Genetic modulation of drug response can cause serious, potentially life-threatening adverse drug reactions, can increase susceptibility to drug-drug interactions, and can diminish or enhance therapeutic efficacy. As a result, the clinical implementation of pharmacogenomics at the bedside could make it possible to avoid adverse drug reactions and maximize drug efficacy. Ideally, selection of medications based on the genetic profile of individual patients will produce optimal effects for specific indications. Over the past decade, a large number of pharmacogenomic variants with demonstrated clinical utility have been identified and incorporated into drug labels by the US Food and Drug Administration (FDA).1 Recognizing that pharmacogenomics has the potential to immediately impact the care of patients in a clinically meaningful fashion, President Obama highlighted the importance of “delivering the right treatments, at the right time, every time to the right person” in his Precision Medicine Initiative Remarks.2 However, the incorporation of pharmacogenomics into clinical practice has proved slow and challenging. Some of those challenges include: 1) delay in the initiation of therapy due to the use of traditional reactive, as opposed to preemptive, ordering of pharmacogenomic testing, 2) lack of support for commercial electronic health record (EHR) systems to integrate genomic data and enable automated clinical decision support at the point of care, 3) limited evidence of the benefit for preemptive testing for efficacy as opposed to testing to avoid rare toxicities, 4) complexity of interpretation of the genotype data, and 5) limited coverage and reimbursement of pharmacogenomic testing by health insurance companies.3 Given these challenges, early implementation of large scale preemptive pharmacogenomics has been restricted to large medical centers with substantial institutional support to cover the costs.4-6 Expansion of these efforts to patient populations willing to pay out of pocket to defray costs could facilitate wider adoption. However, the types of patients willing to pay out of pocket and the amount these patients would be willing to pay for pharmacogenomic testing are unknown.
Direct-to-consumer (DTC) genetic testing companies provide some clues as to what patients might be willing to pay for pharmacogenomic testing. 23andMe is the most widely used DTC genetic testing company that provides health-related information as well as ancestry information to over 1,000,000 customers.7 Cost of testing was initially $999 in 2007, was lowered to $99 in 2012, and was raised to $199 in 2015 with the introduction of a new FDA compliant report (www.23andMe.com). Customers have access to their raw genetic data, but the health-related portion of the interpreted report includes only a small set of results for rare disease carrier status. Further limiting the utility of these results is the lack of integration with individual patient's EHR for use in current and future medical care. Clearly, large numbers of consumers are willing to pay for DTC genetic testing; however, it remains to be determined what types of patients would be willing to incur out of pocket costs to have their health care provider order genetic testing, and what amount would these patients be willing to pay out of pocket. It may be that in the actual health care context, patients have the expectation that the costs of genetic testing should be covered by their health insurance plan. To address these questions, we surveyed patients participating in the Mayo Clinic Right Drug, Right Dose, Right Time Protocol (RIGHT Protocol) study.4 The RIGHT Protocol is tasked with extending pharmacogenomics implementation beyond “reactive genotyping,” to include “preemptive sequencing,” with integration of clinically actionable pharmacogenomic variants into the EHR in the Mayo Clinic and Mayo Clinic Health Systems. The RIGHT Protocol recruited 1013 patients and identified variants in CYP2C19, CYP2C9, VKORC1, SLCO1B1, and CYP2D6 that could cause drug-gene interactions with warfarin, clopidogrel, simvastatin, tamoxifen, opioid analgesics, and selective serotonin reuptake inhibitors. Variant information was added to the EHR for each patient, and parallel point of care clinical decision support for drug-gene interactions was implemented. This study was conducted according to the principles of the Declaration of Helsinki, and informed consent was obtained from all participants. The Mayo Clinic institutional review board approved the study.
In December 2014, all 1010 living RIGHT participants were sent a survey assessing their experiences with and understanding of pharmacogenomics along with their CYP2D6 results and pharmacogenomics educational materials. Patient characteristics for the 869 (86%) who completed the survey are included in Table 1. The average age of the RIGHT patients was 59 years and nearly 40% of patients had a clinically actionable CYP2D6 result (i.e., drug or dose change is recommended). Of those who provided an answer to individual questions, 77% were taking one or more prescription medications, 70% self-reported excellent or very good health, and about 70% strongly or somewhat agreed that they would ask for additional pharmacogenetic tests if it were to become available. Financial strain was assessed with the question “How would you describe your household's financial situation right now?” to account for the wide variation in access to financial resources in people with similar incomes.8 Overall, 65% of patients indicated that after paying the bills, they still had enough money for special things. In contrast, roughly 8% indicated they had to cut back in order to pay bills or were having difficulty paying the bills. We assessed the patient's willingness (or unwillingness) to pay for pharmacogenomic testing by the following question: “If pharmacogenomic tests were available, what would be the maximum out of pocket cost you would be willing to pay?” with responses of $100, $250, $500, $1000, $2000, $5000, and “I would not pay for this unless completely covered by insurance.” Finally, we assessed the patients' perceived benefit of pharmacogenomics testing by assessing whether they would undergo addition pharmacogenomic testing if available and ascertaining their level of confidence that their test results would 1) be used by their health care provider, 2) reduce side effects, and 3) improve chances of getting a medication and a dose that is right for them.
Table 1. Characteristics of the RIGHT Protocol Participant Survey Respondents.
| Characteristics | |
|---|---|
| n | 869 |
| Age, mean years ± standard deviation | 59 ± 5.5 |
| Female, n (%) | 475 (55) |
| Number of prescription medications used in the past month, n (%) (n=797 responded) | |
| 0 | 183 (23) |
| 1-2 | 342 (43) |
| 3+ | 272 (34) |
| Drug/dose change recommended based on CYP2D6 phenotype, n (%)† | |
| No | 530 (61) |
| Yes | 339 (39) |
| Self-reported health status, n (%) (n=858 responded) | |
| Excellent | 153 (18) |
| Very good | 447 (52) |
| Good | 216 (25) |
| Fair | 37 (4) |
| Poor | 5 (1) |
| If more pharmacogenomic tests were to become available, I would ask my health care provider to order these for me, n (%) (n=804 responded) | |
| Strongly agree | 305 (38) |
| Somewhat agree | 253 (31) |
| Neither agree nor disagree | 216 (27) |
| Somewhat disagree | 23 (3) |
| Strongly disagree | 7 (1) |
| Self-reported financial status, n (%) (n=823 responded) | |
| After paying the bills, I still have enough money for special things that I want | 538 (65) |
| I have enough money to pay the bills, but little spare money to buy extra or special things | 216 (26) |
| I have enough money to pay the bills, but only because I have cut back on things | 51 (6) |
| I am having difficulty paying the bills, no matter what I do | 18 (2) |
Phenotypes categorized as drug/dose change recommended are ultra-rapid, extensive to ultra-rapid, poor to intermediate, and poor. Phenotypes categorized as no drug/dose change recommended are extensive, intermediate to extensive, and intermediate.
The figure illustrates the relationship between available financial strain and willingness to pay for testing. Overall, 42% of the patients were not willing to incur out of pocket costs for pharmacogenomic testing. This proportion was highest in those patients with the most financial strain (88%), but a significant proportion of those with less financial strain were also unwilling to pay any amount of money for future testing (56% of those who have to cut back, 47% of those with little spare money, and 36% of those with enough money for special things). Using logistic regression, we assessed other factors that might be associated with a willingness to incur out of pocket costs including age, sex, number of prescription medications, self-reported health, and education. Results were reported as odds ratios (OR), 95% confidence intervals (CI), and associated P values. After adjusting for all other variables, increasing financial strain remained strongly associated with being unwilling to incur any out of pocket costs for pharmacogenomic testing (little spare money OR = 1.4; cut back to pay bills OR = 2.2; difficulty paying bills OR = 15.3, P for trend < 0.001). Likewise, after adjusting for other variables, older patients (OR per 10-year increase in age = 1.48, 95% CI = 1.09-2.01, P = 0.01) and women (OR = 1.36, 95% CI = 0.98-1.90, P = 0.06) were less likely to be willing to incur out of pocket costs. Patients who perceived a low value to pharmacogenomics testing were associated with being less willing to incur out of pocket costs (OR = 2.61, 95% CI = 1.81- 3.77, P < 0.001). Finally, of those patients willing to incur out of pocket costs, an overwhelming majority (87%) would not pay more than $250, with most (58%) indicating that $100 was their maximum.
Figure.
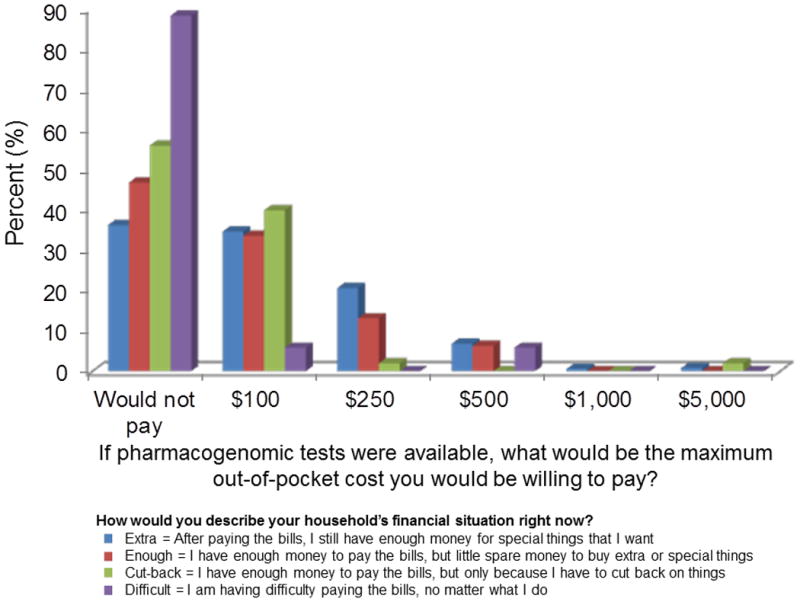
Our results indicate that while a majority of patients might be interested in and value pharmacogenomic testing, a large portion would only do so if it was completely reimbursed by insurance. This finding is particularly compelling since participants had first-hand knowledge of the potential benefits of pharmacogenomic testing with 21% of patients exposed to a drug acting through CYP2D6 in the prior 2 years and over 50% exposed during the past decade. Therefore, these results may reflect a best case scenario and indicate an informed ambivalence to the value of this testing. Or, it may be that the offering of genetic testing in a clinical setting – versus ordering a DTC genetic test on one's own volition – activates the notion that such testing should be a health insurance-covered expense. The impacts of these forces on testing uptake and willingness to pay may be compounded by the fact that it has been challenging to clearly establish the value of preemptive pharmacogenomic testing to the healthcare system despite the recognized value to individualized care.9-11 Cost-effectiveness, cost-utility, and cost-benefit analyses in the area of pharmacogenomics are needed to guide further discussion on this topic and better educate healthcare payers and patients. To facilitate these important research areas, the Mayo Clinic Center of Individualized Medicine is expanding the RIGHT Protocol study to include an additional 10,000 patients.
We observed that a patient's financial situation is a key driver of a willingness to incur any out of pocket costs for pharmacogenomics. These findings add empiric evidence to the concern that future benefits of pharmacogenomics testing may be disproportionately allocated to patients with more financial resources.12, 13 These results support the need for a national conversation about pharmacogenomic testing reimbursement given that advances in clinical medicine often result in widening health disparities among patients with social and economic vulnerability.14
Alternatively, the variable levels of willingness to pay across socioeconomic lines may reflect a differential ambivalence regarding the utility of genetic data. Qualitative work has demonstrated that members of underserved communities may not be particularly interested in the added value of personalized genetic information relative to more significant social and environmental factors contributing to health inequities.15 Although we were unable to access differences among underserved racial/ethnic minority patients, pharmacogenomics hold the promise to reduce health disparities through actionable observations of variable drug responses across ethnic groups for certain conditions.16 However, these advances concomitantly hold the risk to widen disparities through reification of race as a genetic, rather than social construct, with implications for exacerbated discrimination.17-20
In conclusion, strategies to implement pharmacogenomic testing outside of large academic medical centers, where testing can be supported by grant or institutional funding, are needed to address infrastructure and financial barriers. Personalizing treatment for patients based on their individual genetic variation will only improve the health of those who have access to them, further worsening the gap in health indicators in specific populations. Any undertaking of pharmacogenomics will need to include strategies to address the existing gap in healthcare disparities and take measures to ensure populations with economic vulnerability benefit from this new knowledge.
Acknowledgments
This work was supported in part by Mayo Clinic Center for Individualized Medicine, Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery, National Institutes of Health grants U19 GM61388 (The Pharmacogenomics Research Network), R01 GM28157, U01 HG005137, R01 CA138461, R01 AG034676 (The Rochester Epidemiology Project), and U01 HG06379 and U01 HG06379 Supplement (The Electronic Medical Record and Genomics (eMERGE) Network).
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.U.S. Food and Drug Administration Table of Pharmacogenomic Biomarkers in Drug Labels. http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- 2.Obama B. Remarks by the President on Precision Medicine. Office of the Press Secretary 2015 [Google Scholar]
- 3.Meckley LM, Neumann PJ. Personalized medicine: factors influencing reimbursement. Health Policy. 2010;94(2):91–100. doi: 10.1016/j.healthpol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther. 2014;96(4):482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Power of One Million. [2/16/2016]; http://Blog.23andme.com/news/one-in-a-million.
- 8.Szanton SL, Allen JK, Thorpe RJ, Jr, Seeman T, Bandeen-Roche K, Fried LP. Effect of financial strain on mortality in community-dwelling older women. J Gerontol B Psychol Sci Soc Sci. 2008;63(6):S369–374. doi: 10.1093/geronb/63.6.s369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wedlund PJ, de Leon J. Pharmacogenomic testing: the cost factor. Pharmacogenomics J. 2001;1(3):171–174. doi: 10.1038/sj.tpj.6500033. [DOI] [PubMed] [Google Scholar]
- 10.Wong WB, Carlson JJ, Thariani R, Veenstra DL. Cost effectiveness of pharmacogenomics: a critical and systematic review. Pharmacoeconomics. 2010;28(11):1001–1013. doi: 10.2165/11537410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 11.Monaco AD, Beretta M, Pugliese S, Valente D, Francia RD. Early outline evaluation of genotyping costs of pharmacogenomics. J Pharmacogenomics Pharmacoproteomics. 2014;5:123. [Google Scholar]
- 12.Brothers KB, Rothstein MA. Ethical, legal and social implications of incorporating personalized medicine into healthcare. Personalized medicine. 2015;12(1):43–51. doi: 10.2217/pme.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedgecoe A. The Politics of Personalised Medicine - Pharmacogenetics in the Clinic. Cambridge University Press; Cambridge, UK: 2005. [Google Scholar]
- 14.Tehranifar P, Neugut AI, Phelan JC, Link BG, Liao Y, Desai M, et al. Medical advances and racial/ethnic disparities in cancer survival. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2701–2708. doi: 10.1158/1055-9965.EPI-09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg AJ, Hartmann CD, Morello L, Brooks S, Colon-Zimmermann K, Marshall PA. Gene-environment interactions and health inequalities: views of underserved communities. J Community Genet. 2013;4(4):425–434. doi: 10.1007/s12687-013-0143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotimi C, Shriner D, Adeyemo A. Genome science and health disparities: a growing success story? Genome Med. 2013;5(7):61. doi: 10.1186/gm465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn J. How a drug becomes “ethnic”: law, commerce, and the production of racial categories in medicine. Yale J Health Policy Law Ethics. 2004;4(1):1–46. [PubMed] [Google Scholar]
- 18.Rothstein MA, Epps PG. Ethical and legal implications of pharmacogenomics. Nat Rev Genet. 2001;2(3):228–231. doi: 10.1038/35056075. [DOI] [PubMed] [Google Scholar]
- 19.Lee SS. Racializing drug design: implications of pharmacogenomics for health disparities. Am J Public Health. 2005;95(12):2133–2138. doi: 10.2105/AJPH.2005.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank R. What to make of it? The (Re)emergence of a biological conceptualization of race in health disparities research. Soc Sci Med. 2007;64(10):1977–1983. doi: 10.1016/j.socscimed.2007.01.010. [DOI] [PubMed] [Google Scholar]


