Abstract
Background
OncotypeDX, a multi-gene expression assay, has been incorporated into clinical practice as a prognostic and predictive tool. However, its use in resource-constrained international healthcare systems is limited. Here we develop and validate a simplified model using clinicopathologic criteria to predict OncotypeDX score.
Methods
Patients with estrogen receptor (ER) and/or progesterone receptor (PR) positive and HER2 negative invasive ductal carcinoma for whom the OncotypeDX test was successfully performed between 09/2008–12/2011 were retrospectively identified. Tumor size, nuclear and histologic grade, lymphovascular invasion, and ER and PR status were extracted from pathology reports. Data were split into a training dataset comprising women tested 09/2008–04/2011, and a validation dataset comprising women tested 04/2011–12/2011. Using the training dataset, linear regression analysis was used to identify factors associated with OncotypeDX score, and to create a simplified risk score and identify risk cutoffs.
Results
Estrogen and progesterone receptors, tumor size, nuclear and histologic grades, and lymphovascular involvement were independently associated with OncotypeDX. The full model explained 39% of the variation in the test data, and the simplified risk score and cutoffs assigned 57% of patients in the test data to the correct risk category (OncotypeDX score <18, 18–30, >30). 41% of patients were predicted to have OncotypeDX score<18; of these, 83%, 16%, and 2% had true scores of <18, 18–30, and >30, respectively.
Conclusions
Awaiting an inexpensive test that is prognostic and predictive, our simplified tool allows clinicians to identify a fairly large group of patients (41%) with very low chance of having high-risk disease (2%).
Keywords: OncotypeDX, breast cancer
Introduction
OncotypeDX (Genomic Health, Redwood City, CA) is a quantitative reverse transcription polymerase chain reaction-based assay shown to have prognostic and predictive value in early-stage estrogen receptor (ER) positive breast cancers. The OncotypeDX recurrence score (RS) is reported on a 0–100 scale, divided into low (<18), intermediate (18–30), or high risk (>30) categories. It is used to predict the benefits of chemotherapy added to adjuvant hormone therapy in ER positive early-stage breast cancer [1-3]. This multi-gene expression assay has been incorporated into the American Society of Clinical Oncology (ASCO) and National Comprehensive Cancer Network (NCCN) guidelines [4-8].
The current cost of OncotypeDX testing in the United States is approximately $4500 per case. The test has been available since 2004, and Medicaid coverage became available in 2007. While the use of OncotypeDX has been validated and its cost effectiveness has been established [9], its expense may be a concern in some health care systems and communities. In addition, it is time and labor intensive, requiring shipment of tissue samples to a single laboratory with an average 10–14 day turnaround time in the United States.
Because clinicians pre-select when to order testing using clinicopathologic parameters routinely available from tumor specimens, a reliable and inexpensive estimator of the OncotypeDX risk score could be extremely useful, particularly in many foreign healthcare systems with limited financial resources, and especially if it accurately reproduces not only the prognostic results, but also the predictive results. In this study, we sought to develop and validate a simplified estimation tool for the OncotypeDX RS. The scoring system sought to use established pathologic parameters to limit the cost of care. Although previous studies have developed models to predict OncotypeDX score based on histopathologic variables [10-15], our study is unique in that we used only variables that we expect would be a part of routine workup in international and resource-limited settings (i.e., we did not use Ki-67, CyclinD1, Nottingham Score or H-scores for ER and progesterone receptor [PR] [16]) and because we created a simplified risk score that can be calculated without a computer, increasing its clinical utility in such settings. In addition, our study was strengthened by the availability of a large sample size for model fitting and independent model validation.
Methods
After obtaining Institutional Review Board approval, women with ER/PR positive, HER2 negative, invasive ductal carcinoma of the breast for whom the OncotypeDX test was successfully performed between 09/2008–12/2011 were retrospectively identified from our institution's prospectively maintained database. The data were split into a model training dataset consisting of 984 women tested between 09/2008–04/2011, and a model validation dataset consisting of 299 women tested between 04/2011–12/2011. Clinicopathologic characteristics of the two groups were summarized using median and range for continuous covariates, and frequency and percentage for categorical covariates.
Our goal was to develop and validate a linear regression model for OncotypeDX RS, a score derived from the OncotypeDX genomic test that is used to assess the risk of recurrence in early-stage breast cancer and to aid in the decision of whether to use chemotherapy. OncotypeDX scores range from 0–100 and are grouped into three categories when reported commercially: low risk (RS <18), intermediate risk (RS 18–30), and high risk (RS >30).
Univariate linear regression models were used to assess the relationship between various pathologic factors and OncotypeDX score using the training dataset. Patients with HER2 IHC result = ‘3+’ were excluded. Patients with equivocal HER2 by IHC were only included if the FISH test confirmed lack of amplification. HER2 IHC result was examined as a dichotomous variable (>0 versus 0). Factors significant on univariate analysis were entered into a multivariable linear regression model, and backward selection (with selection criterion p < .05) was applied to obtain the final multivariable model. Due to missing data on some patients, the final model is fit on a dataset of 766 patients. To assess model performance, true observed OncotypeDX score was plotted against model-predicted score separately for the training and validation datasets, and R-squared values were calculated. In addition, there was interest specifically in the performance of the model to identify whether a patient's tumor was high risk (score >30). This was assessed using the concordance index (C-index), which represents the proportion of pairs where the model-predicted score is higher for the patient who was truly high risk (true OncotypeDX risk score > 30) than for the patient who was truly low/intermediate risk. The C-index can range from 0.5–1, with 1 representing perfect concordance and 0.5 representing a classifier that is no better than random guessing.
In order to create a tool that could be used easily in real time, a simpler OncotypeDX RS estimation tool was developed based on the final model. Continuous covariates (ER and PR % staining) were dichotomized (80 was selected as a cutoff for both based on observed patterns in the training dataset), and a new multivariable linear regression model was fit. In order to obtain a more clinically useful model while preserving the magnitude of the effects estimated by the model, coefficients from this model were rounded to the nearest integer to obtain the number of points associated with each risk factor. Using the simplified risk score, we classified patients into three groups (0–6 points, 7–12 points, and 13–21 points). Point cutoffs were selected on the basis of their performance and utility in the training dataset only, prior to the assessment of performance in the validation dataset. The number of patients in each simplified risk-score group was reported, and the distribution of true OncotypeDX score (low, intermediate, high) in each group was reported. All statistical analysis was performed in SAS 9.3 (SAS Institute, Cary, NC) and R 3.1.1 (R Foundation, Vienna, Austria), and p-values < 0.05 were considered significant.
Results
The OncotypeDX test was successfully performed on 984 women with ER/PR positive, and HER2 negative invasive breast carcinomas between 09/2008–04/2011. These 984 women formed the training dataset. In this dataset, the median age was 58 years (range 24–90, interquartile range [IQR] 49–65) and the median tumor size was 11mm (range 0.2–47, IQR 8–16). Most women (58%) were classified as low risk based on OncotypeDX RS; 34% were intermediate risk and 8% were high risk (Supplementary Table 1).
Factors associated with OncotypeDX score
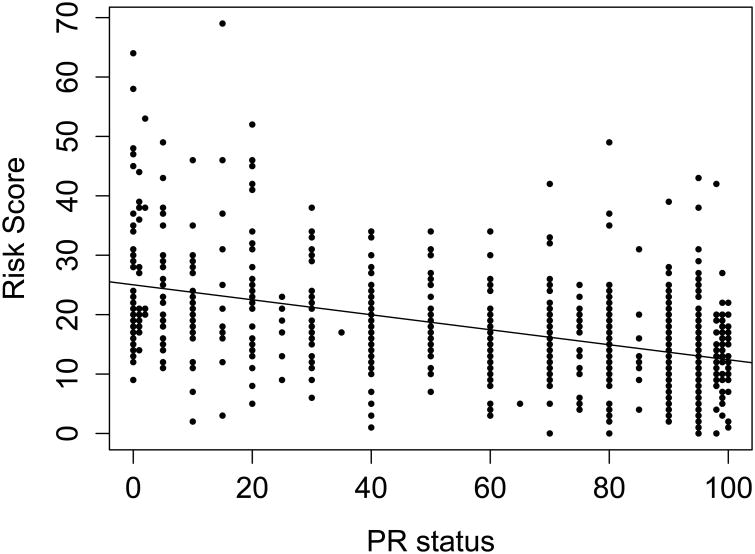
On univariate analysis, ER status, PR status, tumor size >20mm, nuclear grade, histologic grade, lymphovascular invasion (LVI), and HER2 status were all significantly associated with OncotypeDX score (p < .05)(Table 1). The explanatory variable with the largest strength of association (as measured by R2) was PR status, which explained 23% of the variation in OncotypeDX score (R2=0.2347)(Fig. 1). In a multivariable model, LVI and HER2 were no longer significant, and ER status, PR status, tumor size, and nuclear and histologic grade were independently associated with OncotypeDX score (all p<.05)(Table 1). One-unit increases in ER and PR status conferred small (0.11 and 0.21 point) drops in RS. Size >20mm, histologic grade 3, and nuclear grade >1 were associated with moderate (1.3–6.1 point) increases in RS. The multivariable model is estimated on the 766 women with complete data on all model covariates.
Table 1. Linear regression analysis of factors associated with OncotypeDX recurrence score.
| Univariate models | Multivariable model* (n = 766) | Multivariable model with ER and PR as binary** (n=766) | ||||
|---|---|---|---|---|---|---|
| Variable | Effect estimate (standard error) | P-value | Effect estimate (standard error) | P-value | Effect estimate (standard error) | P-value |
| ER | -0.27 (0.02) | < .001 | -0.21 (0.02) | < .0001 | ER ≤ 80: 5.01 (0.93) | < .0001 |
| PR | -0.13 (0.01) | < .001 | -0.11 (0.01) | < .0001 | PR ≤ 80: 5.47 (0.59) | < .0001 |
| Tumor size > 20 mm | 4.13 (0.95) | < .001 | 2.56 (0.80) | 0.0014 | 2.97 (0.91) | 0.0012 |
| Nuclear grade | ||||||
| 2 vs 1 | 3.18 (1.12) | < .001 | 2.36 (0.94) | 0.0122 | 1.98 (1.07) | 0.0644 |
| 3 vs 1 | 10.51 (1.27) | 6.13 (1.13) | < .0001 | 7.00 (1.38) | < .0001 | |
| Histologic grade 3 vs 1/2 | 3.72 (0.60) | < .001 | 1.29 (0.54) | 0.0181 | 1.32 (0.62) | 0.0331 |
| LVI | 1.94 (0.78) | 0.014 | NA | NA | NA | NA |
| HER2 > 0 vs 0 | 1.62 (0.69) | 0.020 | NA | NA | NA | NA |
| Model performance metrics | ||||||
| Multivariable model | Multivariable model with ER and PR as binary | |||||
| Model R2 | ||||||
| Training dataset | 0.42 | 0.25 | ||||
| Validation dataset | 0.39 | 0.27 | ||||
| Model c-index (high RS versus intermediate/low RS) | ||||||
| Training dataset | 0.854 | 0.822 | ||||
| Validation dataset | 0.868 | 0.816 | ||||
Intercept estimate for multivariable model is 37.10. 218 patients were not included in the multivariable model due to missing information for one or more model covariates.
Intercept estimate is 6.57. This model was used to develop the simplified risk score.
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; LVI, lymphovascular invasion
Fig 1.
OncotypeDX risk score versus PR status, with line of best fit (training dataset, n = 915, R2 = 0.2347).
Abbreviations: PR, progesterone receptor
Model performance and external validation
From 04/2011–12/2011, 299 patients met inclusion criteria and were used to validate this OncotypeDX RS prediction tool. The validation cohort had similar clinical characteristics to the training cohort (Supplementary Table 1). Six patients in the validation dataset were missing histologic grade so no model-predicted score could be obtained for them.
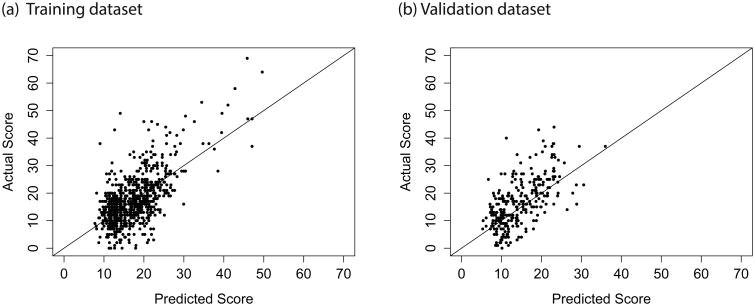
In both the training and validation datasets, we observed correlation between the model-predicted score and the true OncotypeDX score (Fig. 2), with our final model explaining 42% of the variation in score in the training data and 39% of the variation in the validation data (Table 1). Our model had good discrimination between patients with high OncotypeDX score (>30) and low/intermediate OncotypeDX score (training data C-index =0.854, validation data C-index=0.868). The model assigns 68% of the patients in the training data and 71% of the patients in the test data to the correct risk category (OncotypeDX score <18, 18–30, >30).
Fig 2.
Actual OncotypeDX score versus score predicted by the final multivariable model. (a) Training dataset (n = 766, R2 = 0.4184); (b) Validation dataset (n = 293, R2 = 0.3856).
Simplified risk score
A simplified risk score was developed to allow for quick and easy risk prediction (Table 2). The risk score is based on a modified version of the final multivariable model with ER and PR treated as binary covariates (Table 1), The simplified risk score is calculated by summing the points associated with each risk factor that is present: 7 points for nuclear grade 3, 2 points for nuclear grade 2, 5 points for ER ≤80, 5 points for PR ≤80, 3 points for tumor size >20mm, and 1 point for histologic grade 3. In both the training and validation datasets, the association between the simplified score and the true OncotypeDX score was highly significant (p<.0001). R2 was equal to 0.2455 in the training dataset and 0.2759 in the validation dataset.
Table 2.
The simplified risk score is formed by taking the sum of the points assigned to each risk factor that is present. The score ranges from 0 to 21
| Risk factor | Number of points |
|---|---|
| ER ≤ 80 | 5 points |
| PR ≤ 80 | 5 points |
| Tumor size > 20 mm | 3 points |
| Nuclear grade 2 | 2 points |
| Nuclear grade 3 | 7 points |
| Histologic grade 3 | 1 point |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor
As shown in Table 3, in the training dataset, 34% of patients had a simplified risk score between 0–7. In this group, 80% of patients were low risk based on their true OncotypeDX score, 19% were intermediate risk, and only 1% were high risk. Half of the patients had a simplified risk score between 7–12 points, and of the patients in this group, 5% had a high OncotypeDX score. The remaining 16% of patients had a simplified risk score between 13–21. In this group, 27% (33/121) of patients were high risk based on true OncotypeDX score. Performance of the simplified risk score was similar in the validation dataset, although slightly more patients had a simplified risk score between 0 and 7 (41%, compared to 34% in the training dataset)(Table 3). The C-index for the simplified risk score using the outcome OncotypeDX score >30 was 0.808 in both the training and validation datasets. Using the selected cutoffs for the simplified risk score, the score assigns 52% of the patients in the training data and 57% of the patients in the test data to the correct risk category (OncotypeDX score <18, 18–30, >30).
Table 3.
Performance of the simplified risk score in the training data (n = 766) and performance of the simplified risk score in the validation data (n = 293).
| Training Data (n = 766) | |||||
|---|---|---|---|---|---|
| Simplified risk score | Actual OncotypeDX score | Percent of patients in this risk category | |||
| Low risk (< 18) | Intermediate risk (18–30) | High risk (> 30) | Total | ||
| < 7 points | 212 (80%) | 49 (19%) | 3 (1%) | 264 (100%) | 34% |
| 7–12 points | 205 (54%) | 156 (41%) | 20 (5%) | 381 (100%) | 50% |
| > 12 points | 32 (26%) | 56 (46%) | 33 (27%) | 121 (100%) | 16% |
| Validation Data (n = 293) | |||||
| Simplified risk score | Actual OncotypeDX score | Percent of patients in this risk category | |||
| Low risk (< 18) | Intermediate risk (18–30) | High risk (> 30) | Total | ||
| < 7 points | 100 (83%) | 19 (16%) | 2 (2%) | 121 (100%) | 41% |
| 7–12 points | 68 (53%) | 54 (42%) | 6 (5%) | 128 (100%) | 44% |
| > 12 points | 5 (11%) | 26 (59%) | 13 (30%) | 44 (100%) | 15% |
We were additionally interested in whether our simplified model could identify patients with very low OncotypeDX scores, that is, scores from 0–10. In the training data, patients with even the lowest simplified risk score (score=0 or 1) still had a 50% chance of having a true Oncotype score >10.
Discussion
While OncotypeDX is incorporated into ASCO, NCCN, and St. Gallen guidelines as both prognostic for cancer recurrence and predictive of chemotherapy benefit, its use is not universal in all healthcare systems, particularly in countries with limited financial resources. Guth et al examined the use of OncotypeDX in minority and economically disadvantaged women, comparing OncotypeDX testing incidence in an inner-city population at municipal versus tertiary referral centers in the United States. They noted that when Medicaid coverage became available for OncotypeDX in 2007, minority women at tertiary centers began to undergo testing; however, women treated at municipal hospitals were less likely to undergo testing, resulting in a misleading racial disparity driven by care site [17]. In a retrospective observational cohort study by Chen et al, practice characteristics driving OncotypeDX use were analyzed in over 6200 patients using the iKnowMed electronic health record system of McKesson Specialty Health and The US Oncology Network. They found that of all patients eligible, only 29% underwent OncotypeDX testing, and that these patients were younger and had better Eastern Cooperative Oncology Group (ECOG) performance status. They concluded that in many patients, the decision whether to use chemotherapy is oftentimes based on other factors such as tumor type, tumor size, ECOG performance status, patient age, or patient or physician treating preference, rather than OncotypeDX recurrence scores [18]. Bombard et al looked at medical oncologists' views of gene-expression profiling tests such as OncotypeDX by conducting telephone interviews. While many oncologists considered the testing to be a tool that enhanced confidence in their established approach to risk assessments, many had concerns about the cost of testing and the over-reliance on the results within the medical community [19].
Others have attempted to develop an accurate model for OncotypeDX using routine histopathologic variables [10, 11, 13-15]. The Magee equations use Nottingham score, ER H-score, PR H-score, tumor size, and Ki-67 percentage to estimate OncotypeDX score and are available for free online [14]. Our model is unique in that it uses only variables that we expect would be a part of routine workup in international and resource-limited settings, and that, due to its simplicity, it can be calculated instantly without a computer. Our model does not use Ki-67, which may not always be available, especially in resource-limited settings, and it does not use semi-quantitative H-scores, which also may not be part of routine pathologic evaluation. Our model performed similarly to the Magee equations presented in Klein et al in terms of the number of patients assigned to the correct risk category (OncotypeDX score <18, 18–30, >30). In Klein et al, the percent of patients assigned to the correct OncotypeDX RS category ranged from 54.3% to 59.4% across the 4 versions of the equation (original equation and new equations 1, 2, and 3), compared to 57% of patients in our test dataset.
Cuzick et al showed that classical variables plus the 4 IHC markers provided a similar amount of prognostic information as the OncotypeDX score, providing supporting evidence for the idea that a score based on routine histopathologic variables could one day replace OncotypeDX [11]. We could not directly compare our score to theirs, as they measured performance relative to the primary outcome of distant recurrence and we targeted OncotypeDX score itself.
A study exploring the ability of oncologists to predict the RS using standard prognostic criteria revealed that oncologists are able to differentiate between a low or intermediate RS and a high RS using standard prognostic criteria [12]. However, in this study, provision of the actual RS changed the treatment recommendations in nearly 20% of cases, most often in intermediate-risk cases, suggesting that the RS may reduce chemotherapy use [12]. Another study looking at the ability of oncologists to estimate recurrence risk also showed that specialists tend to overestimate the risk of recurrence compared with RS, resulting in potential significant overtreatment of breast cancer patients with adjuvant chemotherapy. Inclusion of actual RS resulted in a 24.9% change in treatment recommendations [20].
In our study, we developed and validated a linear regression model for OncotypeDX RS. Interestingly, PR status was strongly associated with OncotypeDX score and alone explained 23% of the variation in OncotypeDX scores. Our final model explained 42% of the variation in scores in the training data and 39% of the variation in the validation data. Our final model had good discrimination between patients with high OncotypeDX scores and low/intermediate OncotypeDX scores (training data C-index=0.854, validation data C-index=0.868). Furthermore, a simpler OncotypeDX RS estimation tool was developed based on this final model to allow for quick and easy risk prediction. In both training and validation datasets, the simplified risk score had good discrimination between patients with high and low/intermediate OncotypeDX scores (0.808 in both the training and validation datasets). Despite its high discrimination, the simplified risk score using the selected cutoffs has a high misclassification rate, assigning 48% of the patients in the training data and 43% of the patients in the test data to the wrong risk category. Additionally, when we tried to use the tool to identify the very low-risk group defined by Sparano et al (OncotypeDX score 0–10), we found that even among patients with the lowest simplified risk scores, 50% had true OncotypeDX scores >10 [21]. Though this limits its utility, we believe the tool may be useful in health care systems where OncotypeDX testing is not readily available or commonly utilized by showing doctors the distribution of OncotypeDX score for patients in the same simplified risk-score category as their patients (Table 3). The tool allows the identification of a fairly large group of patients (41%) who have a very small chance of being high risk (2%).
One of the limitations of this model is that it was developed using data from a defined set of patients—those who received OncotypeDX testing—hence the possibility of selection bias. Furthermore, the model was developed and validated in the same center. Testing the ability of this simplified tool to predict OncotypeDX RS, in a set of patients at a different center, may further validate its potential.
Within our study design limits, we developed and attempted to validate an estimation tool of OncotypeDX RS. When cost and time are a consideration in utilizing OncotypeDX as a diagnostic tool, our model provides an estimation of risk at lower cost. Our model is not intended to replace the Oncotype RS or other models, but rather to provide a very simple method to calculate risk in settings where other risk estimation methods may not be available. While waiting for a less-expensive test than OncotypeDX that can reduce the overuse of chemotherapy, our model provides a simplified, available technique with the potential to assist physicians and patients in a healthcare system with limited financial resources. The model uses a minimal set of clinical and pathologic variables, and can be calculated instantly without a computer. This model may be useful in low- and middle-income countries, particularly when the financial burden falls on the patients.
Supplementary Material
Supplementary Table 1 Characteristics of the training and validation datasets
Synopsis.
Here we develop and validate a simplified model using clinicopathologic criteria to predict OncotypeDX score. We find our simplified tool allows clinicians to identify a fairly large group of patients with very low chance of having high-risk disease.
Acknowledgments
Funding: This study was funded in part by NIH/NCI Cancer Center Support Grant No. P30 CA008748.
Footnotes
Disclosures: Presented in part at the 2012 Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 1–5, 2012, and the 2013 Annual Meeting of the American Society of Breast Surgeons, Chicago, IL, May 3–5, 2013.
Conflict of interest: The authors have no conflicts of interest to declare.
Compliance with Ethical Standards: Informed consent: This was a retrospective study without patient identifiers.
References
- 1.Bao T, Davidson NE. Gene expression profiling of breast cancer. Adv Surg. 2008;42:249–260. doi: 10.1016/j.yasu.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross JS, Hatzis C, Symmans WF, Pusztai L, Hortobagyi GN. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13(5):477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 3.Turaga K, Acs G, Laronga C. Gene expression profiling in breast cancer. Cancer Control. 2010;17(3):177–182. doi: 10.1177/107327481001700306. [DOI] [PubMed] [Google Scholar]
- 4.Dowsett M, Cuzick J, Wale C, Forbes J, Mallon EA, Salter J, Quinn E, Dunbier A, Baum M, Buzdar A, Howell A, Bugarini R, Baehner FL, Shak S. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J Clin Oncol. 2010;28(11):1829–1834. doi: 10.1200/JCO.2009.24.4798. [DOI] [PubMed] [Google Scholar]
- 5.Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, Baker J, Walker M, Watson D, Hackett J, Blick NT, Greenberg D, Fehrenbacher L, Langholz B, Quesenberry CP. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8(3):R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC, Jr, American Society of Clinical O. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network (NCCN) NCCN Clinical Practice Guidelines in Oncology: Breast Cancer V.2. 2008;2008 [Google Scholar]
- 8.Toi M, Iwata H, Yamanaka T, Masuda N, Ohno S, Nakamura S, Nakayama T, Kashiwaba M, Kamigaki S, Kuroi K Japan Breast Cancer Research Group-Translational Research G. Clinical significance of the 21-gene signature (Oncotype DX) in hormone receptor-positive early stage primary breast cancer in the Japanese population. Cancer. 2010;116(13):3112–3118. doi: 10.1002/cncr.25206. [DOI] [PubMed] [Google Scholar]
- 9.Kondo M, Hoshi SL, Yamanaka T, Ishiguro H, Toi M. Economic evaluation of the 21-gene signature (Oncotype DX) in lymph node-negative/positive, hormone receptor-positive early-stage breast cancer based on Japanese validation study (JBCRG-TR03) Breast Cancer Res Treat. 2011;127(3):739–749. doi: 10.1007/s10549-010-1243-y. [DOI] [PubMed] [Google Scholar]
- 10.Allison KH, Kandalaft PL, Sitlani CM, Dintzis SM, Gown AM. Routine pathologic parameters can predict Oncotype DX recurrence scores in subsets of ER positive patients: who does not always need testing? Breast Cancer Res Treat. 2012;131(2):413–424. doi: 10.1007/s10549-011-1416-3. [DOI] [PubMed] [Google Scholar]
- 11.Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, Zabaglo L, Mallon E, Green AR, Ellis IO, Howell A, Buzdar AU, Forbes JF. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29(32):4273–4278. doi: 10.1200/JCO.2010.31.2835. [DOI] [PubMed] [Google Scholar]
- 12.Kamal AH, Loprinzi CL, Reynolds C, Dueck AC, Geiger XJ, Ingle JN, Carlson RW, Hobday TJ, Winer EP, Goetz MP. Breast medical oncologists' use of standard prognostic factors to predict a 21-gene recurrence score. Oncologist. 2011;16(10):1359–1366. doi: 10.1634/theoncologist.2011-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Umbricht C, Illei PB, Magalhaes MCF, Pesce C, Gage MM, Mylander C, Rosman M, Tafra L, Visvanathan K, Cope L, Wolff AC. An estimation model for Oncotype DX recurrence score using routine histopathologic variables. J Clin Oncol. 2014;32(5s) suppl; abstr 559. [Google Scholar]
- 14.Klein ME, Dabbs DJ, Shuai Y, Brufsky AM, Jankowitz R, Puhalla SL, Bhargava R. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26(5):658–664. doi: 10.1038/modpathol.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner BM, Skinner KA, Tang P, Jackson MC, Soukiazian N, Shayne M, Huston A, Ling M, Hicks DG. Use of modified Magee equations and histologic criteria to predict the Oncotype DX recurrence score. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28(7):921–931. doi: 10.1038/modpathol.2015.50. [DOI] [PubMed] [Google Scholar]
- 16.McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS., Sr Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Archives of pathology & laboratory medicine. 1985;109(8):716–721. [PubMed] [Google Scholar]
- 17.Guth AA, Fineberg S, Fei K, Franco R, Bickell NA. Utilization of Oncotype DX in an Inner City Population: Race or Place? Int J Breast Cancer. 2013;2013:653805. doi: 10.1155/2013/653805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Dhanda R, Tseng WY, Forsyth M, Patt DA. Evaluating use characteristics for the oncotype dx 21-gene recurrence score and concordance with chemotherapy use in early-stage breast cancer. J Oncol Pract. 2013;9(4):182–187. doi: 10.1200/JOP.2012.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bombard Y, Rozmovits L, Trudeau M, Leighl NB, Deal K, Marshall DA. The value of personalizing medicine: medical oncologists' views on gene expression profiling in breast cancer treatment. Oncologist. 2015;20(4):351–356. doi: 10.1634/theoncologist.2014-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joh JE, Esposito NN, Kiluk JV, Laronga C, Lee MC, Loftus L, Soliman H, Boughey JC, Reynolds C, Lawton TJ, Acs PI, Gordan L, Acs G. The effect of Oncotype DX recurrence score on treatment recommendations for patients with estrogen receptor-positive early stage breast cancer and correlation with estimation of recurrence risk by breast cancer specialists. Oncologist. 2011;16(11):1520–1526. doi: 10.1634/theoncologist.2011-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Dees EC, Perez EA, Olson JA, Zujewski J, Lively T, Badve SS, Saphner TJ, Wagner LI, Whelan TJ, Ellis MJ, Paik S, Wood WC, Ravdin P, Keane MM, Gomez Moreno HL, Reddy PS, Goggins TF, Mayer IA, Brufsky AM, Toppmeyer DL, Kaklamani VG, Atkins JN, Berenberg JL, Sledge GW. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. New England Journal of Medicine. 2015;373(21):2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Characteristics of the training and validation datasets