synapsin-1 promoter-driven mCherry tagged with nuclear localization signal (FSW-mCherry-NLS) was used to label all neuronal nuclei in the cultures. Epi-fluorescent live imaging was taken by Leica microscope coupled with digital camera DFC400 (Leica) and analyzed using ImageJ software (National Institutes of Health).
Immunoblotting and protein quantifications
Neurons and cells were lysed in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton-X 100, plus a cocktail protease inhibitors (Roche)), and lysates were analyzed by SDS-PAGE in the presence of DTT (0.1 M). Immunoblotting and quantitative analysis were performed by a dual-channel infrared imaging system with fluorescence-labeled secondary antibodies (800CW and 680LT), an Odyssey Infrared Imager CLX and software Image Studio 5.2.5 (LI-COR Biosciences). Signals were normalized for Tuj1 probed on the same blots as loading controls. Antibodies used were listed in Key Resources Table.
Gene Expression Analyses
To determine the mRNA levels of genes of interest in cultured cells, quantitative RT-PCR measurements were performed on total RNA (isolated with PrepEase RNA Spin Kit, Affymetrix) using TaqMan probes with VeriQuest Probe One-Step qRT-PCR Master Mix (Affymetrix) and an Applied Biosystems 7900HT apparatus. The pre-designed TaqMan primer/probe sets were purchased from Integrated DNA Technologies and tested to show no or minimal cross-species reactivity in pure human neuronal and mouse glial cultures (Fig. S1B, S2A). MAP2 and GAPDH were used as endogenous reference genes.
The assay ID of all used TaqMan primer/probe sets was: human MAP2, Hs.PT.58.20680759; human GAPDH, Hs.PT.58.40035104; human APP, Hs.PT.56a.38768352; human APLP1, Hs.PT.56a.39998510; human APLP2, Hs.PT.56a.23012253.g; human DLK/MAP3K12, Hs.PT.58.38466429; human LZK/MAP3k13, Hs.PT.58.20565862; human MKK7/MAP2K7, Hs.PT.58.40607007; human MBIP, Hs.PT.58.28049660; human BACE1, Hs.PT.58.5050046; human ADAM10, Hs.PT.56a.38403589; human JIP3/MAPK8IP3, Hs.PT.58.776291; human FOS, Hs.PT.58.15540029; human JUN, Hs.PT.58.25094714.g; mouse Map2, Mm.PT.58.10819514; mouse GAPDH, 4352932-0809025; mouse App, Mm.PT.58.10488606; mouse Aplp1, Mm.PT.58.17581301; mouse Aplp2, Mm.PT.15945720; mouse Dlk/Map3k12, Mm.PT.58.7730388; mouse MKK7/Map2k7, Mm.PT.58.28392891.
Luciferase assays of APP promoter reporter plasmids
The regulation of the APP promoter was probed in cultured neurons using the Dual-Luciferase Reporter Assay System (Promega) that allows sequential measurements in a single sample of luminescent signals of firefly luciferase transcribed from the APP reporter and Renilla luciferase transcribed from a control promoter. We inserted the wild-type human APP promoter region (from −554 to +147; PCR-amplified from human genomic DNA extracted from HEK293T cells) into the KpnI and BglII sites of the promoter-less firefly luciferase vector pGL3-Basic (Promega), and obtained a matching mutant APP promoter construct with an AP-1 site substitution (changing the TGATTC sequence to TAATTA) by site-directed mutagenesis. The wild-type or mutant APP reporter plasmids (500 ng) were introduced at D9 together with a control plasmid (pRL-TK; Promega) encoding Renilla luciferase transcribed from a the herpes simplex virus thymidine kinase (HSV-TK) promoter (50 ng) into cultured human neurons in 24 well plates by liposome transfection (Lipofectamin 2000, Life Technologies). The co-transfection of the control plasmid which constitutively expresses Renilla luciferase with the test plasmid enables normalization of the transfection efficiency and recovery during the cellular extraction for each sample in the reporter assays. Each reporter experiment included extracts from sham-transfected cells as a reference control.
At D10 (24 hrs after transfection), cultured neurons were treated for 48 hrs with control solution or ApoE2, ApoE3, or ApoE4 (10 µg/ml). Cell lysates were collected at D12 in passive lysis buffer (Promega), and dual luciferase reporter assays were performed according to manufacturer instructions using a plate-reading luminometer with reagent auto-injectors Apollo LB915 (Berthold Technologies). Samples were compared by subtracting the background activity of the reference control, and then normalizing the firefly luciferase activity of each sample to its Renilla luciferase activity.
APP gene sequence analyses
Human promoter sequences for APP, APLP1, and APLP2 were aligned using CLUSTAL multiple alignment algorithms and the MUSCLE (3.8) program. Nucleotides shared by all three sequences are indicated by an asterisk. The AP-1 consensus binding sequence is boxed, and the transcription start site (+1) marked in bold and boxed. The sequences targeted by sgRNAs (sg1–6) for APP CRISPRi are underlined. APP promoter mutations associated with AD (Theuns et al., 2006) are marked with shading and boxed: −118C-->A, −369C-->G, and −534G-->A. The sequence alignment used is as follows:
Enzyme-linked immunosorbent assay (ELISA) of Aβ
All Aβ measurements were performed using ELISA kits listed in Key Resources Table according to the manufacturer’s instructions. Cultured neurons were washed 3 times on D10 with NBA/B27 to remove existing Aβ, and treated with control solution or ApoE2, ApoE3, or ApoE4 (10 µg/ml in NBA/B27, 0.5 ml per well on a 24-well plate) for 48 hours. The culture medium was harvested at D12 to examine the concentration of secreted Aβ variants. The cultured neurons were then washed for 3 times with PBS, and the wash buffer from the last wash was collected and analyzed for complete removal of ApoE and soluble Aβ by immunoblotting of ApoE and Aβ ELISA. After the washes, lysis buffer (TBS, 1% Triton X-100, 50 µl per well) was added, and the levels of cellular Aβ extracted in cell lysates were measured by ELISA. The HEK293 ApoE proteins (medium supernatants from ApoE-expressing HEK293-F cultures; Methods, Recombinant ApoE and RAP proteins, A), media from mouse glial and cortical cultures were also subjected to ELISA and confirmed to be free of human Aβ.
Non-radioactive in vitro kinase assay
Recombinant human ERK2/MAPK1 were produced in E. coli bacteria (BL21 strain), described as above. The cDNA encoding human ERK2 (NM_002745.4) was synthesized as a gene fragment (gBlock, Integrated DNA Technologies), cloned into pGEX-KG vector, and expressed in bacteria as GST fusion proteins with a linker peptide sequence recognized by rhinovirus 3C protease. At the end of aforementioned purification process, GST cleavage by rhinovirus 3C protease was carried out in kinase buffer: 25 mM Tris-HCl (pH 7.5), 5 mM beta-glycerophosphate, 2mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2. The yield was around 2–5 mg from half a liter culture of E. coli BL21 harboring pGEX-ERK2.
Flag-tagged human MKK7 proteins were expressed in pure human neurons (iN cells) by lentiviral transduction (day 4 to day 10), and activated by ApoE stimulation (10 µg/ml for 2 hours on day 10) as described earlier. The gene fragment of human MKK7 (NM_001297555.1) was synthesized and cloned into AgeI and EcoRI sites on FUGW vector (Addgene plasmid # 14883). After mock or ApoE stimulation, human neurons were lysed with non-denaturing cell lysis buffer and brief sonication. Cell lysis buffer: 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin, and 1 mM PMSF added immediately before use. The immunoprecipitation of unactivated or ApoE-activated Flag-MKK7 was performed by incubating cell lysates of mock- or ApoE-stimulated human neurons with Flag-beads (Sigma A2220, mouse) or HA-beads (control, Sigma A2095, mouse) in cell lysis buffer, overnight at 4°C. For each condition, 50 µl of 50% flurry beads were first washed twice with cell lysis buffer and incubated with neuronal cell lysate collected from a whole 6-well plate of cultured human neurons (~ 4 million cells). After pull-down, the beads were washed twice with cell lysis buffer and once with kinase buffer, and then reconstituted as 50% flurry with kinase buffer. 10 µl of immunoprecipitated beads were eluted by adding 10 µl of 2X SDS-PAGE buffer. The ApoE-induced MKK7 phosphorylation and MKK7 pull-down efficiency were analyzed by immunoblotting with p-MKK7 (Cell Signaling 4171, rabbit)
For in vitro kinase assays determining whether ApoE-activated MKK7 phosphorylates ERK2, 200 ng of recombinant human ERK2 was added into immunoprecipitated beads pelleted from 40 µl 50% flurry: Flag-beads pulling down unactivated or ApoE-activated Flag-MKK7, or control HA-beads. The reactions were done in 40 µl kinase buffer supplemented with 0.1 mM ATP at 30°C for 30 minutes, without or with U0126 50 µM. The reaction were terminated by pelleting down beads and adding 10 µl 5X SDS-PAGE buffer into the harvested supernatants (~40 µl). The phosphorylation of ERK2 was analyzed by running 20 µl of each reaction mixture supernatants for SDS-PAGE and immunoblotting using anti-ERK (Cell Signaling 4695) and anti-p-ERK (Cell Signaling 9106) antibodies.
Cycloheximide chase assay to determine DLK protein stability
The degradation rate of DLK protein was analyzed by quantitative immunoblotting after cycloheximide chase assay. Cycloheximide treatments were performed by adding cycloheximide (0.1 g/l; Sigma) without or together with ApoE3 (10 µg/ml) to cultured human neurons on MEFs at D10. At the indicated time points, neurons were lysed and immunoblotted for DLK protein levels. The data of DLK relative intensity at 0h, 2h, 4h and 6h, after normalization to loading control Tuj1, were fit to an exponential decay function (first-order kinetics, A=A0ekt) to estimate the degradation rate constant (k, hour−1). The half-life time (t1/2) is further calculated from the constant (t1/2 = ln(2)/k) to determine the time required for 50% degradation of DLK protein without or with ApoE3.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative data are presented as means ± SEM. All experiments were independently repeated at least three times. Statistical analyses were conducted using Prism 6 software (GraphPad Software, Inc.). Statistical comparisons between groups were analyzed for significance by one-way analysis of variance (ANOVA) with Tukey’s post-hoc test and Student’s t-test.
Supplementary Material
Figure S1: General experimental strategy for producing human neurons (A), measurements of ApoE mRNA levels in mouse glia, human neurons, and mouse embryonic fibroblasts (MEFs) showing that only glia abundantly express ApoE (B), production of recombinant ApoE2, ApoE3, and ApoE4 in transfected HEK293 cells and bacteria (C), measurements of cellular total Ab, Aβ40, and Aβ42 in human neurons as a function of glial co-cultures and of ApoE2, ApoE3, and ApoE4 added to MEF co-cultures (D), effect of ApoE’s added to human neuron/MEF co-cultures on the expression of α- and β-secretases (E), reproduction of the differential stimulation of Aβ secretion from human neurons by ApoE2, ApoE3, and ApoE4 using neurons differentiated from two different lines of iPS cells (F, G), effect of ApoE’s on Aβ secretion from human neurons cultured on mouse glia (H), and control experiments for measurements of the effects of recombinant ApoE2, ApoE3, and ApoE4 on Aβ-production in and Aβ-secretion from human neurons (I and J) (related to Figure 1)
(A) Diagram illustrating the basic protocol for generating pure human neurons (iN cells) from ES and iPS cells. Top, schematic drawing of the two expression cassettes in lentiviruses used for forced tetracycline-dependent expression of Ngn2; bottom, flow diagram of the overall procedure; see Zhang et al., 2013 for details. After induction of neuronal differentiation, neurons are cultured either on cultured mouse glia (which secrete copious amounts of ApoE and other signaling factors), mouse embryonic fibroblasts (MEFs), or matrigel alone (see also Fig. 1A). The age of neuronal cultures is given in days after induction of differentiation by initiating Ngn2 expression with tetracycline. Human neurons induced from ES cells (H1 line) were used as the standard model throughout the article, and human neurons induced from iPS cells (SKC line and EB line) were only used to study ApoE’s effect on Aβ secretion (Fig. S1F, S1G) and APP transcription (Fig. S3A, S3B).
(B) ApoE is abundantly expressed only in mouse glia but not in MEFs or human neurons. Data show quantitative RT-PCR measurements of ApoE mRNA levels as percent of GAPDH mRNA levels in MEFs, human neurons, and glia, as measured with both mouse-specific (left) and human-specific primers (right) to illustrate RT-PCR assay specificity (n.d., not detectable). For human neurons, measurements were performed at day 10 after induction (D10) with neurons cultured on matrigel. Data are means ± SEM; n≥3 independent experiments for all bar graphs.
(C) SDS-PAGE analysis of recombinant ApoE2, ApoE3, and ApoE4 proteins produced in transfected HEK293 cells and in E. coli. Image shows a representative stain-free SDS-polyacrylamide gel in which proteins are visualized via trihalo compounds included in the gel that react with tryptophan residues in a UV-induced reaction to produce fluorescence. Bovine serum albumin (BSA) was analyzed on the same gel at different concentrations as a protein standard to calculate ApoE concentrations. HEK293 ApoE proteins were prepared from the medium of HEK293-F (FreeStyle) cell suspension cultures grown in serum-free condition and transfected to express human ApoE isoforms. The concentrations of HEK293 ApoE in the harvested medium are typically 500-800 µg/ml (50-80X). The control medium was harvested from transfected HEK293-F cells expressing GFP and used in the experiments at the same dilution (50-80X) as the HEK293-F cell ApoE-containing medium. ApoE proteins were quantified on SDS-Page gels using serum bovine albumin as a standard. Bacterial ApoE proteins were expressed in E. coli BL21 strain as GST fusion proteins, with a linker peptide sequence recognized by rhinovirus 3C protease, under the control of an inducible tac promoter. Protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.05 mM for 6 h) when transformed E. coli had grown to an OD650 of 0.5 at room temperature. E. coli ApoE proteins were purified from bacterial lysates by glutathione beads and collected by cleavage of GST tags by rhinovirus 3C protease (yield; 2-6 mg/l culture).
(D) Glia co-culture dramatically increases the cellular Aβ content in human neurons compared to MEF co-culture; addition of ApoE2, ApoE3, and ApoE4 to human neurons cultured on MEFs that produce only little Aβ differentially elevates their cellular Aβ content with an ApoE4>ApoE3>ApoE2 potency rank order, but does not change the Aβ40/Aβ42 ratio. Total Aβ, Aβ40, and Aβ42 concentrations were analyzed by ELISA in lysates harvested from cultured human neurons as described for Fig. 1E after collection of the culture medium for the measurements, as described in detail for panel H (left). Relative levels of total Aβ (left), Aβ40 (left center), Aβ42 (right middle) and Aβ42/Aβ40 ratio (right) were measured by ELISA at D12 and normalized to those of cultures without ApoE because it is difficult to standardize cellular concentrations in absolute terms.
(E) ApoE2, ApoE3, and ApoE4 do not alter expression of α- and β-secretases. mRNA levels of BACE1 and ADAM10 (the β- and α-secretases that proteolyze APP proteins into Aβ and non-Aβ peptides, respectively) were measured in human neurons on MEFs cultures at D12 after 48 h stimulation by ApoEs (10 µg/ml). Experiments suggest that ApoE does not stimulate Aβ production by increasing β-secretase or decreasing α-secretase expression.
(F & G) ApoE2, ApoE3 and ApoE4 also differentially stimulate Aβ secretion from human neurons derived from two independent iPS-cell lines as previously characterized (Zhang et al., 2013). The differential efficacy of ApoE isoforms exhibited the same rank order of ApoE4>ApoE3>ApoE2 as for H1 ES-cell derived neurons. iPSC-derived human neurons (SKC line and EB line) were cultured and stimulated with ApoE as described for Fig. 1E and (A) above.
(H) ApoE2, ApoE3, and ApoE4 have no effect on total Aβ levels when human neurons are cultured on mouse glia, presumably because glia produces saturating amounts of ApoE (see panel B) and of other factors (Fig. 1D) which occlude any additional effect of exogenous ApoE on the co-cultured neurons. Neuronal Aβ measurements were performed on human neurons co-cultured with mouse glia by a human-specific ELISA that detects total Aβ. As described below in panel F, cultured neurons were washed 3 times at D10 to remove existing Aβ, and treated with control solution or ApoE2, ApoE3, or ApoE4 (10 µg/ml, 0.5 ml per well on a 24-well plate) for 48 h. At D12, the culture medium was collected to examine the concentration of secreted human Aβ (left, Medium). Neurons were then washed for 3 times to remove ApoE and secreted Aβ. After the washes, lysis buffer (50 µl per well) was added, and the Aβ extracted in cell lysates was measured by ELISA and normalized to control condition with no ApoE (right, Cells). Data (means ± SEM) are shown as absolute levels for the medium and as relative levels normalized to the control for cellular Aβ because of variability in the cell extractions.
(I) Control experiments demonstrating that medium containing ApoE2, ApoE3, and ApoE4 contained from transfected HEK293 cells lacks detectable human Aβ, and that medium from cultures of mouse glia or of mixtures of mouse cortical neurons and glia also lack detectable human Aβ. Supernatants harvested from HEK293 cells transfected with plasmids expressing ApoE2, ApoE3 or ApoE4 were analyzed to show that no contamination of human Aβ is carried from the HEK293 to human neurons treated with the ApoE’s. Note that these supernatants from transfected HEK293 cells expressing the ApoE isoforms were used as stock solutions for HEK293 ApoE and were employed for experiments after quantification and dilution (typically 1:50-1:80). The control for all experiments was supernatant harvested from HEK293 cells transfected with control plasmids. Media from mouse glial and cortical cultures were harvested at DIV12. Data are means ± SEM; n≥3 independent experiments for all bar graphs.
(J) Control experiments demonstrating that washing human neurons three times fully removes all Aβ and ApoE from the extracellular space. The flow diagram on the left depicts the experimental protocol whereby human neurons cultured on MEFs were incubated with ApoE2, ApoE3, or ApoE4 starting at 10 days (D10) after induction of neuronal differentiation. At D12, the cell culture medium was collected and analyzed for total Aβ, the cells were washed three times, and the third wash was analyzed again for Aβ and for ApoE. The summary graph in the center depicts the results of ELISA measurements of the levels of total human Aβ in the third wash (means ± SEM), demonstrating that Aβ was undetectable. The panel on the right depicts an immunoblot for ApoE of both the initial medium and the third wash, showing that ApoE2, ApoE3, and ApoE4 were only detectable in the initial medium but not the third wash.
Data in B, and D-I are means ± SEM (n≥3 independent experiments for all samples); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons. Non-significant comparisons are not identified, nor are more significant comparisons (e.g., comparisons of the ApoE3 or ApoE4 samples vs. control).
Figure S2: Control experiments for molecular definition of the neuronal MAP-kinase signaling pathway activated by ApoE in human neurons. Data show that ApoE3-induced ERK1/2 phosphorylation is inhibited by RAP and MEK antagonists but not by PI3-kinase or src inhibitors (A, B), that human neurons express high levels of DLK and MKK7 (C, D), that ApoE3-induced ERK1/2 phosphorylation declines rapidly after cycloheximide addition (E), that manipulations of DLK expression or function are effective as intended but do not affect expression of MBIP or LZK (F), that manipulations of DLK or MKK7 function do not impair neuronal survival (G), that MKK7 CRISPR and LZK shRNAs are highly effective (F, J), and that LZK shRNAs have no effect on ApoE3-induced changes in DLK protein and ERK1/2 phosphorylation levels (I) (related to Fig. 2 and 3)
(A) Representative immunoblots for the experiment shown in Fig. 2B demonstrating that ApoE3-induced ERK1/2-phosphorylation is abolished by the ApoE-receptor blocker RAP (50 µg/ml) and by inhibitors of MEKs (PD98059 and U0126; both 50 µM), but not by inhibitors of JNK (SP600125; 25 µM), PI3K kinase (Wortmannin; 0.1 µM), or Src kinase (PP2; 10 µM). Drugs were applied 30 min before a 2 h ApoE3 incubation at D10.
(B) Independent repetition of the experiments shown in Fig. 2B and panel A above without the ApoE3-minus control for most treatments.
(C) mRNA measurements show that DLK (dual leucine-zipper kinase, a.k.a. MAP3K12) and MKK7 (MAP-kinase kinase 7, a.k.a. MAP2K7) are abundantly expressed in human neurons but only modestly expressed in mouse glia and not detectable (n.d.) in MEFs. Data show quantitative RT-PCR measurements of both mouse and human DLK and MKK7 mRNA abundance in percent of GAPDH abundance (means ± SEM), as measured separately with mouse- and human-specific primers in the three cell types shown to monitor both mRNA abundance and assay specificity.
(D) Confocal images of human neurons cultured on mouse glia and immunostained for DLK and MKK7 as indicated to illustrate neuronal enrichment of DLK and MKK7.
(E) Analysis of effect of ApoE3 on phospho-ERK protein stability based on the quantitative immunoblotting shown in Fig. 2E-2F. The phospho-ERK intensity at the given time points of cycloheximide treatment was normalized to internal control Tuj1 intensity and plotted relative to the 0 h time point. Data were fit to an exponential decay curve for the ApoE3 condition (red line; not possible for the control condition [blue line] due to a lack of change). The cycloheximide chase assay is described in detail in the methods.
(F) ApoE has no effect on the mRNA levels for human DLK, MBIP, and LZK (a.k.a. MAP3K13) in human neurons cultured on MEFs, but DLK shRNAs efficiently suppress, and DLK overexpressions during rescue manipulations massively increase, respectively, DLK mRNA levels; MBIP overexpression, in contrast, enhances MBIP mRNA levels but has no effect on DLK mRNA levels since MBIP inhibits DLK protein, not DLK synthesis; none of these manipulations, finally, alters LZK mRNA levels. Human neurons cultured on MEFs were infected at D4 with lentiviruses expressing (i) a control (EGFP), (ii) DLK shRNAs together with EGFP; (iii) DLK shRNAs together with EGFP and an shRNA-independent DLK rescue protein producing overexpression; and (iv) MBIP. Neurons were treated with ApoE3 at D10-12, and analyzed by quantitative RT-PCR for DLK, MBIP, and LZK expression at D12. mRNA levels are normalized to the control and to MAP2 as an internal standard to control for neuronal differentiation because MAP2 mRNA levels increase in human neurons during differentiation.
(G) Survival of human neurons cultured on MEFs is not impaired by various DLK and MKK7 manipulations (DLK knockdown by shRNAs, DLK rescue overexpression, DLK inhibition by MBIP overexpression, MKK7 inhibition by CRISPR, and MKK7 overexpression). Neurons were subjected to shRNA-mediated knockdown or overexpression of DLK, to MBIP overexpression or CRISPR/Cas9-mediated knockdown or overexpression of MKK7 at D4, treated with control or ApoE3-containing medium from D10-D12, and examined by quantification of neuronal cell density at D12.
(H) LZK shRNAs efficiently suppress LZK mRNA levels but have no effect on DLK mRNA levels. Human neurons cultured on MEFs were infected with control lentivirus or a lentivirus expressing the LZK shRNA at D4, treated with a control solution or 10 µg/ml ApoE3 at D10-D12, and LZK and DLK mRNA levels were measured by quantitative RT-PCR at D12. mRNA levels are normalized to the control condition without ApoE3. mRNA levels are normalized to the control and to MAP2 as an internal standard.
(I) LZK shRNA has no significant effect on the ApoE3-induced increase in DLK protein levels or on the ApoE3-induced enhancement of ERK phosphorylation. Human neurons cultured on MEFs with and without knockdown of LZK and/or ApoE3 treatment were obtained as described for D, and analyzed by quantitative immunoblotting (left, representative immunoblots; right, summary graph of protein levels normalized to Tuj1 as an internal standard).
(J) MKK7 mRNA levels in human neurons cultured on MEFs are not altered by ApoE3, but can be efficiently suppressed via CRISPR/Cas9-mediated MKK7 deletion or increased by MKK7 overexpression. MKK7 mRNA levels were measured by quantitative RT-PCR. Neurons were infected at D4 with lentiviruses expressing Cas9 and a control sgRNA or an MKK7-specific sgRNA targeting exon 1, or with a lentivirus overexpressing MKK7, were treated without or with ApoE3 (10 µg/ml) at D10-D12, and were analyzed at D12. mRNA levels are normalized to the control and to MAP2 as an internal standard.
Data in summary graphs are presented as means ± SEM; n ≥ 3 independent experiments for all bar graphs; statistical significance was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons, comparing the test conditions to the control condition (neurons without ApoE, with control lentiviral manipulations, or without pharmacological treatment; *, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S3: Demonstration that ApoE2, ApoE3, and ApoE4 specifically and differentially increase APP expression also in human neurons that were derived from SKC iPS cells (A) or EB iPS cells (B) and cultured on MEFs; measurements of APP, APLP1, and APLP2 expression in human neurons, mouse glia, and MEFs (C); time course of APP mRNA induction by ApoE’s in human neurons cultured on MEFs (D); and lack of an effect of ApoE’s on APP, APLP1, or APLP2 expression in mouse glia (E) (related to Fig. 4)
(A & B) ApoE2, ApoE3 and ApoE4 differentially increase APP mRNA levels in human neurons that were derived from two different iPSC lines (A, SKC; B, EB) and cultured on MEFs, but do not affect APLP1 or APLP2 mRNA levels. Experiments were carried out as described in Fig. 4C.
(C) Human neurons abundantly express human APP and APLP1 but not APLP2, whereas mouse glia express mouse App and Aplp2 but not Aplp1, and MEFs express only low levels of mouse App. mRNA levels were measured by quantitative human- or mouse-specific RT-PCR assays and normalized for GAPDH mRNA as an internal standard. Measurements are from mouse glia, human neurons cultured on matrigel at D10 after neuronal induction, and MEFs.
(D) Concentration-dependence (left) and time-dependence (right) of the effects of ApoE2, ApoE3, and ApoE4 on APP mRNA levels in human neurons cultured on MEFs. Neurons were cultured on MEFs and treated with ApoE3 at days D10-12 after neuronal induction for the dose-response curve (left), or at the indicated hours before D12 for the time course (right). APP RT-PCR assays were performed at D12 and detected only human transcripts; MAP2 was used as an internal control that relates the APP mRNA levels to those of an internal neuronal marker to correct for the increasing degree of neuronal differentiation during the course of the experiment. mRNA levels were normalized to the untreated control (left) or the 0 h time point (right).
(E) Control experiments demonstrating that addition of ApoE2, ApoE3, and ApoE4 does not alter App, Aplp1, or Aplp2 mRNA levels in cultured mouse glia. Glia were cultured from cortices of newborn mice and passaged for at least three times to remove mouse neurons before ApoE treatment. ApoE2, ApoE3, and ApoE4 (10 µg/ml) were added to mouse glia at 80-90% confluency. Two days later, mRNA levels of App, Aplp1 and Aplp2 were assayed by mouse-specific qRT-PCR and normalized to Gapdh mRNA levels. Note that if there was any effect of the ApoE proteins on App transcription in glia, this would likely be occluded by the copious amounts of endogenous ApoE produced and secreted by the glia (see Fig. S1B). Summary graphs are plotted relative to the levels observed in the absence of ApoE (Ctrl. condition). Data are presented as means ± SEM; n ≥ 3 independent experiments.
Data in summary graphs and plots are presented as means ± SEM (n≥3 independent experiments). Statistical significance was evaluated by one-way ANOVA and Tukey’s post hoc multiple comparisons to control (*, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S4: Further demonstration that those glial factors that were identified in the glial factor screen (Fig. 1D) to enhance Aβ levels also enhance APP transcription (A, B), demonstration that JIP3 shRNAs are effective (C), and characterization of ApoE-dependent stimulation of APP transcription via activation of the DLK-MKK7-ERK MAP-kinase signaling pathway (D-F) (related to Fig. 4)
(A) Multiple glial factors other than ApoE that increase Aβ synthesis in human neurons cultured in the absence of glia (Fig. 1D) are also active in elevating neuronal APP protein levels. Data show cellular APP levels determined by quantitative immunoblotting under the conditions described in Fig. 1D (left, representative immunoblot; right, summary graph of APP protein levels normalized to the internal loading control Tuj1). Note that conditioned glia medium (CGM) robustly increases neuronal (human) APP protein level (yellow bar), and that this increase is replicated by addition of recombinant human ApoE3 (brown bar) and IGF2 and IGFBP2 (black bars). The production and application of CGM and selected glia factors are described in detail in the methods.
(B) Same as A, except that the effect of conditioned glia medium (CGM) and of various factors on the APP mRNA was tested.
(C) The shRNA-mediated knockdown of JIP3 is efficacious. JIP3 mRNA levels were measured in human neurons expressing control or JIP3 shRNAs to document efficacy of the RNAi for the experiments shown in Fig. 4F and S6C. Neurons were infected with lentiviruses expressing shRNAs at D4, and treated at D10 for 4 h with ApoE3 (10 µg/ml) and MG132 (10 µM) before analysis by qRT-PCR.
(D) Quantification of MKK7 and ERK1/2 phosphorylation during the ApoE-induced increase in APP expression to further demonstrate that the ApoE-induced APP increase is mediated by activation of a canonical DLK-dependent MAP-kinase pathway. Summary graphs show quantifications of immunoblots performed in the experiments shown in Fig. 4E from four independent replicates.
(E) APP mRNA levels measured in experiments analogous to those described for Figs. 4E and (A) above. Human neurons cultured on MEFs were subjected at D4 to the indicated molecular manipulations by lentiviral infection, treated at D10 with control or ApoE3-containing (10 µg/ml) medium, and analyzed at D12 by quantitative mRNA RT-PCR. Data confirm that ApoE3 increases APP expression by elevating the APP mRNA levels dependent on DLK and MKK7 activation, whereas DLK or MKK7 overexpression constitutively activates APP expression.
(F) DLK knockdown blocks ApoE-induced increases in APP mRNA levels. Cultures of human neurons grown on MEFs were infected with lentiviruses expressing control shRNA (Ctrl) or DLK shRNA at D4, stimulated with ApoE2, ApoE3, and ApoE4 (10 µg/ml) at D10-12, and assayed for mRNA levels of APP and DLK (to show efficiency of DLK shRNA).
Data in summary graphs and plots are presented as means ± SEM (n ≥ 3 independent experiments). Statistical significance was evaluated by one-way ANOVA and Tukey’s post hoc multiple comparisons to Ctrl. (A, B, D-F), and Student’s t test (C) (*, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S5: Demonstration that the glial factors that activate APP and Aβ synthesis in human neurons co-cultured with mouse glia occlude further effects of exogenous ApoE (A) and act, at least in part, by activating the same MAP-kinase signaling cascade as ApoE (B, C) (related to Fig. 4)
(A) Demonstration that exogenous ApoE3 has no effect on the very high levels of human APP and DLK proteins expressed in human neurons co-cultured with mouse glia (presumably because glial factors already robustly activate DLK and APP levels [yellow bars]) and has no effect on glia or MEFs alone (in which human DLK and APP protein are not detectable (n.d.) under our conditions), but dramatically increases APP and DLK levels in human neurons co-cultured with MEFs (light blue/brown bars), or cultured on matrigel alone (blue bars). Cells cultured under the indicated conditions were treated with ApoE3 (10 µg/ml) from D10-12, harvested, and analyzed by immunoblotting. Left, representative immunoblots; right, summary graphs of protein levels normalized to Tuj1 in conditions containing human neurons, and plotted relative to the levels observed in neurons cultured on MEFs without ApoE3 (light blue bar). The glial marker GFAP was only detected in co-cultures of human neurons on mouse glia and in pure cultures of mouse glia.
(B) APP synthesis in human neurons cultured on mouse glia are insensitive to ApoE owing to copious glial ApoE secretion, but are regulated by the same DLK-dependent MAP kinase signaling pathway as the pathway that is activated by ApoE in the absence of glia. Human neurons co-cultured with glia were transduced with control lentiviruses or lentiviruses expressing DLK shRNAs without or with a DLK overexpression cassette, or expressing the DLK inhibitory protein MBIP at D4. Cells were treated with or without ApoE3 (10 µg/ml) from D10-12, and analyzed at D12 by quantitative immunoblotting for APP and DLK, using Tuji1 as a loading control and GFAP as a control for the glial co-culture (left, representative immunoblot; right, summary graphs of APP and DLK levels). Note that even in the presence of glia, APP levels can be upregulated by additional increases in DLK levels.
(C) Similar to APP synthesis (see B), Aβ40 and Aβ42 levels in human neurons cultured on mouse glia are insensitive to ApoE owing to copious glial ApoE secretion, but are regulated by the same DLK-dependent MAP kinase signaling pathway as the pathway that is activated by ApoE in the absence of glia. Experiments were performed as described for B, except that the concentrations of human Aβ40 and Aβ42 were measured by ELISA in the medium as described in Fig. S1.
Data are presented as means ± SEM; n ≥ 3 independent experiments for all bar graphs; statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and selected Tukey’s post-hoc comparisons, comparing test conditions to control. Non-significant comparisons are not identified.
Figure S6: ApoE is internalized into human neurons but not transported into nucleus (A), expression of BFP-dCas9 and mCherry during CRISPRi experiments are independent of the co-expressed guide RNAs (B), CRISPRi inhibition of the AP-1 binding site in the APP promoter decreases Aβ secretion from neurons even when neurons are co-cultured with glia (C), and ApoE3 increases levels of both APP and cFos mRNAs in a manner that is independent of the JNK-scaffold JIP3 (D)(related to Figure 5)
(A) ApoE is internalized in human neurons into endosomes in a RAP-inhibited manner without being transported into the nucleus. Confocal images of human neurons cultured alone on matrigel, under control conditions (left) or after incubation with ApoE3 (10 µg/ml at D10-12; center) in the absence or presence of the ApoE-receptor inhibitor RAP (50 µg/ml added 30 min prior to ApoE3 addition; right). Neurons were fixed, permeabilzed, and stained for ApoE and NeuN at D12. Top panels show the ApoE signal in gray scale (note background signal of antibody in the absence of ApoE3 addition); bottom panels show merged images of ApoE (green) and NeuN signals (red).
(B) Quantitative immunoblotting reveals that BFP-dCas9 and mCherry (which is co-expressed with the control or the APP sgRNAs) are produced at equal levels during various conditions of the experiments using CRISPRi to target the human APP promoter. Lysates from neurons grown on MEFs and infected at D4 with the various test and control lentiviruses for the CRISPRi experiments were analyzed by immunoblotting for GFP (to probe for BFP-dCas9 since antibodies to GFP cross-react with BFP) and mCherry (to monitor the level of sgRNA expression) at D12 after incubation with control solution or 10 µg/ml ApoE3 at D10-12. Representative immunoblots are shown in Fig. 5C.
(C) Blockage by CRISPRi of the APP promoter AP-1 binding site in human neurons cultured on mouse glia decreases constitutive Aβ42 secretion; this experiment extends the data from Fig. S5 demonstrating that the same signaling pathway which mediates stimulation of APP and Aβ synthesis by ApoE also regulates Aβ synthesis in co-cultures of human neurons with glia. Experiments were carried out on medium of human neurons cultured on glia with CRISPRi lentivirus infection (dCas9 and control sgRNA or APP sg2) and ApoE treatment (see panel A).
(D) ApoE induces both APP and cFos transcription leading to their increased mRNA levels, but has no effect on cJun transcription; these actions of ApoE do not require JIP3, whereas MG132-induced cellular stress enhances cJun transcription but not APP or cFos transcription, and the induction of cJun transcription by MG132 requires JIP3, presumably because it is mediated by JNKs (see Fig. 2D and 4F). Knockdown of JIP3 and treatments with ApoE3 or MG132 treatments are described in Fig. S4C.
Figure S7: DLK pathway and AP-1 transcription factor signaling regulate App transcription and Aβ-synthesis in mouse neurons in culture and in vivo (related to Fig. 6 and 7)
(A) ApoE isoforms do not affect Aβ-synthesis in dissociated mixed cultures of mouse hippocampal neurons and glia, presumably because glia secretes copious amounts of factors. Cultures were treated with control solution or ApoE2, ApoE3, or ApoE4 (all 10 µg/ml) from DIV7 to DIV14, and Aβ40 (left panel) and Aβ42 (middle panel) levels in the culture medium were measured by ELISA at DIV14.
(B) ApoE3 does not affect Aβ42 synthesis in dissociated mixed cultures of mouse cortical neurons and glia, again presumably because glia secretes copious amounts of factors. Cultures were treated with control solution or ApoE3 (10 µg/ml) from DIV7 to DIV12, and Aβ42 levels in the culture medium were measured by ELISA at DIV12.
(C) Demonstration that mouse DLK shRNAs are efficient in mouse neurons, and quantification of the overexpression of human DLK or MKK7 in experiments validating the reagents for the mechanistic studies of Fig. 6. Dissociated cultures of mouse hippocampal neurons and glia were infected at DIV4 with lentiviruses expressing (i) a control shRNA, (ii) DLK shRNA alone, and (iii) DLK shRNA together with human DLK rescue, or (iv) DLK shRNA together with human MKK7 rescue proteins. Note that the primers used for quantitative RT-PCR assays (performed at DIV14) of DLK in the left panel detect both mouse and human DLK, whereas the primers used for human DLK in the middle panel and for human MKK7 in the right panel detect only the human mRNAs. mRNA levels are normalized to the control and to mouse GAPDH as an internal standard.
(D) No change in the Aβ40/42 ratio as a function of various manipulations of the DLK signaling pathway in dissociated cultures of mouse hippocampal neurons and glia primary cultures as described for panel C; data complement those shown in Fig. 6C.
(E) Aβ42-synthesis in dissociated cultures of mouse cortical neurons and glia is decreased by DLK inhibition using MBIP overexpression (left panel), and to interference with the AP-1 binding site in the mouse App-gene promoter by CRISPRi (App AP-1; right panel). Cultures were infected at DIV4 with lentiviruses expressing control proteins or MBIP (left panel) or the control or test CRISPRi components (right panel), and Aβ42 levels were measured by ELISA in the medium at DIV12.
(F) Expression of sgRNAs and dCas9 is efficient in CRISPRi experiments of dissociated cultures of mouse hippocampal neurons and glia (Fig. 6). Expression of mCherry (co-expressed with the control or App AP-1 site sgRNA) and BFP-tagged dCas9 (recognized by GFP antibodies) was analyzed by immunoblotting (representative blots are shown in Fig. 6F). Cultures were infected with lentiviruses expressing dCas9 and sgRNAs (Ctrl. or App AP-1) at DIV4 and harvested at DIV14 for quantitative immunoblotting of APP, mCherry and BFP-dCas9, with Tuj1 as an internal loading control.
(G) Inhibition of AP-1-mediated App transcription does not change Aβ40/Aβ42 ratio based on the analysis of ELISA results from Fig. 6G.
(H) Illustration of high-efficiency in vivo AAV transduction in mouse brain by stereotactic injections into newborn mice, and analysis at P7-9. The types of AAVs injected are listed on the left, and a representative fluorescence image of the mouse brain at P9 viewed under illumination with blue light (440-460 nm) is shown on the right to demonstrate the broad region of infection. Fluorescence is derived from the EGFP co-expressed by the AAVs.
(I) Representative immunoblots complementing the protein expression experiments described in Fig. 7B and 7C.
(J) Quantifications of APP mRNA levels in brain after in vivo application of DN-cFos or CRISPRi to the AP-1 binding sequence in the mouse App promoter. Samples analyzed for protein levels in Figure 7A-7C were used for quantitative RT-PCR; levels are normalized for Tuji1 (tubulin-β3) used as an internal standard.
Data are presented as means ± SEM; n≥3 independent experiments for all bar graphs; statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated by one-way ANOVA and comparing test conditions to control with Tukey’s post-hoc multiple comparisons (A-E) and Student’s t-test (G-I). Non-significant comparisons are not identified.
HIGHLIGHTS.
Human ApoE robustly stimulates APP transcription and Aβ production in human neurons
ApoE4 is more potent and ApoE2 less potent than ApoE3 in stimulating Aβ production
ApoE activates a non-canonical MAP-kinase pathway involving DLK, MKK7, and ERK1/2
ApoE activated MAP-kinases phosphorylate cFos to induce APP and Aβ production
A systematic investigation of how the three ApoE isoforms contribute to Alzheimer’s Disease pathogenesis reveals a direct role for ApoE in driving levels of APP and amyloid-β levels.
Acknowledgments
We thank Dr. J. Herz for the RAP expression vector and ApoE antibodies. This study was supported by grants from the NIH (AG04813101 to M.W. and T.C.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y.A.H. and T.C.S. planned all experiments, analyzed the data, and wrote the paper; Y.A.H. and B.Z. performed all experiments; M.W. provided advice on data analyses and the manuscript.
REFERENCES
- Brouwers N, Sleegers K, Engelborghs S, Bogaerts V, Serneels S, Kamali K, Corsmit E, De Leenheir E, Martin JJ, De Deyn PP, et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer’s disease. Brain. 2006;129:2984–2991. doi: 10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Maloney JA, Kallop DY, Atwal JK, Tam SJ, Baer K, Kissel H, Kaminker JS, Lewcock JW, Weimer RM, et al. Spatially coordinated kinase signaling regulates local axon degeneration. J. Neurosci. 2012;32:13439–13453. doi: 10.1523/JNEUROSCI.2039-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ A, Christa A, Kur E, Lioubinski O, Bachmann S, Willnow TE, Hammes A. LRP2 is an auxiliary SHH receptor required to condition the forebrain ventral midline for inductive signals. Dev. Cell. 2012;22:268–278. doi: 10.1016/j.devcel.2011.11.023. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;12:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet M, Besson FL, Gonneaud, La Joie JR, Chételat G. Imaging brain effects of APOE4 in cognitively normal individuals across the lifespan. Neuropsychol. Rev. 24:290–299. doi: 10.1007/s11065-014-9263-8. [DOI] [PubMed] [Google Scholar]
- Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J. Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama K, Yoshida M, Yamashita A, Deyama T, Baba M, Suzuki A, Mohri H, Ikezawa Z, Nakajima H, Hirai S, et al. MAPK upstream kinase (MUK)-binding inhibitory protein, a negative regulator of MUK/dual leucine zipper-bearing kinase/leucine zipper protein kinase. J. Biol. Chem. 2000;275:21247–21254. doi: 10.1074/jbc.M001488200. [DOI] [PubMed] [Google Scholar]
- Goedert M. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015;161:161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J. Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J. Biol. Chem. 2000;275:25616–25624. doi: 10.1074/jbc.M000955200. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, Mitani A, Joyner D, Thyssen DH, Bacskai BJ, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid β peptide. J. Neurosci. 2012;32:15181–15192. doi: 10.1523/JNEUROSCI.1542-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Goldstein JL, Strickland DK, Ho YK, Brown MS. 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. J. Biol. Chem. 1991;266:21232–21238. [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Xu H, Bu G. ApoE and Aβ in Alzheimer’s disease: accidental encounters or partners? Neuron. 2014;81:740–754. doi: 10.1016/j.neuron.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;14:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Ge YW, Maloney B, Wavrant-De Vrièze F, Hardy J. Characterization of two APP gene promoter polymorphisms that appear to influence risk of late-onset Alzheimer’s disease. Neurobiol. Aging. 2005;26:1329–1341. doi: 10.1016/j.neurobiolaging.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Belin S, He Z. Signaling regulations of neuronal regenerative ability. Curr. Opin. Neurobiol. 2014;27:135–142. doi: 10.1016/j.conb.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch DH, Nägler K, Schumacher S, Göritz C, Müller EC, Otto A, Pfrieger FW. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- Merritt SE, Mata M, Nihalani D, Zhu C, Hu X, Holzman LB. The mixed lineage kinase DLK utilizes MKK7 and not MKK4 as substrate. J. Biol. Chem. 1999;274:10195–10202. doi: 10.1074/jbc.274.15.10195. [DOI] [PubMed] [Google Scholar]
- Monje P, Marinissen PM, Gutkind JS. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol. 2003;23:7030–7043. doi: 10.1128/MCB.23.19.7030-7043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012;4:a011254. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo N, Mitsuda N, Tamatani M, Yamaguchi A, Lee YD, Ogihara T, Vitek MP, Tohyama M. Apolipoprotein E4 stimulates cAMP response element-binding protein transcriptional activity through the extracellular signal-regulated kinase pathway. J. Biol Chem. 2001;276:3046–3053. doi: 10.1074/jbc.M005070200. [DOI] [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 1997;272:18586–94. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, Yi F, Wernig M, Südhof TC. Human Neuropsychiatric Disease Modeling using Conditional Deletion Reveals Synaptic Transmission Defects Caused by Heterozygous Mutations in NRXN1. Cell Stem Cell. 2015;3:316–328. doi: 10.1016/j.stem.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke C, Acuna C, Giam LR, Wernig M, Südhof TC. Conditional deletion of L1CAM in human neurons impairs both axonal and dendritic arborization and action potential generation. J. Exp. Med. 2016;213:499–515. doi: 10.1084/jem.20150951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimplikar SW, Nixon RA, Robakis NK, Shen J, Tsai LH. Amyloid-independent mechanisms in Alzheimer’s disease pathogenesis. J. Neurosci. 2010;30:14946–14954. doi: 10.1523/JNEUROSCI.4305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Hyman BT, Rebeck GW. Apolipoprotein E receptors mediate neurite outgrowth through activation of p44/42 mitogen-activated protein kinase in primary neurons. J. Biol. Chem. 2004;279:34948–34956. doi: 10.1074/jbc.M401055200. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Scheinfeld MH, Roncarati R, Vito P, Lopez PA, Abdallah M, D’Adamio L. Jun NH2-terminal kinase (JNK) interacting protein 1 (JIP1) binds the cytoplasmic domain of the Alzheimer’s beta-amyloid precursor protein (APP) J. Biol. Chem. 2002;277:3767–3775. doi: 10.1074/jbc.M108357200. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Bradke F. The DLK signalling pathway--a double-edged sword in neural development and regeneration. EMBO Rep. 2013;14:605–614. doi: 10.1038/embor.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns J, Brouwers N, Engelborghs S, Sleegers K, Bogaerts V, Corsmit E, De Pooter T, van Duijn CM, De Deyn PP, et al. Promoter Mutations That Increase Amyloid Precursor-Protein Expression Are Associated with Alzheimer Disease. Am. J. Hum. Genet. 2006;78:936–946. doi: 10.1086/504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 2015;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade EJ, Klucher KM, Spector DH. An AP-1 binding site is the predominant cis-acting regulatory element in the 1.2-kilobase early RNA promoter of human cytomegalovirus. J. Virol. 1992;66:2407–17. doi: 10.1128/jvi.66.4.2407-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Eckel RH. What are lipoproteins doing in the brain? Trends Endocrinol. Metab. 2014;25:8–14. doi: 10.1016/j.tem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: General experimental strategy for producing human neurons (A), measurements of ApoE mRNA levels in mouse glia, human neurons, and mouse embryonic fibroblasts (MEFs) showing that only glia abundantly express ApoE (B), production of recombinant ApoE2, ApoE3, and ApoE4 in transfected HEK293 cells and bacteria (C), measurements of cellular total Ab, Aβ40, and Aβ42 in human neurons as a function of glial co-cultures and of ApoE2, ApoE3, and ApoE4 added to MEF co-cultures (D), effect of ApoE’s added to human neuron/MEF co-cultures on the expression of α- and β-secretases (E), reproduction of the differential stimulation of Aβ secretion from human neurons by ApoE2, ApoE3, and ApoE4 using neurons differentiated from two different lines of iPS cells (F, G), effect of ApoE’s on Aβ secretion from human neurons cultured on mouse glia (H), and control experiments for measurements of the effects of recombinant ApoE2, ApoE3, and ApoE4 on Aβ-production in and Aβ-secretion from human neurons (I and J) (related to Figure 1)
(A) Diagram illustrating the basic protocol for generating pure human neurons (iN cells) from ES and iPS cells. Top, schematic drawing of the two expression cassettes in lentiviruses used for forced tetracycline-dependent expression of Ngn2; bottom, flow diagram of the overall procedure; see Zhang et al., 2013 for details. After induction of neuronal differentiation, neurons are cultured either on cultured mouse glia (which secrete copious amounts of ApoE and other signaling factors), mouse embryonic fibroblasts (MEFs), or matrigel alone (see also Fig. 1A). The age of neuronal cultures is given in days after induction of differentiation by initiating Ngn2 expression with tetracycline. Human neurons induced from ES cells (H1 line) were used as the standard model throughout the article, and human neurons induced from iPS cells (SKC line and EB line) were only used to study ApoE’s effect on Aβ secretion (Fig. S1F, S1G) and APP transcription (Fig. S3A, S3B).
(B) ApoE is abundantly expressed only in mouse glia but not in MEFs or human neurons. Data show quantitative RT-PCR measurements of ApoE mRNA levels as percent of GAPDH mRNA levels in MEFs, human neurons, and glia, as measured with both mouse-specific (left) and human-specific primers (right) to illustrate RT-PCR assay specificity (n.d., not detectable). For human neurons, measurements were performed at day 10 after induction (D10) with neurons cultured on matrigel. Data are means ± SEM; n≥3 independent experiments for all bar graphs.
(C) SDS-PAGE analysis of recombinant ApoE2, ApoE3, and ApoE4 proteins produced in transfected HEK293 cells and in E. coli. Image shows a representative stain-free SDS-polyacrylamide gel in which proteins are visualized via trihalo compounds included in the gel that react with tryptophan residues in a UV-induced reaction to produce fluorescence. Bovine serum albumin (BSA) was analyzed on the same gel at different concentrations as a protein standard to calculate ApoE concentrations. HEK293 ApoE proteins were prepared from the medium of HEK293-F (FreeStyle) cell suspension cultures grown in serum-free condition and transfected to express human ApoE isoforms. The concentrations of HEK293 ApoE in the harvested medium are typically 500-800 µg/ml (50-80X). The control medium was harvested from transfected HEK293-F cells expressing GFP and used in the experiments at the same dilution (50-80X) as the HEK293-F cell ApoE-containing medium. ApoE proteins were quantified on SDS-Page gels using serum bovine albumin as a standard. Bacterial ApoE proteins were expressed in E. coli BL21 strain as GST fusion proteins, with a linker peptide sequence recognized by rhinovirus 3C protease, under the control of an inducible tac promoter. Protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.05 mM for 6 h) when transformed E. coli had grown to an OD650 of 0.5 at room temperature. E. coli ApoE proteins were purified from bacterial lysates by glutathione beads and collected by cleavage of GST tags by rhinovirus 3C protease (yield; 2-6 mg/l culture).
(D) Glia co-culture dramatically increases the cellular Aβ content in human neurons compared to MEF co-culture; addition of ApoE2, ApoE3, and ApoE4 to human neurons cultured on MEFs that produce only little Aβ differentially elevates their cellular Aβ content with an ApoE4>ApoE3>ApoE2 potency rank order, but does not change the Aβ40/Aβ42 ratio. Total Aβ, Aβ40, and Aβ42 concentrations were analyzed by ELISA in lysates harvested from cultured human neurons as described for Fig. 1E after collection of the culture medium for the measurements, as described in detail for panel H (left). Relative levels of total Aβ (left), Aβ40 (left center), Aβ42 (right middle) and Aβ42/Aβ40 ratio (right) were measured by ELISA at D12 and normalized to those of cultures without ApoE because it is difficult to standardize cellular concentrations in absolute terms.
(E) ApoE2, ApoE3, and ApoE4 do not alter expression of α- and β-secretases. mRNA levels of BACE1 and ADAM10 (the β- and α-secretases that proteolyze APP proteins into Aβ and non-Aβ peptides, respectively) were measured in human neurons on MEFs cultures at D12 after 48 h stimulation by ApoEs (10 µg/ml). Experiments suggest that ApoE does not stimulate Aβ production by increasing β-secretase or decreasing α-secretase expression.
(F & G) ApoE2, ApoE3 and ApoE4 also differentially stimulate Aβ secretion from human neurons derived from two independent iPS-cell lines as previously characterized (Zhang et al., 2013). The differential efficacy of ApoE isoforms exhibited the same rank order of ApoE4>ApoE3>ApoE2 as for H1 ES-cell derived neurons. iPSC-derived human neurons (SKC line and EB line) were cultured and stimulated with ApoE as described for Fig. 1E and (A) above.
(H) ApoE2, ApoE3, and ApoE4 have no effect on total Aβ levels when human neurons are cultured on mouse glia, presumably because glia produces saturating amounts of ApoE (see panel B) and of other factors (Fig. 1D) which occlude any additional effect of exogenous ApoE on the co-cultured neurons. Neuronal Aβ measurements were performed on human neurons co-cultured with mouse glia by a human-specific ELISA that detects total Aβ. As described below in panel F, cultured neurons were washed 3 times at D10 to remove existing Aβ, and treated with control solution or ApoE2, ApoE3, or ApoE4 (10 µg/ml, 0.5 ml per well on a 24-well plate) for 48 h. At D12, the culture medium was collected to examine the concentration of secreted human Aβ (left, Medium). Neurons were then washed for 3 times to remove ApoE and secreted Aβ. After the washes, lysis buffer (50 µl per well) was added, and the Aβ extracted in cell lysates was measured by ELISA and normalized to control condition with no ApoE (right, Cells). Data (means ± SEM) are shown as absolute levels for the medium and as relative levels normalized to the control for cellular Aβ because of variability in the cell extractions.
(I) Control experiments demonstrating that medium containing ApoE2, ApoE3, and ApoE4 contained from transfected HEK293 cells lacks detectable human Aβ, and that medium from cultures of mouse glia or of mixtures of mouse cortical neurons and glia also lack detectable human Aβ. Supernatants harvested from HEK293 cells transfected with plasmids expressing ApoE2, ApoE3 or ApoE4 were analyzed to show that no contamination of human Aβ is carried from the HEK293 to human neurons treated with the ApoE’s. Note that these supernatants from transfected HEK293 cells expressing the ApoE isoforms were used as stock solutions for HEK293 ApoE and were employed for experiments after quantification and dilution (typically 1:50-1:80). The control for all experiments was supernatant harvested from HEK293 cells transfected with control plasmids. Media from mouse glial and cortical cultures were harvested at DIV12. Data are means ± SEM; n≥3 independent experiments for all bar graphs.
(J) Control experiments demonstrating that washing human neurons three times fully removes all Aβ and ApoE from the extracellular space. The flow diagram on the left depicts the experimental protocol whereby human neurons cultured on MEFs were incubated with ApoE2, ApoE3, or ApoE4 starting at 10 days (D10) after induction of neuronal differentiation. At D12, the cell culture medium was collected and analyzed for total Aβ, the cells were washed three times, and the third wash was analyzed again for Aβ and for ApoE. The summary graph in the center depicts the results of ELISA measurements of the levels of total human Aβ in the third wash (means ± SEM), demonstrating that Aβ was undetectable. The panel on the right depicts an immunoblot for ApoE of both the initial medium and the third wash, showing that ApoE2, ApoE3, and ApoE4 were only detectable in the initial medium but not the third wash.
Data in B, and D-I are means ± SEM (n≥3 independent experiments for all samples); statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons. Non-significant comparisons are not identified, nor are more significant comparisons (e.g., comparisons of the ApoE3 or ApoE4 samples vs. control).
Figure S2: Control experiments for molecular definition of the neuronal MAP-kinase signaling pathway activated by ApoE in human neurons. Data show that ApoE3-induced ERK1/2 phosphorylation is inhibited by RAP and MEK antagonists but not by PI3-kinase or src inhibitors (A, B), that human neurons express high levels of DLK and MKK7 (C, D), that ApoE3-induced ERK1/2 phosphorylation declines rapidly after cycloheximide addition (E), that manipulations of DLK expression or function are effective as intended but do not affect expression of MBIP or LZK (F), that manipulations of DLK or MKK7 function do not impair neuronal survival (G), that MKK7 CRISPR and LZK shRNAs are highly effective (F, J), and that LZK shRNAs have no effect on ApoE3-induced changes in DLK protein and ERK1/2 phosphorylation levels (I) (related to Fig. 2 and 3)
(A) Representative immunoblots for the experiment shown in Fig. 2B demonstrating that ApoE3-induced ERK1/2-phosphorylation is abolished by the ApoE-receptor blocker RAP (50 µg/ml) and by inhibitors of MEKs (PD98059 and U0126; both 50 µM), but not by inhibitors of JNK (SP600125; 25 µM), PI3K kinase (Wortmannin; 0.1 µM), or Src kinase (PP2; 10 µM). Drugs were applied 30 min before a 2 h ApoE3 incubation at D10.
(B) Independent repetition of the experiments shown in Fig. 2B and panel A above without the ApoE3-minus control for most treatments.
(C) mRNA measurements show that DLK (dual leucine-zipper kinase, a.k.a. MAP3K12) and MKK7 (MAP-kinase kinase 7, a.k.a. MAP2K7) are abundantly expressed in human neurons but only modestly expressed in mouse glia and not detectable (n.d.) in MEFs. Data show quantitative RT-PCR measurements of both mouse and human DLK and MKK7 mRNA abundance in percent of GAPDH abundance (means ± SEM), as measured separately with mouse- and human-specific primers in the three cell types shown to monitor both mRNA abundance and assay specificity.
(D) Confocal images of human neurons cultured on mouse glia and immunostained for DLK and MKK7 as indicated to illustrate neuronal enrichment of DLK and MKK7.
(E) Analysis of effect of ApoE3 on phospho-ERK protein stability based on the quantitative immunoblotting shown in Fig. 2E-2F. The phospho-ERK intensity at the given time points of cycloheximide treatment was normalized to internal control Tuj1 intensity and plotted relative to the 0 h time point. Data were fit to an exponential decay curve for the ApoE3 condition (red line; not possible for the control condition [blue line] due to a lack of change). The cycloheximide chase assay is described in detail in the methods.
(F) ApoE has no effect on the mRNA levels for human DLK, MBIP, and LZK (a.k.a. MAP3K13) in human neurons cultured on MEFs, but DLK shRNAs efficiently suppress, and DLK overexpressions during rescue manipulations massively increase, respectively, DLK mRNA levels; MBIP overexpression, in contrast, enhances MBIP mRNA levels but has no effect on DLK mRNA levels since MBIP inhibits DLK protein, not DLK synthesis; none of these manipulations, finally, alters LZK mRNA levels. Human neurons cultured on MEFs were infected at D4 with lentiviruses expressing (i) a control (EGFP), (ii) DLK shRNAs together with EGFP; (iii) DLK shRNAs together with EGFP and an shRNA-independent DLK rescue protein producing overexpression; and (iv) MBIP. Neurons were treated with ApoE3 at D10-12, and analyzed by quantitative RT-PCR for DLK, MBIP, and LZK expression at D12. mRNA levels are normalized to the control and to MAP2 as an internal standard to control for neuronal differentiation because MAP2 mRNA levels increase in human neurons during differentiation.
(G) Survival of human neurons cultured on MEFs is not impaired by various DLK and MKK7 manipulations (DLK knockdown by shRNAs, DLK rescue overexpression, DLK inhibition by MBIP overexpression, MKK7 inhibition by CRISPR, and MKK7 overexpression). Neurons were subjected to shRNA-mediated knockdown or overexpression of DLK, to MBIP overexpression or CRISPR/Cas9-mediated knockdown or overexpression of MKK7 at D4, treated with control or ApoE3-containing medium from D10-D12, and examined by quantification of neuronal cell density at D12.
(H) LZK shRNAs efficiently suppress LZK mRNA levels but have no effect on DLK mRNA levels. Human neurons cultured on MEFs were infected with control lentivirus or a lentivirus expressing the LZK shRNA at D4, treated with a control solution or 10 µg/ml ApoE3 at D10-D12, and LZK and DLK mRNA levels were measured by quantitative RT-PCR at D12. mRNA levels are normalized to the control condition without ApoE3. mRNA levels are normalized to the control and to MAP2 as an internal standard.
(I) LZK shRNA has no significant effect on the ApoE3-induced increase in DLK protein levels or on the ApoE3-induced enhancement of ERK phosphorylation. Human neurons cultured on MEFs with and without knockdown of LZK and/or ApoE3 treatment were obtained as described for D, and analyzed by quantitative immunoblotting (left, representative immunoblots; right, summary graph of protein levels normalized to Tuj1 as an internal standard).
(J) MKK7 mRNA levels in human neurons cultured on MEFs are not altered by ApoE3, but can be efficiently suppressed via CRISPR/Cas9-mediated MKK7 deletion or increased by MKK7 overexpression. MKK7 mRNA levels were measured by quantitative RT-PCR. Neurons were infected at D4 with lentiviruses expressing Cas9 and a control sgRNA or an MKK7-specific sgRNA targeting exon 1, or with a lentivirus overexpressing MKK7, were treated without or with ApoE3 (10 µg/ml) at D10-D12, and were analyzed at D12. mRNA levels are normalized to the control and to MAP2 as an internal standard.
Data in summary graphs are presented as means ± SEM; n ≥ 3 independent experiments for all bar graphs; statistical significance was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons, comparing the test conditions to the control condition (neurons without ApoE, with control lentiviral manipulations, or without pharmacological treatment; *, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S3: Demonstration that ApoE2, ApoE3, and ApoE4 specifically and differentially increase APP expression also in human neurons that were derived from SKC iPS cells (A) or EB iPS cells (B) and cultured on MEFs; measurements of APP, APLP1, and APLP2 expression in human neurons, mouse glia, and MEFs (C); time course of APP mRNA induction by ApoE’s in human neurons cultured on MEFs (D); and lack of an effect of ApoE’s on APP, APLP1, or APLP2 expression in mouse glia (E) (related to Fig. 4)
(A & B) ApoE2, ApoE3 and ApoE4 differentially increase APP mRNA levels in human neurons that were derived from two different iPSC lines (A, SKC; B, EB) and cultured on MEFs, but do not affect APLP1 or APLP2 mRNA levels. Experiments were carried out as described in Fig. 4C.
(C) Human neurons abundantly express human APP and APLP1 but not APLP2, whereas mouse glia express mouse App and Aplp2 but not Aplp1, and MEFs express only low levels of mouse App. mRNA levels were measured by quantitative human- or mouse-specific RT-PCR assays and normalized for GAPDH mRNA as an internal standard. Measurements are from mouse glia, human neurons cultured on matrigel at D10 after neuronal induction, and MEFs.
(D) Concentration-dependence (left) and time-dependence (right) of the effects of ApoE2, ApoE3, and ApoE4 on APP mRNA levels in human neurons cultured on MEFs. Neurons were cultured on MEFs and treated with ApoE3 at days D10-12 after neuronal induction for the dose-response curve (left), or at the indicated hours before D12 for the time course (right). APP RT-PCR assays were performed at D12 and detected only human transcripts; MAP2 was used as an internal control that relates the APP mRNA levels to those of an internal neuronal marker to correct for the increasing degree of neuronal differentiation during the course of the experiment. mRNA levels were normalized to the untreated control (left) or the 0 h time point (right).
(E) Control experiments demonstrating that addition of ApoE2, ApoE3, and ApoE4 does not alter App, Aplp1, or Aplp2 mRNA levels in cultured mouse glia. Glia were cultured from cortices of newborn mice and passaged for at least three times to remove mouse neurons before ApoE treatment. ApoE2, ApoE3, and ApoE4 (10 µg/ml) were added to mouse glia at 80-90% confluency. Two days later, mRNA levels of App, Aplp1 and Aplp2 were assayed by mouse-specific qRT-PCR and normalized to Gapdh mRNA levels. Note that if there was any effect of the ApoE proteins on App transcription in glia, this would likely be occluded by the copious amounts of endogenous ApoE produced and secreted by the glia (see Fig. S1B). Summary graphs are plotted relative to the levels observed in the absence of ApoE (Ctrl. condition). Data are presented as means ± SEM; n ≥ 3 independent experiments.
Data in summary graphs and plots are presented as means ± SEM (n≥3 independent experiments). Statistical significance was evaluated by one-way ANOVA and Tukey’s post hoc multiple comparisons to control (*, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S4: Further demonstration that those glial factors that were identified in the glial factor screen (Fig. 1D) to enhance Aβ levels also enhance APP transcription (A, B), demonstration that JIP3 shRNAs are effective (C), and characterization of ApoE-dependent stimulation of APP transcription via activation of the DLK-MKK7-ERK MAP-kinase signaling pathway (D-F) (related to Fig. 4)
(A) Multiple glial factors other than ApoE that increase Aβ synthesis in human neurons cultured in the absence of glia (Fig. 1D) are also active in elevating neuronal APP protein levels. Data show cellular APP levels determined by quantitative immunoblotting under the conditions described in Fig. 1D (left, representative immunoblot; right, summary graph of APP protein levels normalized to the internal loading control Tuj1). Note that conditioned glia medium (CGM) robustly increases neuronal (human) APP protein level (yellow bar), and that this increase is replicated by addition of recombinant human ApoE3 (brown bar) and IGF2 and IGFBP2 (black bars). The production and application of CGM and selected glia factors are described in detail in the methods.
(B) Same as A, except that the effect of conditioned glia medium (CGM) and of various factors on the APP mRNA was tested.
(C) The shRNA-mediated knockdown of JIP3 is efficacious. JIP3 mRNA levels were measured in human neurons expressing control or JIP3 shRNAs to document efficacy of the RNAi for the experiments shown in Fig. 4F and S6C. Neurons were infected with lentiviruses expressing shRNAs at D4, and treated at D10 for 4 h with ApoE3 (10 µg/ml) and MG132 (10 µM) before analysis by qRT-PCR.
(D) Quantification of MKK7 and ERK1/2 phosphorylation during the ApoE-induced increase in APP expression to further demonstrate that the ApoE-induced APP increase is mediated by activation of a canonical DLK-dependent MAP-kinase pathway. Summary graphs show quantifications of immunoblots performed in the experiments shown in Fig. 4E from four independent replicates.
(E) APP mRNA levels measured in experiments analogous to those described for Figs. 4E and (A) above. Human neurons cultured on MEFs were subjected at D4 to the indicated molecular manipulations by lentiviral infection, treated at D10 with control or ApoE3-containing (10 µg/ml) medium, and analyzed at D12 by quantitative mRNA RT-PCR. Data confirm that ApoE3 increases APP expression by elevating the APP mRNA levels dependent on DLK and MKK7 activation, whereas DLK or MKK7 overexpression constitutively activates APP expression.
(F) DLK knockdown blocks ApoE-induced increases in APP mRNA levels. Cultures of human neurons grown on MEFs were infected with lentiviruses expressing control shRNA (Ctrl) or DLK shRNA at D4, stimulated with ApoE2, ApoE3, and ApoE4 (10 µg/ml) at D10-12, and assayed for mRNA levels of APP and DLK (to show efficiency of DLK shRNA).
Data in summary graphs and plots are presented as means ± SEM (n ≥ 3 independent experiments). Statistical significance was evaluated by one-way ANOVA and Tukey’s post hoc multiple comparisons to Ctrl. (A, B, D-F), and Student’s t test (C) (*, p<0.05, **, p<0.01; ***, p<0.001). Non-significant comparisons are not identified.
Figure S5: Demonstration that the glial factors that activate APP and Aβ synthesis in human neurons co-cultured with mouse glia occlude further effects of exogenous ApoE (A) and act, at least in part, by activating the same MAP-kinase signaling cascade as ApoE (B, C) (related to Fig. 4)
(A) Demonstration that exogenous ApoE3 has no effect on the very high levels of human APP and DLK proteins expressed in human neurons co-cultured with mouse glia (presumably because glial factors already robustly activate DLK and APP levels [yellow bars]) and has no effect on glia or MEFs alone (in which human DLK and APP protein are not detectable (n.d.) under our conditions), but dramatically increases APP and DLK levels in human neurons co-cultured with MEFs (light blue/brown bars), or cultured on matrigel alone (blue bars). Cells cultured under the indicated conditions were treated with ApoE3 (10 µg/ml) from D10-12, harvested, and analyzed by immunoblotting. Left, representative immunoblots; right, summary graphs of protein levels normalized to Tuj1 in conditions containing human neurons, and plotted relative to the levels observed in neurons cultured on MEFs without ApoE3 (light blue bar). The glial marker GFAP was only detected in co-cultures of human neurons on mouse glia and in pure cultures of mouse glia.
(B) APP synthesis in human neurons cultured on mouse glia are insensitive to ApoE owing to copious glial ApoE secretion, but are regulated by the same DLK-dependent MAP kinase signaling pathway as the pathway that is activated by ApoE in the absence of glia. Human neurons co-cultured with glia were transduced with control lentiviruses or lentiviruses expressing DLK shRNAs without or with a DLK overexpression cassette, or expressing the DLK inhibitory protein MBIP at D4. Cells were treated with or without ApoE3 (10 µg/ml) from D10-12, and analyzed at D12 by quantitative immunoblotting for APP and DLK, using Tuji1 as a loading control and GFAP as a control for the glial co-culture (left, representative immunoblot; right, summary graphs of APP and DLK levels). Note that even in the presence of glia, APP levels can be upregulated by additional increases in DLK levels.
(C) Similar to APP synthesis (see B), Aβ40 and Aβ42 levels in human neurons cultured on mouse glia are insensitive to ApoE owing to copious glial ApoE secretion, but are regulated by the same DLK-dependent MAP kinase signaling pathway as the pathway that is activated by ApoE in the absence of glia. Experiments were performed as described for B, except that the concentrations of human Aβ40 and Aβ42 were measured by ELISA in the medium as described in Fig. S1.
Data are presented as means ± SEM; n ≥ 3 independent experiments for all bar graphs; statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated with one-way ANOVA and selected Tukey’s post-hoc comparisons, comparing test conditions to control. Non-significant comparisons are not identified.
Figure S6: ApoE is internalized into human neurons but not transported into nucleus (A), expression of BFP-dCas9 and mCherry during CRISPRi experiments are independent of the co-expressed guide RNAs (B), CRISPRi inhibition of the AP-1 binding site in the APP promoter decreases Aβ secretion from neurons even when neurons are co-cultured with glia (C), and ApoE3 increases levels of both APP and cFos mRNAs in a manner that is independent of the JNK-scaffold JIP3 (D)(related to Figure 5)
(A) ApoE is internalized in human neurons into endosomes in a RAP-inhibited manner without being transported into the nucleus. Confocal images of human neurons cultured alone on matrigel, under control conditions (left) or after incubation with ApoE3 (10 µg/ml at D10-12; center) in the absence or presence of the ApoE-receptor inhibitor RAP (50 µg/ml added 30 min prior to ApoE3 addition; right). Neurons were fixed, permeabilzed, and stained for ApoE and NeuN at D12. Top panels show the ApoE signal in gray scale (note background signal of antibody in the absence of ApoE3 addition); bottom panels show merged images of ApoE (green) and NeuN signals (red).
(B) Quantitative immunoblotting reveals that BFP-dCas9 and mCherry (which is co-expressed with the control or the APP sgRNAs) are produced at equal levels during various conditions of the experiments using CRISPRi to target the human APP promoter. Lysates from neurons grown on MEFs and infected at D4 with the various test and control lentiviruses for the CRISPRi experiments were analyzed by immunoblotting for GFP (to probe for BFP-dCas9 since antibodies to GFP cross-react with BFP) and mCherry (to monitor the level of sgRNA expression) at D12 after incubation with control solution or 10 µg/ml ApoE3 at D10-12. Representative immunoblots are shown in Fig. 5C.
(C) Blockage by CRISPRi of the APP promoter AP-1 binding site in human neurons cultured on mouse glia decreases constitutive Aβ42 secretion; this experiment extends the data from Fig. S5 demonstrating that the same signaling pathway which mediates stimulation of APP and Aβ synthesis by ApoE also regulates Aβ synthesis in co-cultures of human neurons with glia. Experiments were carried out on medium of human neurons cultured on glia with CRISPRi lentivirus infection (dCas9 and control sgRNA or APP sg2) and ApoE treatment (see panel A).
(D) ApoE induces both APP and cFos transcription leading to their increased mRNA levels, but has no effect on cJun transcription; these actions of ApoE do not require JIP3, whereas MG132-induced cellular stress enhances cJun transcription but not APP or cFos transcription, and the induction of cJun transcription by MG132 requires JIP3, presumably because it is mediated by JNKs (see Fig. 2D and 4F). Knockdown of JIP3 and treatments with ApoE3 or MG132 treatments are described in Fig. S4C.
Figure S7: DLK pathway and AP-1 transcription factor signaling regulate App transcription and Aβ-synthesis in mouse neurons in culture and in vivo (related to Fig. 6 and 7)
(A) ApoE isoforms do not affect Aβ-synthesis in dissociated mixed cultures of mouse hippocampal neurons and glia, presumably because glia secretes copious amounts of factors. Cultures were treated with control solution or ApoE2, ApoE3, or ApoE4 (all 10 µg/ml) from DIV7 to DIV14, and Aβ40 (left panel) and Aβ42 (middle panel) levels in the culture medium were measured by ELISA at DIV14.
(B) ApoE3 does not affect Aβ42 synthesis in dissociated mixed cultures of mouse cortical neurons and glia, again presumably because glia secretes copious amounts of factors. Cultures were treated with control solution or ApoE3 (10 µg/ml) from DIV7 to DIV12, and Aβ42 levels in the culture medium were measured by ELISA at DIV12.
(C) Demonstration that mouse DLK shRNAs are efficient in mouse neurons, and quantification of the overexpression of human DLK or MKK7 in experiments validating the reagents for the mechanistic studies of Fig. 6. Dissociated cultures of mouse hippocampal neurons and glia were infected at DIV4 with lentiviruses expressing (i) a control shRNA, (ii) DLK shRNA alone, and (iii) DLK shRNA together with human DLK rescue, or (iv) DLK shRNA together with human MKK7 rescue proteins. Note that the primers used for quantitative RT-PCR assays (performed at DIV14) of DLK in the left panel detect both mouse and human DLK, whereas the primers used for human DLK in the middle panel and for human MKK7 in the right panel detect only the human mRNAs. mRNA levels are normalized to the control and to mouse GAPDH as an internal standard.
(D) No change in the Aβ40/42 ratio as a function of various manipulations of the DLK signaling pathway in dissociated cultures of mouse hippocampal neurons and glia primary cultures as described for panel C; data complement those shown in Fig. 6C.
(E) Aβ42-synthesis in dissociated cultures of mouse cortical neurons and glia is decreased by DLK inhibition using MBIP overexpression (left panel), and to interference with the AP-1 binding site in the mouse App-gene promoter by CRISPRi (App AP-1; right panel). Cultures were infected at DIV4 with lentiviruses expressing control proteins or MBIP (left panel) or the control or test CRISPRi components (right panel), and Aβ42 levels were measured by ELISA in the medium at DIV12.
(F) Expression of sgRNAs and dCas9 is efficient in CRISPRi experiments of dissociated cultures of mouse hippocampal neurons and glia (Fig. 6). Expression of mCherry (co-expressed with the control or App AP-1 site sgRNA) and BFP-tagged dCas9 (recognized by GFP antibodies) was analyzed by immunoblotting (representative blots are shown in Fig. 6F). Cultures were infected with lentiviruses expressing dCas9 and sgRNAs (Ctrl. or App AP-1) at DIV4 and harvested at DIV14 for quantitative immunoblotting of APP, mCherry and BFP-dCas9, with Tuj1 as an internal loading control.
(G) Inhibition of AP-1-mediated App transcription does not change Aβ40/Aβ42 ratio based on the analysis of ELISA results from Fig. 6G.
(H) Illustration of high-efficiency in vivo AAV transduction in mouse brain by stereotactic injections into newborn mice, and analysis at P7-9. The types of AAVs injected are listed on the left, and a representative fluorescence image of the mouse brain at P9 viewed under illumination with blue light (440-460 nm) is shown on the right to demonstrate the broad region of infection. Fluorescence is derived from the EGFP co-expressed by the AAVs.
(I) Representative immunoblots complementing the protein expression experiments described in Fig. 7B and 7C.
(J) Quantifications of APP mRNA levels in brain after in vivo application of DN-cFos or CRISPRi to the AP-1 binding sequence in the mouse App promoter. Samples analyzed for protein levels in Figure 7A-7C were used for quantitative RT-PCR; levels are normalized for Tuji1 (tubulin-β3) used as an internal standard.
Data are presented as means ± SEM; n≥3 independent experiments for all bar graphs; statistical significance (*, p<0.05, **, p<0.01; ***, p<0.001) was evaluated by one-way ANOVA and comparing test conditions to control with Tukey’s post-hoc multiple comparisons (A-E) and Student’s t-test (G-I). Non-significant comparisons are not identified.