ABSTRACT
Inflammation is a significant risk factor for colon cancer. Recent work has demonstrated essential roles for several infiltrating immune populations in the metaplastic progression following inflammation. Hypoxia and stabilization of hypoxia-inducible factors (HIFs) are hallmark features of inflammation and solid tumors. Previously, we demonstrated an important role for tumor epithelial HIF-2α in colon tumors; however, the function of epithelial HIF-2α as a critical link in the progression of inflammation to cancer has not been elucidated. In colitis-associated colon cancer models, epithelial HIF-2α was essential in tumor growth. Concurrently, epithelial disruption of HIF-2α significantly decreased neutrophils in the colon tumor microenvironment. Intestinal epithelial HIF-2α-overexpressing mice demonstrated that neutrophil recruitment was a direct response to increased epithelial HIF-2α signaling. High-throughput RNA sequencing (RNA-seq) analysis of HIF-2α-overexpressing mice in conjunction with data mining from the Cancer Genome Atlas showed that the neutrophil chemokine CXCL1 gene was highly upregulated in colon tumor epithelium in a HIF-2α-dependent manner. Using selective peptide inhibitors of the CXCL1-CXCR2 signaling axis identified HIF-2α-dependent neutrophil recruitment as an essential mechanism to increase colon carcinogenesis. These studies demonstrate that HIF-2α is a novel regulator of neutrophil recruitment to colon tumors and that it is essential in shaping the protumorigenic inflammatory microenvironment in colon cancer.
KEYWORDS: HIF-2α, inflammation, colon cancer, cancer, colon, hypoxia, neutrophils
INTRODUCTION
Colon cancer remains a significant public health concern and is the second leading cause of cancer-associated deaths in the United States (1). Patients with chronic inflammation associated with inflammatory bowel disease (IBD), comprising ulcerative colitis and Crohn's disease, are at an increased lifetime risk of developing colon cancer; these tumors are termed colitis-associated cancers (CAC) (2). The genetic changes of sporadic colon cancer have been well defined, and a comprehensive genetic analysis of CAC was recently reported (3). In contrast to sporadic colon cancer, CAC are associated with early loss of the TP53 tumor suppressor and less frequent inactivation of adenomatous polyposis coli (APC) (4). Inflammation is an important component of the progression of sporadic cancer, and the inflammatory response is essential in the initiation and progression of CAC (5). The precise mechanisms that initiate the protumorigenic response following inflammation remain unknown.
Hypoxia is a characteristic feature of IBD and nearly all solid tumors, including those of the colon (6). Hypoxia promotes activation of the hypoxia-inducible factors (HIFs). HIFs consist of a heterodimer of an O2-labile α-subunit (HIF-1α, HIF-2α, and HIF-3α) and an O2-stable β-subunit (ARNT) (7). HIFs regulate transcription of target genes that mediate cellular responses to hypoxic microenvironments. HIFs are also essential factors promoting tumorigenesis and regulate several neoplastic processes, including growth, evasion of apoptosis, and chemoresistance (8). Previously, we showed that overexpression of intestinal epithelial HIF-2α but not HIF-1α can increase colon tumor progression in mouse models of sporadic colon tumorigenesis (9, 10). The essential role of HIF-2α and the mechanisms by which it regulates CAC have not been defined.
Inflammation is a critical component of the colon tumor microenvironment, and colon tumors are highly infiltrated with cells of both the innate and adaptive immune systems (5). Neutrophils are granulocytic myeloid cells with a critical role in the innate immune response (11) and are highly prevalent in the colon tumor microenvironment (12), but the function of neutrophils in the initiation and progression of cancer remains controversial. Previous studies have shown that neutrophils can be polarized into antitumorigenic (N1) and protumorigenic (N2) types (13). N2 neutrophils promote tumorigenesis through suppression of antitumor immunity, activation of oncogenic signaling through secretion of neutrophil elastase, and activation of angiogenesis (14–16). On the other hand, antitumorigenic N1 neutrophils can suppress tumorigenesis through direct tumor cytotoxicity and activation of antitumor immunity (17). Neutrophil recruitment into tumors can be regulated by tumor-derived secretion of a variety of chemokines and cytokines. However, the precise mechanisms mediating recruitment of neutrophils into colon tumors are not well defined.
In the present study, we show that the colon epithelial hypoxic response through activation of HIF-2α is essential in colon tumorigenesis in mouse models of CAC. Mechanistically, intestinal epithelial HIF-2α is a critical mediator of neutrophil recruitment to colon tumors through direct transcriptional regulation of the potent neutrophil chemokine CXCL1 in colon tumors. Taken together, these studies provide novel insights into hypoxic inflammatory responses in the progression of colon tumors and suggest a rationale for the targeting of HIF-2α in colon tumors.
RESULTS
HIF-2α is essential in inflammation-induced colon tumorigenesis.
One of the most commonly utilized models to study intestinal tumorigenesis is the ApcMin/+ mouse model. ApcMin/+ mice harbor a germ line truncation mutation of the Apc gene and spontaneously develop intestinal adenomas (18). However, this model does not completely recapitulate human colon tumorigenesis, as the vast majority of the tumors develop in the small intestine, with few colon tumors observed. Moreover, few of the tumors progress beyond adenoma, and they rarely become invasive. Inflammation is an essential component of the colon tumor microenvironment, and previous studies have shown that acute colonic inflammation induced by dextran sulfate sodium (DSS) can increase the incidence of colon tumorigenesis in the ApcMin/+ mouse (19). To directly determine the functional role of HIF-2α expression in inflammation-induced colon tumorigenesis, mice with intestine epithelium-specific disruption of HIF-2α by villin-cre-mediated recombination were crossed to ApcMin/+ mice (Hif-2αΔIE/ApcMin/+) and compared to littermate controls with intact HIF-2α expression (Hif-2αF/F/ApcMin/+). The Hif-2αΔIE/ApcMin/+ mice had significantly reduced colon tumor numbers and reduced tumor burdens (Fig. 1A to D). Furthermore, tumors from Hif-2αΔIE/ApcMin/+ mice had significantly increased apoptosis as measured by terminal deoxynucleotidyltransferase (TdT) dUTP nick end labeling (TUNEL) staining (Fig. 1E and F). These results show that colon epithelial HIF-2α is important in inflammation-driven colon tumorigenesis.
FIG 1.
HIF-2α is essential for inflammation-induced colon tumorigenesis. (A) Hif-2α+/+/ApcMin/+ (n = 5) and Hif-2αΔIE/ApcMin/+ (n = 7) mice were treated for 5 days with DSS and analyzed 28 days following the final day of DSS administration. (B to D) Tumor numbers (B), tumor burdens (C), and gross images (D) of the colons of Hif-2α+/+/ApcMin/+ and Hif-2αΔIE/ApcMin/+ mice 25 days following DSS administration. (E and F) Representative images of hematoxylin and eosin (H&E) analysis and TUNEL staining (E) and quantification of TUNEL-positive cells (F) in tumor tissue from Hif-2α+/+/ApcMin/+ and Hif-2αΔIE/ApcMin/+ mice. (G and H) qPCR expression analysis of myeloid cell markers (G) and S100a8 in tumors and/or adjacent normal tissue (H) from Hif-2α+/+/ApcMin/+ and Hif-2αΔIE/ApcMin/+ mice. *, P < 0.05, **, P < 0.01, and ***, P < 0.001 compared to Hif-2α+/+/ApcMin/+mice; ##, P < 0.01, and ###, P < 0.001 compared to normal tissue. Statistical analysis was performed with Student's t test or two-way ANOVA, followed by Sidak's multiple-comparison test. The error bars indicate standard deviations.
Epithelial expression of HIF-2α can promote inflammatory responses (10) and can modulate the immune environment in tumors. Colon tumors from Hif-2αF/F/ApcMin/+ mice had a significant increase in the panmyeloid cell marker CD11b compared to adjacent normal tissue. However, tumors from Hif-2αΔIE/ApcMin/+ mice had a significant reduction in tumor CD11b compared to those from Hif-2αF/F/ApcMin/+ mice, suggesting tumor epithelial expression of HIF-2α regulates myeloid cell influx into tumors. To determine the precise myeloid cell type that was decreased by disruption of Hif-2α, quantitative-PCR (qPCR) analysis was conducted for monocyte, macrophage, and neutrophil markers. Significantly less expression of the neutrophil marker gene Ly6g and the myeloperoxidase gene (Mpo) was observed in Hif-2αΔIE/ApcMin/+ than in Hif-2αF/F/ApcMin/+ colon tumors, whereas no changes in expression of the monocyte marker CD68 or macrophage marker genes Emr1, iNos, and Arg1 were observed (Fig. 1G). Moreover, the calcium binding protein S100a8, which is abundantly expressed by neutrophils, is significantly reduced in both normal and tumor tissue from Hif-2αΔIE/ApcMin/+ mice (Fig. 1H) (20). These data suggest epithelial cell expression of HIF-2α is essential for the presence of neutrophils in colon tumors.
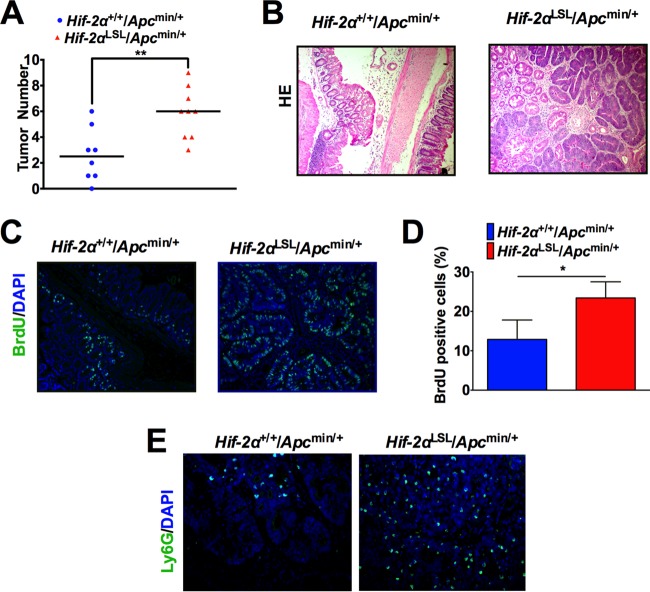
To assess if HIF-2α is sufficient to drive colon tumorigenesis, mice with overexpression of O2-stable HIF-2α downstream of a loxP-stop-loxP (LSL) cassette knocked into the Rosa26 allele (10) were crossed to ApcMin/+ mice to generate Hif-2αLSL/ApcMin/+ mice. These mice develop significantly more colon tumors than age-matched HIF-2α wild-type (WT) controls (Hif-2α+/+/ApcMin/+) at 3 months (Fig. 2A and B). Moreover, the tumors have a significantly higher proportion of proliferating cells, as shown by incorporation of bromodeoxyuridine (BrdU) (Fig. 2C and D). Concurrent with increased tumorigenesis, tumors from Hif-2αLSL/ApcMin/+ mice have a higher presence of neutrophils than Hif-2α+/+/ApcMin/+ mice by Ly6G immunofluorescence (Fig. 2E).
FIG 2.
HIF-2α increases inflammation-induced colon tumorigenesis. (A) Tumor numbers from colon tissue of Hif-2α+/+/ApcMin/+ (n = 8) and Hif-2αLSL/ApcMin/+ mice (n = 9). **, P < 0.01. (B) Representative images of H&E analysis. (C) Representative images of BrdU incorporation in tumors from Hif-2α+/+/ApcMin/+ and Hif-2αLSL/ApcMin/+ mice. DAPI, 4′,6-diamidino-2-phenylindole. (D) Quantification of the data in panel C. *, P < 0.05. (E) Representative images of Ly6G immunofluorescence in tumors from Hif-2α+/+/ApcMin/+ and Hif-2αLSL/ApcMin/+ mice. Statistical analysis was performed with Student's t test. The error bars indicate standard deviations.
HIF-2α regulates intratumoral neutrophils in colitis-associated colon cancer.
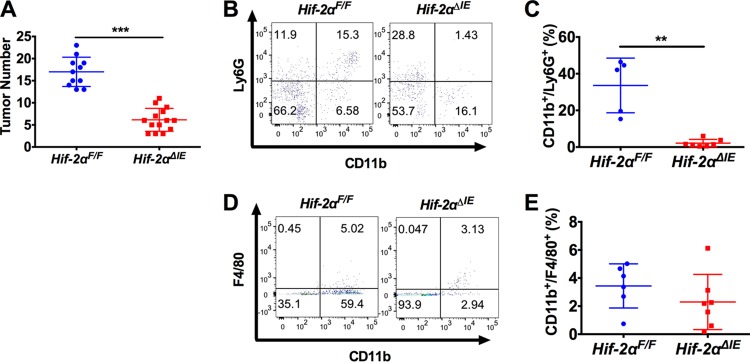
In order to appropriately model the role of HIF-2α in the inflammatory progression of colon cancer, we used the azoxymethane (AOM)/DSS model of CAC in mice with intestine epithelium-specific deletion of HIF-2α (Hif-2αΔIE) (21). Hif-2αΔIE and littermate control mice (Hif-2αF/F) were injected intraperitoneally (i.p.) with AOM (10 mg/kg of body weight) on day 0 and treated with DSS (1.5% [wt/vol]) in their drinking water beginning on day 5 for 5 days and changed back to regular drinking water for 2 weeks for three cycles. Consistent with the inflammation-induced ApcMin/+ model, Hif-2αΔIE mice had significantly reduced colon tumor numbers compared to littermate controls, suggesting HIF-2α expression is essential for inflammation-induced colon tumorigenesis (Fig. 3A). To analyze neutrophil infiltration in tumors, flow cytometry was performed on individual colon tumors. Colon tumors from Hif-2αΔIE mice had a significant reduction of CD11b+ Ly6G+ neutrophils from the colon tumor microenvironment compared to Hif-2αF/F colon tumors (Fig. 3B and C). No changes in tumor macrophages (CD11b+ F4/80+) were observed (Fig. 3D and E). These data suggest epithelial HIF-2α modulates the colon tumor microenvironment by regulating infiltration of tumor-associated neutrophils.
FIG 3.
Disruption of intestinal epithelial HIF-2α decreases colon tumors and intratumoral neutrophils in a CAC model. (A) Tumor numbers from colons of Hif-2αF/F (n = 11) and Hif-2αΔIE (n = 14) mice following AOM/DSS-induced CAC. (B and D) Flow cytometry analysis of CD11b/Ly6G double-positive cells (B) or CD11b/F4/80 double-positive cells (D) gated from CD45+ cells in tumors from Hif-2αF/F and Hif-2αΔIE mice. (C and E) Quantification of flow cytometry data in tumors. **, P < 0.01, and ***, P < 0.001 compared to Hif-2α+/+ mice. Statistical analysis was performed with Student's t test. The error bars indicate standard deviations.
HIF-1α does not impact colon tumorigenesis or neutrophil recruitment.
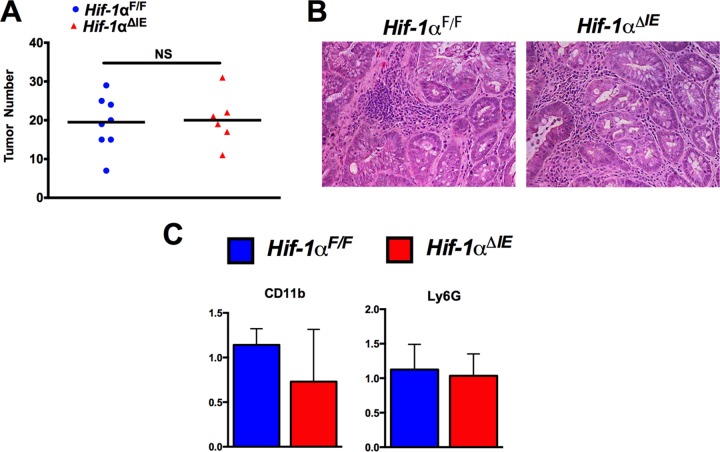
Our studies suggest that HIF-2α is a major regulator of colitis-associated colon tumorigenesis through recruitment of protumorigenic neutrophils. To address overlap in the functions of HIF-2α and HIF-1α in colon tumorigenesis, we assessed the role of HIF-1α using mice with intestine epithelial disruption of HIF-1α (Hif-1αΔIE). In the AOM/DSS model of colitis-associated colon cancer, no difference in tumorigenesis was observed in Hif-1αΔIE mice compared to WT controls (Hif-1αF/F) (Fig. 4A and B). Concurrently, no changes in expression of neutrophil markers (Ly6g and Cd11b) were observed in Hif-1αF/F colon tumor tissue relative to Hif-1αΔIE colon tumor tissue (Fig. 4C).
FIG 4.
HIF-1α does not impact colon tumorigenesis or neutrophil recruitment. (A) Tumor numbers from colons of Hif-1αF/F (n = 8) and Hif-1αΔIE (n = 6) mice following AOM/DSS-induced CAC. (B) Representative images of H&E staining from Hif-1αF/F and Hif-1αΔIE tumor tissue. (C) qPCR of Cd11b and Ly6g expression in tumor tissue from panel A. Statistical analysis was performed with Student's t test. NS, not significant. The error bars indicate standard deviations.
Colon epithelial HIF-2α regulates neutrophil chemotaxis.
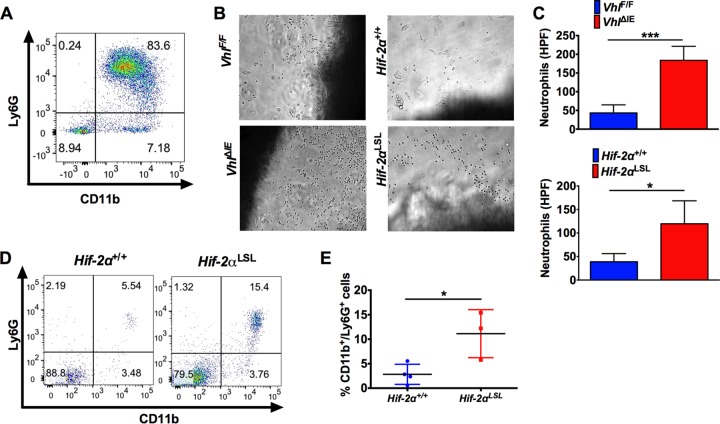
To determine if epithelial HIF-2α can regulate neutrophil chemotaxis, we used a transwell assay. Neutrophils were isolated from bone marrow and were shown to be highly pure (over 80%) (Fig. 5A). The isolated colon explants used were from mice with epithelial deletion of the von Hippel-Lindau gene (VhlΔIE). Previous work has shown that intestinal epithelial stabilization of HIF-2α is promoted in these mice under normoxic conditions (22). A dramatic increase in neutrophil migration through the Transwell toward colon explants from VhlΔIE mice compared to WT (VhlF/F) colon tissue explants was observed (Fig. 5B and C). To more directly assess the effects of HIF-2α, the medium was conditioned with colon tissue from HIF-2α-overexpressing mice (Hif-2αLSL), and this led to a significant increase in neutrophil Transwell chemotaxis compared to colon tissues explanted from wild-type littermate mice (Hif-2α+/+) (Fig. 5B and C). These data demonstrate that HIF-2α is important in neutrophil recruitment in vitro. Flow cytometry analysis of normal colon tissue from mice overexpressing HIF-2α in the intestinal epithelium (Hif-2αLSL) showed a significant increase in intracolonic neutrophils compared to wild-type littermate mice (Fig. 5D and E). Together, these data demonstrate an essential and sufficient role of epithelial HIF-2α in neutrophil recruitment in the colon.
FIG 5.
Intestinal HIF-2α activation promotes recruitment of neutrophils to the colon. (A) Flow cytometric analysis of CD11b/Ly6G staining of neutrophils isolated from bone marrow. (B and C) Representative images of neutrophils (B) and quantification of numbers of cells that migrated into the bottom well of a Transwell toward VhlF/F, VhlΔIE, Hif-2α+/+, and Hif-2αLSL colon tissue explants (C). HPF, high-power field. (D and E) Flow cytometry analysis (D) and quantification (E) of CD11b/Ly6G double-positive cells gated from CD45+ cells in colon tissue from Hif-2αLSL mice compared to Hif-2α+/+ mice. *, P < 0.05, and ***, P < 0.001 compared to VhlF/F or Hif-2α+/+ mice. Statistical analysis was performed with Student's t test. The error bars indicate standard deviations.
CXCL1 is highly induced by intestinal epithelial HIF-2α.
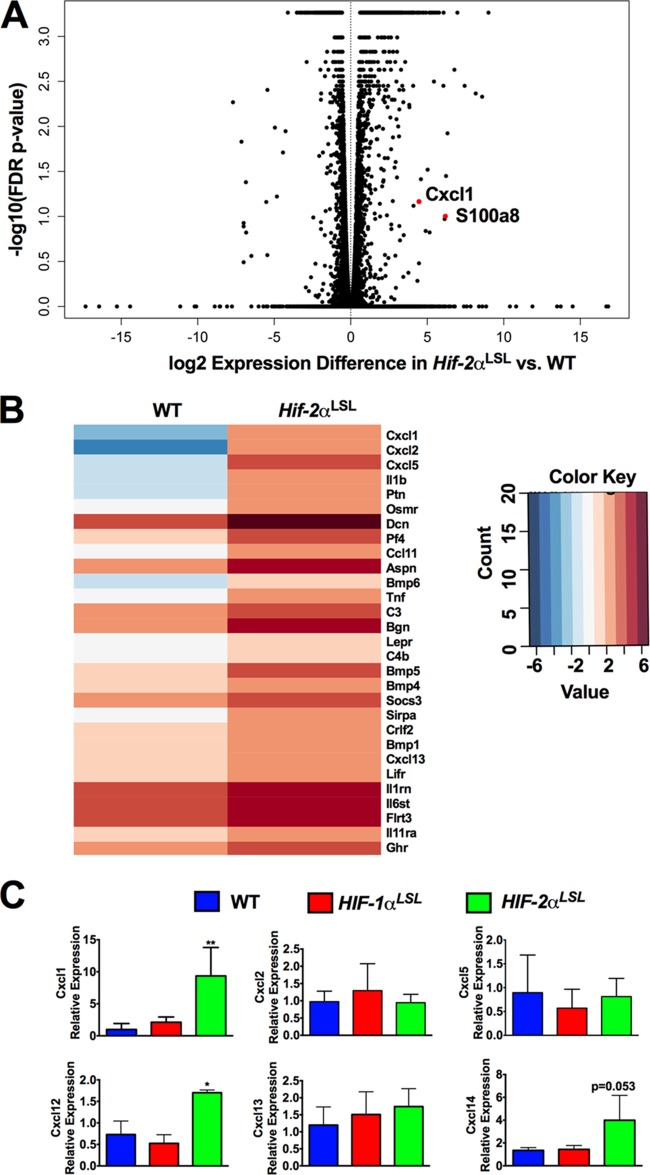
Neutrophils are recruited to solid tumors through tumor secretion of cytokines and chemokines (23). To determine the precise mechanism by which epithelial HIF-2α recruits neutrophils into the intestine, high-throughput RNA sequencing (RNA-seq) analysis was performed on colon tissues from Hif-2αLS) and WT (Hif-2α+/+) mice. Pathway analysis showed that genes encoding neutrophil-attractive chemokines, such as Cxcl1, Cxcl2, and Cxcl5, and neutrophil markers, such as S100a8, were highly increased in the colon tissues from Hif-2αLSL mice (Fig. 6A and B) (a full gene list is provided in Table S1 in the supplemental material). The expression of several members of the CXC family of chemokines was examined by qPCR of colon tissue from HIF-1α-overexpressing mice (Hif-1αLSL), Hif-2αLSL mice, and littermate controls. Only Cxcl1 expression was robustly increased (P < 0.001) in colon tissue in Hif-2αLSL mice compared to WT and Hif-1αLSL mice (Fig. 6C). These data suggest that activated epithelial HIF-2α in colon tumors may recruit neutrophils through secretion of cytokines and chemokines.
FIG 6.
Activation of intestinal epithelial HIF-2α increases CXCL1 expression. (A) Volcano plot of RNA-seq analysis in colon tissues from Hif-2α+/+ (n = 6) and Hif-2αLSL (n = 6) mice. FDR, false-discovery rate. (B) Heat map of genes related to cytokine activity enriched by Panther gene ontology analysis from Hif-2α+/+ and Hif-2αLSL colon tissues. (C) qPCR analysis of CXC family chemokines in colon tissue from WT, Hif-1αLSL, and Hif-2αLSL mice. *, P < 0.05, and **, P < 0.001 compared to normal colon tissues. Statistical analysis was performed by one-way ANOVA, followed by Dunnett's multiple-comparison test. The error bars indicate standard deviations.
HIF-2α is an essential regulator of CXCL1 expression in colon tumors.
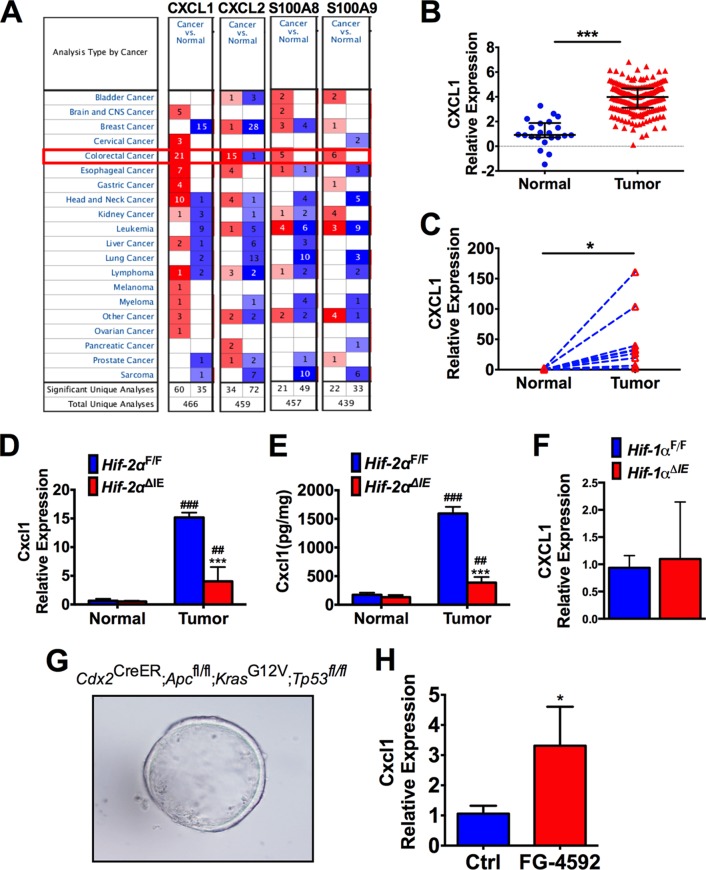
Oncomine data analysis indicated that CXCL1 was greatly increased in human colon tumors compared to normal colon tissues (Fig. 7A and B). We further confirmed by qPCR that CXCL1 was significantly increased in a set of colon tumor tissues compared to the adjacent normal colon tissues (Fig. 7C). CXCL1 expression is induced in colon tumors, but the major regulators of CXCL1 expression in colon tumors are currently unknown. Consistent with patient tumor analysis, CXCL1 expression was highly induced in AOM/DSS-induced colon tumors in Hif-2αF/F mice compared to normal adjacent tissue. This increase was significantly attenuated in tumors isolated from Hif-2αΔIE mice (Fig. 7D and E). No difference in Cxcl1 expression was observed in tumors from Hif-1αF/F or Hif-1αΔIE mice (Fig. 7F).
FIG 7.
Epithelial HIF-2α is essential for CXCL1 expression in colon tumors. (A) Oncomine database analysis of CXCL1, CXCL2, and neutrophil markers in several independent microarray analyses from colon cancer and normal tissues. (B and C) CXCL1 gene expression in the Cancer Genome Atlas data set (B) and a set of 8 pairs of colon tumors and adjacent normal tissues collected at the University of Michigan (C). (D and E) qPCR analysis of Cxcl1 mRNA (D) and enzyme-linked immunosorbent assay (ELISA) analysis of CXCL1 protein (E) in CAC tumor tissue and adjacent normal tissue from Hif-2αF/F and Hif-2αΔIE mice. (F) qPCR analysis of Cxcl1 expression in tumors from Hif-1αF/F and Hif-1αΔIE mice. (G) Representative image of colon enteroids following deletion of Apc and TP53 and activation of Kras. (H) qPCR of Cxcl1 expression in colon enteroids treated with FG-4592 or vehicle control (Ctrl). *, P < 0.05, and ***, P < 0.001 compared to WT, Hif-2αF/F, or Ctrl. ##, P < 0.01, and ###, P < 0.001 compared to normal tissue. Expression was normalized to β-actin. Statistical analysis was performed with Student's t test, a paired t test, or two-way ANOVA, followed by Sidak's multiple-comparison test. The error bars indicate standard deviations.
The tumor microenvironment is a complex milieu of tumor epithelial cells, immune cells, and stromal cells. Previous genetic analysis suggested that in tumor xenograft models, greater than 99% of the Cxcl1 transcripts are expressed directly by the tumor epithelial cells relative to tumor stromal cells (24). To evaluate if epithelial hypoxia signaling directly regulates Cxcl1 expression, we generated colon enteroids from mice with colon epithelium-specific deletion of Apc, activation of the oncogene Kras, and loss of the Tp53 tumor suppressor gene, which are commonly observed mutations in human colon tumors (Fig. 7F). Colon enteroids are an ideal model to mechanistically study colorectal cancer, because they maintain cell polarization and tight junctions in three dimensions and the cultures can be generated from primary colon epithelial tissue harboring mutations that are most commonly selected for in human colon tumors (25). To activate hypoxia signaling, the enteroids were treated with the potent PHD inhibitor FG-4592, which stabilizes HIF under normoxic conditions (26). Compared to untreated enteroids, activation of hypoxia significantly induced Cxcl1 expression, demonstrating that epithelial hypoxia signaling is sufficient to activate Cxcl1 expression (Fig. 7G). Taken together, our data suggest epithelial HIF-2α is a master regulator of CXCL1 expression in colon tumors.
HIF-2α regulates the Cxcl1 promoter through HRE- and Myc-associated zinc finger (MAZ)-dependent mechanisms.
HIFs activate target gene transcription by binding to hypoxia response elements (HREs), which are defined as 5′-RCGTG-3′, in promoter and enhancer regions. Analysis of the CXCL1-proximal promoter identified six canonical HREs clustered at distal and proximal sites (Fig. 8A). The proximal promoter region of Cxcl1 was cloned into the pGL3-luciferase reporter construct. Using cotransfection in HCT116 cells, overexpression of oxygen-stable HIF-2α was shown to directly activate the Cxcl1 promoter, similar to HIF-2α activation of the well-characterized HREs of the enolase gene promoter (P2.1) (Fig. 8B). To evaluate the dependence of these HREs on HIF-2α-mediated CXCL1 induction, a series of deletion constructs to disrupt the HREs were generated. HIF-2α activation of the Cxcl1 promoter was attenuated when the distal HREs were deleted and completely ameliorated when both the distal and proximal HREs were removed. These data demonstrate that the HREs are essential for HIF-2α-mediated Cxcl1 promoter induction (Fig. 8C).
FIG 8.

HIF-2α and MAZ are essential for CXCL1 activation. (A) Analysis of the Cxcl1-proximal promoter showing there are six HREs present (I to VI) in proximal and distal areas. (B) Cxcl1 or enolase (P2.1) promoter luciferase activity assays in HCT116 cells expressing HIF-2α. (C) Cxcl1 promoter luciferase activity assays with deletion constructs in HCT116 cells expressing HIF-2α. (D) qPCR and Western blot analysis of MAZ knockdown efficiency in HCT116 cells expressing MAZ targeting shRNAs (MAZ sh1 and MAZ sh2). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E) CXCL1 promoter luciferase activity assay in HCT116 cells expressing MAZ targeting shRNAs. **, P < 0.01, and ***, P < 0.001 compared to EV or Ctrl. Statistical analysis was performed by two-way ANOVA, followed by Sidak's multiple-comparison test. The error bars indicate standard deviations. EV, empty vector.
It has been suggested that target gene specificity for HIF-1α and HIF-2α is mediated by interactions with other transcription cofactors (27). Previously, our work has shown that HIF-2α inflammatory target gene activation is dependent upon interaction with an essential cofactor, Myc-associated zinc finger (10, 28, 29). MAZ is a Cys2-His2-type zinc finger transcription factor that is highly upregulated in several human cancers and regulates tumor growth (30). To determine if MAZ is essential for HIF-2α-dependent CXCL1 promoter induction, we used two targeting short hairpin RNAs (shRNAs) to generate stable knockdowns of MAZ expression in HCT116 cells (MAZ sh1 and MAZ sh2) (Fig. 8D). Compared to cells stably expressing scrambled shRNA (control [Ctrl] cells), MAZ sh1 and MAZ sh2 significantly attenuated CXCL1 promoter activation in response to HIF-2α (Fig. 8E).
CXCR2 inhibition reduces HIF-2α-driven colon tumorigenesis.
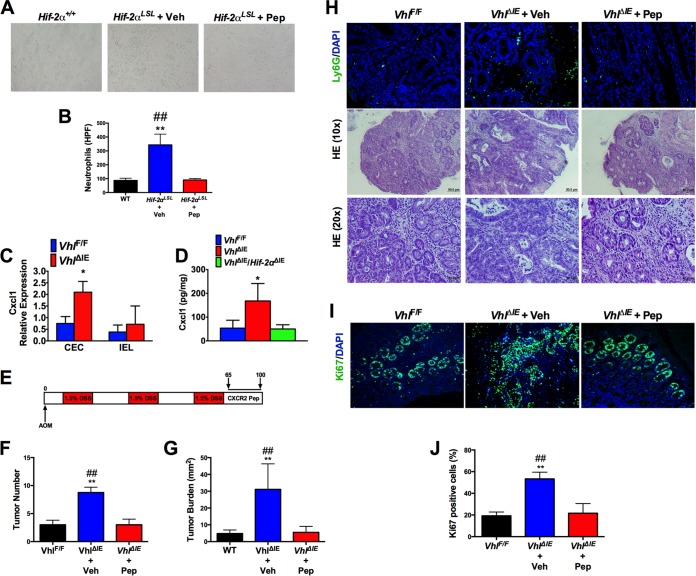
CXCL1 induces neutrophil recruitment by binding its cognate receptor, CXCR2, expressed on the surfaces of neutrophils. To determine if CXCL1 signaling through CXCR2 is the major mechanism by which epithelial HIF-2α mediates neutrophil recruitment, we used a well-characterized CXCR2-blocking peptide mimetic, CXCR2 pepducin (Pep). CXCR2 is a G protein-coupled receptor, and pepducins block CXCR2 signaling and decrease neutrophil influx into sites of inflammation and tumors (31, 32). Bone marrow-derived neutrophils were isolated and treated with the CXCR2-Pep or control pepducin (Veh). Blocking CXCR2 completely attenuated HIF-2α-induced neutrophil migration (Fig. 9A and B).
FIG 9.
HIF-2α-mediated neutrophil recruitment is essential for colon tumorigenesis. (A and B) Representative images of neutrophils (A) and quantification (B) of Ctrl pepducin (Veh)- or CXCR2 pepducin (PEP)-treated cells that migrated into the bottom well of a Transwell toward Hif-2α+/+ and Hif-2αLSL colon tissue explants. (C) qPCR analysis of Cxcl1 expression in CECs compared to IELs of VhlF/F and VhlΔIE mice. (D) ELISA analysis of CXCL1 protein in colon tissues of VhlF/F (n = 6), VhlΔIE (n = 3), and VhlΔIE/Hif-2αΔIE (n = 4) mice. (E) Schematic diagram of AOM/DSS-induced CAC and CXCR2-pepducin treatment protocol. (F to J) Tumor counting (F) and tumor burden analysis (G), representative images of H&E staining and Ly6G staining (H) and Ki67 staining (I), and quantification of Ki67 staining (J) in colon tissue from VhlF/F (n = 4), VhlΔIE plus Veh (n = 4), and VhlΔIE plus PEP (n = 3) groups. *, P < 0.05, and **, P < 0.01 compared to VhlF/F; ##, P < 0.01 compared to VhlΔIE plus CXCR2-pepducin. Statistical analysis was performed by one-way ANOVA, followed by Dunnett's multiple-comparison test. The error bars indicate standard deviations.
We next assessed the functional role of neutrophil recruitment in HIF-2α-driven colon tumorigenesis. To investigate this axis, we used the AOM/DSS model of CAC in VhlΔIE mice. Previous work had shown that these mice have a higher propensity to develop colon tumors in a HIF-2α-dependent manner (9). Unlike the Hif-2αLSL mice, VhlΔIE mice can survive 3 cycles of DSS. To confirm that epithelial deletion of Vhl increases Cxcl1 expression, we isolated purified colon epithelial cells (CECs) and intraepithelial lymphocytes (IELs) (33) from VhlΔIE and VhlF/F mice. Cxcl1 expression is significantly increased in CECs of VhlΔIE mice compared to those of VhlF/F mice but not in IELs (Fig. 9C). Moreover, CXCL1 expression is induced in VhlΔIE mice in a HIF-2α-dependent manner using mice with double disruption of VHL and HIF-2α (VhlΔIE/Hif-2αΔIE) (Fig. 9D). To address the role of CXCR2-mediated neutrophil recruitment to colon tumors, VhlΔIE mice were randomized to treatment with Ctrl-pepducin (Veh) or CXCR2-pepducin once daily for days 65 to 100 by subcutaneous injection following the third cycle of DSS (Fig. 9E). Compared to littermate control VhlF/F mice, VhlΔIE mice treated with Ctrl-pepducin developed significantly more colon tumors and had higher tumor burdens and increased neutrophil influx (Fig. 9F to H). The VhlΔIE mice treated with CXCR2-pepducin had significantly reduced HIF-2α-driven colon tumorigenesis and neutrophil infiltration. In addition, tumors from VhlΔIE mice had a significant increase in tumor cell proliferation as measured by Ki67 immunofluorescence staining, which was attenuated in VhlΔIE mice treated with CXCR2-pepducin (Fig. 9I and J). Taken together, our studies suggest that HIF-2α-mediated neutrophil recruitment through the CXCL1-CXCR2 axis is essential for its role in colon tumorigenesis.
DISCUSSION
Inflammation and hypoxia are intimately linked, and hypoxia has been previously shown to regulate the inflammatory microenvironments of many tumor types. Hypoxia increases ovarian cancer tumor growth through secretion of CCL28, which facilitates recruitment of immune-suppressive T regulatory (Treg) cells to promote tumor growth (34). In pancreatic ductal adenocarcinoma, HIF-1α is a tumor suppressor through blockade of protumorigenic B cell recruitment to tumors (35). Interestingly, our data show that epithelial expression of HIF-2α can modulate the inflammatory milieu of colon tumors by regulating the recruitment of intratumoral neutrophils. Hypoxic regulation of cytokines and chemokine secretion from tumor cells can modulate neutrophil recruitment to promote hepatocellular carcinoma (36). Additionally, neutrophils tend to be localized to hypoxic zones within uterine tumors (37). Mechanistically, we have discovered a novel HIF-2α target gene, the CXCL1 gene. CXCL1 is a member of the CXC family of chemokines and is a potent neutrophil chemoattractant to sites of inflammation or tumors by binding its cognate receptor, CXCR2 (38). Consistent with our work showing that epithelial CXCL1 is induced by HIF-2α, previous studies using xenograft models demonstrated that the vast majority (>99%) of Cxcl1 transcripts in colon tumors are derived from tumor epithelial cells (24). Our studies clearly demonstrate that epithelial HIF-2α can regulate Cxcl1 induction in colon tumors.
The functional role of neutrophils in the progression of tumors is not clear, as both antitumorigenic (N1) and protumorigenic (N2) neutrophils have been described (13). Neutrophils expressing the hepatocyte growth factor (HGF) receptor, c-MET, have been shown to be largely antitumorigenic in colon tumors (28). However, large-scale meta-analysis studies have shown that neutrophils are highly correlated with adverse outcomes across more than 25 different tumor types (39). Neutrophils are critical mediators of metastasis in murine models of breast cancer (33). It has also been suggested that neutrophils are essential in the inflammatory progression of colon tumors, as depletion of neutrophils with anti-Ly6G antibody significantly reduced colon tumors (40). Moreover, a high neutrophil-to-lymphocyte ratio portends a poor prognosis for colon cancer patients (41). In our study, we show that HIF-2α-driven colon tumorigenesis is dependent upon neutrophil influx into colon tumors through the neutrophil CXCL1 receptor, CXCR2. A critical role for CXCR2 in the initiation and progression of colon cancer and pancreatic ductal adenocarcinoma has been described (42, 43). Inhibition of neutrophil influx via CXCR2 decreased HIF-2α-driven colon tumorigenesis, progression, and proliferation. These studies demonstrate mechanistically how hypoxic inflammatory responses can modulate the colon tumor immune microenvironment to promote tumor growth. More work is needed to determine the precise mechanisms by which neutrophils promote colon tumorigenesis.
The studies reported here suggest that that epithelial HIF-2α, but not HIF-1α, selectively modulates neutrophil recruitment into tumors without affecting other myeloid cell populations. Neutrophils are critical to set up an oxygen gradient in the intestine (44). These oxygen gradients promote tissue repair in a HIF-1α-dependent manner. HIF-1α is highly active in intestinal inflammation, and genetic deletion of intestinal epithelial HIF-1α exacerbates colitis (45). HIF-1α is an essential regulator of the expression of intestinal-barrier-protective genes, such as the intestinal trefoil factor gene (Itf), CD73, and multidrug resistance gene 1 (Mdr-1) (45, 46). The bidirectional signaling of hypoxia and neutrophils may be a feed forward mechanism mediated by HIF-2α that is critical to establish an oxygen gradient during acute inflammation for HIF-1α-dependent injury repair (44). However, our data suggest that in chronic inflammation, this mechanism exacerbates tumorigenesis.
Previously, we have shown that activation of HIF-2α can promote colon tumor cell growth in a cell-autonomous manner. Epithelial expression of HIF-2α is a potent activator of inflammatory responses and increases the progression of intestinal inflammation (10). Additionally, HIF-2α is a transcriptional regulator of proinflammatory cyclooxygenase 2 (COX2) and microsomal prostaglandin E synthase (mPGES) to increase tumor inflammation, and treatment of HIF-2α-overexpressing mice with the anti-inflammatory nimesulide can reduce colon tumorigenesis (47). Interestingly, HIF-1α activation has no effect on colon tumorigenesis (48). Similarly, we found that HIF-1α has no effect on the expression of Cxcl1. Previous studies have highlighted the dichotomous role of HIF-1α and HIF-2α in several cancer models. For example, in renal cell carcinoma, HIF-2α is essential for tumor cell growth whereas HIF-1α decreases cell growth (49). In pancreatic cancer, mouse genetic models demonstrated that HIF-2α is essential for tumorigenesis whereas HIF-1α decreases tumorigenesis through repression of infiltrating protumorigenic B cells (35, 50). However, in lung cancer, it has been shown that HIF-2α exerts a tumor-suppressive effect (51). These studies demonstrate the need to carefully evaluate the tumor-specific roles of HIF-1α and HIF-2α for therapeutic targeting. HIF-2α-specific inhibitors have been developed that target a novel ligand-binding pocket that is located within the PAS-B domain of HIF-2α but not in HIF-1α (52). These novel tools may provide an exciting therapeutic avenue to decrease tumor cell proliferation, as well as to decrease tumor-promoting inflammatory responses in colon cancer.
MATERIALS AND METHODS
Animals.
VhlF/F, VhlΔIE, Hif-1αΔIE, Hif-1αF/F, Hif-2αΔIE, Hif-2αF/F, Hif-2αLSL, and Hif-2α+/+ mice were described previously (10, 22). To evaluate HIF-2α in colon tumorigenesis, Hif-2αΔIE and Hif-2αLSL mice were crossed to ApcMin/+ mice. To induce colon tumorigenesis in Hif-2αΔIE/ApcMin/+ mice, the animals were treated with 2% DSS in their drinking water for 5 days and then placed back on regular drinking water for 28 days. For AOM/DSS experiments, animals were injected i.p. with azoxymethane (10 mg/kg) and then cycled on 1.5% DSS in their drinking water for 5 days, followed by regular drinking water for 2 weeks for three cycles. For the CXCR2-pepducin experiment, following the third cycle, VhlΔIE mice were treated with CXCR2-pepducin (palmitoyl [pal]-RTLFKAMGQKHR) or control peptide (pal-TRFLAKMHQGHKR) (Genscript) for 35 consecutive days (2.5 mg/kg subcutaneously [s.c.]) (53). All the animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Flow cytometry.
Single-cell suspensions from fresh normal colon or colon tumor tissue were prepared by finely mincing the tissue and incubating it with collagenase type II (Sigma-Aldrich; 1 mg/ml) at 37οC for 1 h and then passed through a 40-μm cell strainer. The single-cell suspensions were stained in Hanks' balanced salt solution (HBSS)-2% fetal bovine serum (FBS) with eFluor780-conjugated anti-CD45 (eBioscience), phycoerythrin (PE)-conjugated anti-Ly6G (BD), allophycocyanin (APC)-conjugated anti-Cd11b (eBioscience), and eFluor450-conjugated anti-F4/80 (eBioscience). Flow cytometry was performed using an LSR Fortessa (BD). Flow cytometry data were analyzed using FlowJo software.
Neutrophil isolation and transwell assay.
Bone marrow cells were suspended in HBSS buffer supplemented with 20 mM HEPES and 0.5% FBS. The isolated bone marrow was disaggregated through an 18-gauge (18-G) needle. To lyse the residual red blood cells (RBCs), 0.2% NaCl was added to the cells for 45 s, and then the reaction was stopped with 1.2% NaCl. The cells were resuspended in HBSS buffer and carefully layered over 62% Percoll. Centrifugation was performed at 2,200 rpm for 30 min. The cell pellet was washed twice with PBS and used for antibody staining to confirm the purity. The antibodies used were peridinin chlorophyll protein (PerCP)-Cy5.5-conjugated anti-CD45 (eBioscience), APC-conjugated anti-CD11b (eBioscience), PE-conjugated anti-Ly6G (BD), and eFluor450-conjugated anti-F4/80 (eBioscience) antibodies. Debris (FSC-A/SSC-A) and doublets (FSC-A/FSC-H) were excluded, and CD45+ cells were then subgated on CD11b+ and Ly6G+ neutrophils. The numbers flow cytometry-gated populations indicate the relative percentages of each population. For transwell assays, 1 × 106 neutrophils were cultured in the top well in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% antibiotic/antimycotic, and fresh colon explants were plated in the bottom well. Migration was assessed 2 hours after plating.
Isolation of colon epithelial cells and intraepithelial lymphocytes.
Isolation of CECs and intraepithelial lymphocytes was performed as previously described (10). Briefly, colon tissue was isolated and incubated with EDTA and dithiothreitol (DTT) in HBSS. To separate CECs and IELs, the tissue was passed through a cell strainer and exposed to 67% to 44% Percoll gradient separation. CECs were collected from the top, and IELs were collected at the interface of the Percoll gradient.
Histology.
Colon tissue and tumors were excised, fixed, sectioned, and stained as previously described (9). The antibodies for immunofluorescence assays were as follows: BrdU (eBioscience), Ki67 (Vector Laboratories), Ly6G (BD), and Alexa Fluor 488–goat anti-mouse IgG (Molecular Probes Inc.).
RNA isolation, qPCR analysis, and RNA-seq.
RNA was isolated, and qPCR analysis was conducted as previously described (9). The primers are listed in Table 1. RNA sequencing libraries were prepared using the TruSeq RNA library prep kit v2 (Illumina) following the manufacturer's recommended protocol. The libraries were sequenced using single-end 50-cycle reads on a HiSeq 2500 sequencer (Illumina) at the University of Michigan DNA Sequencing Core Facility.
TABLE 1.
Primers

a F, forward; R, reverse.
RNA-seq data analysis.
Raw sequencing read quality was assessed utilizing FastQC. Reads were aligned to the reference mouse transcriptome (UCSC mm10) using Bowtie v 2.1.0.0 (54) and TopHat v 2.0.9 (55). Default parameters were used for the alignment, with the exception of “-b2-very-sensitive,” “-no-coverage-search,” and “-no-novel-juncs.” Mate inner pair distances were estimated by TopHat, and the values were used in the alignment. Expression quantification and differential expression analysis between Hif-2αLSL and Hif-2α+/+ mice were conducted using CuffDiff v 2.1.1 (56) with the parameter settings “-multi-read-correct,” “-compatible-hits-norm,” and “-upper-quartile-norm” for normalization of expression calculations across samples. For the CuffDiff analysis, we used UCSC mm10.fa as the reference genome and UCSC mm10.gtf as the reference transcriptome. Genes were considered differentially expressed between conditions at a false-discovery rate-adjusted P value of <0.05 (57).
Pathway analyses.
A directional analysis was conducted on all genes by including the P value of the differential-expression test as a measure of the effect size and the log2 fold difference in expression as a measure of the effect direction using iPathways (Advaita). Differentially expressed pathways were identified utilizing the Panther classification system (http://pantherdb.org/). KEGG biological pathways and gene ontology biological processes were considered differentially expressed at a P value of <0.05.
CXCL1 luciferase reporter activity.
The Cxcl1 promoter was cloned using primers listed in Table 1. The Cxcl1 promoter fragments were subsequently cloned into the pGL3-basic vector (Promega). Luciferase activity assays were performed as previously described and normalized to β-galactosidase activity (47). HCT116 cells expressing MAZ targeting shRNAs were generated as previously described (10).
Enteroid culture.
Enteroids were generated from colon tissue from mice with inducible, colon epithelium-specific deletion of Apc, activation of Kras, and loss of Tp53 (Cdx2CreER; Apcfl/fl; KrasG12V; Tp53fl/fl). The mice were sacrificed, and the colon was cut open longitudinally. All plasticware was precoated with 0.1% bovine serum albumin (BSA), and all steps were carried out on ice unless otherwise specified. The tissue was incubated for 15 min at room temperature in 2.5 μg/ml amphotericin B (Fungizone; ThermoFisher) in Dulbecco's phosphate-buffered saline containing 25 μg/ml gentamicin (Gibco) and 50 μg/ml normocin (InvivoGen) (DPBSgn). The colon tissue was cut into lengthwise strips (approximately 3 mm by 5 mm). The tissue was incubated in 10 mM DTT for 15 min at room temperature, changing to fresh DTT every 5 min. The tissue was rinsed in DPBSgn, rinsed once with 8 mM EDTA, and then incubated/rotated in 8 mM EDTA at 4°C for 75 min. The EDTA was removed, and the tissue was washed three times with DPBSgn. The tissue was then “snap-shaken” 10 times to manually separate colon crypts. The crypt-containing supernatant was immediately added to 1.5 ml of cold FBS in a BSA-coated 50-ml tube, and the shaking step was repeated twice more. The crypts were spun at 40 × g for 2 min at 4°C. The pellet was washed in DPBSgn and spun again at 40 × g for 2 min at 4°C. The pellet was resuspended in a solution of 66% Matrigel (Corning), 33% KGMG medium (KGMG Bullet kit; Lonza), and 10 μM Rock inhibitor (Y27632; Miltenyi) at a concentration of 2 crypts/μl, accounting for 250 μl per well in a 6-well plate. Four diagonal strips of 60 μl of the culture were added to each well of a prewarmed cell culture plate using a cut tip. After 30 min, medium containing 10 μM Y27632 was added. The medium was changed daily for 3 days. On the fourth day, the cultures were treated with either vehicle or 100 μM the PHD inhibitor FG-4592 (Cayman Chemicals) overnight and then lysed directly in TRIzol for qPCR analysis.
Data analysis.
Error bars in the figures represent standard deviations (37). P values were calculated by independent t test, paired t test, one-way analysis of variance (ANOVA), Dunnett's t test, and two-way ANOVA. Immunofluorescence staining and Western blot analysis were quantified with ImageJ.
Supplementary Material
ACKNOWLEDGMENTS
We declare that we have no conflict of interest.
This work was supported by NIH grants (CA148828 and DK095201 to Y.M.S.), the University of Michigan Gastrointestinal Peptide Center (Y.M.S.), a pilot grant from the University of Michigan GI Spore (CA130810 to Y.M.S.), the Crohn's Colitis Foundation of America (grant number 276556 to X.X.), a research grant to J.A.C. from the University of Michigan Comprehensive Cancer Center (which receives funding from NCI grant P30CA046592), funding from the Ravitz Foundation (J.A.C.), and a Research Scholar Award from the American Gastroenterological Association (to X.X.). D.T. was supported by a T32 training grant (T32 DK 094775; Training in Basic and Translational Digestive Sciences).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00481-16.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin 66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kim ER, Chang DK. 2014. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol 20:9872–9881. doi: 10.3748/wjg.v20.i29.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER. 2011. Molecular genetics of colorectal cancer. Annu Rev Pathol 6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 4.Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, Lauwers GY, Selaru FM, Popoli M, Pittman ME, Ke X, Hruban RH, Meltzer SJ, Kinzler KW, Vogelstein B, Harris CC, Papadopoulos N. 2016. Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology 150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terzić J, Grivennikov S, Karin E, Karin M. 2010. Inflammation and colon cancer. Gastroenterology 138:2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 6.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. 2000. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol 157:411–421. doi: 10.1016/S0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL. 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semenza GL. 2012. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue X, Taylor M, Anderson E, Hao C, Qu A, Greenson JK, Zimmermann EM, Gonzalez FJ, Shah YM. 2012. Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res 72:2285–2293. doi: 10.1158/0008-5472.CAN-11-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, Shah YM. 2013. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology 145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridlender ZG, Albelda SM. 2012. Tumor-associated neutrophils: friend or foe? Carcinogenesis 33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 12.Rao H-L, Chen J-W, Li M, Xiao Y-B, Fu J, Zeng Y-X, Cai M-Y, Xie D. 2012. Increased intratumoral neutrophil in colorectal carcinomas correlates closely with malignant phenotype and predicts patients' adverse prognosis. PLoS One 7:e30806. doi: 10.1371/journal.pone.0030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM. 2009. Polarization of tumor-associated neutrophil (TAN) phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houghton AM, Rzymkiewicz DM, Ji H, Gregory AD, Egea EE, Metz HE, Stolz DB, Land SR, Marconcini LA, Kliment CR, Jenkins KM, Beaulieu KA, Mouded M, Frank SJ, Wong KK, Shapiro SD. 2010. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nat Med 16:219–223. doi: 10.1038/nm.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nozawa H, Chiu C, Hanahan D. 2006. Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc Natl Acad Sci U S A 103:12493–12498. doi: 10.1073/pnas.0601807103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishalian I, Bayuh R, Eruslanov E, Michaeli J, Levy L, Zolotarov L, Singhal S, Albelda SM, Granot Z, Fridlender ZG. 2014. Neutrophils recruit regulatory T-cells into tumors via secretion of CCL17—a new mechanism of impaired antitumor immunity. Int J Cancer 135:1178–1186. doi: 10.1002/ijc.28770. [DOI] [PubMed] [Google Scholar]
- 17.Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, Stephen TL, Ranganathan A, Deshpande C, Akimova T, Vachani A, Litzky L, Hancock WW, Conejo-Garcia JR, Feldman M, Albelda SM, Singhal S. 2014. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest 124:5466–5480. doi: 10.1172/JCI77053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taketo MM, Edelmann W. 2009. Mouse models of colon cancer. Gastroenterology 136:780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka T, Kohno H, Suzuki R, Hata K, Sugie S, Niho N, Sakano K, Takahashi M, Wakabayashi K. 2006. Dextran sodium sulfate strongly promotes colorectal carcinogenesis in ApcMin/+ mice: inflammatory stimuli by dextran sodium sulfate results in development of multiple colonic neoplasms. Int J Cancer 118:25–34. doi: 10.1002/ijc.21282. [DOI] [PubMed] [Google Scholar]
- 20.Pruenster M, Kurz ARM, Chung K-J, Cao-Ehlker X, Bieber S, Nussbaum CF, Bierschenk S, Eggersmann TK, Rohwedder I, Heinig K, Immler R, Moser M, Koedel U, Gran S, McEver RP, Vestweber D, Verschoor A, Leanderson T, Chavakis T, Roth J, Vogl T, Sperandio M. 2015. Extracellular MRP8/14 is a regulator of β2 integrin-dependent neutrophil slow rolling and adhesion. Nat Commun 6:6915. doi: 10.1038/ncomms7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor M, Qu A, Anderson ER, Matsubara T, Martin A, Gonzalez FJ, Shah YM. 2011. Hypoxia-inducible factor-2α mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology 140:2044–2055. doi: 10.1053/j.gastro.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah YM, Ito S, Morimura K, Chen C, Yim S-H, Haase VH, Gonzalez FJ. 2008. Hypoxia-inducible factor augments experimental colitis through a MIF-dependent inflammatory signaling cascade. Gastroenterology 134:2036–2048. doi: 10.1053/j.gastro.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell DR, Huttenlocher A. 2016. Neutrophils in the tumor microenvironment. Trends Immunol 37:41–52. doi: 10.1016/j.it.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, Inghirami G, Trusolino L, Fekete Z, De Ridder M, Cassoni P, Storme G, Bertotti A, Medico E. 2015. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 25.Dame MK, Jiang Y, Appelman HD, Copley KD, McClintock SD, Aslam MN, Attili D, Elmunzer BJ, Brenner DE, Varani J, Turgeon DK. 2014. Human colonic crypts in culture: segregation of immunochemical markers in normal versus adenoma-derived. Lab Invest 94:222–234. doi: 10.1038/labinvest.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramakrishnan SK, Anderson ER, Martin A, Centofanti B, Shah YM. 2015. Maternal intestinal HIF-2α is necessary for sensing iron demands of lactation in mice. Proc Natl Acad Sci U S A 112:E3738–E3747. doi: 10.1073/pnas.1504891112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C-J, Sataur A, Wang L, Chen H, Simon MC. 2007. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1α and HIF-2α. Mol Biol Cell 18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finisguerra V, Di Conza G, Di Matteo M, Serneels J, Costa S, Thompson AAR, Wauters E, Walmsley S, Prenen H, Granot Z, Casazza A, Mazzone M. 2015. MET is required for the recruitment of anti-tumoural neutrophils. Nature 522:349–353. doi: 10.1038/nature14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L, Xue X, Taylor M, Ramakrishnan SK, Nagaoka K, Hao C, Gonzalez FJ, Shah YM. 2014. Hypoxia-inducible factor/MAZ-dependent induction of caveolin-1 regulates colon permeability through suppression of occludin, leading to hypoxia-induced inflammation. Mol Cell Biol 34:3013–3023. doi: 10.1128/MCB.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smits M, Wurdinger T, van het Hof B, Drexhage JAR, Geerts D, Wesseling P, Noske DP, Vandertop WP, de Vries HE, Reijerkerk A. 2012. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J 26:2639–2647. doi: 10.1096/fj.11-202820. [DOI] [PubMed] [Google Scholar]
- 31.Kaneider NC, Agarwal A, Leger AJ, Kuliopulos A. 2005. Reversing systemic inflammatory response syndrome with chemokine receptor pepducins. Nat Med 11:661–665. doi: 10.1038/nm1245. [DOI] [PubMed] [Google Scholar]
- 32.Steele CW, Karim SA, Foth M, Rishi L, Leach JDG, Porter RJ, Nixon C, Jeffry Evans TR, Carter CR, Nibbs RJB, Sansom OJ, Morton JP. 2015. CXCR2 inhibition suppresses acute and chronic pancreatic inflammation. J Pathol 237:85–97. doi: 10.1002/path.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau C-S, Verstegen NJM, Ciampricotti M, Hawinkels LJAC, Jonkers J, de Visser KE. 2015. IL17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang L-P, Gimotty PA, Gilks CB, Lal P, Zhang L, Coukos G. 2011. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature 475:226–230. doi: 10.1038/nature10169. [DOI] [PubMed] [Google Scholar]
- 35.Lee KE, Spata M, Bayne LJ, Buza EL, Durham AC, Allman D, Vonderheide RH, Simon MC. 2016. Hif1α deletion reveals pro-neoplastic function of B cells in pancreatic neoplasia. Cancer Discov 6:256–269. doi: 10.1158/2159-8290.CD-15-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou S-L, Zhou Z-J, Hu Z-Q, Huang X-W, Wang Z, Chen E-B, Fan J, Cao Y, Dai Z, Zhou J. 2016. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology 150:1646–1658. doi: 10.1053/j.gastro.2016.02.040. [DOI] [PubMed] [Google Scholar]
- 37.Blaisdell A, Crequer A, Columbus D, Daikoku T, Mittal K, Dey Sudhansu K, Erlebacher A. 2015. Neutrophils oppose uterine epithelial carcinogenesis via debridement of hypoxic tumor cells. Cancer Cell 28:785–799. doi: 10.1016/j.ccell.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zlotnik A, Yoshie O. 2000. Chemokines: a new classification system and their role in immunity. Immunity 12:121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 39.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang K, Bai Y-P, Wang C, Wang Z, Gu H-Y, Du X, Zhou X-Y, Zheng C-L, Chi Y-Y, Mukaida N, Li Y-Y. 2012. Crucial involvement of tumor-associated neutrophils in the regulation of chronic colitis-associated carcinogenesis in mice. PLoS One 7:e51848. doi: 10.1371/journal.pone.0051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pine JK, Morris E, Hutchins GG, West NP, Jayne DG, Quirke P, Prasad KR. 2015. Systemic neutrophil-to-lymphocyte ratio in colorectal cancer: the relationship to patient survival, tumour biology and local lymphocytic response to tumour. Br J Cancer 113:204–211. doi: 10.1038/bjc.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele Colin W, Karim Saadia A, Leach Joshua DG, Bailey P, Upstill-Goddard R, Rishi L, Foth M, Bryson S, McDaid K, Wilson Z, Eberlein C, Candido Juliana B, Clarke M, Nixon C, Connelly J, Jamieson N, Carter CR, Balkwill F, Chang DK, Evans TRJ, Strathdee D, Biankin AV, Nibbs RJB, Barry ST, Sansom OJ, Morton JP. 2016. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell 29:832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, DuBois RN. 2013. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, Bayless AJ, Scully M, Saeedi BJ, Golden-Mason L, Ehrentraut SF, Curtis VF, Burgess A, Garvey JF, Sorensen A, Nemenoff R, Jedlicka P, Taylor CT, Kominsky DJ, Colgan SP.. 2014. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity 40:66–77. doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. 2004. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest 114:1098–1106. doi: 10.1172/JCI200421086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuta GT, Turner JR, Taylor CT, Hershberg RM, Comerford K, Narravula S, Podolsky DK, Colgan SP.. 2001. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med 193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue X, Shah YM. 2013. Hypoxia-inducible factor-2α is essential in activating the COX2/mPGES-1/PGE(2) signaling axis in colon cancer. Carcinogenesis 34:163–169. doi: 10.1093/carcin/bgs313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue X, Ramakrishnan SK, Shah YM. 2014. Activation of HIF-1α does not increase intestinal tumorigenesis. Am J Physiol 307:G187–G195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raval RR, Lau KW, Tran MGB, Sowter HM, Mandriota SJ, Li J-L, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. 2005. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol 25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Criscimanna A, Duan L-J, Rhodes JA, Fendrich V, Wickline E, Hartman DJ, Monga SPS, Lotze MT, Gittes GK, Fong G-H, Esni F. 2013. PanIN-specific regulation of Wnt signaling by HIF2α during early pancreatic tumorigenesis. Cancer Res 73:4781–4790. doi: 10.1158/0008-5472.CAN-13-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, Vachani A, Jacks T, Chodosh LA, Kissil JL, Simon MC, Keith B. 2010. HIF-2α deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A 107:14182–14187. doi: 10.1073/pnas.1001296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheuermann TH, Li Q, Ma H-W, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, Tambar UK, Gardner KH, Bruick RK. 2013. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol 9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamieson T, Clarke M, Steele CW, Samuel MS, Neumann J, Jung A, Huels D, Olson MF, Das S, Nibbs RJB, Sansom OJ. 2012. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest 122:3127–3144. doi: 10.1172/JCI61067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.