Abstract
Hemin linked to hexa(ethylene glycol)bishydrazide was patterned by inkjet printing into periodic microarrays, and evaluated for their ability to capture bacterial pathogens expressing various hemin receptors. Bacterial adhesion was imaged under darkfield conditions with Fourier analysis, supporting a label-free method of pathogen detection. Hemin microarrays were screened against a panel of sixteen bacteria and found capable of capturing multiple species, some with limits of detection as low as 103 cfu/mL. Several Gram-positive strains including Staphylococcus aureus and Bacillus anthracis also exhibited rapid adhesion, enabling pattern recognition within minutes of exposure. This can be attributed to differences in hemin acquisition systems: aggressively adherent bacteria express cell-surface hemin receptors (CSHRs) that enable direct hemin binding and uptake, whereas other types of bacteria including most Gram-negative strains rely on the secretion and recapture of soluble proteins (hemophores) for hemin acquisition, with consequently longer times for ligand binding and detection.
Keywords: bacteria, heme, hydrazide, microarray, optical sensing, pathogen detection
Graphical Abstract
INTRODUCTION
The frequency of pathogenic bacteria outbreaks has been increasing at an alarming rate. Drug-resistant strains of bacteria are on the rise, creating concerns of a “post-antibiotic era” with no viable countermeasures against a pandemic of incurable infectious diseases.1,2,3 The threat of drug-resistant bacteria is particularly grave with respect to hospital-acquired infections, the most severe of which are grouped by the acronym ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species).4 Pathogens in the Enterobacteriaceae are also a concern and a major cause of gastrointestinal illnesses, and easily transmitted by food and water contamination, unsanitary conditions in crowded spaces, or interspecies contact. Another threat to biosecurity comes from the deliberate release of weaponized pathogens such as Bacillus anthracis, which have been used in past acts of bioterrorism.5 For each setting, the risk of infection can be reduced by implementing technologies enabling the rapid and label-free detection and screening of bacteria, as part of an early warning system. In the most extreme cases, early and rapid detection may be the best defense against further spread of disease.
Rapid pathogen detection methods are also needed in point-of-care settings for medical treatment.6 In the absence of a convenient diagnostic, physicians often take a “scorched earth” approach by prescribing a variety of broad-spectrum antibiotics to treat a potential infection. It is now recognized that such a practice has the dire consequence of creating conditions that select for antibiotic-resistant bacteria, whose burden on the U.S. healthcare system and associated societal costs has been estimated to be well over $50 billion a year.7,8 In the meantime, current practices in pathogen analysis continue to rely on the submission of samples to clinical laboratories for culturing in selective media.9,10 Culture-based methods remain the gold standard for reliable analysis but can take up to several days for verification, and are poorly suited for situations where rapid detection is urgent. Many clinical laboratories are also equipped with DNA amplification techniques based on the polymerase chain reaction (PCR), which is an excellent method for establishing the presence of specific bacteria but is susceptible to sample contamination and misinterpretation.11 Practical alternatives to these laboratory methods remain to be established; efforts toward point-of-care technologies for rapid pathogen analysis include lateral-flow immunoassays, immunomagnetic separations, DNA microarray analysis, or combinations thereof.9,10,12 While all of these methods are faster than traditional culturing, they vary with respect to sensitivity, specificity, and reliability. Moreover, nearly all of these methods are reliant upon purified bacteria samples and trained personnel, and are vulnerable to nonfunctional mutations of bacterial antigens.
As an alternative to immunological and nucleic acid-based methods for pathogen identification, we have been investigating low molecular-weight ligands that target immutable functions or recognition processes that correlate strongly with bacterial virulence or survival.13,14,15 These ligands typically have high affinity for the bacteria’s cognate receptors, which are expressed constitutively on the outer wall or membrane or presented on fimbriae or pili during their search for host cells. Immutable recognition ligands can be displayed as periodic microarrays on substrates and exposed to suspensions of active bacteria, then rinsed and examined under quasi-darkfield conditions for rapid and label-free pathogen detection, with reproducible limits of detection (LOD) of 103 colony-forming units per milliliter (cfu/mL).13–15 This is achieved by using a fast Fourier transform (FFT) algorithm to convert periodic signals into peak frequencies, with subsequent reduction in background noise. Two great benefits of using FFT-based readouts are (i) reliable signal recovery from heterogeneous sample images, created by nonspecific binding events or uneven lighting conditions, and (ii) a higher degree of fault tolerance relative to single-point measurements, as the Fourier signal strength is based on the overall occupancy (fill factor) of the microarray elements, with detection still possible at an array occupancy as low as 1%.14 Last but far from least, small-molecule conjugates can circumvent many of the limitations intrinsic to antibody-based detection strategies, such as chemical and thermal stability or invalidation caused by nonfunctional mutations.
One such immutable function involves the acquisition of hemin, the oxidized form of heme and a primary source of iron which many bacteria require for survival. Approximately 95% of the bioavailable iron in mammals is associated with hemin or heme-containing proteins.16 However, the concentration of free hemin in blood is extremely low (ca. 10−18 M),17 and is tightly regulated by host organisms to prevent oxidative damage as well as its acquisition by opportunistic pathogens. To overcome this, bloodborne pathogens have evolved sophisticated and diverse mechanisms for acquiring and metabolizing hemin.18,19,20,21 Some bacteria secrete extracellular proteins known as hemophores to scavenge hemin from damaged cells, with subsequent delivery to TonB-dependent transport proteins on the bacterial cell surface,21,22 while other species employ surface-bound receptors capable of direct hemin acquisition. A well-known example of the latter is the iron-regulated surface determinant (Isd), a family of receptors expressed on the exterior of several Gram-positive pathogens for hemin acquisition.23,24
Here we show that pathogens with the capacity to metabolize heme can be captured and detected in a label-free manner using periodic microarrays of a synthetic hemin conjugate (1). Hemin microarrays were evaluated against a panel of sixteen Gram-positive and -negative bacteria, several with LODs as low as 103 cfu/mL. For certain species, the time to detection can also be rapid, sometimes within minutes of exposure, depending on the pathogen’s mechanism of hemin acquisition. In particular, our studies point to a novel mechanism for the rapid detection of bacteria that express cell-surface hemin receptors (CSHRs), with discrimination from other species which rely on soluble hemophores for hemin acquisition. In this regard, we note that the direct acquisition of hemin has never been explicitly characterized for any pathogen species, although the existence of such pathways has at times been assumed.25
RESULTS AND DISCUSSION
Synthesis and purification of hemin–bishydrazide 1
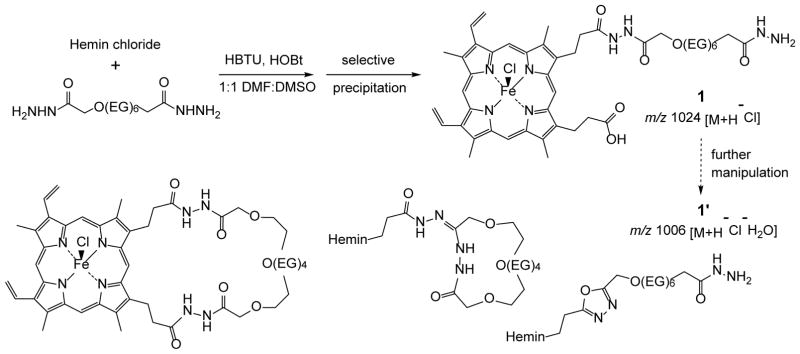
The protoporphyrin-IX ring of hemin has two carboxylic acid groups that are reactive toward standard peptide coupling reagents, but with sufficient selectivity to produce monofunctionalized derivatives as major products.26,27 Performing such conjugations in the presence of coordinated Fe(III) is less common, given its potential for oxidative side reactions. Nevertheless, we found that hemin chloride reacted smoothly with hexa(ethylene glycol)bishydrazide13 on a gram scale using HBTU/HOBt coupling conditions (Scheme 1).
Scheme 1.
Synthesis of hemin–EG6-bishydrazide 1 and dehydration byproduct(s) 1′, with possible structures below (EG =-CH2CH2O-).
While the synthesis of hemin–EG6-bishydrazide 1 was facile, its isolation with acceptable purity proved much more challenging. Although the hemin–bishydrazide adduct appears to be stable in solution, especially under acidic conditions (pH < 4), concentrating the reaction mixture by rotary evaporation produces one or more dehydration byproducts (1′) as determined by electrospray ionization mass spectrometry (ESI-MS). Increasing the basicity of the reaction mixture also promotes the apparent dehydration of 1. Possible dehydration products include macrocyclic condensation of the ω-hydrazide with the carboxylate or a hydrazide carbonyl, or condensation of the acylhydrazide into a 1,2,4-oxadiazole; however, attempts to separate 1 and 1′ by chromatography with silica gel, alumina, Fluorisil, or by reverse-phase (RP) HPLC were all detrimental to purity and mass recovery, frustrating attempts at further characterization.28
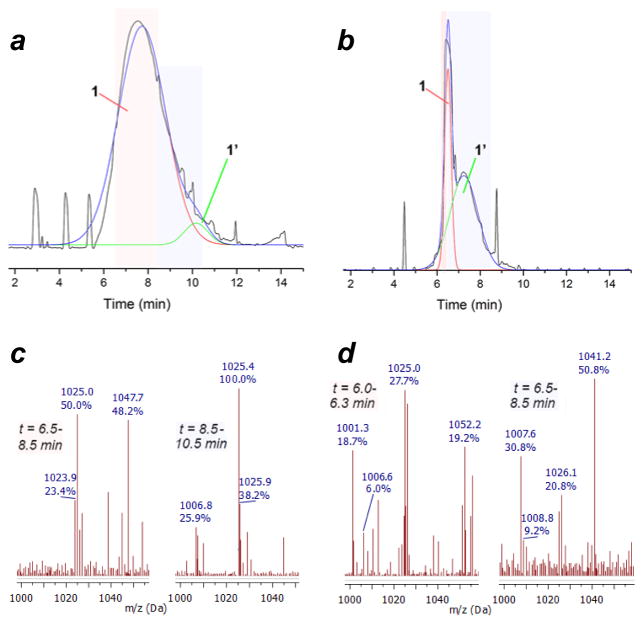
After extensive experimentation, we found that trituration of the reaction mixture with ether, redissolution in acetonitrile, addition of 16 v/v% trifluoroacetic acid for selective precipitation of unreacted hemin, and retrituration with ether allowed us to obtain hemin–bishydrazide 1 as a dark solid in practical yields (60% mass recovery) and over 95% purity, as determined by RP-HPLC analysis (Figure 1a). Isocratic elution of 1 redissolved in acetonitrile with 5% TFA produced a nearly Gaussian peakshape, with confirmation of structure by ESI-MS. To confirm the sensitivity of conjugate 1 to dehydration, isolated fractions with pure product were concentrated then resubjected to RP-HPLC analysis, which yielded a dramatic increase in dehydration product 1′ (65% by peak deconvolution; Figure 1b).
Figure 1.
HPLC chromatogram and ESI-MS data for hemin–bishydrazide 1 ([M(FeII)+H–Cl] = 1025) and its dehydration byproduct 1′ (m/z 1007) after elution with 95:5 CH3CN:CF3CO2H on a reverse-phase C18 silica column. (a) Compound 1 was precipitated from acidic CH3CN then redissolved and subjected to RP-HPLC to produce a nearly Gaussian lineshape (>95% purity based on peak deconvolution). (b) Concentrating and resubjecting this sample to RP-HPLC produced an altered distribution with a much larger secondary peak. (c) ESI-MS of the HPLC fractions from the original sample indicated the major product to be conjugate 1, with traces of dehydration byproduct 1′ eluting after 8.5 min. (d) ESI-MS of the HPLC fractions from resubjected sample indicated the major product to be byproduct 1′ with a large reduction in 1.
Hemin microarrays by inkjet printing
In previous studies, immutable ligands were presented as polyvalent conjugates on bovine serum albumin (BSA), and patterned onto activated glass slides by microcontact or inkjet printing.13–15 However, albumin can interfere with hemin presentation by occlusion within a hydrophobic binding pocket.29 We reasoned that the EG6-bishydrazide linker would be of sufficient length to project hemin conjugates tethered on passivated slides and enable the binding of bacterial hemin receptors. Furthermore, the propionyl (−CH2CH2CO2H) substituents were deemed unessential for binding: X-ray crystal structures of two heme-binding domains (HasR and Isd) have shown hemin to be bound with propionyl groups extending outside the binding pocket with minimum interactions.19,30
Hemin microarrays were generated with a piezoelectric inkjet printer, which deposited 10-pL droplets of buffered solutions containing 1 onto epoxysilane-coated glass slides. Printer parameters were optimized to generate dot-matrix arrays with periodicities of 80 μm in the x-direction and 150 μm in the y-direction, whose quality was confirmed by darkfield optical imaging (Figure 2). Slides were blocked with 0.1% BSA followed by copious washing prior to storage in the dried state. These microarrays could be stored in the dark under ambient conditions for at least 6 months without loss of activity.
Figure 2.
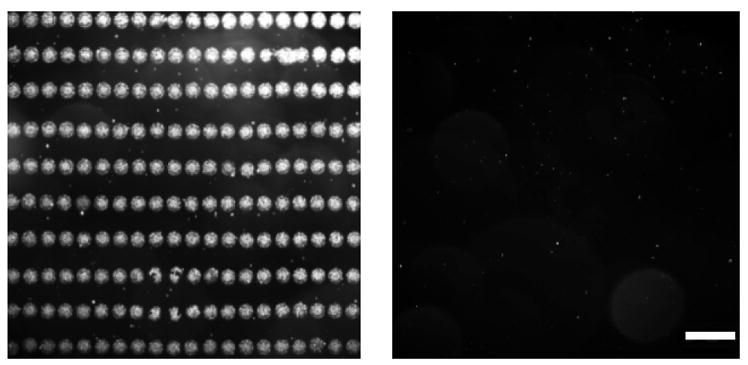
Microarrays of 1 prepared by piezoelectric inkjet printing. Left, darkfield image of freshly printed microarray, prior to blocking or washing; right, microarray after post-printing treatment, prior to bacterial exposure. Bar = 100 μm.
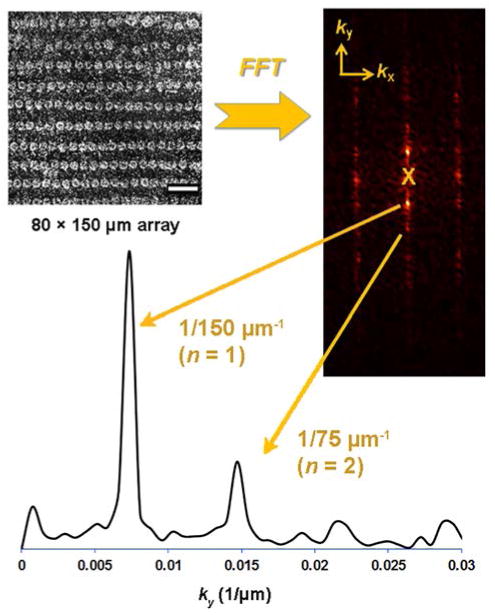
Bacterial capture assays were performed as previously described (see Experimental Section for details).14 Briefly, bacteria were cultured in media at 37 °C until a concentration of 108 cfu/mL was achieved, then subjected to serial dilution and cast onto ligand microarrays at ambient temperature for a period of 15–60 min. The microarray slides were gently rinsed and dried, then examined by darkfield optical microscopy at 10 × magnification (Figure 3). Darkfield images were cropped and scaled to approximately 1 pixel/μm, then subjected to fast Fourier transform (FFT) image analysis with conversion into reciprocal lattice units (μm−1). Fourier spectra were generated from linescans in the x- or y-direction, using the primary reciprocal lattice peak (n = 1; k = 1/a) to characterize the limits of pathogen detection. This approach is tolerant to noise generated by nonspecific binding (see below), and produces fewer false positives than single-point detection methods with reproducible LODs of 103 cfu/mL.14
Figure 3.
Darkfield image of bacterial capture by hemin microarray (80 × 150 μm) is converted into its reciprocal lattice by FFT, and replotted as a linescan for analysis of the periodic signal (n=1). Signal-to-noise ratios (S/N) above 3 indicate positive detection; data at the origin (X) is removed prior to analysis.
The hemin microarrays were evaluated against a panel of sixteen Gram-positive and - negative bacteria including ESKAPE representatives, four of which exhibited positive affinity (Table 1). The lowest LODs (103 cfu/mL) were observed with Staphylococcus aureus, Yersinia enterocolitica, and Escherichia coli O157:H7, the first being an ESKAPE bacteria and the latter two being pathogens in the Enterobacteriaceae. To test for binding specificity toward EG6-tethered hemin, several high-affinity strains were incubated with slides printed with unmodified hemin or EG6-bishydrazide linker alone. These resulted in weak and aperiodic capture pattern even at the highest bacteria concentrations, confirming the importance of ligand presentation. Gram-positive and -negative strains with no known affinity for hemin (e.g. Bacillus megaterium, Salmonella enterica)36 were also included as negative controls, and further confirmed the specificity of hemin recognition in pathogen capture.
Table 1.
Bacteria evaluated for hemin microarray capture and their acquisition systems
| Bacterial strain (source) | LOD in normal or low-Fe media (cfu/mL)a | Hemin acquisition system (receptor proteins)b,c |
|---|---|---|
| Gram positive: | ||
| Bacillus anthracis (Ames 35) | Fe+: 1E4 Fe−: 1E5 |
Isd(C,E,X1,X2); Bsl(K); Hal |
| Bacillus anthracis (Stern Vaccine) | Fe+: n.d. Fe−: 1E5 |
|
| Bacillus megaterium (BCRC 10608) | Fe+/−: n.d. | n/a |
| Enterococcus faecium (VRE) | Fe+/−: n.d. | n/a |
| Listeria monocytogenes (J0161) | Fe+: 1E5 Fe−: 1E6 |
Hup(C); Hbp2/Svp(A) |
| Staphylococcus aureus (PCI 1203) | Fe+: 1E4 Fe−: 1E3 |
Isd(A,B,C,E,H) |
| Staphylococcus epidermidis (AMC 263) | Fe+: 1E6 Fe−: 1E5 |
Isd(A,B,C,E,H) |
| Streptococcus pneumoniae (CDC CS111) | Fe+: 1E5 Fe−: 1E6 |
unassigned31,32 |
| Gram negative: | ||
| Acinetobacter baumanii (DSM 6974) | Fe+: 1E6 Fe−: 1E6 |
unassigned33,34 |
| Escherichia coli O104:H21 (CDC 1994-3024) | Fe+: 1E5 Fe−: n.d. |
n/a |
| Escherichia coli O157:H7 (CDC EDL 933) | Fe+: 1E3 Fe−: 1E5 |
Chu(A); Hma; Shu(A) |
| Enterobacter aerogenes (NCDC 819-56) | Fe+/−: n.d. | n/a |
| Klebsiella pneumoniae (S 389) | Fe+: 1E6 Fe−: 1E5 |
unidentified35 |
| Pseudomonas aeruginosa (PAO1-LAC) | Fe+: n.d. Fe−: 1E6 |
Has(A,R); Phu(R) |
| Salmonella enterica, ssp. typhimurium (LT2) | Fe+/−: n.d. | n/a |
| Yersinia enterocolitica (WA-314) | Fe+: 1E6 Fe−: 1E3 |
Hem(R) |
LOD based on strength of 1/a peak, with S/N > 3; n.d. = not detected (S/N < 3).
Abbreviations: Bsl, B. anthracis S-layer; Chu, E. coli heme utilization; Hal, heme-acquisition leucine-rich; Has, heme acquisition system; Hbp, hemin binding protein; Hem, hemin; Hma, heme acquisition protein; Hup, heme uptake; Isd, Iron-regulated surface determinant; Phu, Pseudomonas heme uptake system; Shu, Shigella heme uptake; Svp, surface virulence-associated protein.
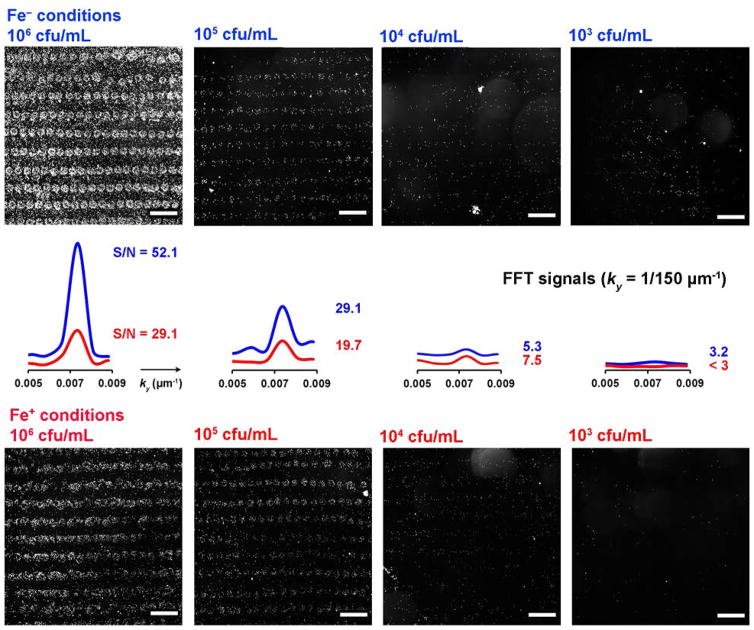
In some cases, hemin affinity could be increased by culturing bacteria in iron-deficient media, with a corresponding enhancement in detection sensitivity. This is attributable to the induced expression of hemin acquisition proteins by an iron-dependent ferric uptake regulator (fur).37,38 For example, S. aureus were grown in standard and iron-deficient tryptic soy broth (the latter with 3 mM 2,2′-bipyridine), then exposed to hemin microarrays at different concentrations for a 60-min period. S. aureus cultured in iron-deficient media demonstrated a LOD at 103 cfu/mL, while the same bacteria raised in standard media had a higher LOD at 104 cfu/mL (Figure 4). However, growth under iron-deficient conditions did not universally lead to increased sensitivity, and sometimes even caused an apparent decrease in binding avidity. Overall, we find that the detection of hemin-acquiring bacteria does not require pre-conditioning under iron-challenged conditions.37
Figure 4.
Darkfield images of S. aureus captured by hemin microarrays using different concentrations and culturing media (standard (Fe+) or iron-deficient (Fe−)), with 60-min. exposure times. FFT signals of respective images indicate a LOD of 103 cfu/mL for S. aureus grown in iron-challenged media, versus 104 cfu/mL for bacteria grown in standard media.
Discrimination of hemin-binding bacteria based on time to detection
In the course of this work, it became evident that hemin microarrays were capable of capturing certain bacteria more rapidly than others at similar concentrations, independent of their LODs. This can be illustrated clearly by comparing the minimum time to detection (TTD) for Staphylococcus aureus and Yersinia enterocolitica. Both of these species can be detected by the hemin microarrays at the most sensitive level (103 cfu/mL) within 1 hour, although they have very different mechanisms for acquiring hemin. Staphylococcus expresses surface-bound receptors (IsdB, IsdH) that capture and extract hemin from hemoproteins, which is then relayed to downstream receptors (IsdA, IsdC, IsdE) and a cell-wall transporter for uptake (see Table 1).21,38,39 In contrast, Gram-negative bacteria are known to secrete soluble hemophores that scavenge for hemin or hemoproteins in the extracellular milieu, with subsequent transfer to a TonB-dependent outer-membrane transporter, presumably by a diffusion-controlled process.36,37 With regard to Yersinia enterocolitica, a TonB-dependent hemin transporter (HemR) has been characterized,40 although its cognate hemophore has yet to be identified.
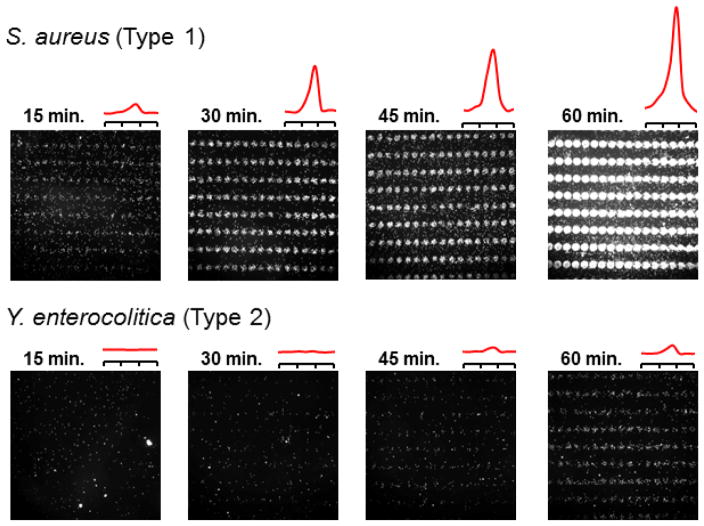
S. aureus and Y. enterocolitica were cultured under iron-challenged conditions then diluted to 106 cfu/mL and incubated with hemin microarrays, with one substrate removed every 15 minutes over a 60-minute period followed by darkfield imaging and FFT analysis (Figure 5). A marked difference in capture rate and TTD was clearly observed: For S. aureus, patterned bacterial adhesion could be detected within the first 15 minutes of exposure with a steady increase in signal over time, whereas Y. enterocolitica adhesion could not be reliably confirmed until 45 minutes after initial exposure.
Figure 5.
Time-dependent capture and detection of S. aureus and Y. enterocolitica by hemin microarrays, exposed to 106 cfu/mL. FFT analysis of darkfield images (ky = 150 μm−1) indicates a TTD of 15 minutes or less for S. aureus, and over 30 minutes for Y. enterocolitica.
The large difference in TTD suggests an original approach for typing and discriminating bacterial pathogens according to their mechanisms of hemin acquisition. Bacteria that express high-affinity hemin receptors on their outer walls or membranes are capable of capturing hemin directly from nearby sources (Type 1), whereas bacteria that express soluble hemophores can harvest hemin or hemoproteins over larger distances (Type 2). However, the recapture of hemophores by Type 2 bacteria requires more time, with a delay in hemin acquisition. In the context of pathogen detection, Type 1 bacteria can be rapidly immobilized onto hemin microarrays in a single step, whereas Type 2 bacteria would be delayed by an induction period involving the secretion of hemophores and their diffusion toward the substrate, followed by bacterial capture by microarray elements coated with surface-bound hemophores (Figure 6).
Figure 6.
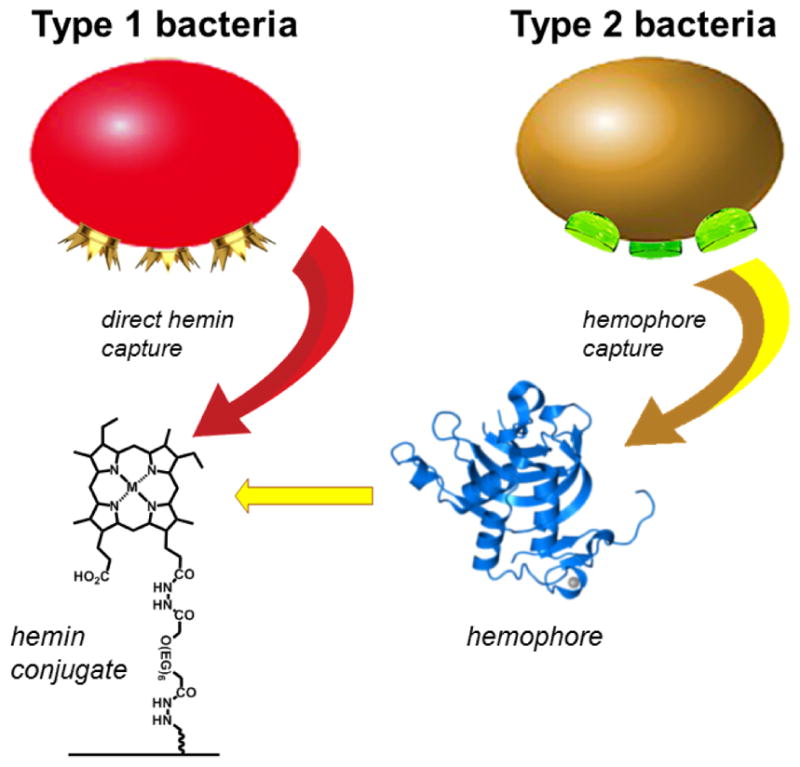
Rapid detection of Type 1 bacteria is enabled by direct hemin capture, whereas detection of Type 2 bacteria is delayed by the intermediacy of hemophore release and recapture.
There is strong evidence that many Gram-positive bacteria have cell-surface hemin receptors (CSHRs) capable of direct hemin uptake. For example, staphylococci and Bacillus anthracis are known to express multiple Isd proteins on their outer cell walls (Table 1).21,23,41 On the other hand, within the limits of current knowledge, Gram-negative bacteria do not appear to express CSHRs but rely instead on hemophore harvesting and recapture for hemin acquisition. Bacteria with TonB-dependent hemin transporters are also thought to express these proteins with low density,19,21,22 which further reduces their rate of adhesion to hemin-coated substrates.
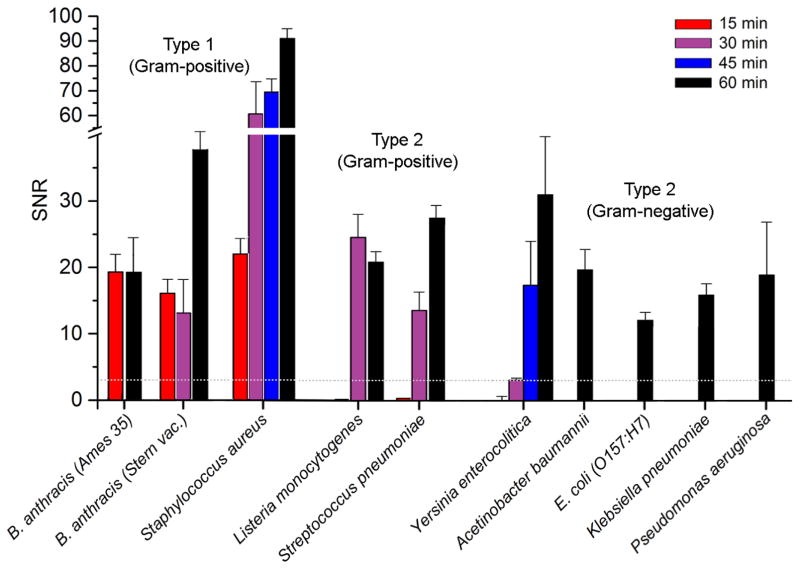
To determine whether other Gram-positive species besides Staphylococcus might also be amenable of rapid detection using hemin microarrays, we evaluated strains of Bacillus anthracis (Ames 35 and Stern vaccine), Listeria monocytogenes (J0161), and Streptococcus pneumoniae (CDC CS111) in a time-dependent manner at 106 cfu/mL. Like S. aureus, the patterned adhesion of Bacillus anthracis strains could be detected in under 15 minutes; for Listeria monocytogenes and Streptococcus pneumoniae, TTD was achieved within 30 minutes (Figure 7). We did not evaluate Gram-negative bacteria other than Yersinia enterocolitica in a time-dependent manner; however, many of the bacteria listed in Table 1 were recently screened for their uptake of Ga(III)-protoporphyrin IX (Ga-PpIX), a fluorescent hemin analog, with the observation that all Gram-negative species experienced significant delays in Ga-PpIX uptake relative to B. anthracis, S. aureus, and S. pneumoniae.42 We thus consider time-dependent hemin capture to be a valid mechanism for discriminating CSHR-expressing bacteria from those that rely on hemophore-based harvesting systems.
Figure 7.
Time-dependent detection of select bacteria at 106 cfu/mL, based on FFT analysis of their patterned adhesion on hemin microarrays. All analyses were performed in triplicate; mean S/N values presented with standard deviations. TTD (S/N > 3; above dotted grey line) is less than 15 min for Type 1 Gram-positive bacteria (B. anthracis, S. aureus) and 30 min for Type 2 bacteria (L. monocytogenes, S. pneumoniae). Type 2 Gram-negative strains such as Yersinia enterocolitica are at the LOD after 30 min, but produce strong signals at 45 and 60 min; other Gram-negative species exhibit similar delays.
It is worthwhile to revisit the relative advantages of using darkfield imaging of hemin microarrays (and immutable ligand microarrays in general) for label-free pathogen detection.13–15 A recent review on optical biosensors for pathogen detection has highlighted several factors that are important in the development of practical sensors for point-of-care applications 43 In addition to speed and sensitivity, sensors should be inexpensive, easy to use, portable, require minimum sample preparation, and permit integration with multiplexing strategies. Our approach to reagentless pathogen detection is well aligned with these criteria: darkfield microarray imaging is performed with simple collection optics for detecting intact bacteria, without cell lysis or amplification steps.
Among the many technologies being developed for whole-bacteria detection, those best meeting the conditions above include lateral-flow immunoassays (LFIAs) with prepackaged reagents, and surface plasmon resonance (SPR) and its variants using functionalized glass substrates. Commercial LFIAs have been optimized for rapid detection (within 10 minutes) but have rather poor sensitivity and often require sample enrichment. Recent versions of FLIA have been reported to detect bacteria with LODs of 300–500 cfu/mL, albeit at the expense of a short TTD.44,45 SPR methods using antibody- or bacteriophage-coated substrates offer better performance, with LODs of 10–100 cfu/mL in 25 min or less.46,47 In comparison, darkfield microarray imaging provides reproducible LODs on the order of 1000 cfu/mL and, in the case of Type 1 bacteria, TTDs of 15 min or less. This is in addition to the aforementioned benefits of FFT-based readouts for high fault tolerance, and the robust chemical recognition and stability offered by the immutable ligand approach.
Finally, it should be emphasized that FFT analysis of pathogen capture by hemin microarrays provides an experimentally robust approach for detecting bacteria that form biofilms or networks, whose presence can create signal interference. Two key advantages of using a Fourier-based approach for signal readout from periodic microarrays are (i) the large reduction in noise created by nonspecific binding events, and (ii) a more reliable analysis of signal quality based on multiple array elements, relative to intensity measurements of single data points. We have previously used simulations to demonstrate that FFT analysis of microarrays with partially occupied matrix elements or high levels of background noise (with relative intensities up to one third of that generated by specific binding) can still reveal distinct peaks at the fundamental harmonic (k=1/a).14
Here we validate the FFT readout method by applying it toward the specific detection of Bacillus anthracis, following a 15-min exposure to hemin microarrays. B. anthracis cultured in solution generally exhibits planktonic behavior but can become highly self-associative during its growth phase, often forming chain networks.48 This is evident even during the initial stages of surface adhesion: bacteria bound to the microarray elements serve as nucleation sites for network growth within minutes (Figure 8). Despite this the Fourier signal is still discernible, which confirms the active role of the hemin conjugates in patterned adhesion.
Figure 8.
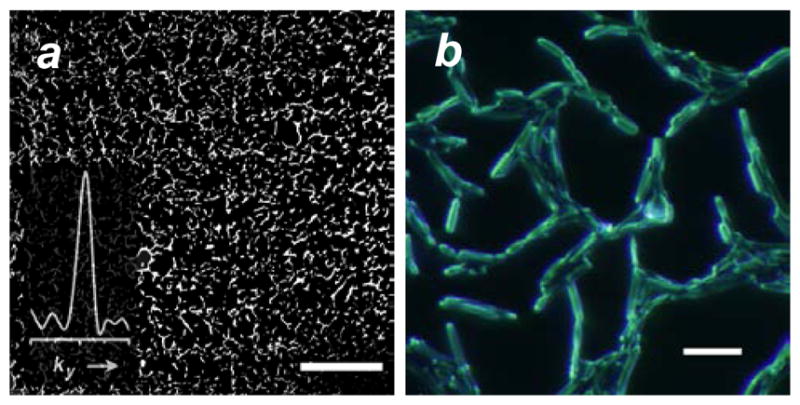
(a) Microarray-initiated growth of Bacillus anthracis network, after 15-min exposure at 106 cfu/mL (bar = 300 μm); FFT image analysis yields a peak signal (ky = 1/a) with a S/N of 14.5. (b) Magnification of B. anthracis network showing individual bacteria (bar = 10 μm).
CONCLUSIONS
Hemin microarrays prepared with EG6-bishydrazide conjugate 1 can support a rapid, label-free method for detecting bacterial pathogens with active hemin acquisition systems, with limits of detection as low as 103 cfu/mL in several cases. Rapid detection (15 minutes or less) is possible with Gram-positive bacteria that express CSHRs capable of direct hemin binding, offering a practical method for their discrimination against other heme-utilizing bacteria. The microarray-based detection platform offers a robust approach toward point-of-care detection technologies that are quick and cost-effective without requiring specialized equipment or training. Finally, we note that the practical synthesis of 1 enables the development of molecular conjugates, for targeted labeling or delivery of antibiotics to hemin-utilizing pathogens.
MATERIALS AND METHODS
General procedures
All starting materials and reagents were obtained from commercial sources, and used as received unless otherwise noted. Dimethylformamide (DMF) was obtained in anhydrous form; dimethyl sulfoxide (DMSO) was distilled over CaH2 under reduced pressure. Infrared (IR) spectra were acquired with a Nexus 670 spectrometer (Thermo) equipped with a grazing-angle attenuated total reflectance module (GATR, Harrick), with samples deposited directly on a Ge window; UV–visible spectra were collected on a Varian Cary50 spectrometer in quartz cuvettes. Label-free optical images were acquired at 10 × magnification using an upright microscope (Olympus BX60) equipped with a darkfield condenser and a charge-coupled device camera (Rolera EM-C2, QImaging).
Synthesis
Hexa(ethylene glycol)bishydrazide was synthesized as previously described,14 with a slight modification in the purification procedure: the crude product was concentrated to a dry solid (1.0 g), resuspended in anhydrous methanol (20 mL) and filtered to remove yellow precipitate, then concentrated again to a clear colorless oil (0.70 g). HPLC analysis of the EG6-bishydrazide (Phenomenex C18 column, 0–30% aq. CH3CN gradient elution) indicated >95% purity based on peak area integration.
Hemin–EG6-bishydrazide (1) was prepared on a gram scale using O-(benzotriazole-1-yl)tetramethyluronium hexafluorophosphate (HBTU)-mediated coupling conditions. Hemin chloride (1 g, 1.5 mmol), HBTU (1.14 g, 3.0 mmol), and hydroxybenzotriazole (HOBt, 459 mg, 3.0 mmol) were dissolved in anhydrous 1:1 DMF:DMSO (16 mL) and stirred for 2 h at 0 °C. The reaction mixture was treated by the dropwise addition of EG6-bishydrazide (1.28 g, 3.0 mmol) dissolved in 2 mL anhydrous DMF, stirred for 20 min at 0 °C, then diluted tenfold with diethyl ether to precipitate hemin conjugate 1 as well as unreacted hemin. This was filtered, washed with cold ether, dried under reduced pressure, then resuspended in 5:1 CH3CN:CF3CO2H (10 mL) and filtered through Celite to remove unreacted hemin. Compound 1 was obtained by precipitation with ether (100 mL) at –20 °C and collected as a brown solid (0.97 g, 60% yield). HPLC analysis of 1 (Sunfire C18 column, 95:5 CH3CN:CF3CO2H) indicated lower and upper purity limits of 88 and >90% based on peak area integration. UV–vis (DMSO): λmax 404 nm, ε 1.05 × 105 L mol−1 cm−1. IR (cm−1): 3236, 2935, 1684, 1662, 1456, 1178, 1086, 841. HRESI-MS: m/z calcd for C50H65N8O12Fe [M–Cl]+ 1024.3994; found 1024.3979. No NMR spectra were not acquired due to the presence of Fe(III).
Microbiological culture conditions
All bacteria strains were obtained from ATCC, BEI Resources, or Microbiologics, and cultured at 37 °C under standard environmental conditions unless otherwise specified. Acinetobacter baumannii (DSM 6974), Bacillus megaterium (BCRC 10608), Enterobacter aerogenes (NCDC 819-56), and Klebsiella pneumoniae (S 389) were grown in nutrient broth; Pseudomonas aeruginosa (PAO1-LAC) and Yersinia enterocolita (WA-314) were grown in Luria–Burtani broth; Bacillus anthracis (Stern vaccine and Ames 35 strains), Escherichia coli O157:H7 (CDC EDL 933) and O104:H21 strains (CDC 1994-3024), Listeria monocytogenes (J0161), Salmonella enterica typhimurium (LT2), Staphylococcus aureus (PCI 1203), Staphylococcus epidermidis (AMC 263), and vancomycin-resistant Enterococcus faecium (VRE) were grown in tryptic soy broth; Streptococcus pneumoniae (CDC CS111) was grown in brain–heart infusion broth. Iron-deficient conditions were generally met by first growing the bacteria in standard media, then subculturing in medium containing 3 mM 2,2′-bipyridine; B. anthracis was subcultured in media containing 0.5 mM bipyridine. Tryptic soy broth was used in all cases to achieve growth in iron-challenged media except E. aerogenes and Y. enterocolitica, which used standard media listed above with 3 mM 2,2′-bipyridine. In some cases (Listeria monocytogenes and Enterococcus faecium), iron was further depleted by adding ethylenediaminetetraacetic acid (EDTA) or ethylenediamine-di(o-hydroxyphenyl)acetic acid (EDDHA) to induce expression of hemin-harvesting systems, however no increases in expression could be determined.
Bacterial suspensions were typically incubated for 16 hours at 37 °C in capped culture tubes under a 5% CO2 atmosphere, until an optical density of 1 was achieved (λ = 600 nm). Bacterial counts were estimated as colony-forming units per milliliter (cfu/mL) by plating serial dilutions onto agar plates, followed by incubation for 16 hours at 37 °C. For pathogen detection studies, bacteria were isolated from growth media by three rounds of centrifugation (5200 g, 5 min) and resuspension in phosphate buffered saline (PBS, pH 7.4).
Selection of hemin-binding bacteria
S. aureus was subject to a selection process to optimize its native affinity for hemin for positive control studies. Glass slides with periodic microarrays of 1 was exposed for 1 hour to a 0.25-mL suspension of S. aureus in PBS (107 cfu/mL), then carefully rinsed and dried under a stream of nitrogen. The slide was placed face-down onto mannitol salt agar plates then lifted after several seconds, followed by incubation for up to 72 hours at 37 °C. Colonies were harvested with a sterile inoculation loop and cultured in tryptic soy broth for 16 hours, then subcultured in broth with 3 mM 2,2′-bipyridine for another 16 hours. The process was repeated twice more using increasingly thinner populations of S. aureus at 106 and 105 cfu/mL.
Microarray printing
Dot-matrix arrays with 80 and 150 μm periodicities along the x- and y-directions respectively were prepared with a piezoelectric inkjet printer (DMP–2800, Fujifilm Dimatix), using a monopolar waveform with a voltage of 25–30 V. Hemin conjugate 1 was dissolved in 50:50 DMSO:PBS (2 mg/mL) and jetted onto epoxy-activated glass slides (Nexterion E, Schott) cut to 1 × 1 cm. The microarrays were incubated for 12 hours at room temperature in 75% relative humidity, then gently rinsed by spraying the back face of the chips with water, PBS containing 0.005% Tween-20, then water again before drying under a nitrogen stream. The patterned chips were blocked by immersion in 0.1% BSA for one hour, followed by gentle rinsing with water and drying as described above.
Pathogen detection by label-free optical imaging
Hemin microarrays were exposed to live bacteria cultures for 15–60 minutes at 106–103 cfu/mL, then gently rinsed with water and imaged under darkfield conditions with a resolution of 1 pixel/μm2. FFT image analysis was performed using commercial software (WSxM) as previously described;14 signal quality was based on the intensity of the principal harmonic peak in reciprocal lattice space (ky = 1/150 μm−1) relative to background noise, using S/N = 3 as the LOD threshold. Background noise (N) was quantified as the standard deviation of spectral data between 1/300 and 1/75 μm−1, not including principal or secondary harmonic peaks. In some cases, the limit of detection was statistically strengthened by combining the data from three darkfield images of the same experimental sample prior to FFT analysis, which effectively increased the sampling population.
Acknowledgments
The authors acknowledge financial support from the Department of Defense (W911SR-08-C-0001) through the U.S. Army RDECOM (Edgewood Contracting Division), the National Science Foundation (CMMI-1449358), the Birck Nanotechnology Center at Purdue University, the Purdue University Center for Cancer Research (P30 CA023168), and startup funds from Purdue University. We thank BEI Resources at NIAID for providing several of the bacterial strains used in this study, and Phil Low and Ron Reifenberger for helpful discussions over the years.
References
- 1.Alanis AJ. Resistance to antibiotics: Are we in the post-antibiotic era? Arch Med Res. 2005;36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR 1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Antimicrobial Resistance: WHO Global Report on Surveillance. World Health Organization Press; Geneva: 2014. [Google Scholar]
- 4.Rice LB. Progress and Challenges in Implementing the Research on ESKAPE Pathogens. Infect Control Hosp Epidemiol. 2010;31:S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 5.Binkley CE, Cinti S, Simeone DM, Colletti LM. Bacillus Anthracis as an Agent of Bioterrorism: A Review Emphasizing Surgical Treatment. Ann Surg. 2002;236:9–16. doi: 10.1097/00000658-200207000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, et al. Better tests, better care: Improved diagnostics for infectious diseases. Clin Infect Dis. 2013;57:S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gracia JF, Onnela J-P, Barnett ML, Eguíluz VM, Christakis NA. Spread of pathogens in the patient transfer network of US hospitals. 2015 arXiv 1504.08343 [physics.soc-ph] [Google Scholar]
- 8.Centers for Disease Control and Prevention, U.S. Department of Health and Human Services. Antimicrobial Resistance Posing Growing Health Threat. 2011 http://www.cdc.gov/media/releases/2011/p0407_antimicrobialresistance.html (downloaded 5/4/2016)
- 9.Dwivedi HP, Jaykus LA. Detection of pathogens in foods: The current state-of-the-art and future directions. Crit Rev Microbiol. 2011;37:40–63. doi: 10.3109/1040841X.2010.506430. [DOI] [PubMed] [Google Scholar]
- 10.López-Campos G, Martínez-Suárez J, Aguado-Urda M, López-Alonso V. Microarray Detection and Characterization of Bacterial Foodborne Pathogens. Springer; US: 2012. Detection, Identification, and Analysis of Foodborne Pathogens; pp. 13–32. [Google Scholar]
- 11.Kim J, Lim J, Lee C. Quantitative real-time PCR approaches for microbial community studies in wastewater treatment systems: Applications and considerations. Biotechnol Adv. 2013;31:1358–1373. doi: 10.1016/j.biotechadv.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Su W, Gao X, Jiang L, Qin J. Microfluidic platform towards point-of-care diagnostics in infectious diseases. J Chromatogr A. 2015;1377:13–26. doi: 10.1016/j.chroma.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Adak AK, Leonov AP, Ding N, Thundimadathil J, Kularatne S, Low PS, Wei A. Bishydrazide glycoconjugates for recognition and capture of bacterial pathogens. Bioconjugate Chem. 2010;21:2065–2075. doi: 10.1021/bc100288c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adak AK, Boley JW, Lyvers DP, Chiu GT, Low PS, Reifenberger R, Wei A. Label-Free Detection of Staphylococcus aureus Captured on Immutable Ligand Microarrays. ACS Appl Mater Interfaces. 2013;5:6404–6411. doi: 10.1021/am4016236. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Lyvers DP, Wei A, Reifenberger RG, Low PS. Label-Free Detection of a Bacterial Pathogen Using an Immobilized Siderophore, Deferoxamine. Lab Chip. 2012;12:971–976. doi: 10.1039/c2lc20904g. [DOI] [PubMed] [Google Scholar]
- 16.Otto BR, Verweij-van Vught AM, MacLaren DM. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 17.Nobles CL, Maresso AW. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics. 2011;3:788–796. doi: 10.1039/c1mt00047k. [DOI] [PubMed] [Google Scholar]
- 18.Wilks A, Barker KD. Mechanisms of Heme Uptake and Utilization in Bacterial Pathogens. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science. Vol. 15. World Scientific Publishing; Singapore: 2010. pp. 357–398. [Google Scholar]
- 19.Benson D, Rivera M. Heme Uptake and Metabolism in Bacteria. In: Banci L, editor. Metallomics and the Cell. Springer; Berlin: 2013. pp. 279–332. [DOI] [PubMed] [Google Scholar]
- 20.Clarke TE, Tari LW, Vogel HJ. Structural biology of bacterial iron uptake systems. Curr Top Med Chem. 2001;1:7–30. doi: 10.2174/1568026013395623. [DOI] [PubMed] [Google Scholar]
- 21.Farrand AJ, Skaar EP. Heme and Infectious Diseases. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science. Vol. 26. World Scientific Publishing; Singapore: 2013. pp. 317–377. [Google Scholar]
- 22.Mayfield JA, Dehner CA, DuBois JL. Recent advances in bacterial heme protein biochemistry. Curr Op Chem Biol. 2011;15:260–266. doi: 10.1016/j.cbpa.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microb Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Sheldon JR, Heinrichs DE. Recent developments in understanding the iron acquisition strategies of Gram-positive pathogens. FEMS Microbiol Rev. 2015;39:592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- 25.Mason HY, Lloyd C, Dice M, Sinclair R, Ellis W, Jr, Powers L. Taxonomic identification of microorganisms by capture and intrinsic fluorescence detection. Biosens Bioelectr. 2003;18:521–527. doi: 10.1016/s0956-5663(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 26.Giuntini F, Boyle R, Sibrian-Vazquez M, Vicente MGH. Porphyrin Conjugates for Cancer Therapy. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science. Vol. 27. World Scientific Publishing; Singapore: 2010. pp. 303–416. [Google Scholar]
- 27.Azad BB, Cho CF, Lewis JD, Luyt LG. Synthesis, radiometal labeling and in vitro evaluation of a targeted PPIX derivative. Appl Radiat Isotop. 2012;70:505–511. doi: 10.1016/j.apradiso.2011.11.054. [DOI] [PubMed] [Google Scholar]
- 28.NMR analysis of 1′ could not be performed due to the presence of paramagnetic Fe(III).
- 29.Zunszain PA, Ghuman J, Komatsu T, Tsuchida E, Curry S. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct Biol. 2003;3:6. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriwaki Y, Caaveiro JMM, Tanaka Y, Tsutsumi H, Hamachi I, Tsumoto K. Molecular Basis of Recognition of Antibacterial Porphyrins by Heme-Transporter IsdHNEAT3 of Staphylococcus aureus. Biochemistry 2011. 2011;50:7311–7320. doi: 10.1021/bi200493h. [DOI] [PubMed] [Google Scholar]
- 31.Tai SS, Lee CJ, Winter RE. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect Immun. 1993;61:5401–5405. doi: 10.1128/iai.61.12.5401-5405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero-Espejel ME, Gonzalez-Lopez MA, de Jesus Olivares-Trejo J. Streptococcus pneumoniae requires iron for its viability and expresses two membrane proteins that bind haemoglobin and haem. Metallomics. 2013;5:384–389. doi: 10.1039/c3mt20244e. [DOI] [PubMed] [Google Scholar]
- 33.McConnell MJ, Actis L, Pachon J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 34.Zimbler D, Penwell W, Gaddy J, Menke S, Tomaras A, Connerly P, Actis L. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. BioMetals. 2009;22:23–32. doi: 10.1007/s10534-008-9202-3. [DOI] [PubMed] [Google Scholar]
- 35.Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, Badger JH, Durkin AS, Huot H, Shrivastava S, Kothari S, et al. Complete Genome Sequence of the N2-Fixing Broad Host Range Endophyte Klebsiella pneumoniae 342 and Virulence Predictions Verified in Mice. PLoS Genetics. 2008;4:e1000141. doi: 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crosa JH, Mey AR, Payne SM. Iron Transport in Bacteria. American Society of Microbiology Press; 2004. [Google Scholar]
- 37.Runyen-Janecky LJ. Role and regulation of heme iron acquisition in Gram-negative pathogens. Front Cell Infect Microbiol. 2013;3:55. doi: 10.3389/fcimb.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammer ND, Skaar EP. Molecular Mechanisms of Staphylococcus aureus Iron Acquisition. Annu Rev Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, Stillman MJ. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283:28125–28136. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maltais TR, Morales-de-Echegaray AV, Younis W, Kadasala NR, Dutta S, Seleem MN, Wei A. Manuscript submitted 2016 [Google Scholar]
- 43.Yoo SM, Lee SY. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016;34:7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Li CZ, Vandenberg K, Prabhulkar S, Zhu X, Schneper L, Methee K, Rosser CJ, Almeide E. Paper based point-of-care testing disc for multiplex whole cell bacteria analysis. Biosens Bioelectron. 2011;26:4342–4348. doi: 10.1016/j.bios.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 45.Bruno JG. Application of DNA aptamers and quantum dots to lateral flow test strips for detection of foodborne pathogens with improved sensitivity versus colloidal gold. Pathogens. 2014;3:341–355. doi: 10.3390/pathogens3020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh A, et al. Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst. 2011;136:4780–4786. doi: 10.1039/c1an15547d. [DOI] [PubMed] [Google Scholar]
- 47.Tawil N, Mouawad F, Lévesque S, Sacher E, Mandeville R, Meunier M. The differential detection of methicillin-resistant, methicillin-susceptible and borderline oxacillin-resistant Staphylococcus aureus by surface plasmon resonance. Biosens Bioelectron. 2013;49:334–340. doi: 10.1016/j.bios.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 48.Koehler TM. Bacillus anthracis physiology and genetics. Mol Aspects Med. 2009;30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]