ABSTRACT
One major concern over the clinical application of embryonic stem cell (ESC)-derived cells is the potentiation of latent tumorigenicity by residual undifferentiated cells. Despite the use of intensive methodological approaches to eliminate residual undifferentiated cells, the properties of these cells remain elusive. Here, we show that under a serum-free neural differentiation condition, residual undifferentiated cells markedly delay progression of their cell cycle without compromising their pluripotency. This dormant pluripotency was maintained during reculture of the cells under a serum-free condition, whereas upon serum stimulation, the cells exited the dormant state and restarted proliferation and differentiation into all three germ layers. Microarray analysis revealed a set of genes that is significantly upregulated in the dormant ESCs compared with their levels of regulation in proliferating ESCs. Among them, we identified the transcription factor Forkhead box O3 (FoxO3) to be an essential regulator of the maintenance of pluripotency in dormant ESCs. Our study demonstrates that the transition into the dormant state endows residual undifferentiated cells with FoxO3-dependent and leukemia inhibitory factor/serum-independent pluripotency.
KEYWORDS: FoxO3, embryonic stem cells, pluripotency, regenerative medicine, stem cell quiescence
INTRODUCTION
Embryonic stem cells (ESCs) are characterized by infinite proliferation, self-renewal, and pluripotency. These features are maintained ex vivo in mouse ESCs (mESCs) using leukemia inhibitory factor (LIF), a cytokine capable of activating STAT signaling, in concert with either serum or bone morphogenic protein (BMP) to induce inhibitor-of-differentiation (Id) proteins (1–3). mESCs secrete fibroblast growth factor 4 (FGF4), which induces autocrine stimulation of mitogen-activated protein kinase (MAPK) and promotes differentiation; therefore, inhibition of the MAPK signaling cascade promotes the self-renewal of ESCs (4). Additional inhibition of glycogen synthase kinase 3 (GSK3) is sufficient for maintenance of mESC pluripotency in the defined culture regime, commonly referred to as “2i,” which includes a Mek inhibitor to block the MAPK cascade and a GSK3 inhibitor (5). In the absence of differentiation-inhibitory signals, FGF4-mediated autocrine signals are the dominant trigger of ESC differentiation, such that the cells are no longer able to maintain pluripotency.
mESCs commit to a neuronal lineage when LIF and serum-derived signals are minimized under differentiation culture conditions (6–9). Although the majority of cells differentiate into neuronal cells, a subpopulation remains in an undifferentiated state (10–12), which complicates the clinical application of ESC-derived cells. Methodological approaches to the elimination of undifferentiated cells from cultures have been developed by taking advantage of differences between pluripotent and differentiated cells, such as metabolism, gene expression profiles, and cell surface antigens, or by extending the differentiation period of ESCs (12–17). However, the properties of residual undifferentiated cells are largely unclear, with even less being known about how cells maintain an undifferentiated state in the absence of LIF and serum/BMP signals.
In the study described in this report, we found that during in vitro neural differentiation of mESCs, a subpopulation of cells transits into a dormant state without compromising their pluripotency. These cells manifest gene expression profiles that are distinct from those of proliferating mESCs. However, upon serum stimulation, these cells can exit from the dormant state and restart proliferation and differentiation into all three germ layers. We identified Forkhead box O3 (FoxO3), a member of the Forkhead family of transcription factors, to be an essential regulator of the maintenance of pluripotency in the dormant mESCs. This study demonstrates a novel property of mESCs that may account for residual undifferentiated cells in the absence of LIF and serum/BMP signals.
RESULTS
Residual undifferentiated cells express pluripotent genes after neural differentiation of mESCs by SFEBq.
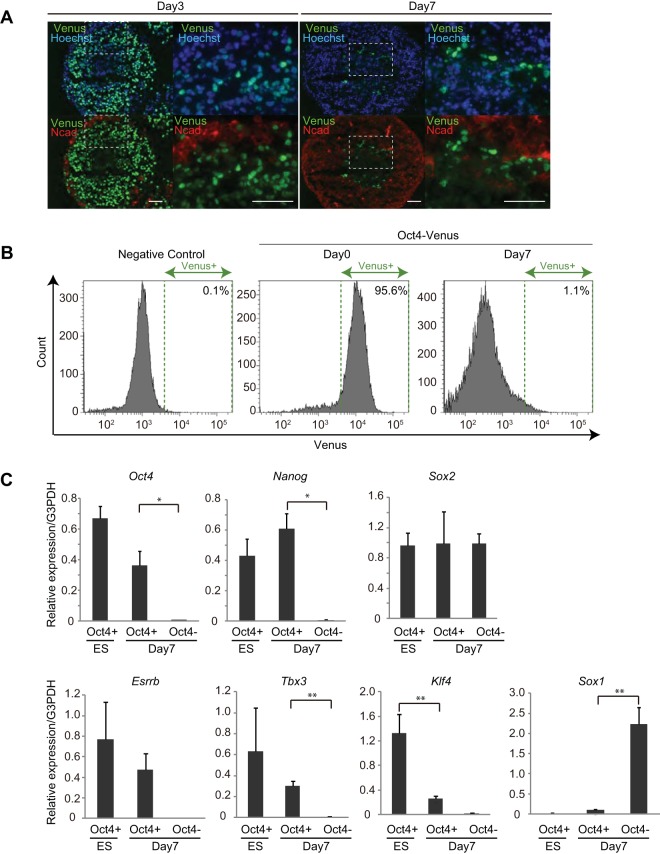
To detect residual undifferentiated cells after in vitro differentiation, we used the previously established mESC line in which Oct4-Venus was knocked in at the locus of Oct4, a gene essential for pluripotency (Oct4-Venus mESCs) (18). A single colony was picked up and clonally expanded for use in the experiments to ensure the homogeneity of the cell population. For in vitro neural differentiation, we cultured Oct4-Venus mESCs under serum-free floating culture of embryoid body-like aggregates with quick reaggregation (SFEBq) culture conditions, a highly efficient neural differentiation system (9). Consistent with the findings of previous studies, the majority (98.5% ± 1.38%) of Oct4-Venus mESCs lost Oct4 expression and expressed both a neural progenitor-specific gene, Sox1, and a neuronal marker protein, N-cadherin, during SFEBq culture (1 to 7 days) (Fig. 1A and B; see also Fig. S1A and B in the supplemental material). However, a small population (1.52% ± 1.32%) of cells reproducibly retained Oct4-Venus expression even after the cells were cultured in SFEBq medium for 7 days (Fig. 1A and B). We isolated Venus-positive and -negative cells by fluorescence-activated cells sorting (FACS) on day 7 of SFEBq culture (here referred to as day 7 Oct4+ and day 7 Oct4− cells, respectively) and analyzed the expression profiles of mESC pluripotent genes. Day 7 Oct4− cells expressed Sox1; however, mESCs and day 7 Oct4+ cells did not, confirming the isolation of undifferentiated cells by the FACS method employed (Fig. 1C, Sox1). We also found that Klf4 expression was significantly decreased in day 7 Oct4+ cells, whereas the level of expression of Oct4, Nanog, Sox2, Esrrb, and Tbx3 in these cells was only partially decreased or even increased compared with that in undifferentiated mESCs (Fig. 1C). These results indicate that a population of undifferentiated cells on day 7 of SFEBq culture continues to express a subset of pluripotent genes.
FIG 1.
Residual undifferentiated cells express pluripotent genes after neural differentiation of mESCs by SFEBq. (A) Images of Oct4-Venus (green), N-cadherin (red), and Hoechst (blue) in cryosections of cell spheres of SFEBq-cultured Oct4-Venus ESCs on day 3 and day 7. Magnified images of the boxed areas in the left panels are shown on the right. Bars = 50 μm. (B) FACS analysis for detecting Venus signals in EB5 mESCs (negative control), undifferentiated Oct4-Venus mESCs (Oct4-Venus, day 0), and SFEBq-cultured Oct4-Venus mESCs on day 7 (Oct4-Venus, day 7). (C) RT-qPCR analysis for expression of the indicated genes in FACS-isolated undifferentiated Oct4-Venus mESCs (ES, Oct4+), SFEBq-cultured day 7 Oct4+ cells (day 7, Oct4+), and SFEBq-cultured day 7 Oct4− cells (day 7, Oct4−). Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test; **, P < 0.01, analyzed by t test. G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
Day 7 Oct4+ cells retain self-renewal ability and pluripotency.
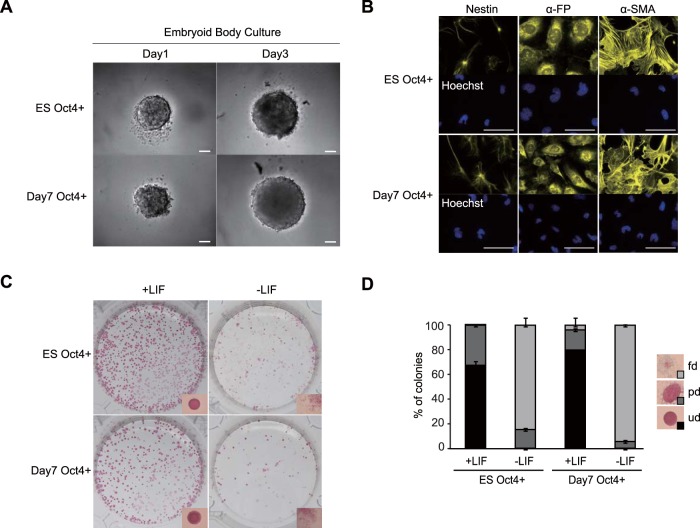
As day 7 Oct4+ cells expressed pluripotent genes, we examined whether these cells also retained pluripotency and a self-renewal ability. We found that in serum-containing embryoid body (EB) culture medium, day 7 Oct4+ cells robustly proliferated (Fig. 2A) and showed the capacity to differentiate into all three germ layers, including nestin-expressing ectodermal cells, α-fetoprotein (α-FP)-expressing endodermal cells, and α-smooth muscle actin (α-SMA)-expressing mesodermal cells (Fig. 2B), indicating that day 7 Oct4+ cells retain pluripotency. Moreover, in LIF-conditioned mESC maintenance medium containing serum, day 7 Oct4+ cells formed colonies expressing alkaline phosphatase (AP), a marker for undifferentiated cells (Fig. 2C). The ratio of undifferentiated colonies was similar in day 7 Oct4+ cells (79.6% ± 0.41%) and mESCs (67.4% ± 3.46%) (Fig. 2D). In contrast to the viability of day 7 Oct4+ cells, day 7 Oct4− cells were inviable in EB culture medium (Fig. S2A) and barely formed AP-expressing colonies with crystal violet (CV) staining in LIF-conditioned mESC maintenance medium (Fig. S2B and C), confirming distinct cell properties between day 7 Oct4+ and day 7 Oct4− cells. Collectively, these results show that day 7 Oct4+ cells retain both pluripotency and a self-renewal ability. Notably, day 7 Oct4+ cells hardly self-renewed when LIF was removed from the mESC maintenance medium, suggesting that, similar to mESCs, these cells require LIF for their self-renewal (Fig. 2C and D).
FIG 2.
Day 7 Oct4+ cells retain self-renewal ability and pluripotency. (A) Phase-contrast images of EBs derived from FACS-isolated undifferentiated Oct4-Venus mESCs (ES Oct4+) and SFEBq-cultured day 7 Oct4+ cells (day 7 Oct4+) on day 1 and day 3. Bars = 100 μm. (B) Images of nestin, α-FP, and α-SMA in differentiated EBs derived from FACS-isolated undifferentiated Oct4-Venus mESCs (ES Oct4+) and SFEBq-cultured day 7 Oct4+ cells (day 7 Oct4+). Bars = 50 μm. (C) Images of the AP assay for detection of the self-renewal ability of FACS-isolated undifferentiated Oct4-Venus mESCs (ES Oct4+) and SFEBq-cultured day 7 Oct4+ cells (day 7 Oct4+). Cells were cultured in LIF-conditioned medium (+LIF) or non-LIF-conditioned medium (LIF−). Magnified representative images of colonies are shown at the bottom of each panel. (D) Analysis of the differentiation status of colonies in the AP assay whose results are shown in panel C. fd, fully differentiated; pd, partially differentiated; ud, undifferentiated.
Day 7 Oct4+ cells remain in a dormant state.
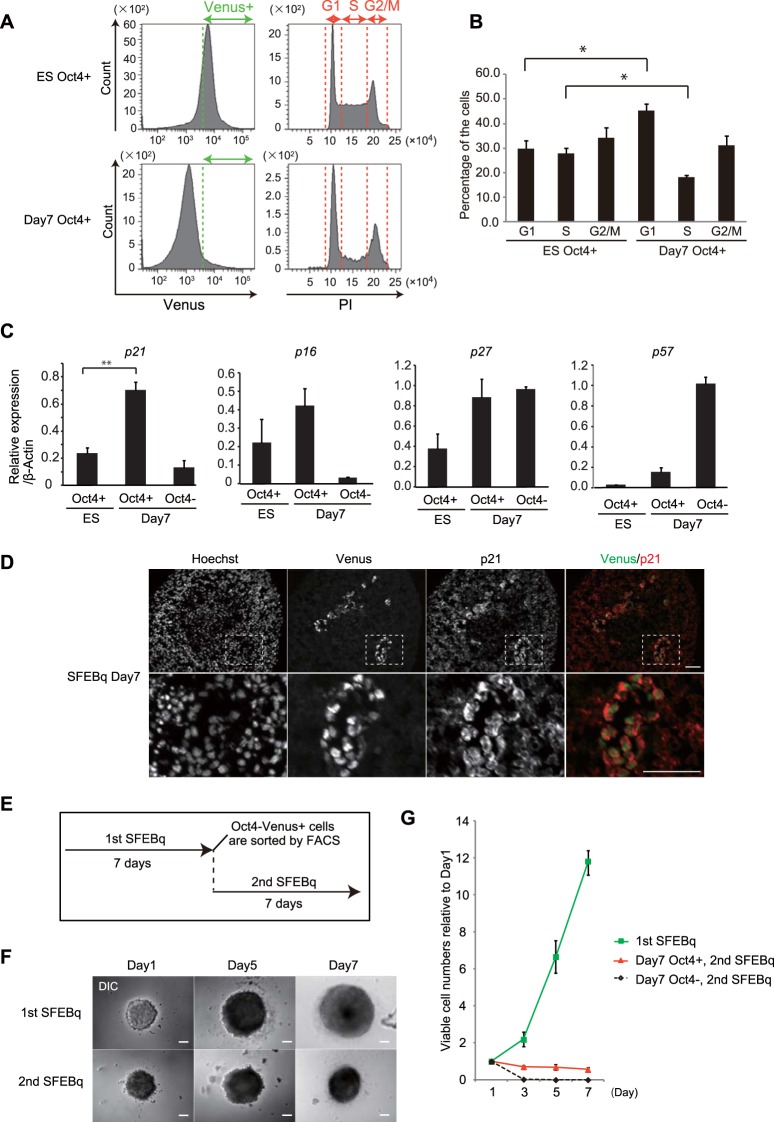
Next, we investigated the cell cycle profiles of day 7 Oct4+ cells. FACS analysis of the DNA content within each cell showed that the population of G1-phase cells was significantly increased in day 7 Oct4+ cells compared with that in mESCs (Fig. 3A and B). Conversely, the population of S-phase cells was significantly decreased in day 7 Oct4+ cells, whereas no significant difference in the G2/M-phase population was observed (Fig. 3A and B). These results indicate the delay or arrest of cell cycle progression in G1 phase in day 7 Oct4+ cells. Consistent with these observations, we found that, among a set of cyclin-dependent kinase (CDK) inhibitors, expression of p21 was significantly upregulated in day 7 Oct4+ cells compared with its level of expression in mESCs and day 7 Oct4− cells (Fig. 3C). In addition, p21 protein was expressed in Oct4+ cells in SFEBq cell aggregates on day 7 (Fig. 3D). Furthermore, when day 7 Oct4+ cells were isolated by FACS and reaggregated under SFEBq culture conditions (Fig. 3E), cell aggregates rarely grew to a large size during an additional 7 days of culture (Fig. 3F, 2nd SFEBq) and the cell proliferation rate was significantly suppressed compared with that of undifferentiated mESC-derived cells (Fig. 3G). These results indicate that day 7 Oct4+ cells markedly delay their cell cycle progression and exhibit a dormant-like phenotype.
FIG 3.
Day 7 Oct4+ cells arrest their cell cycle progression. (A) FACS analysis for the cell cycle profiles of undifferentiated Oct4-Venus mESCs (ES Oct4+) and SFEBq-cultured cells on day 7 (day 7 Oct4+). DNA contents were analyzed by PI staining (right) in the Venus+ cell fraction (left). (B) Quantification of the cell cycle profiles shown in panel A. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by the Bonferroni multiple-comparison test. (C) RT-qPCR analysis for the expression of CDK inhibitors in FACS-isolated undifferentiated Oct4-Venus mESCs (ES, Oct4+), SFEBq-cultured day 7 Oct4+ cells (day 7, Oct4+), and SFEBq-cultured day 7 Oct4− cells (day 7, Oct4−). Data represent the mean ± SEM (n = 3). **, P < 0.01, analyzed by t test. (D) Images of Oct4-Venus, p21, and Hoechst in cryosections of SFEBq-cultured Oct4-Venus mESCs on day 7. Magnified images of the boxed areas in the top panels are shown in the bottom panels. Bars = 50 μm. (E) Experimental schemes for SFEBq reculture assay. (F) Phase-contrast images of cell spheres in the first and second SFEBq cultures on days 1, 3, and 7. DIC, differential inference contrast. Bars = 100 μm. (G) Proliferation curve of cells during primary and secondary SFEBq culture of FACS-isolated undifferentiated Oct4-Venus mESCs (1st SFEBq), SFEBq-cultured day 7 Oct4+ cells (day 7 Oct4+, 2nd SFEBq), and SFEBq-cultured day 7 Oct4− cells (day 7 Oct4−, 2nd SFEBq). Data represent the mean ± SEM (n = 3).
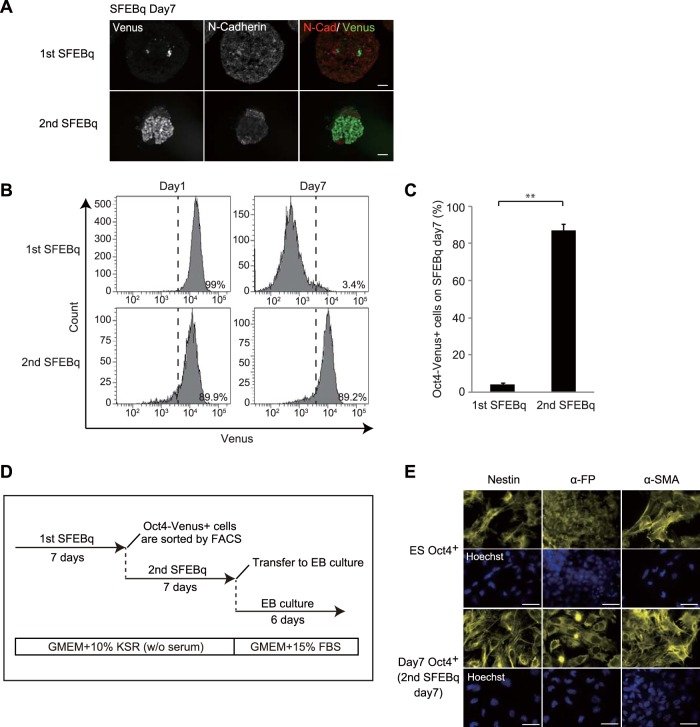
In contrast to cell aggregates from primary SFEBq cultures, aggregates from secondary SFEBq cultures displayed a majority of cells persistently expressing Oct4-Venus on day 7 (Fig. 4A). FACS analysis confirmed sustained expression of Oct4-Venus in a number of cells in aggregates from secondary SFEBq cultures on day 7 (87.1% ± 3.4%) compared with that in aggregates from primary SFEBq cultures on day 7 (4.2% ± 0.7%) (Fig. 4B and C). Furthermore, secondary day 7 Oct4+ cells differentiated into all three germ layers during EB culture, indicating that these cells retain pluripotency (Fig. 4D and E). Notably, when secondary day 7 Oct4+ cells were cultured in LIF-conditioned mESC maintenance medium containing serum for 2 weeks (Fig. S3A), the cells formed dome-shaped undifferentiated colonies (Fig. S3B) expressing mESC pluripotent genes at a level comparable to that for undifferentiated mESCs (Fig. S3C). In addition, these cells proliferated and differentiated in SFEBq culture at a rate equivalent to that for undifferentiated mESCs (Fig. S3D to F), implying that the dormant pluripotent state is reversible. Collectively, these data demonstrate that on day 7 of SFEBq culture a population of undifferentiated cells remains in a dormant state, whereby cells arrest their cell cycle progression without compromising their properties of pluripotency and plasticity.
FIG 4.
Day 7 Oct4+ cells sustain pluripotency in the 2nd SFEBq culture. (A) Images of Oct4-Venus (green) and N-cadherin (N-Cad; red) in cryosections of cell spheres in the first and second SFEBq cultures on day 7. Bars = 50 μm. (B) FACS analysis for detecting the Oct4-Venus signals of cells in the primary and secondary SFEBq cultures on day 7. (C) Quantification of the Oct4-Venus-positive cells for which the results are shown in panel B. Data represent the mean ± SEM (n = 3). **, P < 0.01, analyzed by t test. (D) Experimental schemes for SFEBq reculture assay, followed by EB assay. (E) Images of nestin, α-FP, and α-SMA in differentiated EBs derived from FACS-isolated undifferentiated Oct4-Venus mESCs (ES Oct4+) and secondary SFEBq-cultured day 7 Oct4+ cells (day 7 Oct4+). Bars = 100 μm.
Dormant pluripotent cells emerge during the course of differentiation.
As ESCs display heterogeneous properties (18–22), it is possible that dormant ESCs exist within the original ESC population. To investigate this possibility, we performed a dye dilution assay using the CellTrace reagent, an analog of the 5(6)-carboxyfluorescein N-hydroxysuccinimidyl ester, which covalently binds to intracellular proteins. Once the reagent is incorporated into cells, it is diluted with every cell division, such that only the dormant cell subpopulation retains the dye after several rounds of cell division. We incorporated this reagent into Oct4-Venus mESCs, cultured them in either mESC maintenance or SFEBq medium, and then analyzed changes in dye intensity by FACS every other day for 7 days. Our results show that the dye intensity decreased synchronously without any dye-positive cells being retained after 7 days of culture in mESC maintenance medium (Fig. S4A, middle row), whereas under the SFEBq culture condition, a subpopulation of dye-retaining cells emerged on day 5 and day 7 of differentiation (Fig. S4A, bottom row). In addition, under the SFEBq culture condition, the cell cycle profiles of the Oct4-Venus-positive cell population were almost equal on days 0, 1, 3, and 5, whereas the population of S-phase cells was markedly decreased on day 7 (Fig. S4B), suggesting that the majority of the Oct4-Venus-positive cells was dividing until day 5 and then shifted to a slow cycling/cell cycle arrest phase by day 7. These results support the notion that dormant pluripotent cells do not exist within the original ESC population but, rather, emerge during the course of differentiation.
Expression of FoxO3 is upregulated in day 7 Oct4+ cells.
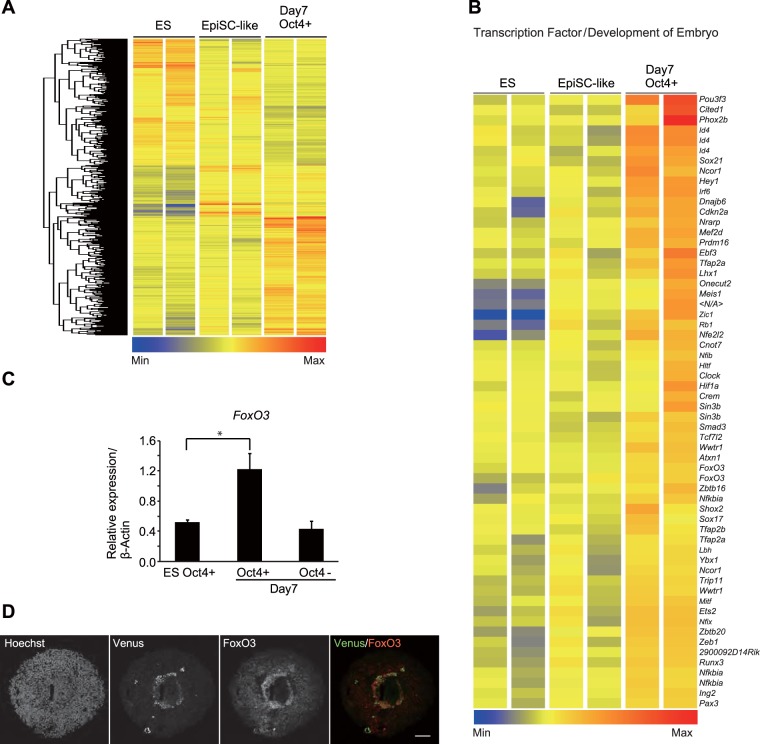
To identify genes accounting for the dormant pluripotent phenotype of day 7 Oct4+ cells, we examined their gene expression profiles using DNA microarray analysis. Epiblast-derived stem cells (EpiSCs), characterized by increased and reduced levels of expression of Fgf5 and the ESC surface antigen Pecam1, respectively (23, 24), are pluripotent but have cellular properties and gene expression profiles different from those of ESCs (23, 25). Consistent with the findings of a previous study (26), we observed the increased expression of Fgf5 at about day 3 of SFEBq culture (Fig. S5A). In addition, we noticed that on day 3 of SFEBq culture, a subpopulation of Oct4-Venus-positive cells showed reduced Pecam1 expression (Fig. S5B). FACS-isolated Venus-positive (Venus+)/Pecam1-negative (Pecam1−) cells recapitulated several notable properties of EpiSCs, including reduced expression of mESC-specific genes (Klf4 and Rex1), increased expression of the EpiSC-specific gene Fgf5 (Fig. S5C), and reduced self-renewability in LIF-conditioned medium, all of which occurred without compromising their pluripotent potential (Fig. S5D to G). Therefore, we defined the FACS-isolated Venus+/Pecam1-positive (Pecam1+) population of mESCs and the Venus+/Pecam1− population of SFEBq cell aggregates on day 3 to represent undifferentiated mESCs and EpiSC-like cells, respectively, for use as controls in our DNA microarray analysis of dormant pluripotent cells. Microarray and gene ontology analysis revealed that genes involved in developmental processes, chromatin organization, and gene expression were significantly upregulated in day 7 Oct4+ cells compared with their levels of regulation in undifferentiated mESCs and EpiSC-like cells (Fig. 5A and Table S2). In addition, we found that transcription factors involved in embryonic development were significantly upregulated in day 7 Oct4+ cells (Fig. 5B). Among them, we confirmed increased expression of FoxO3, a well-known transcription factor governing various biological processes, including cell cycle arrest, autophagy, metabolic regulation, and cellular stress responses (27–29), in day 7 Oct4+ cells by quantitative PCR (qPCR) (Fig. 5C). Moreover, FoxO3 protein was expressed in Oct4+ cells of SFEBq cell aggregates on day 7 (Fig. 5D). In addition to FoxO3, FoxO1, FoxO4, and FoxO6 were also partially upregulated in day 7 Oct4+ cells at the transcriptional level (Fig. S6A). We found that the FoxO1 and FoxO6 proteins but not the FoxO4 protein were expressed in Oct4+ cells of SFEBq cell aggregates on day 7 (Fig. S6B), suggesting the involvement of FoxO1 and FoxO6 but not FoxO4 in the dormant pluripotent phenotype of day 7 Oct4+ cells. The expression levels of FoxO3, FoxO1, and FoxO6 were decreased to levels comparable to those in undifferentiated mESCs after 2 weeks of culture in LIF-conditioned mESC maintenance medium containing serum (Fig. S3C), confirming the specific upregulation of these genes in the dormant pluripotent cells.
FIG 5.
Expression of FoxO3 is upregulated in day 7 Oct4+ cells. (A) Unsupervised hierarchical clustering and heat map display of genes differentially expressed (greater than or less than 1.4-fold) in FACS-isolated SFEBq day 7 Oct4+ cells compared with their expression in undifferentiated mESCs (ES) and EpiSC-like cells. (B) Heat map display of transcription factors involved in embryonic development. Transcription factors upregulated in SFEBq day 7 Oct4+ cells (>1.4-fold) are shown. Min, minimum; Max, maximum. (C) RT-qPCR analysis for the expression of FoxO3 in FACS-isolated undifferentiated Oct4-Venus mESCs (ES Oct4+), SFEBq-cultured day 7 Oct4+ cells (day 7, Oct4+), and SFEBq-cultured day 7 Oct4− cells (day 7, Oct4−). Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test. (D) Images of Oct4-Venus (green), FoxO3 (red), and Hoechst (blue) in cryosections of cell spheres of SFEBq cultures on day 7. Bar = 75 μm.
FoxO3 governs the pluripotency of day 7 Oct4+ cells.
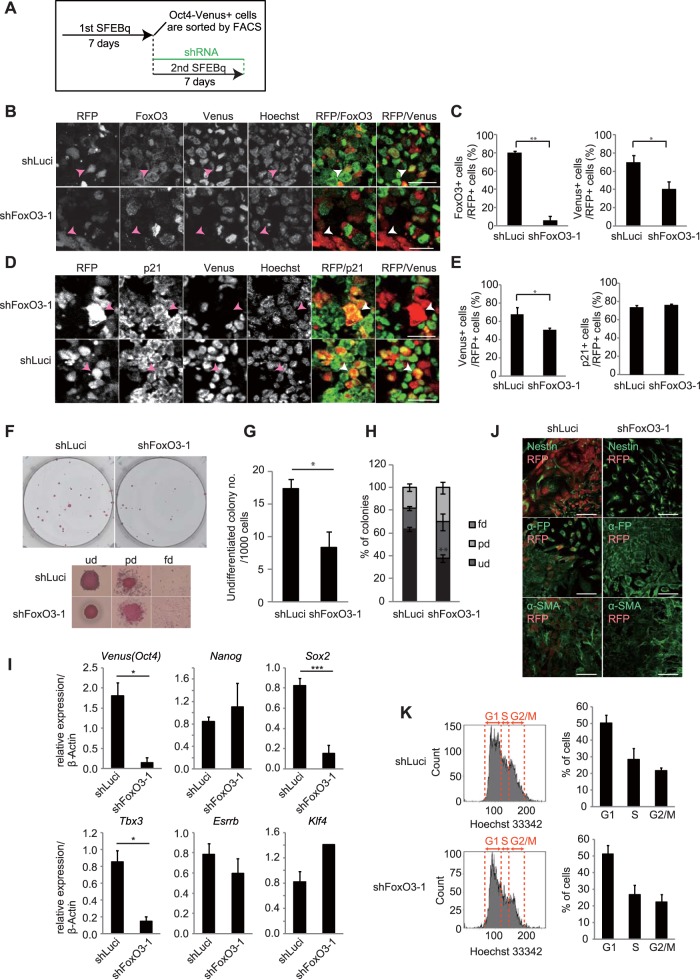
Because FoxO3 is known to regulate the quiescence of adult stem cells, including muscle stem cells, hematopoietic stem cells (HSCs), and neural stem cells (NSCs) (30–34), we interrogated whether FoxO3 also regulates the dormant phenotype of day 7 Oct4+ cells. To address this possibility, we knocked down FoxO3 using lentivirus-mediated short hairpin RNA (shRNA) in day 7 Oct4+ cells during secondary SFEBq culture (Fig. 6A), where shRNA-infected cells could be monitored by their red fluorescent protein (RFP) expression. The shRNA effectively decreased the protein levels of ectopically expressed FoxO3 in HEK293T cells (Fig. S6C, shFoxO3-1). On day 7, we confirmed that the expression of endogenous FoxO3 protein was significantly decreased in cells infected with the virus encoding shRNA against FoxO3 (shFoxO3) compared with that in control cells infected with the virus encoding shRNA against luciferase (Luci) (shLuci) (Fig. 6B and C). In infected cells, we observed that the depletion of FoxO3 resulted in a significant decrease in Oct4-Venus expression (Fig. 6B and C), whereas expression of p21 was unchanged (Fig. 6D and E). FACS-isolated shFoxO3-infected cells manifested reduced self-renewal ability (Fig. 6F and G) with a decreased ratio of undifferentiated cells (Fig. 6H) in LIF-conditioned mESC maintenance medium compared with the findings for shLuci-infected cells. In addition, expression of Oct4 and Sox2 was significantly decreased in the FoxO3-specific shRNA-infected cells compared to that in Luci-specific shRNA-infected cells (Fig. 6I). Similar results were obtained by using an independent shRNA against FoxO3 (Fig. S6C, shFoxO3-2, and D), excluding the possibility that the shRNAs used had off-target effects. We also observed a significant decrease in the level of expression of Tbx3 in the FoxO3-specific shRNA-infected cells (Fig. 6I). However, this is unlikely the result of FoxO3 depletion, because Tbx3 expression was not decreased in the cells depleted of FoxO3 by another independent FoxO3-specific shRNA (Fig. S6D, shFoxO3-2). We further found that the FoxO3-specific shRNA-infected cells showed a compromised capacity for differentiation into three germ layers in EB culture compared with that of Luci-specific shRNA-infected cells (Fig. 6J), indicating a requirement for FoxO3 for the maintenance of differentiation potential in day 7 Oct4+ cells. Although the depletion of FoxO3 results in compromised pluripotency in day 7 Oct4+ cells, it barely altered the cell cycle profiles of these cells (Fig. 6K). This result is consistent with the observation that p21 expression is unchanged in these cells (see Fig. 6D and E), suggesting that the dormancy of these cells is maintained in a manner that is independent of FoxO3. Collectively, these results demonstrate that FoxO3 governs the pluripotency of undifferentiated dormant cells emerging during the course of differentiation of mESCs. In contrast to depletion of FoxO3, depletion of FoxO1 by shRNA (Fig. S6C) barely altered the expression profiles of mESC pluripotent genes in day 7 Oct4+ cells (Fig. S6E), suggesting that FoxO1 plays a minor role in pluripotency maintenance in these cells.
FIG 6.
FoxO3 is responsible for maintenance of pluripotency in day 7 Oct4+ cells. (A) Experimental schemes for the FoxO3a knockdown assay during secondary SFEBq culture. (B) Images of RFP, Oct4-Venus, FoxO3, and Hoechst in cryosections of secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or control luciferase (Luci)-specific shRNA. The merge images of RFP (red)/FoxO3 (green) and RFP (red)/Oct4-Venus (green) are shown in the right two panels. RFP represents shRNA-infected cells. Arrowheads indicate representative RFP+ cells. Bars = 25 μm. (C) Quantification of FoxO3-positive (left) and Oct4-Venus-positive (right) cells in the shRNA-infected cells shown in panel B. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test; **, P < 0.01, analyzed by t test. (D) Images of RFP, Oct4-Venus, p21, and Hoechst in cryosections of secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or control luciferase-specific shRNA. The merge images of RFP (red)/p21 (green) and RFP (red)/Oct4-Venus (green) are shown in the right two panels. Arrowheads indicate representative RFP+ cells. Bars = 25 μm. (E) Quantification of p21-positive (right) and Oct4-Venus-positive (left) cells in the shRNA-infected cells shown in panel D. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test. (F) Images of AP assay for detection of the self-renewal ability of FACS-isolated (RFP-positive [RFP+] cell fractions) secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or Luci-specific shRNA. Magnified representative images of colonies in each differentiation status are shown at the bottom. (G) Average number of AP-positive undifferentiated colonies in panel F. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test. (H) Analysis of the differentiation status of colonies in the AP assay for which the results are shown in panel F. **, P < 0.01, analyzed by t test. (I) RT-qPCR analysis for expression of the indicated genes in FACS-isolated (RFP+ cell fractions) secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or Luci-specific shRNA. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test; ***, P < 0.001, analyzed by t test. (J) Images of nestin, α-FP, and α-SMA in differentiated EBs derived from secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or Luci-specific shRNA. For RFP+/nestin-positive cells, 26.8% were positive for Luci-specific shRNA and 4.6% were positive for FoxO3-specific shRNA-1; for RFP+/α-FP-positive cells, 24% were positive for Luci-specific shRNA and 0.5% were positive for FoxO3-specific shRNA-1; and for RFP+/α-SMA-positive cells, 40.8% were positive for Luci-specific shRNA and 3.8% were positive for FoxO3-specific shRNA-1. These data are for >1,000 cells. (K) FACS analysis for the cell cycle profiles of FACS-isolated (RFP+ cell fractions) secondary day 7 SFEBq-cultured cells infected with FoxO3-specific shRNA-1 or Luci-specific shRNA. (Left) DNA contents were analyzed by Hoechst staining; (right) quantification of cell cycle profiles. Data represent the mean ± SEM (n = 3).
Next, we interrogated whether FoxO3-dependent residual undifferentiated cells emerge during EB culture. Similar to the results for the SFEBq culture on day 7, a subpopulation of cells retained Oct4-Venus expression in EB culture on day 6 (Fig. S7A). Furthermore, the expression of FoxO3 was significantly upregulated in day 6 Oct4+ cells in EB culture compared with the level of regulation in undifferentiated mESCs (Fig. S7B). To evaluate the requirement for FoxO3 in these cells, we established Oct4-Venus mESC lines which stably express FoxO3-specific shRNA or control Luci-specific shRNA (Fig. S7C). The capacity of the mESC lines expressing shFoxO3 to differentiate into all three germ layers was comparable to that of the mESC lines expressing shLuci (Fig. S7D). Under this condition, we found that a population of Oct4+ cells in EB culture on day 6 was significantly decreased in mESCs expressing shFoxO3 compared with that in mESCs expressing shLuci (Fig. S7E and F). These results suggest that FoxO3-mediated dormant pluripotency mechanisms function not only in SFEBq culture but also under other differentiation conditions.
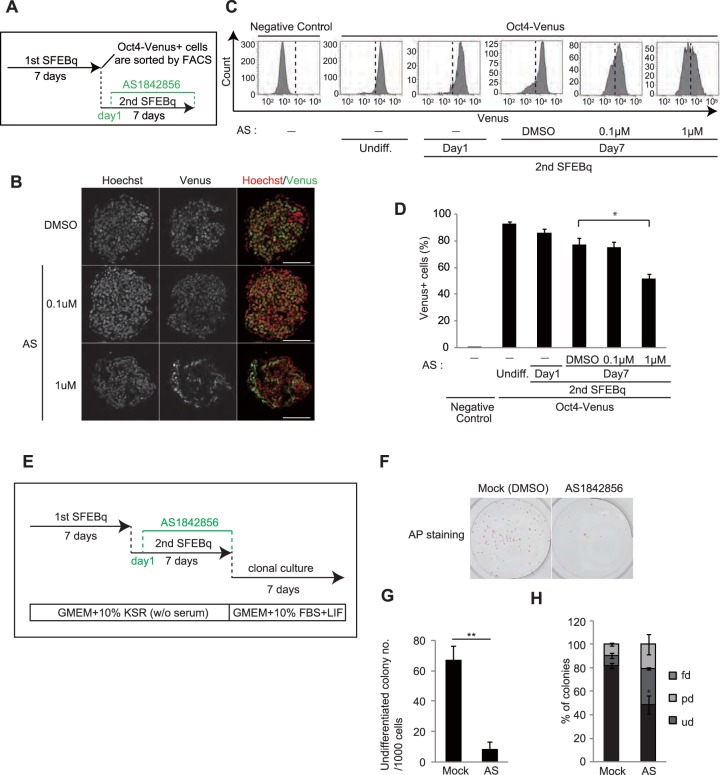
It has been reported that AS1842856, an inhibitor of FoxO1, also partially suppresses FoxO3 (35). We found that treatment of day 7 Oct4+ cells with AS1842856 during secondary SFEBq culture (Fig. 7A) resulted in a decreased population of cells expressing Oct4-Venus (Fig. 7B to D), whereas AS1842856 treatment barely altered the expression level of Oct4-Venus in self-renewing mESCs cultured in mESC maintenance medium (Fig. S8A and B). Furthermore, AS1842856 significantly compromised the self-renewal ability of day 7 Oct4+ cells during secondary SFEBq culture (Fig. 7E to H). These results imply the possible application of AS1842856 to eliminate residual undifferentiated cells from ESC differentiation culture.
FIG 7.
AS1842856 compromised pluripotency maintenance in day 7 Oct4+ cells. (A) Experimental scheme for the AS1842856 (AS) treatment assay. (B) Images of Oct4-Venus (green) and Hoechst (blue) in cryosections of secondary day 7 SFEBq-cultured cells treated with AS1842856 or the control (dimethyl sulfoxide [DMSO]). Bars = 75 μm. (C) FACS analysis for detecting Oct4-Venus signals in EB5 mESCs (negative control), undifferentiated Oct4-Venus mESCs (Undiff.), secondary SFEBq-cultured Oct4-Venus mESCs on day 1 (2nd SFEBq, day 1), and secondary SFEBq-cultured Oct4-Venus mESCs on day 7 with or without AS1842856 treatment (2nd SFEBq, day 7). (D) Quantification of the Oct4-Venus-positive cells shown in panel C. Data represent the mean ± SEM (n = 3). *, P < 0.05, analyzed by t test. (E) Experimental scheme for AS1842856 treatment, followed by the colony formation assay with AP staining. (F) Images of the AP assay for detection of the self-renewal ability of the secondary day 7 SFEBq-cultured cells treated with AS1842856 or the control (dimethyl sulfoxide). (G) Average number of AP-positive undifferentiated colonies in panel F. Data represent the mean ± SEM (n = 3). **, P < 0.01. (H) Analysis of the differentiation status of colonies in the AP assay shown in panel G. *, P < 0.05, analyzed by t test.
DISCUSSION
In this report, we describe a subpopulation of mESCs that transit into a dormant state during neural differentiation without compromising pluripotency. Our results show that dormant cells maintain their pluripotency in a FoxO3-dependent manner. In human ESCs, FoxO1 directly binds to the promoter of the Oct4 and Sox2 genes to regulate pluripotency (36). FoxO binding motifs also exist within the promoter region of the mouse Oct4 and Sox2 genes (36); however, whether the FoxO family is involved in the pluripotency of mESCs or induced pluripotent stem cells (iPSCs) has been controversial (36, 37). Our result showing that shRNA against FoxO3 but not shRNA against FoxO1 compromised the expression of Oct4 and Sox2 in dormant pluripotent cells (Fig. 6I; see also Fig. S6D and E in the supplemental material) suggests that the pluripotency of dormant cells is mainly reliant on FoxO3.
How dormant pluripotent cells emerge remains an open question. FoxO3 is positively regulated by AMP-activated protein kinase (AMPK) and negatively regulated by Akt (38–41). Our results show that serum-derived factors drive dormant pluripotent cells into a proliferative state. Serum-derived growth factors activate Ras-MAPK signal transduction, leading to inhibition of the liver kinase B1 (LKB1)-AMPK pathway, a key metabolism-sensing signaling pathway (42–45). In addition, insulin or insulin-like growth factors in serum activate the phosphatidylinositol 3-kinase (PI3K)-Akt pathway (39–41). As Oct4-Venus-positive cells often sit in the center of SFEBq cell aggregates on day 7, a conceivable hypothesis is that energy restriction or growth factor limitation may trigger activation of the LKB1-AMPK pathway and/or inhibition of the PI3K-Akt pathway, eliciting the emergence of pluripotent dormant cells. Supporting this idea, AMPK agonist 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) enhanced the dormant phenotype of mESCs and induced FoxO3 expression in SFEBq culture (data not shown), which is consistent with the previous report demonstrating that AICAR inhibits the proliferation of mESCs in EB culture medium and enhances their pluripotency in mESC maintenance medium (46, 47). In our experiments, the dormant state is maintained under serum-free conditions; thus, once cells are stimulated with serum-derived growth factors, they may exit from the dormant state and lose the characteristic of FoxO3-dependent pluripotency.
During the preparation of the manuscript, Scognamiglio and colleagues reported that Myc-knockout ESCs display a dormant phenotype (48). Interestingly, our microarray analysis revealed that Myc expression was decreased in day 7 Oct4+ cells. It has previously been reported that FoxO3 suppresses Myc-dependent transcription and decreases the expression of Myc via microRNA (49–51). In addition, the CDK inhibitor p21 is a direct target of Myc (52); therefore, it is possible that FoxO3 induces the downregulation of Myc in pluripotent dormant cells. Although our results show that while depletion of FoxO3 leads to a loss of Oct4-Venus expression, it did not affect p21 expression in pluripotent dormant cells or their cell cycle profiles (Fig. 6D, E, and K). This suggests that FoxO3-independent pathways may potentially be involved in the mechanisms of cell cycle arrest occurring in pluripotent dormant cells on day 7.
As the existence of residual undifferentiated cells is an obstacle to the clinical application of ESCs/iPSCs, various methods to eliminate these cells have been reported. Conventionally, these methods take advantages of the differences in properties between self-renewing ESCs and differentiated cell types. Our study shows that a subpopulation of residual undifferentiated cells sits in a dormant state, with their properties being distinct from those of self-renewing ESCs. These results raise the possibility that a subpopulation of residual undifferentiated cells cannot be eliminated by conventional methods. In this context, cancer stem cells also remain in a dormant state and then start to expand in response to extracellular signals (53). Intriguingly, the dormancy observed in several types of cancer stem cells is reportedly maintained by FoxO3 (54, 55). Therefore, FoxO3-mediated regulation of stem cell dormancy is likely a common event among cancer stem cells and pluripotent dormant cells. Our results imply that FoxO3-mediated dormancy maintenance mechanisms are a valid target for strategies to eliminate residual undifferentiated cells for ESC- and iPSC-based therapies.
MATERIALS AND METHODS
Cell culture.
Mouse ESCs (EB5 cells; catalog number AES0151; Riken BioResource Center [BRC]) (56, 57) were maintained in Glasgow minimum essential medium (G-MEM) supplemented with 10% (vol/vol) KnockOut serum replacement (KSR; Gibco), 1% (vol/vol) fetal bovine serum (FBS; Gibco), 0.1 mM nonessential amino acids (NEAA; Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), and LIF (Wako) on a 0.1% (wt/vol) gelatin-coated dish. Oct4-Venus mESCs (catalog number AES0153; Riken BRC) (18) were cultured in G-MEM supplemented with 15% FBS, 1 mM sodium pyruvate, 0.1 mM NEAA, 0.1 mM 2-mercaptoethanol, and LIF on a 0.1% (wt/vol) gelatin-coated dish. A single colony was picked up and grown in the medium for clonal expansion. All the data were obtained by using the single cell clone.
mESC differentiation.
Neural differentiation of mESCs by the serum-free floating culture of embryoid body-like aggregates with quick reaggregation (SFEBq) culture was performed as previously described (9). Briefly, Oct4-Venus mESCs were promptly reaggregated in a low-adhesion 96-well plate (Sumilon spheroid plates; Sumitomo) at a density of 3,000 cells per well in a 150-μl volume and then cultured in SFEBq medium (G-MEM supplemented with 10% KSR, 90 μM NEAA, 0.9 mM sodium pyruvate, and 44 μM 2-mercaptoethanol) for up to 7 days. The culture medium was replenished every other day. In the reculture assay, Oct4-Venus-positive mESCs were isolated by fluorescence-activated cell sorting (FACS) on day 7 of SFEBq culture and then reaggregated in a low-adhesion 96-well plate, as described above. In the FoxO inhibition assay, AS1842856 (Calbiochem) was added to recultured mESCs on days 1, 3, and 5 at a concentration of either 0.1 or 1 μM. For the embryoid body (EB) assay, mESCs were dissociated with 0.25% (vol/vol) trypsin–1 mM EDTA, reaggregated as described above, and then cultured in EB medium (G-MEM supplemented with 15% FBS, 1 mM sodium pyruvate, 0.1 mM NEAA, and 0.1 mM 2-mercaptoethanol). On day 3, cell aggregates were transferred to a glass dish coated with 0.1% (wt/vol) gelatin and cultured for another 3 days.
Lentivirus-based shRNA expression.
The shRNA target sequences are listed in Table S1 in the supplemental material. The oligonucleotides encoding each shRNA target sequence were cloned into the lentiviral vector CSII-EF-mRFP1-RfA (Riken BioResource Center). HEK293T cells were cotransfected with the shRNA vectors together with the pGAG/Pol and pVSV-G plasmids by a calcium phosphate-based method. The cell culture supernatant containing viral particles was concentrated and stored at −80°C. The viral solutions were mixed with polybrane (8 μg/ml), the mixture was added to Oct4-Venus-positive cells on day 1 of the SFEBq reculture assay, and the culture was incubated for 7 days. Unless indicated otherwise, the cell aggregates were dissociated into single cells with phosphate-buffered saline (PBS) containing 1 mM EDTA and resuspended in PBS containing 2% (vol/vol) KSR, and the RFP-positive cell fractions were isolated by FACS to purify the cells infected with lentivirus expressing shRNA.
FoxO3-specific shRNA- and Luci-specific shRNA-stable mESC lines.
Oct4-Venus mESCs were infected with lentivirus encoding FoxO3-specific shRNA-1 or luciferase-specific shRNAs as described above. RFP-positive cells were isolated by FACS with a FACSAria II cell sorter and incubated in mESC maintenance medium, followed by selection of a single colony for clonal expansion. Undifferentiated colonies were selected by morphology. The proper differentiation potential of each cell line was confirmed by the SFEBq culture method.
RT-qPCR assays.
Total RNA was isolated using an RNeasy minikit (Qiagen). cDNA was synthesized from 400 ng of RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen). For gene expression analysis of the lentivirus-based shRNA-infected mESCs on day 7 of the SFEBq reculture assay (Fig. 6I and S6D and E), cDNA obtained from the FACS-purified RFP-positive cells was amplified using a CellAmp total RNA amplification kit (real time; version 2; TaKaRa), according to the manufacturer's protocol. Reverse transcription-quantitative PCR (RT-qPCR) was performed using a Kapa SYBR Fast universal kit (Kapa Biosystems) on an ABI 7500 cycler (Applied Biosystems). Data were analyzed by ABI 7500 SDS software (version 1.2). The primers used are listed in Table S1.
Flow cytometry.
mESCs and SFEBq day 7 cell aggregates were dissociated with 0.25% (vol/vol) trypsin–1 mM EDTA and PBS containing 1 mM EDTA, respectively. Venus-positive and -negative cell fractions were sorted and analyzed using a FACS FACSAria II flow cytometer with FACSDiva software (BD Biosciences). The EB5 cell line was used as a negative control. To isolate mESC populations and epiblast stem cell-like (EpiSC-like) cells for microarray analysis, Oct4-Venus mESCs cultured in the maintenance medium and SFEBq day 3 cell aggregates were dissociated with 0.25% (vol/vol) trypsin–1 mM EDTA, incubated for 1 h in SFEBq medium to recover surface antigens, washed with 2% (vol/vol) FBS in PBS, and stained with allophycocyanin-conjugated CD31 antibody (catalog number 551262; BD Pharmingen) or control rat IgG2a (catalog number 557690; BD Pharmingen) at 4°C for 45 min. After staining, the cells were washed with 2% (vol/vol) FBS in PBS and sorted by FACS with a FACSAria II cell sorter. On day 3, the Venus+ CD31+ subpopulation of Oct4-Venus mESCs and the Venus+ CD31− subpopulation of SFEBq cell aggregates were defined as mESCs and EpiSC-like cells, respectively. For propidium iodide (PI) staining, cells were fixed with 1% (wt/vol) paraformaldehyde (PFA) on ice for 30 min, followed by incubation with 70% (vol/vol) ethanol on ice for 2 h. Samples were then treated with 200 μg/ml RNase at 37°C for 30 min and incubated with 50 μg/ml PI at room temperature for 10 min. For Hoechst staining, cells were incubated with 10 μg/ml Hoechst 33342 (BD Pharmingen) in SFEBq medium at 37°C for 1 h. The cells were then washed with PBS three times and analyzed by FACS with a FACSAria III cells sorter and FACSDiva software.
Immunostaining.
Immunostaining of SFEBq cell aggregates was performed as previously described (9). Briefly, cell aggregates were fixed in 4% (wt/vol) PFA and embedded in optimal cutting temperature compound (Tissue-Tek). Samples were sectioned using a cryostat (catalog number CM1850; Leica) and subjected to immunostaining. For p21 staining, sectioned samples were permeabilized with 100% methanol at −20°C for 30 min. For staining of Venus, N-cadherin, FoxO1, FoxO4, FoxO3, and FoxO6, sectioned samples were permeabilized with 0.3% (vol/vol) Triton X-100 in PBS at room temperature for 30 min. Samples were then blocked with 2% (wt/vol) skim milk in PBS at room temperature for an additional 30 min, treated with primary antibodies at 4°C overnight, washed with 0.05% (vol/vol) Tween 20 in PBS, and incubated with secondary antibodies at room temperature for 1 h. For staining of EB cultures, cells were fixed with 4% (wt/vol) PFA at room temperature for 10 min, permeabilized with 0.1% (vol/vol) Triton X-100, and stained with primary antibodies in 1% bovine serum albumin at 4°C overnight. The antibodies used included anti-green fluorescent protein (anti-GFP; catalog number GF090R; Nacalai Tesque), anti-N-cadherin (catalog number 610920; BD Transduction Laboratory), anti-FoxO1 (catalog number C29H4; Cell Signaling), anti-FoxO3a (catalog number 75D8; Cell Signaling), anti-FoxO4 (catalog number ab63254; Abcam), anti-FoxO6 (catalog number 19122-1-AP; Proteintech), anti-p21 (catalog number sc-397; Santa Cruz), antinestin (catalog number sc-58813; Santa Cruz), anti-α-smooth muscle actin (catalog number M0851; Dako), and anti-α-fetoprotein (catalog number MAB1368; R&D Systems).
Cell tracing assay.
Oct4-Venus mESCs were stained with CellTrace violet reagents (Life Technologies) according to the manufacturer's instructions and cultured in either mESC maintenance medium or SFEBq culture medium. Cells were dissociated on day 1, 3, 5, or 7 and analyzed by FACS with a FACSAria II cell sorter and FACSDiva software.
Colony-forming assay.
Cells were dissociated and seeded onto a 0.1% (wt/vol) gelatin-coated 6-well plate (3,000 cells per well) and cultured in mESC maintenance medium with or without LIF for 7 days. For the alkaline phosphatase (AP) staining assay, cell samples were stained using a leukocyte alkaline phosphatase kit (catalog number 86R-1KT; Sigma-Aldrich) according to the manufacturer's protocol. For quantifications, 100 colonies were analyzed for their morphology and staining intensity. For the crystal violet (CV) staining assay, AP-stained cells were incubated in 0.1% (wt/vol) crystal violet (catalog number 09803-62; Nacalai Tesque) dissolved in methanol at room temperature for 5 min and gently rinsed with water.
Cell proliferation assay.
Oct4-Venus mESCs cultured in mESC maintenance medium and SFEBq day 7 cell aggregates were dissociated and the Venus-positive or -negative cells were sorted by FACS with a FACSAria II cell sorter. The cells were then incubated in SFEBq culture medium for 1, 3, 5, or 7 days. Cells were dissociated and stained with trypan blue solution (Nacalai Tesque) to quantify the number of viable cells. The cell proliferation rate was calculated as the average number of viable cells relative to that on day 1 of culture.
Microarray analysis.
Total RNA was isolated from FACS-sorted mESCs, EpiSC-like cells, and Venus-positive cells from SFEBq day 7 cell aggregates using an RNeasy minikit in accordance with the manufacturer's protocol. RNA quality was confirmed by use of a bioanalyzer (Agilent Technology). Reverse transcription, labeling, and hybridization were performed using a Mouse Genome 430 (version 2.0) array kit (Affymetrix). Statistical analysis of gene expression was performed using GeneSpring GX software (Agilent Technologies), and differentially expressed genes were identified using t statistics with an adjusted P value of <0.05. Genes upregulated more than 1.4-fold in Venus-positive cells from SFEBq day 7 cell aggregates were subjected to gene ontology analysis. Heat maps were generated using GeneSpring GX software.
Statistics.
All data are representative of those from at least three independent experiments. Statistical analyses were performed by t test or the Bonferroni multiple-comparison test. Data are presented as mean ± standard error of the mean (SEM).
Accession number(s).
The microarray data are available in the Gene Expression Omnibus (GEO) database under accession number GSE89912.
Supplementary Material
ACKNOWLEDGMENTS
We thank N. Takata and E. Sakakura (Riken CDB) and M. Maekawa and T. Yamamoto (Kyoto University) for technical guidance, Y. Komatsuzaki (Kyoto University) for technical assistance, and H. Harada (Kyoto University) for critical scientific discussions.
M.I. and F.T. designed the experiments. M.I. performed the experiments. M.I. and F.T. analyzed the data. M.I and F.T. wrote the manuscript.
We have no conflicts of interest to declare.
This work was supported by JSPS KAKENHI grant number 15J05582 (to M.I.), MEXT KAKENHI grant number 26116712 (to F.T.), and the Naito Foundation (to F.T.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00417-16.
REFERENCES
- 1.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 2.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 3.Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281–292. doi: 10.1016/S0092-8674(03)00847-X. [DOI] [PubMed] [Google Scholar]
- 4.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. 2007. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 5.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. 2000. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron 28:31–40. doi: 10.1016/S0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. 2005. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci 8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 8.Smukler SR, Runciman SB, Xu S, van der Kooy D. 2006. Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J Cell Biol 172:79–90. doi: 10.1083/jcb.200508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda H, Takahashi J, Watanabe K, Hayashi H, Morizane A, Koyanagi M, Sasai Y, Hashimoto N. 2006. Fluorescence-activated cell sorting-based purification of embryonic stem cell-derived neural precursors averts tumor formation after transplantation. Stem Cells 24:763–771. doi: 10.1634/stemcells.2005-0137. [DOI] [PubMed] [Google Scholar]
- 12.Doi D, Morizane A, Kikuchi T, Onoe H, Hayashi T, Kawasaki T, Motono M, Sasai Y, Saiki H, Gomi M, Yoshikawa T, Hayashi H, Shinoyama M, Refaat MM, Suemori H, Miyamoto S, Takahashi J. 2012. Prolonged maturation culture favors a reduction in the tumorigenicity and the dopaminergic function of human ESC-derived neural cells in a primate model of Parkinson's disease. Stem Cells 30:935–945. doi: 10.1002/stem.1060. [DOI] [PubMed] [Google Scholar]
- 13.Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M, Perlingeiro RC. 2008. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med 14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 14.Endo K, Hayashi K, Saito H. 2016. High-resolution identification and separation of living cell types by multiple microRNA-responsive synthetic mRNAs. Sci Rep 6:21991. doi: 10.1038/srep21991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K. 2013. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Tomizawa M, Shinozaki F, Sugiyama T, Yamamoto S, Sueishi M, Yoshida T. 2013. Survival of primary human hepatocytes and death of induced pluripotent stem cells in media lacking glucose and arginine. PLoS One 8:e71897. doi: 10.1371/journal.pone.0071897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, Benvenisty N. 2013. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell 12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. 2008. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 19.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 20.Niwa H, Ogawa K, Shimosato D, Adachi K. 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 21.Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. 2009. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol 7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacArthur BD, Sevilla A, Lenz M, Muller FJ, Schuldt BM, Schuppert AA, Ridden SJ, Stumpf PS, Fidalgo M, Ma'ayan A, Wang J, Lemischka IR. 2012. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat Cell Biol 14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 24.Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, Ignatchenko V, McDonald AC, Garner J, Gramolini AO, Rossant J, Kislinger T. 2012. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell 22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 26.Eiraku M, Sasai Y. 2012. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr Opin Neurobiol 22:768–777. doi: 10.1016/j.conb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Seoane J, Le HV, Shen L, Anderson SA, Massague J. 2004. Integration of Smad and Forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 28.Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. 2013. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A. 2007. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, Hiller D, Jiang S, Protopopov A, Wong WH, Chin L, Ligon KL, DePinho RA. 2009. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopinath SD, Webb AE, Brunet A, Rando TA. 2014. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Rep 2:414–426. doi: 10.1016/j.stemcr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, Palmer TD, Butte AJ, Brunet A. 2009. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. 2010. Discovery of novel Forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol 78:961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, Mungamuri SK, Rimmele P, Kennedy M, Sellers R, Landthaler M, Tuschl T, Chi NW, Lemischka I, Keller G, Ghaffari S. 2011. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 13:1092–1099. doi: 10.1038/ncb2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Tian C, Zheng JC. 2013. FoxO3a contributes to the reprogramming process and the differentiation of induced pluripotent stem cells. Stem Cells Dev 22:2954–2963. doi: 10.1089/scd.2013.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutzner N, Kalbacher H, Krones-Herzig A, Rosl F. 2012. FOXO3 is a glucocorticoid receptor target and regulates LKB1 and its own expression based on cellular AMP levels via a positive autoregulatory loop. PLoS One 7:e42166. doi: 10.1371/journal.pone.0042166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 40.Lin K, Dorman JB, Rodan A, Kenyon C. 1997. daf-16: an HNF-3/Forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 41.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 42.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. 2003. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 44.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A 101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, Cantley LC. 2009. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell 33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chae HD, Lee MR, Broxmeyer HE. 2012. 5-Aminoimidazole-4-carboxyamide ribonucleoside induces G(1)/S arrest and Nanog downregulation via p53 and enhances erythroid differentiation. Stem Cells 30:140–149. doi: 10.1002/stem.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi X, Wu Y, Ai Z, Liu X, Yang L, Du J, Shao J, Guo Z, Zhang Y. 2013. AICAR sustains J1 mouse embryonic stem cell self-renewal and pluripotency by regulating transcription factor and epigenetic modulator expression. Cell Physiol Biochem 32:459–475. doi: 10.1159/000354451. [DOI] [PubMed] [Google Scholar]
- 48.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast AM, Baumgartner D, Carnevalli LS, Atzberger A, Haas S, von Paleske L, Boroviak T, Worsdorfer P, Essers MA, Kloz U, Eisenman RN, Edenhofer F, Bertone P, Huber W, van der Hoeven F, Smith A, Trumpp A. 2016. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell 164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A. 2007. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol 27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, Chang M, Zheng H, Wang YA, Kwiatkowski DJ, Kaelin WG Jr, Signoretti S, DePinho RA. 2010. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kress TR, Cannell IG, Brenkman AB, Samans B, Gaestel M, Roepman P, Burgering BM, Bushell M, Rosenwald A, Eilers M. 2011. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol Cell 41:445–457. doi: 10.1016/j.molcel.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 52.Seoane J, Le HV, Massague J. 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 53.Giancotti FG. 2013. Mechanisms governing metastatic dormancy and reactivation. Cell 155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. 2010. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature 463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 55.Sunayama J, Sato A, Matsuda K, Tachibana K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Sakurada K, Kayama T, Tomiyama A, Kitanaka C. 2011. FoxO3a functions as a key integrator of cellular signals that control glioblastoma stem-like cell differentiation and tumorigenicity. Stem Cells 29:1327–1337. doi: 10.1002/stem.696. [DOI] [PubMed] [Google Scholar]
- 56.Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J. 2002. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol 22:1526–1536. doi: 10.1128/MCB.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa K, Matsui H, Ohtsuka S, Niwa H. 2004. A novel mechanism for regulating clonal propagation of mouse ES cells. Genes Cells 9:471–477. doi: 10.1111/j.1356-9597.2004.00736.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.