Abstract
Inhibition of bromodomain and extraterminal motif (BET) proteins such as BRD4 bears great promise for cancer treatment and its efficacy has been frequently attributed to Myc downregulation. Here, we use B-cell tumors as a model to address the mechanism of action of JQ1, a widely used BET inhibitor. Although JQ1 led to widespread eviction of BRD4 from chromatin, its effect on gene transcription was limited to a restricted set of genes. This was unlinked to Myc downregulation or its chromatin association. Yet, JQ1-sensitive genes were enriched for Myc and E2F targets, were expressed at high levels, and showed high promoter occupancy by RNAPol2, BRD4, Myc and E2F. Their marked decrease in transcriptional elongation upon JQ1 treatment, indicated that BRD4-dependent promoter clearance was rate limiting for transcription. At JQ1-insensitive genes the drop in transcriptional elongation still occurred, but was compensated by enhanced RNAPol2 recruitment. Similar results were obtained with other inhibitors of transcriptional elongation. Thus, the selective transcriptional effects following JQ1 treatment are linked to the inability of JQ1-sensitive genes to sustain compensatory RNAPol2 recruitment to promoters. These observations highlight the role of BET proteins in supporting transcriptional elongation and rationalize how a general suppression of elongation may selectively affects transcription.
Introduction
The c-Myc gene encodes for a basic helix-loop-helix leucine zipper transcription factor that pleiotropically regulates the expression of genes linked to cell cycle, cell growth and cellular metabolism.1 In normal cells, the expression of c-Myc is tightly regulated by upstream mitogenic signals to ensure time- and context-dependent transcriptional activation and prevent unscheduled cellular proliferation.2 The c-Myc proto-oncogene is frequently deregulated in hematological cancers following chromosomal rearrangements leading to its constitutive overexpression.3, 4, 5 In solid tumors, c-Myc and its paralogues are found amplified or upregulated by upstream oncogenic lesions activating the WNT, RAS and Notch pathways.6 Upregulation of Myc in tumors supports the high proliferative and metabolic activity of cancer cells leading to their addiction and reliance on continuous Myc expression for their proliferation and survival.7, 8, 9, 10 As the c-Myc protein, as a transcription factor, is resilient to small molecule inhibition, several alternative venues have been explored in order to target its activity and expression in cancer cells. One of the most promising approaches comes from the use of chemical inhibitors of BRD4,11 a chromatin reader that acts as a positive regulator of transcription. BRD4 belongs to the bromodomain and extraterminal motif (BET) family of bromodomain containing proteins, which also includes BRD2, BRD3 and BRDT. These proteins are characterized by two N-terminal bromodomains (BRD), which mediate the binding to acetylated chromatin12 and one extraterminal domain (ET), which is required for protein–protein interactions.13 The use of competitive inhibitors such as JQ1, designed to target the bromodomain binding pocket,14, 15 has demonstrated efficacy and selectivity in targeting tumor cells, particularly in hematological tumors where their efficacy was linked to Myc downregulation.11, 15, 16, 17 Indeed, in multiple myelomas bearing chromosomal rearrangements that bring the coding region of c-Myc under the transcriptional control of the IgH locus, BRD4 inhibition leads to the selective eviction of BRD4 from the IgH enhancers, thus shutting off the expression of the translocated c-Myc.17 Similarly, BRD4 inhibition in myeloid leukemia specifically impairs Myc-deregulated expression orchestrated by the MLL/AF9 fusion protein.15
Here, we follow-up on these observations and investigate the mechanism underlying the efficacy of BET inhibitors in Myc-driven tumors by carrying out a detailed analysis based on genome-wide mRNA expression and ChIPseq experiments. We provide evidences that Myc activity can be targeted by BRD4 inhibitors even in the absence of either its downregulation or its eviction from chromatin. BRD4 inhibition, despite broadly targeting transcriptional elongation, results in defined transcriptional changes affecting a subset of expressed cellular genes. These genes are characterized by high levels of promoter-associated chromatin marks, such as H3K4me3 and H3K27Ac, which pair with strong enrichment of promoter-associated RNAPol2, BRD4 and transcription factors such as Myc and E2F.
This is linked to the high expression level of such genes, reflecting a general strategy to support robust gene expression by maximizing the recruitment of transcription factors and RNAPol2 on promoters. This efficient recruitment of positive transcription factors represents a liability that makes the expression of such genes ‘limited' by BRD4-dependent promoter clearance. Indeed, upon BRD4 inhibition, although the majority of expressed genes can compensate for the drop in transcriptional elongation by enhancing the recruitment of RNAPol2 to their promoters, JQ1-sensitive genes cannot, consequently their expression levels will markedly decrease. Our results highlight how the targeting of a housekeeping cellular function such as transcriptional elongation may result in the selective alteration of defined transcriptional programs. These observations provide a strong rationale for the pharmacological targeting of transcriptional elongation to selectively eradicate cancer cells.
Materials and methods
Cell culture
Burkitt's lymphoma (BL-2, BL-28, DAUDI, P3HR1, RAJI and RAMOS) and acute myeloid leukemia (MV4.11 and THP.1) cell lines were purchased from ATCC (Manassas, VA, USA). The multiple myeloma cell lines were kindly provided by Dr G. Tonon. The Eμ-Myc lymphomas were derived from Eμ-Myc mice.18 Eμ-Myc lymphoma cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) and Iscove's Modified Dulbecco's Medium (ratio 1:1) supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 1% penicillin/streptomycin, 25 μm β-mercaptoethanol, 1% non-essential amino acids. Murine embryonic fibroblasts (MEFs) were derived from 13.5 day post-coitum C57/BL6 or MycER knock-in embryos.19 Burkitt's lymphoma (BL), acute myeloid leukemia (AML) and multiple myeloma (MM) cell lines were cultured in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine and 1% penicillin/streptomycin. MEFs were cultures with DMEM medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 1% penicillin/streptomycin, 25 μm β-mercaptoethanol, 1% non-essential amino acids. All the cells were grown at 37 °C and 5% CO2, except for MEFs that were grown at 37 °C in low oxygen.
Antibodies and primers
A full list of antibodies and primers used in this work is provided as Supplementary Table 2.
Chemicals
Chemicals used: PHA-767491 (Cat no. 217707, Calbiochem, San Diego, CA, USA) and 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB, cat. no D1916, Sigma-Aldrich S.r.l., St Louis, MO, USA) were used to inhibit CDK9. JQ1 was kindly provided by Dr J Bradner.
Cell transfection, viral production and infection
Viral particles were produced as previously described.20 BL cells were infected using spin infection protocol. Briefly, 2 × 106 cells were resuspended in 2 ml of viral supernantant supplemented with 8 μg/ml of polybrene. The cells were spun at 1800 r.p.m. for 1.5 h and then grown at 37 °C for 3 h. The medium was replaced with 2 ml of fresh medium for an overnight recovery. Twenty-four hours post infection, cells were selected with 2.5 μg/ml of puromycin. When doxycycline-inducible vectors were used, transfection, virus production and cell culture were performed using medium supplemented with 10% of fetal bovine serum Tet-free. Induction was performed with 2 μg/μl doxycycline.
Plasmids
LT3GEPIR shREN and RT3GEN shBRD4 were kindly provided by Dr J Zuber.21 LT3GEPIR shBRD4 (602-1817-1838) vectors were obtained subcloning small hairpin RNA (shRNA) targeting BRD4 from RT3GEN to LT3GEPIR, using XhoI and EcoRI restriction enzymes.
Cell growth assay
The cell growth was measured using the CellTiterGlo Luminescent Cell Viability Assay (Promega, Fitchburg, WI, USA). For cells growing in suspension (BL, AML, MM and Eμ-Myc lymphoma cells) 250 000 cells per ml in a total volume of 4 ml were cultured in 6-well plate in presence of the indicated drugs or vehicle dimethyl sulfoxide (DMSO). The assay was performed in triplicate every 24 h using 100 μl of cell suspension and 100 μl of CellTiterGlo. The luminescence was read in a white 96-well plate using a multiwell plate reader (Glomax, Promega). For adherent cells (MEFs), 500 cells were plated in each well of a white 96-well plate, with a total volume of 100 μl. Each condition was plated in triplicate and the luminescence was read after the addition of 100 μl of CellTiterGlo using a multiwell plate reader (Glomax, Promega).
Cell cycle and dead cell discrimination analysis
The cell cycle progression was analyzed by BromodeoxyUridine (BrdU) incorporation. Overall 250 000 cells per ml of BL, AML, MM or Eμ-Myc lymphomas were cultured in a total volume of 15 ml in presence of DMSO or JQ1 (100 nm for BL, AML and MM and 50 nm for Eμ-Myc lymphomas) for 24 h. BrdU (33 μM) was added to the culture 20 min before collecting. Cells were collected and processed as described.20 To assess viability, live cells were washed once with 1 ml of 1% bovine serum albumin. Cells were incubated in the presence of propidium iodide (50 μg/ml in phosphate-buffered saline (PBS)) for 5 min at room temperature and then analyzed by FACS (fluorescence-activated cell sorting).
Western blot
For western blot analysis, 250 000 cells per ml of BL, AML, MM and Eμ-Myc lymphoma cells were cultured in a total volume of 20 ml. Twenty-four hours after plating, different concentrations of JQ1 (0, 50, 100, 250 and 500 nm) were added to the culture for either 6 or 24 h. Cells were collected, washed once in PBS and lysed for 10 min on ice in an adequate volume of lysis buffer (20 mm HEPES pH 7.5, 100 mm NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100) supplemented with MINI-complete Protease Inhibitor Cocktail Tablets (Roche, Indianapolis, IN, USA) and phosphatase inhibition (0.4 mm ortovanadate, 10 mm NaF). The cell lysate was sonicated with Branson sonicator and cleared by centrifugation at full speed at 4 °C. Proteins were quantified by Bradford assay. Proteins (20–30 μg) were boiled at 95 °C with Laemmli sample buffer and loaded on Mini-PROTEAN TGX Gel (Bio-Rad, Hercules, CA, USA). Trans-Blot Turbo Transfer System (Bio-Rad) was used to transfer proteins to Trans-Blot Turbo Nitrocellulose Transfer Packs (Bio-Rad). Blocking was performed with Tris-buffered saline (TBS)+5% of non-fat milk or with TBS+5% of bovine serum albumin. Primary antibody was incubated overnight at 4 °C, whereas secondary antibody was incubated for 1 h at room temperature. The western blots were developed with ECL (Amsharm) using the ChemiDoc System (Bio-Rad).
RNA extraction and expression quantification
For expression analysis, 250 000 cells per ml of BL, AML, MM and Eμ-Myc lymphoma cells (total volume of 20 ml) or 500 000 cells per 10 cm plate of MEFs were treated, 24 h after plating, with different concentrations of JQ1 (0, 50, 100, 250 and 500 nM) for either 6 or 24 h. Cells were collected and washed once in PBS. RNA was extracted using RNeasy columns (Qiagen, Hilden, Germany) performing on-column DNA digestion with DNase (Qiagen). 1 μg of RNA was retrotranscribed using the ImPromII kit (Promega) according to the manufacture's instruction. cDNA (10 ng) were used to perform real-time qPCR using FAST SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA).
RNA for Microarray assay was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) from 107 RAJI cells (250 000 cells per ml) treated, 24 h after the plating, with DMSO or 100 nm of JQ1 for additional 24 h. Total RNA was treated with TurboDNase (Ambion, Waltham, MA, USA) and processed for oligonucleotide microarray profile through Affymetrix Human Gene 1.0 ST arrays platform. Nanostring assay was performed using a codeset containing probes for known genes deregulated by Myc.23 Briefly, 107 cells (250 000 cells per ml) of Eμ-Myc lymphoma (ly9644, ly27805 and ly28514) were cultured, for 24 h and then treated with either DMSO or 50 nm JQ1 for an additional 24 h. Total RNA was extracted using TRIZOL reagent (Invitrogen) according to manufacturer's instructions and DNA digestion was performed using TurboDNase (Ambion). Overall 100 ng of total RNA was used to proceed with the probe hybridization according to manufacturer's instructions.
4-Thiouridine labeling
4-Thiouridine (4-sU) labeling was performed as previously described23 with minor modifications. RAJI (300 000 cells per ml) were cultured in 100 ml of complete medium. Twenty-four hours after plating, cells were treated with either vehicle (DMSO) or JQ1 (100 nm) for 24 h. A pulse of 30 min of 4-sU (300 μm) was performed. After collecting the cells, RNA was extracted with the Qiagen miRNeasy kit according to the manufacturer's instructions and DNase I digestion was performed. Around 40 μg in 100 μl of RNase-free water of total RNA were used for the biotinylation reaction (2 h at 25 °C) with 100 μl of biotinylation buffer (2.53 stock: 25 mM Tris pH 7.4, 2.5 mm EDTA) and 50 μl of EZ-link biotin-HPDP (1 mg/ml in DMF; Pierce/Thermo Scientific 21341). RNA was precipitated and unbound biotin-HPDP was removed by a combination of chloroform/isoamyl alcohol (24:1) precipitation with purification using MaXtract high density tubes from Qiagen. Biotinylated RNA was purified using Dynabeads MyOne Streptavidin T1 (Invitrogen). 50 μL of beads were first washed (twice in washing buffer A (100 mm NaOH, 50 mm NaCl) and once in washing buffer B (100 mm NaCl)) and then resuspended in 100 μl of buffer C (2 m NaCl, 10 mm Tris pH 7.5, 1 mm EDTA, 0.1% Tween-20) to a final concentration of 5 μg/μl. RNA was added in an equal volume to beads and rotated at room temperature for 15 min. Beads were washed three times with washing buffer C. RNA was eluted from the beads in 100 μl of 10 mM EDTA in 95% formamide (65 °C, 10 min). RNA was extracted with the RNeasy MinElute Spin columns from Qiagen according to the manufacturer and eluted in 14 μl of RNase-free water. RNA was retrotranscribed with SuperScript VILO cDNA Synthesis Kit, according to manufacturer's instruction. Real-time qPCR was performed using FAST SYBR Green Master Mix (Applied Biosystems).
Chromatin immunoprecipitation
BL, MM cells (250 000 cells per ml) were plated and DMSO or JQ1 (100 nm for the cell lines, 50 nm for Eμ-Myc lymphomas) were added 24 h after the initial plating. After 24 h of drug treatment, cells were counted and washed once with PBS. Cells (108) were resuspended in 10 ml PBS and fixed. For Myc, Histone Marks, RNA PolII and E2F1 ChIP, cells were fixed using formaldehyde (final concentration of 1%), for BRD4 ChIP cells were fixed using glutaraldehyde (final concentration 1%). The fixation step was carried out at room temperature for 10 min and quenched with 0.125 m glycine for 5 min at room temperature. Cells were washed twice with PBS and resuspended in LB1 buffer (50 mm HEPES pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% Glycerol, 0.5% NP-40, 0.25% Triton X-100) for 10 min on ice. After centrifugation, nuclei were extracted resuspending cells at room temperature for 10 min in LB2 buffer (10 mm Tris-HCl pH 8, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA). The extracted nuclei were finally resuspended in LB3 buffer (10 mm Tris-HCl pH 8, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% Na-Deoxycholate, 0.5% N-lauroylsarcosine) and sonicated in order to obtain DNA fragments of 300–100 bp. For BRD4, Myc, E2F1, total RNAPol2 and RNAPol2-S5p ChIP, the lysate from 50 × 106 cells was incubated with 10 μg of antibody previously bound to protein G Dynabeads (Invitrogen) in PBS+0.5% bovine serum albumin. For Histone Marks ChIP, the DNA from 20 × 106 cells was incubated with 5 μg of the antibody previously bound to protein G Dynabeads (Invitrogen) in PBS+0.5% bovine serum albumin. For RNAPol2-S2p ChIP, the lysate corresponding to 10 × 107 cells was incubated with 60 μl of hybridoma overnight on a rotating wheel at +4 °C. After the incubation with the antibody, beads were collected using the DynaMag magnet, washed six times with 1 ml of RIPA buffer (50 mm HEPES pH 7.5, 500 mm LiCl, 1 mm EDTA, 1% NP-40, 0.7% Na-Deoxycholate) and once with 1 ml of TE 1X+50 mm NaCl. For cells fixed with formaldehyde, de-crosslinking was performed overnight at 65 °C with 150 μl of TE 1X+2% SDS. For cells fixed with glutaraldehyde, de-crosslinking was performed with 150 μl of TE+1% SDS+ 100 mm NaHCO3. Samples were first treated for 1 h with RNaseA at 37 °C, then Proteinase K was added and the de-crosslink reaction was incubated overnight at 65 °C. DNA was purified with PCR Qiaquick columns (Qiagen) and quantified using PicoGreen (Invitrogen) or QUBIT (Invitrogen). For ChIPqPCR, 1 μl of purified was used to perform real-time PCR using FAST SYBR Green Master Mix (Applied Biosystems).
Publicly available data sets analyzed in this work
Data were retrieved from GEO database. MM.1S cell line: GSE31365 (expression data), GSE42355 (ChIPseq data relative to BRD4, RNAPol2, Cdk9 and MED1), GSE42161 (Myc ChIPseq), GSE43743 (RNAPol2 ChIPseq in CDK9i treated cells). OCLY cell line: GSE45630 (expression data) and GSE46663 (ChIPeq).
NGS data filtering and quality assessment
ChIPseq and RNA-Seq reads sequenced with the Illumina HiSeq2000 were filtered using the fastq_masker (setting the options to -Q33 -q 20 -r -N -v -i) and, for ChIPseq reads, also with fastq_quality_trimmer (setting the options to -Q33 -t 20 -l 10 –v -i). These tools are part of the FASTX-Toolkit suite (http://hannonlab.cshl.edu/fastx_toolkit/). Their quality was evaluated and confirmed using the FastQC application (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Analysis of ChIPseq and RNA-Seq data
ChIPseq NGS reads were aligned with the BWA tool.24 Alignment was performed with BWA-MEM and with default settings, using hg19 genome assembly for OCLY and Raji cells and hg18 for MM1.S cells. Peaks were called with the MACS v1.4 software.25 Peaks' P-value threshold was set to 10−9 for MM1.S data and 10−8 for RAJI and OCLY cells, using the R script ‘filterpeaks.R'. FDR (false discovery rate), determined as the ratio between the negative and the positive peaks, was set to 5% for all the data. Negative peaks were found by MACS on the input samples, using the ChIP as reference. Normalized reads count within a genomic region was determined as the number of reads per million of library aligned reads (r.p.m.), that were subtracted by the input normalized reads, ‘compEpiTools' bioconductor R package.26 Peak read density (reads per million of reads per base pair) for a particular region was determined as the ratio between the normalized reads count and the length of the region in base pair. Random Forest is a supervised machine learning algorithm that predicts an output variable from a set of input features. As input features we used gene expression levels in DMSO-treated cells, enrichments of transcription factors and histone marks on the TSSs of the genes. Gene downregulation (that is, JQ1 sensitivity) was set as the output variable. The analysis was carried out using ‘randomForest' R package.27
Definition of promoter, intragenic and intergenic regions and superenhancers
In order to assess if a specific ChIPseq peak mapped to a promoter, a gene body or to an intergenic region, the following criteria were applied: regions that overlap with at least one bp with any promoter (defined as genomic region (−2000; +1000) bp spanning TSSs, transcription start sites), were considered as belonging to promoters; regions that were not promoters but had at least 1 bp overlapping with any gene body were considered intragenic. The remaining regions (that did not overlap either with promoters or gene bodies) were considered intergenic. Annotations were performed with the R annotation packages TxDb.Hsapiens.UCSC.hg19.knownGene for OCLY and RAJI cells and TxDb.Hsapiens.UCSC.hg18.knownGene for MM1.S cells of Bioconductor. Superenhancers were called according to Lovèn et al.28 using BRD4 as factor of interest. Briefly, BRD4 peaks that were close to each other within a distance of 12.5 kb were merged together; they were then ranked according to BRD4 r.p.m. Stitched peaks above the inflection point of the curve where BRD4 peaks where ranked by their enrichment (Supplementary Figure 8) were defined superenhancers (SE). For in silico association of SE and enhancers (E) to neighboring genes, promoters were classified as active if both RNAPol2, BRD4, H3K4me3 and H3K27Ac peaks (in untreated samples) were found within a window of ±5000 bp from annotated TSSs. BRD4-bound enhancers were defined as those stitched BRD4 peaks, not defined as SE, that did not have any overlap with an active promoter. An active gene was considered associated to SE or E if its TSS was within a 50 kb window from their boundaries.
RNAPol2 stalling index
The RNA polymerase II stalling index (SI, also called elongation rate)29 was calculated as SI=Prom/GB; prom refers to the read counts on the promoter (TSS±300 bp interval) and GB to the read counts in the gene body (the interval between TSS +301 and 3,000 bp after the TSS). These values were normalized both to library size (total number of reads) and to the length of the interval, and only genes with GB>600 and with a RNAPol2 ChIPseq peak in the promoter region were considered. RNAPol2 signal in gene bodies was plotted using the same criteria that were used for SI calculation; genes were expanded by 20% upstream and 20% downstream and then divided into 150 bins, for which the input-subtracted reads were counted. Reads were normalized both for library size and gene length, using ‘GRcoverageInbins' function of compEpiTools R package.26
Analysis of microarray data
Microarray raw data (CEL files) were analyzed with Genespring GX 11 with RMA with probe level summarization. Raw data were normalized for the median of the expression between the six samples. P-values were calculated with t-test and adjusted with BH (Benjamini–Hochberg) multiple testing correction. The first quartile of values distribution was eliminated and a threshold of Log2 Fold Change <−0.5 or Log2 Fold Change >0.5 was used to define downregulated or upregulated genes, respectively.
Results
Cell growth inhibition by JQ1 is independent of Myc downregulation
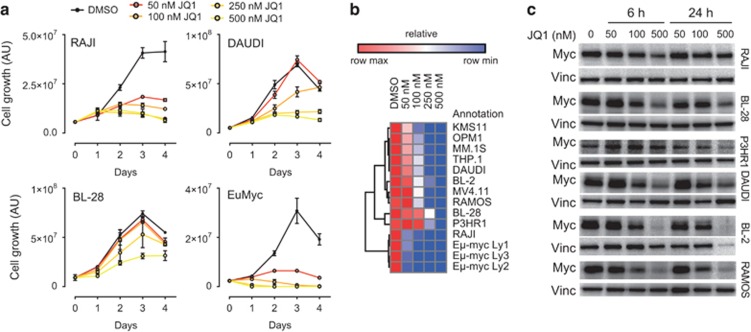
To gain additional insight into the mechanism of action of JQ1 and its relation to Myc expression, we focused our attention on B-cell lymphomas bearing chromosomal translocations involving the c-Myc locus. We selected a panel of Burkitt's lymphomas lines (BL), as human model and Eμ-Myc primary lymphomas as mouse models of poorly differentiated B-cell lymphomas carrying a IgH-Myc chromosomal rearrangement.30 We assessed the sensitivity of BL and Eμ-Myc lymphomas cells to BET inhibition by evaluating cell growth in samples treated with increasing doses of JQ1, ranging from 50 to 500 nM (Figures 1a and b; Supplementary Figures 1 and 2a). As positive controls, we used MM (MM.1 S, OPM1, KMS11) and AML cell lines (MV4.11, THP.1) for which the sensitivity to BET inhibitors has already been reported11, 17 (Supplementary Figures 3a and d). All the BL cell lines (BL-2, BL-28, DAUDI, P3HR1, RAJI and RAMOS) and Eμ-Myc lymphomas tested were responsive to BET inhibition, showing growth arrest in a time- and dose-dependent manner (Figure 1a; Supplementary Figures 1a and 2a). RAJI cells and Eμ-Myc lymphomas were among the most sensitive, showing a marked decrease in cell growth already after 48 h of treatment, at relatively low doses of JQ1 (100 nm and 50 nm, respectively; Figure 1b; Supplementary Figures 1 and 2). We next addressed whether JQ1 treatment would affect Myc levels. As expected, JQ1 effectively downregulated Myc in AML and MM cell lines, with a maximum effect achieved at 500 nm (Supplementary Figures 3b–f). Instead, pharmacological treatment of Burkitt's cell lines gave a composite response in terms of Myc downregulation with RAMOS, DAUDI and BL-2 cells showing a clear dose-dependent decrease of mRNA (Supplementary Figure 1b) and protein levels (Figure 1d), while in P3H1R, BL-28 and RAJI cells, Myc protein and mRNA were downregulated only at the highest concentration of JQ1 (Figure 1c; Supplementary Figure 1b). RAJI cells were already sensitive to JQ1 at low concentrations, at which Myc levels were unaffected (Figures 1a–c). Similarly, in Eμ-Myc lymphomas, the growth inhibitory effect of JQ1 did not associate with Myc downregulation (Supplementary Figures 2b and c). Thus, the anti-proliferative effect of JQ1 was independent from its ability to downregulate Myc.
Figure 1.
BET inhibition is cytostatic in B-cell lymphomas. (a) Cell growth analysis of Burkitt's lymphoma cell lines (RAJI, DAUDI and BL-28) and a representative primary Eμ-Myc lymphoma grown in vitro in the presence of increasing concentrations of JQ1. For each time point, the mean and the s.d. of three technical replicates is reported. (b) Heatmap reporting the relative cell growth of different cell lines exposed to increasing concentration of JQ1 for 48 h. (c) Western blotting analysis of c-Myc level assessed in different BL cell lines at 6 and 24 h post JQ1 administration. Vinculin (vin) was used as a loading control.
BET inhibition affects Myc and E2F-dependent transcriptional programs
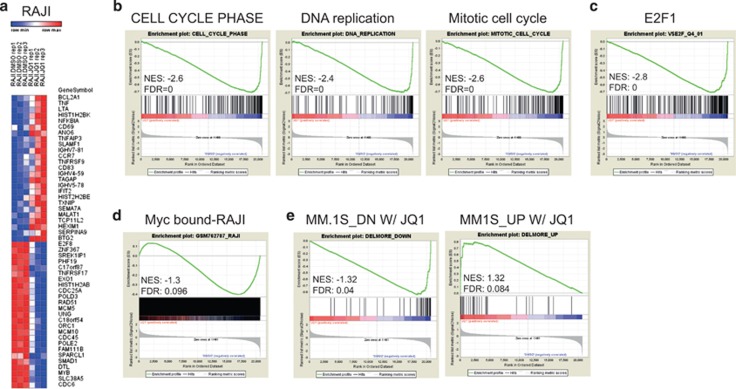
As BET inhibition has been frequently associated to either Myc downregulation11, 16, 17, 31, 32 or selective inhibition of its transcriptional programs,33, 34 we asked whether in those cell lines that showed sensitivity to JQ1 in the absence of Myc downregulation, JQ1 could act by regulating Myc activity rather than its expression. We first assessed the expression levels of selected Myc target genes in BL lines treated with increasing concentrations of JQ1. In all the cell lines analyzed, we observed a dose-dependent inhibition of the expression of NCL and IFRD2, two well-characterized Myc target genes23 (Supplementary Figure 4a). The effect of JQ1 was rather rapid as downregulation of Myc target genes could be appreciated already at 6 h after the addition of JQ1, thus suggesting a direct transcriptional effect. RAJI, P3HR1 and BL-28 showed downregulation of NCL and IRFD2 already at 100 nm, a concentration of JQ1 that did not affect Myc levels in these cell lines (Figure 1c; Supplementary Figure 4a). Next, we performed a genome-wide transcriptional analysis in RAJI cells, chosen as a paradigm for BL lines that showed sensitivity to JQ1 in the absence of Myc downregulation. Microarray analysis resulted in the identification of 1498 differentially expressed genes (DEGs; Figure 2a; Supplementary Table 1): 1017 genes were downregulated by JQ1 (68% of all the DEGs) while 481 genes were upregulated (32% of all the DEGs). The expression of selected genes was verified by RT-qPCR (Supplementary Figure 4b). In line with the observed cytostatic effect of JQ1 (Supplementary Figure 5), downregulated genes were enriched in genes linked to cell cycle control, cell cycle progression and DNA replication (Figure 2b). Accordingly, these genes had promoters enriched for transcription factors binding motifs recognized by E2F1 (Figure 2c). DEG-up genes were less defined from an ontological perspective and, as noted by others,35 contained genes such as HEXIM1 which may represent compensatory transcriptional responses (not shown). GSEA revealed a clear enrichment of Myc bound genes36 in DEG-down (51% were Myc bound) whereas only a slight enrichment in Myc bound genes was noted for the upregulated (24% were Myc bound; Figure 2d). As the ontology of DEGs in RAJI was reminiscent of the ontological annotation of all the differentially expressed genes identified upon BET inhibition in MM cell lines, we also performed GSEA using as a gene set the differentially expressed genes identified in MM.1 S upon JQ1 treatment.17 A significant enrichment score was measured for either up- and down-regulated genes. Thus, although JQ1 treatment in different cell lines may lead to a different outcome in terms of Myc regulation, the downstream transcriptional programs affected by JQ1 were similar (Figure 2e). Similar results were observed when shRNAs targeting BRD4 were used in RAJI cells (Supplementary Figures 6a and b): not only silencing BRD4 had a marked cytostatic effect (Supplementary Figure 6c), but also led to the selective decrease in mRNA levels of KIF2C and MCM2, two DEG-down genes (Supplementary Figure 6d), whereas the expression of either c-Myc or RPL36 (a No-DEG gene) was unaltered (Supplementary Figures 6e and f), thus suggesting that BRD4 is a prominent target of JQ1 in this cell line. We also profiled three independent Eμ-Myc lymphomas for the expression of a subset Myc target genes that have been previously identified as being bound and regulated by Myc in B-cells isolated from Eμ-Myc mice:23 virtually all of the 80 Myc target genes showed significant downregulation upon JQ1 treatment (Supplementary Figure 4c). Thus, despite the lack of Myc downregulation in RAJI and in Eμ-myc lymphomas following JQ1 treatment, alterations at the transcriptional level were associated with low expression of Myc target genes suggesting that BET inhibition may selectively affect Myc activity.
Figure 2.
BET inhibition leads to selective transcriptional alterations. (a) Heatmap of the relative fold change of the top 50 deregulated genes determined by microarray analysis in RAJI cells treated with 100 nm JQ1 for 24 h. (b–e) GSEA plots of differentially expressed genes identified in RAJI cells, following JQ1 treatment (100 nm for 24 h)
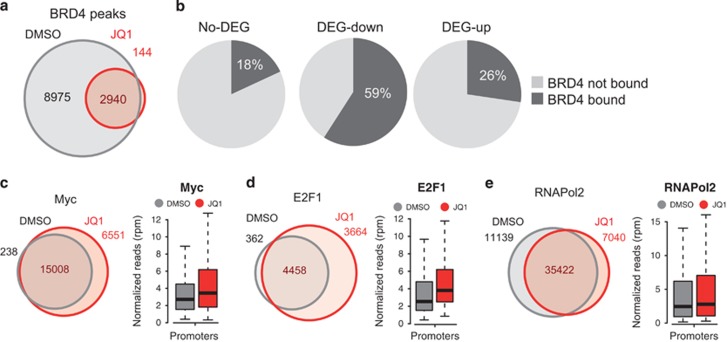
JQ1 causes BRD4 eviction without affecting Myc or E2F binding to chromatin
The observation that JQ1 administration affected the expression of Myc target genes without altering Myc expression lead us to evaluate whether the effect of JQ1 was mediated by selective eviction of BRD4 from specific genomic loci (that is, Myc target genes). We profiled BRD4 genome-wide chromatin association by ChIPseq and identified a total of 11915 BRD4 ChIP peaks in vehicle treated RAJI (Figure 3a), 36% of which were proximal to an annotated promoter (Supplementary Figures 7a and b). BRD4 inhibition led to its widespread eviction from chromatin, with only 3084 peaks detected in cells treated with JQ1 (Figure 3a). Both promoter associated and the intergenic/intragenic peaks were equally reduced in number and enrichment (Supplementary Figure 7b). Thus our genome-wide analysis did not show any evidence for selective eviction of BRD4 from a subset of defined genomic loci, but rather a widespread loss of chromatin associated BRD4. We then focused on the genes that were differentially expressed upon JQ1 treatment, in order to evaluate their association with the ChIPseq data sets. Around 60% of the DEG-down genes had a promoter-associated BRD4 peak, whereas only 18% of the genes not showing relevant expression changes had a proximal BRD4 peak (Figure 3b). There was also a substantial fraction of the DEG-up genes with BRD4 bound at their promoter (Figure 3b). Thus despite the presence of BRD4 on gene promoters was not predictive of the transcriptional response, there was a good association between binding of BRD4 and differential expression. We next mapped SEs, given their relevance in the control of oncogenic programs.11, 34, 37, 38, 39, 40 Based on the BRD4-ChIPseq, we identified 269 SEs in RAJI (Supplementary Figure 8a). These genomic regions were highly enriched for enhancer activation marks as H3K27Ac, which was greater than the enrichment found on promoters or regular enhancers (Supplementary Figure 8b). SEs were also characterized by a high H3K4me1/H3K4me3 ratio a feature typical of enhancers (Supplementary Figure 8c). Annotation of genes associated to SE revealed a slight enrichment for genes affected by JQ1, with 50 genes, out of the 1017 DEGs, found proximal to SE (Supplementary Figure 8d).
Figure 3.
ChIPseq analyses in RAJI cells. RAJI cells were treated with 100 nm JQ1 (gray) for 24 h or mock treated (red). (a) Venn diagram of the number of BRD4 peaks identified in mock-treated or JQ1-treated RAJI cells. (b) Pie charts showing the fraction of differentially expressed genes bound at their promoter by BRD4. (c) ChIPseq analysis of c-Myc. Left: Venn diagram reporting the number of c-Myc peaks identified in mock treated (gray) or JQ1-treated cells (red). Right: the box plot of relative enrichments of the c-Myc peaks mapped on promoters. (d) ChIPseq analysis of E2F1, as in c. (e) ChIPseq analysis of RNAPol2, as in c.
We next evaluated whether BET inhibition would affect the genome distribution of either Myc or E2F. Unexpectedly, ChIPseq analyses revealed that for both transcription factors ChIPseq peaks there was a slight increase in peak numbers and relative enrichment, with a more pronounced effect on peaks annotated to promoters (Figures 3d and e; Supplementary Figures 7a, c and d). Profiling of RNAPol2 by ChIPseq revealed that although its genome-wide distribution was largely unaffected by JQ1, as evidenced by the large overlap of the peaks detected in either control or JQ1 samples, the enrichment of RNAPol2 peaks associated to promoters increased in a slight but significant way (Figure 3f; Supplementary Figures 7a and e). Thus, the selective transcriptional effect exerted by JQ1, did not depend on either Myc or E2F levels neither was due to their failure to localize on chromatin. In line with this, ectopic overexpression of either Myc (Supplementary Figure 9), E2F1 (Supplementary Figures 10a–c) or both (Supplementary Figures 10d and e) failed to rescue the expression of JQ1-sensitive genes.
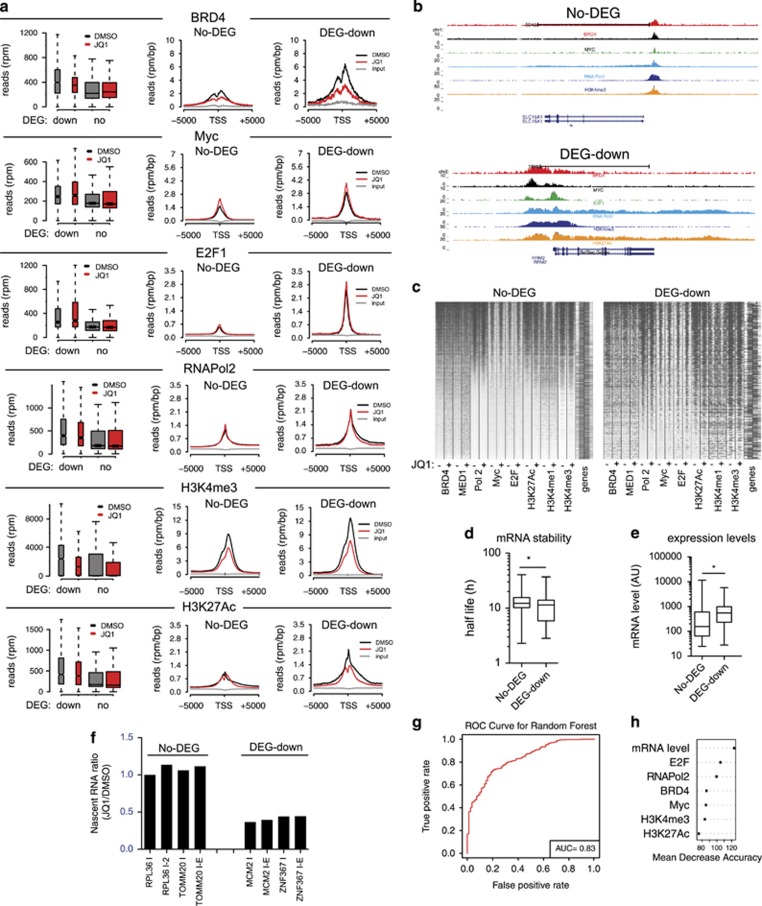
JQ1-sensitive genes are highly expressed and present marked promoter enrichment for RNAPol2 and transcription factors
To gain further insight into the selective transcriptional effect observed upon BRD4 inhibition, we calculated the enrichments of transcription factors and RNAPol2 on promoter of genes that were either unaffected (No-DEG) or downregulated (DEG-down) by JQ1. We observed a marked difference on promoter occupancy of RNAPol2, BRD4, Myc or E2F, with DEG-down genes displaying a robust binding of all these factors (Figures 4a–c). Of note, the higher promoter occupancy associated with high levels of activating chromatin marks such as H3K4me3 and H3K27Ac (Figures 4a–c). This suggested a link between the transcriptional alteration observed upon BET inhibition and promoter occupancy. To verify whether this was a general feature of JQ1-sensitive genes, we analyzed promoter occupancy in MM.1S (a MM cell line) and OCLY (a DLBCL cell line), for which there were publicly available data sets reporting ChIPseq data of cells treated with JQ1.17, 34 Expressed genes were subsetted based on their differential expression upon JQ1 treatment.17, 34 For both cell lines, genes downregulated upon BET inhibition were characterized by higher binding of RNAPol2 and transcription factors (Supplementary Figures 11 and 12), suggesting that this might be a common characteristic of promoters of JQ1-sensitive genes. In order to evaluate whether intrinsic properties of mRNAs may also account for the differential transcriptional responses, we determined whether differential gene expression was associated with general properties of the mRNAs transcribed, such as mRNA levels and relative stability. We failed to observe a clear link between mRNAs stability and their differential expression upon JQ1 treatment, as the two genes subsets, DEG-down and No-DEG, both showed no statistically significant differences in the half-life of their mRNAs (Figure 4d). Instead, a peculiar feature of DEG-down genes was related to their expression levels as genes downregulated by JQ1 were significantly more abundant than No-DEGs (Figure 4e). This feature matched with the promoter composition of these genes that showed high occupancy of RNAPol2, Myc, E2F and BRD4 (Figures 4a–c). Thus, genes downregulated by JQ1 were highly expressed genes with promoters enriched in BRD4, Myc and E2F binding. Nascent mRNA analysis performed by 4-sU labeling confirmed that DEG-down genes were transcriptionally inhibited upon JQ1 treatment, while mRNA synthesis of No-DEGs was unaltered (Figure 4f). Given the association between expression levels and promoter features, we set out to test whether mRNA abundance, transcription factors binding and RNApol2 enrichment would be good predictors for genes that will be differentially expressed upon BRD4 inhibition. We used machine learning techniques based on random forest to evaluate if the features described above would be good classifiers of the differential gene expression observed upon BRD4 inhibition. The ROC curve showed good predicting power of our features (Figure 4g). Ranking our features based on their predictive power revealed that they all contributed significantly to the prediction (Figure 4h).
Figure 4.
Analysis of promoter occupancy in JQ1-sensitive genes. RAJI cells were treated with 100 nm JQ1 or vehicle (DMSO) for 24 h. Genes were subsetted based on their differential expression following JQ1 treatment in DEG-down (genes downregulated) or No-DEG (genes not affected by JQ1). (a) The enrichment of RNAPol2, BRD4, Myc, E2F and selected chromatin marks is reported as a box plot (left) and as cumulative reads distribution (center and right panels) calculated around the TSS. (b) Genome browser views of representative DEG-down or No-DEG genes. (c) Ranked heatmap showing the distribution of transcription factors, RNAPol2 and chromatin marks on the promoter of either DEG-down or No-DEG genes. For each factor the signal relative to mock-treated cells (−) and JQ1-treated cells (+) is shown. (d) Box plot showing the analysis of mRNA stability based on Schwanhäusser et al.63 *P-value <0.05 (Student's t-test). (e) Box plot showing relative expression levels of either DEG-down or No-DEG genes, based on μArray analysis. *P-value <0.05 (Student's t-test). (f) Nascent mRNA analysis of selected DEG-down or No-DEG genes performed by 4-sU labeling. Data are reported as normalized to the values determined for mock-treated cells (g) Receiver operating characteristic (ROC) curve associated to the Random Forest classifier. Area under the curve (AUC) shows that the response to JQ1 treatment (downregulation of a gene) could be predicted using the expression levels and the transcription factors and chromatin marks enrichments on the TSSs. (h) Predictive power of the features used in g. Features were ranked based on their variable importance in the prediction of JQ1 response. All the features contributed significantly to the prediction, with gene expression level, E2F and RNAPol2 enrichments being among the most predictive.
Compensatory RNAPol2 recruitment accounts for selective transcriptional downregulation following BET inhibition
As BRD4 had been previously implicated in the regulation of transcriptional elongation,11, 41, 42, 43, 44 we sought to determine transcriptional dynamics by analyzing RNA polymerase 2 distribution along transcribed genes. Upon BET inhibition, we observed an increase in the RNAPol2 stalling index (that is, the ratio of the enrichment measured at promoter versus the enrichment determined on the gene body) for both DEG-down and No-DEG genes (Supplementary Figure 13a), thus suggesting a general role of BRD4 in regulating RNAPol2 activity. Yet, the increase in stalling index was due to diametrically opposite effects of JQ1 on RNAPol2 distribution in the two gene subsets.
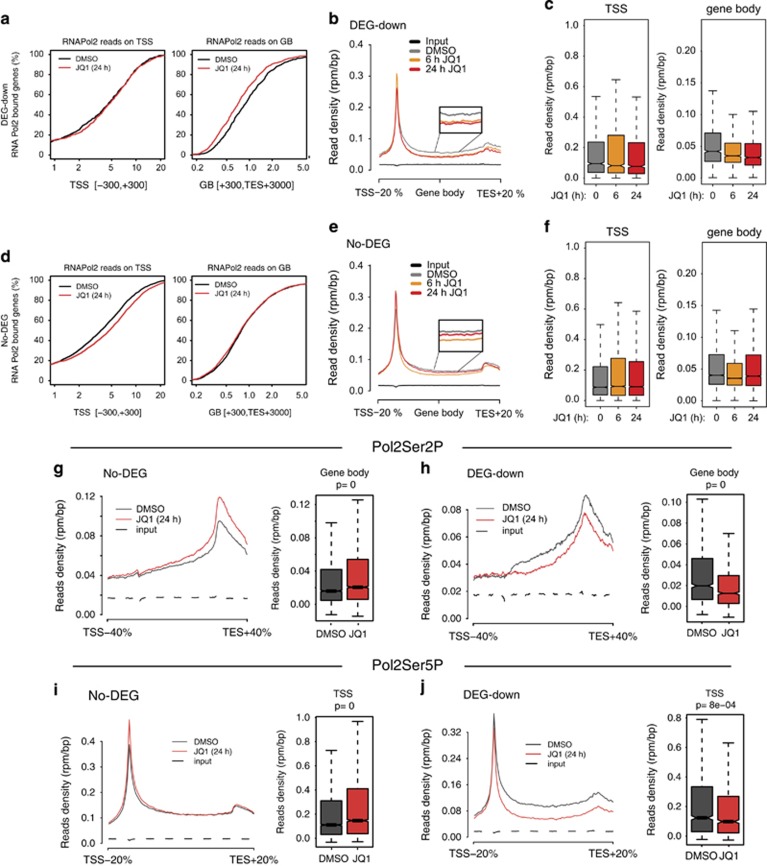
DEG-down genes showed a clear decrease in elongating RNAPol2, shortly after BET inhibition (6 h), which persisted also at longer time points, whereas RNAPol2 occupancy at promoters was unaltered (Figures 5a–c). Accordingly, there was a clear reduction in the elongating form of RNAPol2 (Pol2Ser2P, phosphorylated on Ser2) along gene bodies of DEG-down (Figure 5h; Supplementary Figure 13b) whereas the initiating form of RNAPol2 (Pol2Ser5P, phosphorylated on Ser5) localized at the TSSs, was only slightly reduced (Figure 5j). Thus, while JQ1 treatment clearly affected RNAPol2 elongation in DEG-down genes, at promoter level, these genes did not show any evidence for a compensatory increase in either recruitment or initiation of RNAPol2, thus suggesting that RNAPol2 recruitment was already at near-equilibrium. On the other hand, following BET inhibition, No-DEG genes showed a decrease in elongating RNAPol2 (6 h post-BET inhibition) which was transient and was rescued at longer time points to reach a level comparable to that observed in unchallenged cells. Concomitantly, promoter-associated RNAPol2 increased progressively with time (Figures 5d–f). Profiling of Pol2Ser5P and Pol2Ser2P confirmed the increase in initiating RNAPol2 (Figure 5i; Supplementary Figure 13c) and the rescue of elongating RNAPol2 (Figure 5g; Supplementary Figure 13d) at longer time points. Thus, on No-DEG genes, perturbation of the elongation rate constant (due to inhibitory effect of JQ1) led to a transient increase in the amount of promoter-associated RNAPol2 leading to a new steady state where the increment in promoter-associated RNAPol2, compensated for the lowering of RNAPol2 promoter escape by acting on mass effect (that is, more substrate), thus allowing proficient mRNA transcription. Thus, while No-DEGs can rescue a less efficient elongation by increasing RNAPol2 recruitment and initiation, DEG-down genes, by having maximized the promoter recruitment of RNAPol2 and transcription factors, have little ability to compensate for the drop in elongating RNAPol2. This regulation may reflect the need to support the high expression levels of DEG-down genes with robust transcriptional flux.
Figure 5.
Analysis of RNAPol2 distribution on genes differentially regulated by JQ1. RAJI cells were treated with 100 nm JQ1 (gray) or vehicle (DMSO) for 24 h. Genome-wide distribution of total RNAPol2 (RNAPol2) and its Ser2 (Pol2Ser2P) and Ser5 (Pol2Ser5P) phosphorylated forms were analyzed by ChIPseq. (a, d) Cumulative read count of RNAPol2 localized either on the TSS (transcriptional start site; left panels) or on gene bodies (GB, right panel) of either DEG-down or No-DEG genes. (b, e) Occupancy of RNAPol2 along DEG-down genes (b) or No-DEG genes (e). The insets show an enlarged snapshot of the gene body. (c, f) Box plot showing the read counts at TSS or gene bodies (GB) for either DEG-down genes (c) or No-DEG genes (f) at the indicated times following JQ1 addition. (g, h) Occupancy of Pol2Ser2P along either No-DEG genes (h) or DEG-down genes (g). The box plots showing the read counts on gene bodies are reported on the side of the distribution plots. (i, j) Occupancy of Pol2Ser5P along either No-DEG genes (i) or DEG-down genes (j). The box plots showing the read counts on gene bodies are reported on the side of the distribution plots.
To strengthen our observations, we re-analyzed a published data set describing BRD4 inhibition in MM.1S cells.17 Genes downregulated by JQ1 showed a clear drop in elongating RNAPol2 as evidenced by the reduction in gene body occupancy (Supplementary Figures 14a–d and i), whereas the enrichment of RNAPol2 associated to their promoters was largely unchanged (Supplementary Figures 14a–d). Conversely, No-DEGs showed RNAPol2 promoter stalling with negligible changes in gene body-associated RNAPol2 (Supplementary Figures 14e–h). Similarly to what observed in RAJI, genes downregulated by JQ1 in MM.1S displayed promoters that were strongly enriched for Myc, RNAPol2 and BRD4 binding compared with No-DEGs (Supplementary Figure 11), thus again a selective transcriptional response was associated to genes that displayed features of high promoter occupancy and therefore were intrinsically more susceptible to drops in elongation rates.
Targeting CDK9-dependent elongation selectively affects specific transcriptional programs
Our data suggests that DEG-down genes, due to their scant ability to increase the recruitment of RNAPol2 to promoters, have a limited capacity to compensate gene transcription when drops in transcriptional elongation occur. A consequence of this model will be the prediction that any event leading to impairment of elongation may (i) selectively alter RNApol2 dynamics and (ii) lead to transcriptional alterations that are similar to the one observed upon BRD4 inhibition.
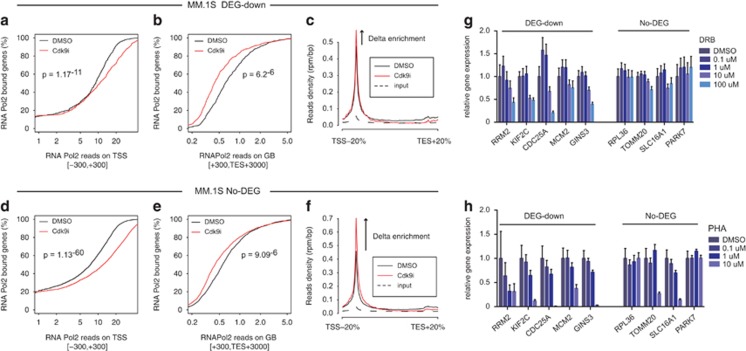
To address the first point, we re-analyzed a published data set reporting RNAPol2 ChIPseq data of MM.1S cells treated with a CDK9i,37 with the aim of verifying whether the DEG-down genes identified upon BRD4 inhibition in MM.1S would show poor promoter stalling also when elongation was impaired by pharmacological inhibition of CDK9. As expected given the high concentration of CDK9i used in this study, we observed a broad effect on the stalling index of either JQ1 DEG-down and JQ1 No-DEG genes (Supplementary Figure 15). This was due to a general reduction in elongation as either JQ1 DEG-down and the No-DEG genes showed a sizable drop in RNAPol2 occupancy on gene bodies (Figures 6b and e). Yet, this decrease in elongation was more pronounced in the JQ1 dependent DEG-down genes as shown in the cumulative graph relative to RNAPol2 enrichment at gene bodies, where the cumulative enrichment of RNAPol2 in CDK9i treated cells (Figure 6b, red curve of the gene body plot) is more right shifted with respect to the control distribution (black line, DMSO), compared to the corresponding curve calculated for No-DEGs (Figure 6e). Accordingly, proximal promoter stalling of RNAPol2 upon Cdk9 inhibition was more pronounced in No-DEGs (Figures 6a, c, d and f), again underscoring their intrinsic flexibility in mounting compensatory responses. This supports the hypothesis that different subsets of cellular genes may be subjected to a differential transcriptional control that depends on either promoter occupancy and efficiency of transcriptional elongation. Highly expressed genes will display highly occupied promoters to support their high transcriptional flux and will be more sensitive to fluctuations in elongation rates (that is, their expression is limited by elongation). This behavior would be relatively independent on the drug used to impair elongation but will be a direct consequence of the degree of gene activity. To experimentally validate this hypothesis, we asked whether CDK9 inhibition in RAJI would selectively affect gene transcription, similarly to what observed following BET inhibition. We treated RAJI cells with increasing doses of two commonly used elongation inhibitors, PHA-767491 and DRB. At each dose tested, we measured the expression of a subset of JQ1 dependent DEG-down and a subset of No-DEGs. At intermediate concentrations of DRB, we observed selective downregulation of the DEG-down genes whereas the No-DEGs were relatively stable (Figure 6g). The relative transcriptional resilience of the No-DEGs was confirmed even at the highest DRB concentration (that is, 100 μm). Similar results were observed with PHA-767491: at intermediate doses DEGs-down expression was generally affected while No-DEGs were relatively insensitive (Figure 6h). Notably, Myc and E2F mRNA levels were affected only at the highest concentration of DRB and PHA-767491, whereas at intermediate concentration, where a selective effect on DEG-down was already observed, their relative levels were comparable to those measured in untreated cells (Supplementary Figures 16a and b).
Figure 6.
Pharmacological inhibition of CDK9-dependent elongation phenocopies BET inhibition. (a–f) ChIPseq analysis of RNAPol2 in MM.1S cells treated with CDK9i, based on published data.37 Genes were subsetted in DEG-down and No-DEG based on their differential gene expression upon JQ1 treatment.17 Cumulative read counts of RNAPol2 on either TSS (a, d) or on gene bodies (GB) (b, e). Occupancy of RNAPol2 along DEG-down genes (c) or No-DEG genes (f) is displayed on the side. (g, h) Expression analysis by RT-qPCR of selected DEG-down and No-DEG genes performed in RAJI treated with either DRB (g) or PHA-767491 (h). Expression values are reported as mean of technical triplicates, normalized to the RPLP0 gene and mock-treated samples.
Discussion
Here, we report an investigation on the mechanism of action of JQ1, a BET inhibitor with potent and broad anti-cancer activity. Although in several blood-borne tumors the efficacy of JQ1 and other BET inhibitors has been ascribed to the selective control of the expression of the Myc oncogene,11, 15, 16, 17 therefore providing a strong rationale for their anti-tumoral activity, we here describe a number of instances where, despite showing robust anti-growth activity, BET inhibition does not lead to Myc downregulation. This was unexpected given that BLs and Eμ-Myc tumors bear rearrangements that have features similar to those reported in MMs, and suggests that the expression of translocated Myc in BLs may depend on different regulatory elements. Indeed, we noticed that although in MMs the IgH enhancers translocated upstream the Myc gene are strongly acetylated and bound by BRD4, in RAJI and other BL cell lines, these enhancers show low levels of BRD4, despite having prominent H3K27Ac (Supplementary Figure 17). Also, despite in RAJI the Myc 5' enhancer and its promoter are bound by BRD4, this binding does not appear to be sensitive to JQ1 (Supplementary Figure 17). These observations may explain why BRD4 inhibition does not affect Myc transcription in some BL tumors (Supplementary Figure 17). Regardless the effect on Myc levels, we noticed that the transcriptional programs altered upon BRD4 inhibition were similar among different hematological tumors, thus suggesting that the selective transcriptional effects observed were not solely due to Myc downregulation but to a more complex effect that led to inhibition of Myc-dependent transcription. Our data shows that BET inhibition targets transcriptional elongation and that BRD4 (and its paralogs) can control elongation on a broad set of genes, as indicated by the large number of promoters bound by BRD4 in our data set, as well as in published ones.17, 45, 46 This is in line with previous data supporting a role for BRD4 in regulating elongation and transcriptional activation mediated by either promoters and/or promoter/enhancer activation.41, 42, 43, 47 Despite this pervasive interaction at promoters of expressed genes, the consequences of BET inhibition on transcription are not global but restricted to a subset of genes that show defined characteristics: these genes are expressed at high levels, have promoters highly enriched for chromatin marks associated with gene activation (that is, H3K27Ac and H3K4me3) and show high promoter occupancy by RNAPol2 and associated transcription factors (Myc, E2F and BRD4). This peculiar promoter configuration likely reflects a general strategy in transcriptional control based on the maximization of the recruitment of RNAPol2 and transcription factors in order to support a robust transcriptional flux. High RNAPol2 promoter occupancy is likely dictated by the chromatin state of such promoters as suggested by their high enrichments in chromatin marks linked to transcriptional activation48 (H3K27Ac and H3K4me3), while BRD4 and Myc seem to be dispensable since neither BRD4 eviction (us) nor Myc downregulation17, 29 impairs RNAPol2 recruitment. This is also supported by recent genome-wide studies showing that in vitro and in vivo Myc progressively invades promoters already pre-marked by RNAPol2.23, 49, 50, 51
Our work is in agreement with previous evidences that link BRD4 to the control of elongation43, 52, 53, 54 and support a model where Myc29, 55, 56, 57 and other transcription factors may be critical in the regulation of transcriptional elongation, with BRD4 laying downstream of Myc in the control of CDK9-dependent RNAPol2 activation. This does not exclude an involvement of distal regulatory elements, indeed BRD441, 47 and other cofactors like JMJD644 have been implicated in the long-range control of promoter proximal paused genes and have been shown to favor enhancer-mediated transcriptional activation. Further work will be needed to assess this relevant point.
The association of high RNAPol2 promoter occupancy, BRD4 and transcriptional flux may also reflect intrinsic and basic principles of genes transcription, as recent evidences suggest that highly transcribed genes rely on BRD4 to efficiently recruit topoisomerase 1 (TOP1) to prevent topological stress due to high transcriptional rates.58 This may be particularly relevant in Myc-driven tumor, where transcriptional amplification of Myc target genes will be predicted to be reliant on BRD4 and TOP1.
Although the recruitment of transcription factors and RNAPol2 to promoters is not diminished by BRD4 inhibition, RNAPol2 distribution along genes is clearly affected at a global level (Figures 5c and f; Supplementary Figure 13a). Yet, the consequences on transcription are selectively observed on those genes, DEG-down, that show high levels of RNAPol2 occupancy at promoters. This differential effect is due to kinetic compensation of RNAPol2 elongation occurring on No-DEG genes. On these genes the amount of promoter-associated RNAPol2 is at the steady state and depends on both RNAPol2 recruitment and initiation (loading on promoters) and its release into the gene body upon its conversion to the elongating form (promoter release). If RNAPol2 promoter release is lowered (that is, upon JQ1 treatment) then the immediate effect will be that the amount of elongating RNAPol2 will decrease, while, as a consequence, the amount of paused RNAPol2 will gradually increase. This increase in paused RNAPol2 will be the substrate of the second arm of the reaction (elongation), thus as promoter-associated RNAPol2 builds up, its conversion to the elongating form will increase as well. At the point in time when the new steady state is reached, there will be an increase in promoter-associated RNAPol2 compared with the starting steady state (unchallenged elongation) and a substantial amount of elongating RNAPol2 (despite the rate constant of the elongation is reduced). This re-adjustment to the new steady state provides an intrinsic compensation to transcription. This is the behavior we observed in JQ1-insensitive genes (No-DEGs), where, upon JQ1 treatment, a clear increase in promoter paused RNAPol2 was observed, while elongation, despite being affected shortly after BET inhibition, was restored at longer time points. On the other hand, the epigenetic state and the high levels of positive transcription factors (BRD4, Myc and E2F) found on promoters of DEG-down genes allows an efficient recruitment of RNAPol2, that is thus at the near-equilibrium, as evidenced by the observation that inhibition of elongation does not lead to a further increase in promoter paused RNAPol2. Therefore, DEG-down genes are (i) rate limited by the release of the promoter-associated RNAPol2 and its conversion to the elongating form and (ii) inherently susceptible to elongation inhibitors given their scarce ability to compensate for elongation drops by enhancing RNAPol2 recruitment at promoters.
This is supported by our observations, the re-analysis of published data set17, 37 and the use of CDK9/DSIF inhibitors, overall suggesting that transcriptional elongation is rate limiting for the control of the expression levels of DEG-down genes. Thus, even in the instances where BET inhibition does not lead to Myc downregulation, the efficacy and cancer selectivity of such compounds is due to their selective targeting of highly transcribed genes such as proliferative and metabolic genes, that, given the prominent role exerted by Myc in rewiring such programs during cell transformation,2 will be enriched in Myc targets. This may also help explaining why targeting general transcriptional regulators such as CDK9 may indeed lead to specific transcriptional alterations and effectively target Myc addicted cancers.59 This link is also confirmed by unbiased approaches as we (Sara Rohban and Stefano Campaner, unpublished) and others60 have noticed that inhibition of components of the basal transcriptional machinery is synthetic lethal with Myc overexpression. Thus, RNAPol2 promoter clearance represents a cancer cell liability that sensitizes cancer cells to the action of elongation inhibitors. This concept may also expand beyond cancer, being RNAPol2 promoter pausing a general mechanism required for selective and quantitative transcriptional regulation of pathways like cell cycle, developmental processes and stress responses.61 Indeed in m-ESC, cell cycle genes have a high pausing index which associates with high levels of promoter-associated RNAPol2.62 Interestingly, also there the alteration of pause elongation was linked to specific transcriptional deregulation since genetic inactivation of the pausing factor NELF primarily deregulated genes involved in cell cycle and signal transduction.62 On the other hand, lowly transcribed genes, which bear ‘unsaturated' promoters, will still have the chance to compensate for drops in elongation efficiency by enhancing RNAPol2 recruitment.
In summary, our work suggests that the high proliferative and metabolic avidity of cancer cells requires a quantitatively robust transcriptional output. This is supported by oncogenic pathways that sustain a strong transcriptional flux by maximizing promoter recruitment of RNAPol2 and transcription factors such as Myc, E2F an BRD4. This renders gene expression rate limited by RNApol2 promoter clearance, thereby exposing cancer cells to the action of elongation inhibitors (such as BET inhibitors). Thus, transcriptional elongation is a pathway that can be targeted for selective cancer treatment.
Accession numbers
RAJI microarray expression data and ChIPseq data have been deposited in NCBI's Gene Expression Omnibus (GEO) (Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository) and are accessible through GEO Series accession number GSE76192 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76192).
Acknowledgments
We thank Dr J Bradner, Dr M Esteban, Dr Giovanni Tonon and Dr J Zuber for kindly sharing reagents. Dr V Bianchi, Dr S De Pretis, G Melloni and P Bora for discussion and suggestions on computational analyses. Salvatore Bianchi, Thelma Capra and Luca Rotta for technical support in NGS sequencing. Dr B Amati for critical reading of this manuscript, continuous support, suggestions and inspiration. This work was supported by the Italian Association for Cancer Research (AIRC, IG grant 13135).
Author contributions
ED performed all the experiments described and contributed to the experimental design and the bioinformatic analyses; HM analyzed the microarray data and performed GSEA; OC performed all the computational and bioinformatic analyses on ChIPseq and expression data; AS contributed to ChIPseq data production and analyses; MM and MP contributed to bioinformatics and computational analysis. SC designed the study and wrote the paper. ED, OC and AS contributed to the discussion of the data and the drafting of the manuscript.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015; 5: 1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Sabo A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 2015; 15: 593–607. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA 1982; 79: 7824–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA 1982; 79: 7837–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S, Barth R, Hood L, Prehn J, Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science 1982; 218: 1319–1321. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8: 976–990. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004; 431: 1112–1117. [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 2002; 297: 102–104. [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 2013; 27: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 2002; 109: 321–334. [DOI] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA 2011; 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012; 149: 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 2011; 31: 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011; 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 2011; 18: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell 2008; 14: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol 2010; 12: 54–59. [DOI] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013; 5: 1704–1713. [DOI] [PubMed] [Google Scholar]
- Liu L, Xu Y, He M, Zhang M, Cui F, Lu L et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell 2014; 15: 574–588. [DOI] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore K, de Pretis S, Lister R, Morelli MJ, Bianchi V, Amati B et al. methylPipe and compEpiTools: a suite of R packages for the integrative analysis of epigenomics data. BMC Bioinformatics 2015; 16: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by random forest. R News 2002; 3: 18–22. [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB et al. c-Myc regulates transcriptional pause release. Cell 2010; 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985; 318: 533–538. [DOI] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 2013; 3: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 2012; 120: 2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff K, Beckers A, Bell E, Nortmeyer M, Thor T, Sprussel A et al. A Cre-conditional MYCN-driven neuroblastoma mouse model as an improved tool for preclinical studies. Oncogene 2015; 34: 3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013; 24: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadury J, Nilsson LM, Muralidharan SV, Green LC, Li Z, Gesner EM et al. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc Natl Acad Sci USA 2014; 111: E2721–E2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz V, Butzhammer P, Hirsch B, Hecht J, Gutgemann I, Ehlers A et al. Deep sequencing of MYC DNA-binding sites in Burkitt lymphoma. PLoS One 2011; 6: e26837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB et al. Genome-wide localization of small molecules. Nat Biotechnol 2014; 32: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 2014; 159: 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 2015; 526: 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 2009; 138: 1122–1136. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 2014; 21: 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19: 535–545. [DOI] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013; 155: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner WA, Van Bortle K, Li L, Ramos E, Takenaka N, Corces VG. Distinct isoforms of the Drosophila Brd4 homologue are present at enhancers, promoters and insulator sites. Nucleic Acids Res 2013; 41: 9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem 2012; 287: 43137–43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol 2007; 9: 932–944. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014; 158: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 2006; 8: 764–770. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012; 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005; 19: 523–534. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009; 138: 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 2003; 22: 5707–5711. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem 2001; 276: 48562–48571. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem 2002; 277: 40156–40162. [DOI] [PubMed] [Google Scholar]
- Gargano B, Amente S, Majello B, Lania L. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle 2007; 6: 2031–2037. [DOI] [PubMed] [Google Scholar]
- Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan-Salis KY et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 2016; 165: 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev 2014; 28: 1800–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci USA 2012; 109: 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A et al. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev 2012; 26: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell 2015; 58: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J et al. Global quantification of mammalian gene expression control. Nature 2011; 473: 337–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.