ABSTRACT
Salmonella is recognized as one of the most important foodborne bacteria and has wide health and socioeconomic impacts worldwide. Fresh pork meat is one of the main sources of Salmonella, and efficient and fast methods for detection are therefore necessary. Current methods for Salmonella detection in fresh meat usually include >16 h of culture enrichment, in a few cases <12 h, thus requiring at least two working shifts. Here, we report a rapid (<5 h) and high-throughput method for screening of Salmonella in samples from fresh pork meat, consisting of a 3-h enrichment in standard buffered peptone water and a real-time PCR-compatible sample preparation method based on filtration, centrifugation, and enzymatic digestion, followed by fast-cycling real-time PCR detection. The method was validated in an unpaired comparative study against the Nordic Committee on Food Analysis (NMKL) reference culture method 187. Pork meat samples (n = 140) were either artificially contaminated with Salmonella at 0, 1 to 10, or 10 to 100 CFU/25 g of meat or naturally contaminated. Cohen's kappa for the degree of agreement between the rapid method and the reference was 0.64, and the relative accuracy, sensitivity, and specificity for the rapid method were 81.4, 95.1, and 97.9%, respectively. The 50% limit of detections (LOD50s) were 8.8 CFU/25 g for the rapid method and 7.7 CFU/25 g for the reference method. Implementation of this method will enable faster release of Salmonella low-risk meat, providing savings for meat producers, and it will help contribute to improved food safety.
IMPORTANCE While the cost of analysis and hands-on time of the presented rapid method were comparable to those of reference culture methods, the fast product release by this method can provide the meat industry with a competitive advantage. Not only will the abattoirs save costs for work hours and cold storage, but consumers and retailers will also benefit from fresher meat with a longer shelf life. Furthermore, the presented sample preparation might be adjusted for application in the detection of other pathogenic bacteria in different sample types.
KEYWORDS: Salmonella, food safety, foodborne pathogens, fresh meat, rapid tests, real-time PCR, sample preparation, validation
INTRODUCTION
Salmonella is a global foodborne pathogen responsible for approximately 30% of foodborne outbreaks in the United States and 23% in the European Union (1, 2). Pork meat remains an important source of human salmonellosis, which is why the absence of Salmonella is confirmed at critical control points at abattoirs (2). However, the reference culture-based methods require several days to obtain a test result (3, 4), thereby postponing the market release of fresh meat.
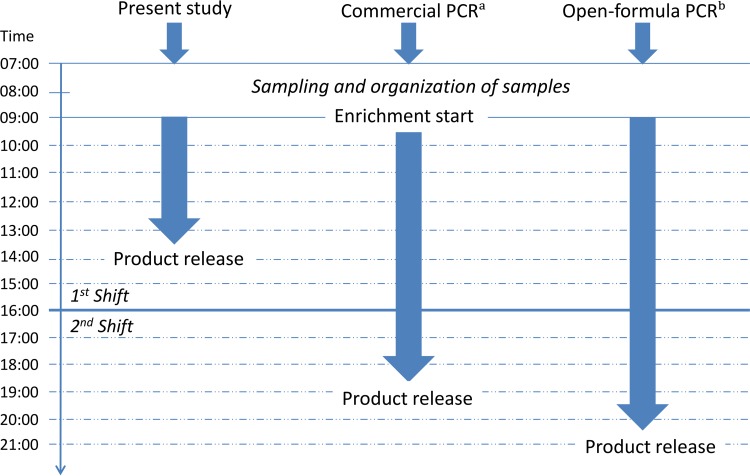
To ensure faster release, rapid methods intended for use at abattoirs must be sensitive, robust, noncomplex, low-cost, and, last but not least, amendable to high throughput (5, 6). Rather than there being a lack of fast detection technologies, lack of user-friendly and noncomplex preanalytical methods is the major bottleneck of rapid testing. In addition, the time to result for the fastest same-day method (7) requires a two-shift laboratory staff. Although rapid and sensitive detection technologies have been reported for the detection of Salmonella, these are mostly limited due to the complexity of the sample preparation. Some methods require multiple washing and centrifugation steps (8), while others are based on advanced filtration systems (9). To our knowledge, there is no publication of a same-shift, sensitive, and high-throughput method specific for the detection of Salmonella in fresh meat. We previously validated a same-day high-throughput PCR method developed for routine use at abattoirs (10). The method, however, requires 12 h of enrichment, corresponding to two working shifts. The requirement for two working shifts is also true for commercial methods, e.g., the fast raw meat protocol iQ-Check Salmonella II (Bio-Rad). With iQ-Check, as well as similar commercial kits (e.g., GeneDisc; Pall Corporation), the detection time for meat samples can be 9 to 10 h. The present study aimed at developing an even faster method, enabling abattoirs to release meat within one working shift, thus extending the shelf-life of fresh meat in the distribution chain and reducing cost of storage (Fig. 1).
FIG 1.
Timeline for the rapid method developed in the present study compared to some of the currently validated PCR-based detection methods. Commercial PCR (denoted by the superscript letter “a”), a typical commercial PCR, e.g., iQ-Check Salmonella II fast protocol for raw meats (Bio-Rad). The open-formula protocol (denoted by the superscript letter “b”) is according to Löfström et al. (10). The timeline is based on two laboratory technicians analyzing 50 samples. The analysis time for a single sample with the presented method is less than 5 h.
In industrialized countries, meat samples, if contaminated, often harbor very low numbers of Salmonella (11, 12); therefore, an enrichment step is an inevitable part of molecular detection methods. Ideally, this enrichment should maximize the number of Salmonella organisms while limiting the background flora. In addition, as Salmonella in meat is often injured from the chilling process, the international standards recommend a two-step enrichment procedure, where the first nonselective preenrichment step is followed by a selective enrichment step. Several attempts to reduce the enrichment time have been reported (13–16). Studies in which shortening of the 16- to 20-h reference enrichment time was investigated reported that at least 8 h of incubation was required for meat samples (15, 16), while a 3-h incubation resulted in the detection of Salmonella in highly contaminated oysters (17). Omitting the time-consuming enrichment step would therefore be optimal, and several direct detection methods have also been published (9, 18–20). However, most of these methods are complex, not high throughput, and have detection limits of >50 CFU/25 g, i.e., above the reported levels of Salmonella in meat (12). Moreover, the legislative demand of absence of 1 CFU/25 g (21) cannot currently be met by the available direct detection technologies, making some sort of short-culture enrichment step necessary.
PCR is a rapid, sensitive, and very specific detection technology. However, when applying PCR-based detection, the whole chain of analysis must be optimized for this, including sampling, sample preparation, and composition of the PCR master mix (22). At abattoirs and meat-cutting plants, relevant sample matrices include carcass swabs, meat cuts, and meat juice (2). These samples are challenging for rapid PCR-based detection, as certain PCR inhibitors are expected and have to be removed prior to detection, e.g., heme from blood (23), myoglobin from tissue (24), and fat (25). PCR inhibition can be relieved by using a DNA polymerase more tolerant to inhibitors (26) or by adding PCR facilitators to the PCR master mix (27), but often a combination with a sample preparation step is needed in order to generate a PCR-compatible sample. Studies have shown that PCR inhibitors can be reduced by physical, biochemical, immunological, and physiological sample preparation methods. These include flotation (18), filtration (9, 20), adsorption and addition of surfactants (19), enzymatic digestion (9), DNA extraction (28), and enrichment in a PCR-compatible medium (15), all of which were considered when designing the described rapid method.
The present study describes the development and comparative validation of the first PCR-based method for detection of Salmonella in pork meat in less than 5 h, with comparable sensitivity to current reference culture methods.
RESULTS
Method development. (i) Preenrichment conditions.
Changing various parameters for the enrichment step showed that preheating of the buffered peptone water (BPW) to 45°C and incubation at 41.5°C led to a smaller initial temperature decrease (45°C ± 1.0°C to 23.0 ± 0.9°C versus 37°C ± 1.0°C to 21.2 ± 1.0°C, respectively) when adding it to the 4°C cold meat (Table 1). Furthermore, a higher BPW temperature (38.6°C versus 32.3°C) was obtained after the 3-h incubation step for the largest sample size tested (10 samples at 25 cm2 [i.e., 10 by 25 cm2]). This resulted in a significant increase in Salmonella growth of 1.7 log CFU/ml (95% confidence interval [CI], 1.5 to 1.9 CFU/ml) 0.7 log CFU/ml (95% CI, 0.6 to 0.8 CFU/ml) for BPW preheating/incubation temperatures of 45°C/41.5°C and 37°C/37°C, respectively. As both the initial temperature decrease and the final BPW temperature were optimal with smaller sample size in larger volumes of BPW, the final sample size for the rapid method was set to 25 g and incubation conditions to 3 h at 41.5°C in 60 ml of BPW preheated to 45°C.
TABLE 1.
Results from optimization studies showing the effect of sample size, volume, and temperature of BPW and temperature of incubation on growth of Salmonella
| Sample sizeb | Vol of BPW (ml) | Temp of BPW (°C) | Temp of incubator (°C) | Temp (°C) ata: |
Salmonella growth (log CFU/ml) | |||
|---|---|---|---|---|---|---|---|---|
| 0 min | 60 min | 120 min | 180 min | |||||
| 10 by 25 cm2 | 100 | 45 | 41.5 | 23.0 ± 0.9 | 29.9 ± 0.7 | 35.7 ± 0.3 | 38.8 ± 0.2 | 1.7 ± 0.2 |
| 10 by 25 cm2 | 100 | 37 | 37 | 21.2 ± 1.0 | 27.3 ± 3.0 | 30.8 ± 2.0 | 32.6 ± 0.5 | 0.7 ± 0.1 |
| 5 by 25 cm2 | 100 | 45 | 41.5 | 26.3 | 35.2 | 38.7 | 40.1 | 2.0 ± 0.2 |
| 1 by 25 cm2 | 100 | 45 | 41.5 | 34.5 | 37.7 | 39.4 | 40.2 | 2.2 ± 0.1 |
| 1 by 25 g | 30 | 37 | 37 | 22.0 ± 0.0 | 34.0 ± 0.9 | 1.2 ± 0.2 | ||
Data are shown as the means and 95% confidence intervals (CIs); if no CI is shown, data were based on a single experiment.
Sample size is presented as the number of samples by the mass. A 25-cm2 sample weighed between approximately 11 and 17 g.
(ii) Filter pore size.
Filter bags with four different pore sizes were tested for their efficiency to filter the sample liquid (Table 2). The smallest pore size (63 μm) was found to retain too much liquid, as fat and proteins from the meat clogged the pores, leading to a low recovery of the added liquid. The largest pore size tested (500 μm) allowed too much meat debris to pass through the filter. A comparison of the four filters showed that they did not yield significantly different numbers of Salmonella per milliliter of filtrate, but the 63-μm-pore size resulted in a significantly lower number of total Salmonella organisms, as the extract volume was considerably lower. In a comparison of the real-time PCR results from the samples filtered through 280-μm- or <250-μm-pore-size filters, no statistically significant difference was found, although a pore size of 280 μm generally resulted in lower threshold cycle (CT) values and thus potentially a better detection limit than the <250-μm-pore filter (data not shown). The pore size of 280 μm was selected for the rapid method.
TABLE 2.
Effects of pore size of filter bag on the recovered number of Salmonella organisms and the volume obtained after filtration
| Pore size of filter bag (μm) | Recovery after filtrationa |
||
|---|---|---|---|
| Salmonella/ml (log CFU/ml) | Vol (ml)b | Total no. of Salmonella (log CFU) | |
| 63 | 3.0 ± 0.1 | 35 ± 7 | 4.6 ± 0.1 |
| <250 | 3.1 ± 0.1 | 65 ± 7 | 4.9 ± 0.1 |
| 280 | 3.0 ± 0.0 | 80 ± 7 | 4.9 ± 0.0 |
| 500 | 2.9 ± 0.1 | 95 ± 7 | 4.9 ± 0.1 |
Data are shown are the means and 95% confidence intervals.
The added volume was 100 ml.
(iii) Selective removal of meat matrix.
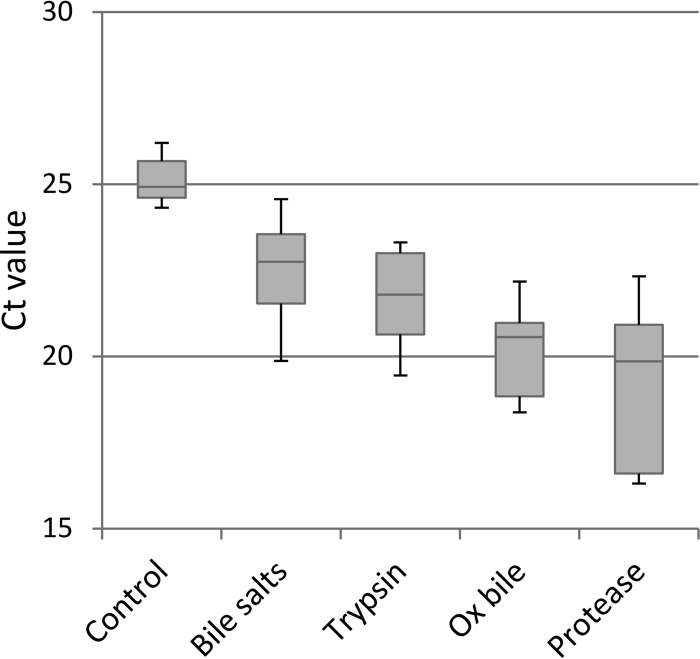
To reduce the PCR inhibitory effect of the meat, different enzymes and bile products were tested for their ability to degrade and remove the meat proteins and fat. The goal was to obtain a pellet that could subsequently be used in downstream analysis with only a simple DNA extraction by boiling. The MolYsis kit was excluded from these tests based on the results from an initial screening (data not shown), where treatment with the MolYsis kit followed by lysis by boiling resulted in higher CT values (i.e., poorer detection limit). Furthermore, a lower number of positive samples was found using the MolYsis kit than with lysis by boiling. As shown in Fig. 2, treatment with bile salts, trypsin, ox bile, and the alkaline protease prior to DNA extraction all resulted in decreased CT values compared to those with the control. Treatment with bile salts did not generate CT values significantly different from the control, whereas treatment with trypsin, ox bile, and protease did. Among the tested reagents, the protease treatment was found to generate the lowest CT values.
FIG 2.
Boxplots illustrating the difference in CT values obtained after treatment of the preenriched meat filtrate with various reagents to reduce the PCR inhibitory effect of meat proteins and fat.
(iv) Manual handling.
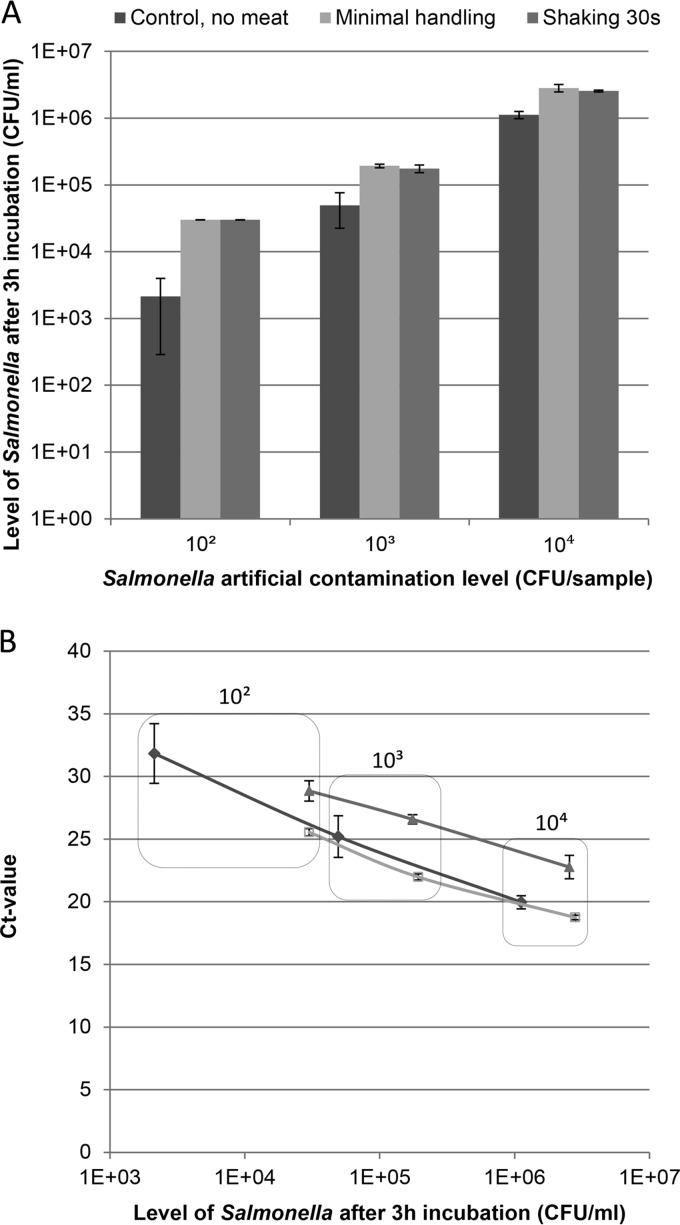
To reduce the time and manual labor for sample treatment and to improve PCR performance, different procedures for handling the meat samples before enrichment were investigated. The results showed that the recovery of Salmonella was not significantly affected by the level of handling (Fig. 3A). However, a significant improvement in CT values was observed when reducing the level of handling from 30 s of shaking to minimal handling that includes gently separating the meat pieces by hand prior to enrichment (Fig. 3B).
FIG 3.
(A) Salmonella levels obtained after 3 h of incubation with artificial contamination levels of 102, 103, and 104 CFU/sample. (B) PCR standard curves based on CT values and plate counts after 3 h of incubation for artificially contaminated meat samples shaken for 30 s (▲), artificially contaminated filter bags without meat (◆), and artificially contaminated meat samples where meat pieces were gently separated (minimal handling) (□).
(v) Comparative validation study.
A comparative study was performed on artificially and naturally contaminated meat samples representing different types of pork cuts (Table 3). The relative specificity of the rapid method was 100%, as neither method generated false-positive results from Salmonella-negative samples. In an analysis of artificially contaminated meat with a low level of Salmonella (1 to 10 CFU), one sample of tenderloin was found to be false positive for Salmonella, according to the NordVal definition, which affected the specificity for this sample type. However, subsequent culture confirmed the result of the rapid method. The relative sensitivity, on the other hand, was affected by samples that were false negative according to the NordVal definition, but culturing confirmed these samples to be true negative. For artificially contaminated meat with high levels of Salmonella (i.e., 10 to 100 CFU), the reference method and the rapid method both detected Salmonella in all samples.
TABLE 3.
Results from the comparative validation study of the developed rapid method against the NMKL 187 reference culture method on artificially and naturally contaminated meat samples
| Sample type | No. of samplesa |
% valueb |
κ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | PA | NA | FNc | TP | FP | AC | SE | SP | ||
| Artificially contaminated | ||||||||||
| Loin cut with rind | 25 | 11 | 10 | 3 | 1 | 0 | 84.0 | 85.7 | 100.0 | 0.68 |
| Tenderloin | 25 | 13 | 10 | 1 | 0 | 1 | 96.0 | 92.9 | 90.9 | 0.92 |
| Shoulder cut | 25 | 14 | 10 | 1 | 0 | 0 | 96.0 | 93.3 | 100.0 | 0.92 |
| Total | 75 | 38 | 30 | 5 | 1 | 1 | 92.0 | 90.7 | 96.8 | 0.84 |
| Naturally contaminated | ||||||||||
| Tongue (paired homogenization) | 50 | 14 | 16 | 10 | 10 | 0 | 60.0 | 100.0 | 100.0 | 0.20 |
| Tongue (pooled homogenization) | 15 | 15 | 0 | 0 | 0 | 0 | 100.0 | 100.0 | 100.0 | 1.00 |
| Total | 65 | 29 | 16 | 10 | 10 | 0 | 69.2 | 100.0 | 100.0 | 0.41 |
| Overall total | 140 | 67 | 46 | 15 | 11 | 1 | 81.4 | 95.1 | 97.9 | 0.64 |
N, total no. of samples per sample type, calculated as PA + NA + FN + TP + FP. PA, positive agreement; NA, negative agreement; FN, false negative (positive result by NMKL 187, negative by rapid method); TP, true positive (confirmed positive result by rapid method, negative by NMKL 187); FP, false positive (negative result by NMKL 187 and positive by rapid method, but not confirmed).
AC, relative accuracy; SE, sensitivity; SP, specificity.
False negative according to NordVal definition, but confirmation showed that with the exception of one tongue sample, all of these were true negatives.
For the naturally contaminated meat samples with paired homogenization, the reference method and the rapid method gave the same number of positive samples but not always on the same samples (data not shown). The confirmation of the negative samples analyzed with the rapid method indicated that this was due to uneven distribution of Salmonella; only one discrepancy between the rapid method and confirmation was observed. In another test round, where the naturally contaminated meat samples were pooled for homogenization and then divided into paired samples, all tested samples were positive for Salmonella with both methods. The LOD50s for the rapid method and the NMKL 187 reference method were found to be 8.8 and 7.7 CFU/sample, respectively.
DISCUSSION
In this study, a rapid method for the detection of Salmonella in pork meat in less than 5 h was developed and validated. To shorten the time of analysis, culture enrichment was initially avoided, and instead, physical and biochemical processes were examined to concentrate Salmonella in the sample and remove the meat matrix. Direct detection without culture enrichment has previously been reported using flotation (18, 29) or filtration (9, 20). However, these methods are either too laborious to be applicable in a routine laboratory or have too high of a detection limit to meet the legislative demands of 1 CFU/25 g of meat. In the present study, it was found that even with the filtration and removal of meat proteins by enzymatic digestion, the targeted number of Salmonella cells was too low to detect without culture enrichment (data not shown). An enrichment step, although short, was therefore needed.
An early study by Bej et al. indicated that 3-h nonselective enrichment in preheated BPW resulted in robust detection of Salmonella in oysters, although in that study, the detection limit was 1 to 10 CFU/g (17), whereas the present study could detect 1 to 10 CFU in 25-g samples. To our knowledge, this is the first publication on the use of elevated incubation temperatures in combination with a shortened enrichment time for the detection of Salmonella in meat. Other studies using short enrichments have found the lowest incubation time for meat samples intended for detection with PCR to be a minimum of 8 h (15, 16), but none of these avoided the initial temperature decrease of the enrichment broth, which was found to have a significant effect on the growth of Salmonella in the present study.
Inclusion of a culture enrichment step allowed for confirmation of not only the presence, but also the viability, of Salmonella. The enrichment step also meant that less manual handling of the meat was required, which was found to have a significantly positive effect on the PCR, with lower and less varied CT values. The minimal handling of samples was introduced after observing that 30 s of shaking released PCR inhibitory meat components into the BPW. These findings are in agreement with those of a previous study (30) reporting that untreated and hand-massaged samples led to higher PCR detection frequencies than those of samples that had been stomached. Minimal handling was also favorable with regard to implementation of the rapid method in a high-throughput routine laboratory.
The use of enzymes and bile products for selective removal of interacting sample material has previously been reported (28, 31). Although those studies were based on samples of whole blood, the challenges are similar to those of the present study, i.e., high levels of PCR inhibitors and very low concentration of the targeted pathogen. It was reported that the MolYsis basic kit combined with selective DNA extraction could improve the detection limit for Staphylococcus aureus in blood samples (28). However, the combination of the MolYsis kit and boiling lysis applied in the present study resulted in poorer CT values and a lower number of detected samples than those with lysis by boiling alone (data not shown). As the samples used for testing the efficiency of the MolYsis kit were artificially contaminated with 103 CFU/25 g of sample, the poor results obtained could reflect a major loss of bacterial cells during treatment or reduced the usefulness of the kit for Gram-negative bacteria, as suggested by Horz et al. (32). It was shown in the present study that ox bile, but not bile salts, could be an alternative to enzymatic digestion. In a study by Zhou and Pollard (31), ox bile treatment in combination with a micrococcal nuclease was applied in selective removal of human DNA in blood samples infected with Salmonella enterica serovar Typhi, and DNA subsequently was extracted by the QIAamp DNA minikit, resulting in improved PCR sensitivity. The present study also tested the use of micrococcal nuclease in combination with enzymes or bile products, but it resulted in poorer CT values or no amplification (data not shown). This difference in results could be caused by the different DNA extraction methods applied. The simple boiling lysis applied in the present study would not have removed leftover bile, resulting in PCR inhibition. The results from the present study showed that the protease could replace more expensive reagents for removal of the meat matrix and reduction of PCR inhibition. To our knowledge, the tested protease has not previously been used for this purpose, although other proteases have been used similarly to degrade the meat matrix (9, 33). An earlier generation of the protease was used to degrade gelatin filters used in air sampling (34), prior to PCR analysis, and the enzyme could also prove useful in PCR-compatible sample preparation for other protein-based matrices.
Challenges due to uneven distribution of Salmonella in the samples were encountered when testing naturally contaminated meat. Before dividing the samples for testing with the rapid or reference method, the meat was cut in smaller pieces and mixed. However, this homogenization was insufficient to produce two equally contaminated samples, affecting the agreement between the methods. This is in accordance with the findings of Jensen et al. (35), where differences in the results from two methods used to analyze naturally contaminated samples were related to very low and unevenly distributed Salmonella contamination, even after thorough homogenization. In the study by Josefsen et al. (15), this problem was solved by using the same sample enrichment for both the PCR test and the NMKL culture reference method. The same approach was not possible in the present study, due to differences in enrichment conditions. This issue is foreseen in the revised ISO 16140-2:2016 protocol, where unpaired validation is allowed (36). In the present study, this issue was addressed by confirming all negative results with the rapid method by culture, according to the reference method. By this approach, all but one sample was found to be true negative. For artificially contaminated samples, the agreement between the rapid method and the reference method was generally higher than that observed for naturally contaminated samples. Due to the well-known difficulties with artificial contamination at low levels (37), subsequent confirmation by culture was needed, which showed that the negative samples were true negatives. However, the NordVal definitions were applied, which declared the samples to be false negatives, thus influencing the relative accuracy, relative sensitivity, relative specificity, and Cohen's kappa for the artificially contaminated samples, which would otherwise have been 98.7%, 102.6%, 97.2%, and 0.97, respectively.
The rapid method developed and validated in this study was designed for single 25-g meat samples. Studies are in progress to evaluate the suitability of the method for five or even 10 composite samples. Sample compositing is a way to improve throughput, although the sensitivity can be impaired due to target dilution (38). Furthermore, efforts are being put into optimizing the various handling steps, rendering the method even more applicable for a high sample throughput. The sample matrix in this study was different cuts of raw pork meat, but the method might also be adapted to other sample types. As with the reference culture method, which has different preenrichment specifications for certain foods (3), a similar approach might be suitable for this rapid method. All included sample preparation steps (enrichment media, enzyme treatment, and DNA extraction) are nonselective for Salmonella. Therefore, the protocol might be adjusted for application to other pathogenic bacteria, or even coupled with metagenomics.
In conclusion, the rapid method developed in this study showed good agreement with the reference culture method NMKL 187, with comparable LODs. Studies are planned for international validation according to the revised ISO 16140-2:2016 protocol (36). The presented rapid method may have the potential for detection of other pathogens and other sample types and have a relatively high throughput compared to other reported rapid methods for the detection of Salmonella in meat. The presented method enables abattoirs to achieve faster market release of fresh meat with longer shelf-life and less cold storage.
MATERIALS AND METHODS
Sample material.
Salmonella-negative pork meat (loin cut with rind, tenderloin, and shoulder cut) was obtained from local supermarkets, while meat from pig carcasses naturally contaminated with Salmonella was provided by Danish meat producers.
Bacterial strains and preparation of inoculum.
Salmonella strains (S. enterica serovar Typhimurium DT 193 [DMRI 4984 PX], S. enterica serovar Dublin [DMRI 4983 PX], and S. enterica serovar Derby [DMRI 4985 PX]) were obtained from the Danish Technological Institute, DMRI (Høje Taastrup, Denmark) and stored in Protect multipurpose microorganism preservation system (Technical Service Consultants Ltd., Lancashire, UK) containing 20% glycerol as a cryo-protectant. Strains were revived on tryptone soy agar with sheep blood (TSASB; Oxoid Microbiology Products, Thermo Fisher Scientific) and isolated on xylose lysine deoxycholate (XLD) agar (Oxoid). The inoculation culture was prepared by transferring 3 to 4 colonies of each of the Salmonella strains into a separate tube containing 4 ml of nutrient broth (8.5 g of sodium chloride [catalog no. A1371,9025; AppliChem], 20 g of nutrient broth [catalog no. 234000; Becton Dickinson], and 1 liter of demineralized water [pH 6.6 to 7.0]) and then incubated at 37.0 ± 1.0°C for 1 to 2 h without shaking. The final inoculum used for artificial contamination was prepared by mixing the three cultures using equal volumes of each strain. This final inoculum was stored at 2 to 5°C, while enumeration was performed by plating 10-fold dilution series in sterile saline (0.9% NaCl) on TSASB plates (Oxoid), with subsequent incubation at 37.0 ± 1.0°C for 18 ± 2 h.
Artificial contamination.
To confirm that meat used for artificial contamination experiments was not naturally contaminated with Salmonella, a 25-g portion was analyzed using a validated real-time PCR method (10). Pork meat samples were cut using sterile scalpels, transferred to sterile filter bags (BagPage model +; Interscience), and inoculated with droplets (approximately 10 μl) of diluted final inoculum directly on the meat. The inoculated meat was stored overnight at 2 to 5°C to simulate the cold stressed conditions encountered during cooling and cold storage in the pork production line.
Method development.
In order to develop the most rapid method, several steps, where optimization was feasible, were identified, including enrichment, sample preparation, and real-time PCR conditions. Experiments for optimization of the method were performed on artificially contaminated pork meat (loin cut with rind, tenderloin, and shoulder cut). Unless otherwise stated, three biological replicates and two PCR replicates per biological replicate were used per variable.
Enrichment conditions.
In the initial experiments, 3 and 4 h of enrichment were evaluated. This enrichment was optimized by changing the temperature of the enrichment medium and incubator. The results were evaluated by determining the differences in lag phase and generation time of Salmonella. Combinations of different meat sample sizes (1 by 25 cm2 to 10 by 25 cm2, corresponding to 11 to 168 g) and 30 to 100 ml of buffered peptone water (BPW; Oxoid) were incubated at 37.0 to 41.5 ± 1.0°C with the BPW preheated to 37.0 or 45.0 ± 1.0°C. The temperature of the enrichment medium was monitored using an Almemo 2290-8 Thermologger (Erich Blichfeld A/S, Kolding, Denmark), with the probes inserted into two noninoculated process controls.
Sample preparation. (i) Filter pore size.
Stomacher bags with internal filter walls (filter bags) were used to separate the inoculated media from the larger meat pieces. Smaller pore sizes were expected to retain more fat and meat debris while allowing the bacteria to be collected in the filtrate, thus improving the detection. The filter bags evaluated for the enrichment were Seward (pore size, 500 μm; Seward Ltd., Worthing, UK), BagPage model + (pore size, 280 μm; Interscience, Saint Nom, France), BagPage model R (pore size, <250 μm; Interscience), and BagPage model F (pore size, 63 μm; Interscience). The effects of pore size on the recovery of Salmonella in saline (0.9%) were evaluated using 5 by 25-cm2-thin slices (thickness, approximately 5 mm) of artificially contaminated meat.
(ii) Selective removal of meat matrix.
For selective removal of meat components, the following reagents were tested: protease (FoodPro alkaline protease, 0.6 U/μg; Danisco), trypsin (from porcine pancreas, approximately 1.5 U/μg; Sigma), bile salt (50% sodium cholate, 50% sodium deoxycholate; Sigma), ox bile (bile bovine; Fluka), and the MolYsis Basic 5 bacterial DNA enrichment kit (Molzym, Bremen, Germany). The enzymes (protease and trypsin) were tested as described in “Removal of meat components” below. For ox bile and bile salt, the 1-ml suspensions of pellet (obtained from the centrifuged enrichment filtrate resuspended in phosphate-buffered saline [PBS], autoclaved, containing 0.31 g of potassium di-hydrogen phosphate [catalog no. 1.04873; Merck], 2.77 g of di-sodium hydrogen phosphate-12-hydrate [catalog no. 0304; J. T. Baker], 8.15 g of sodium chloride [catalog no. A1371,9025; AppliChem], and 1 liter of Milli-Q water) were divided equally into two Eppendorf tubes, and to the contents of each of these tubes was added 500 μl of ox bile (0.5 g suspended in 5 ml of sterile water) or bile salt (86 mg suspended in 5 ml of sterile water) solution, which was then thoroughly mixed and incubated at 37.0 ± 1.0°C for 10 min. The tubes were centrifuged at 3,000 × g and the supernatant discarded before adding 500 μl of PBS to each tube. The content of the two tubes was collected in one tube and centrifuged at 3,000 × g, discarding the supernatant before adding 50 μl of 1 mM Tris-HCl with 10 mM EDTA (1× Tris-EDTA [TE] buffer [pH 8.0]). The bacterial cells were lysed by boiling as described in “DNA extraction,” below. The MolYsis Basic 5 kit was used on the 1-ml pellet suspension transferred to a 2-ml sterile polypropylene tube. A volume of 250 μl of buffer CM was added and mixed thoroughly by vortexing before incubation at 21°C ± 4°C for 5 min. Subsequently, 250 μl of DB1 buffer and 10 μl of MolDNase B were added, and the samples were incubated at 21°C ± 4°C for 15 min. Samples were then centrifuged at 3,000 × g and the supernatant carefully removed by pipetting before 1 ml of RS buffer was added and the samples were vortexed again. Samples were centrifuged at 3,000 × g, and the supernatant was removed. A volume of 50 μl of 1× TE buffer was added, and the DNA was extracted by boiling lysis, as described in “DNA extraction” below.
(iii) Manual handling.
In initial experiments, the meat and enrichment media were manually homogenized by shaking the filter bags for 30 s before enrichment. However, to avoid too much handling of samples and the resulting release of meat components, a minimal handling procedure was tested. In the minimal handling procedure, the meat pieces were separated by gently massaging the outside of the filter bag, after the addition of BPW, and incubating at 37.0°C ± 1.0°C for 3 h without further handling. Following the incubation, enumeration was performed by plating 10-fold dilution series in sterile saline (0.9% NaCl) on TSASB plates (Oxoid), with subsequent incubation at 37.0 ± 1.0°C for 18 ± 2 h and PCR results generated using the “Rapid method protocol” described below.
Real-time PCR conditions.
The real-time PCR for the rapid method was based on a previously validated Salmonella PCR assay (7), using a fast cycling thermal profile as described below.
Rapid method protocol. (i) Enrichment.
BPW (Oxoid) with 0.5% Tween 20 (molecular grade; catalog no. P94 16; Sigma) was preheated to 45°C. A 60-ml volume of this preheated enrichment medium was added to meat samples consisting of 25 ± 1 g of artificially inoculated or naturally contaminated meat. The meat pieces were separated by gently massaging from the outside of the bag to ensure that the entire surface of the meat was exposed to the enrichment medium. The samples were incubated at 41.5°C ± 1°C for 3 h ± 5 min.
(ii) Removal of meat components.
Following the enrichment step, 50 ml of sample liquid was collected by pouring from the filtered side of the bag, centrifuging for 5 min at 3,000 × g, and discarding the supernatant. The pellet was resuspended in 1 ml of PBS and transferred to a 1.5-ml Eppendorf tube, and 100 μl of protease (FoodPro alkaline protease, 0.6 U/μg; Danisco) was added. This suspension was mixed by vortexing for 20 s and incubating at 37.0°C ± 1.0°C for 5 min in a heating block. This vortexing and 5-min incubation were performed twice. The suspension was centrifuged for 5 min at 3,000 × g, and the supernatant was discarded by pipetting. A volume of 50 μl of TE buffer (1×) was added to the pellet, followed by mixing by vortexing until the pellet was fully dissolved.
(iii) DNA extraction.
The pretreated samples were lysed by boiling at 98°C ± 2°C for 15 min in a heating block. The samples were then centrifuged for 1 min at 3,000 × g.
Real-time PCR analysis.
For the specific detection of Salmonella in meat samples, a TaqMan real-time PCR method targeting a region within the ttrRSBCA locus (39) was performed on a StepOnePlus (Life Technologies) using the fast cycling option (20 s initial denaturation at 95°C and 40 cycles of 95°C for 1 s and 60°C for 20 s). Each reaction mixture contained 9 μl of template DNA and 16 μl of PCR master mix, prepared as previously described (15). Fluorescence measurements (i.e., 6-carboxyfluorescein [FAM] for the Salmonella target probe and VIC for the internal amplification control [IAC] probe) were obtained online and analyzed with the StepOne software (version 2.0). The baseline was set manually for each experiment starting around cycle 5 to 6 and ending two cycles before the lowest threshold cycle (CT) value was obtained. Based on an evaluation of multiple experiments, the threshold was assigned to 1,000 ΔRn (normalized fluorescence signal) for the Salmonella target and 300 ΔRn for the IAC. All amplification curves crossing the threshold were considered positive.
In every PCR analysis, an IAC was included in all samples to detect PCR inhibition. Furthermore, a positive DNA control (Salmonella Typhimurium in a concentration of approximately 0.005 ng), a negative DNA control (Escherichia coli in a concentration of approximately 5 ng), as well as a nontemplate control (NTC; containing only the PCR mix and 9 μl of PCR-grade water) were included. Duplicate PCR analysis was performed for each biological replicate. A CT value was considered an outlier if it differed from the other replicates representing the same sample by more than 3 cycles.
Comparative study.
To evaluate the performance of the developed rapid method, it was compared to the reference culture method NMKL 187 (3), which is comparable to the ISO 6579/A1:2007 method (40), on artificially and naturally contaminated meat samples, according to the NordVal (Nordic method validation body) guidelines for validation of alternative methods (41), as previously described (7). For the artificially contaminated samples, three contamination levels were tested (0 CFU/sample [n = 30], 1 to 10 CFU/sample [n = 30], and 10 to 100 CFU/sample [n = 15]). All samples artificially contaminated with 1 to 10 CFU were confirmed by subsequent culture (see below) to register possible false-negative results. This was likewise done for samples giving a negative result by the rapid method. The naturally contaminated samples were cut into smaller pieces and mixed thoroughly to obtain a more even distribution of Salmonella, before the samples were split into parallel subsamples.
Culture confirmation.
For samples inoculated at a low level and samples with negative results from the rapid method, results were confirmed by culture in accordance with the NMKL 187 method (3). A volume of 225 ml of BPW was added to the filter bags containing the meat sample and the remaining 10 ml of BPW from the enrichment step of the rapid method. These mixtures were incubated further at 37.0 ± 1.0°C for 18 ± 1 h, before 100 μl was distributed in three droplets on a modified semisolid Rappaport-Vassiliadis (MSRV) (3) agar plate. Further verification was performed according to NMKL 187 and by real-time PCR, as previously described (10).
Data analysis.
If no overlap was found between confidence intervals (CI) (α = 0.05), the differences between, e.g., CT values, were considered to be statistically significant. Boxplots were constructed by plotting the area between the 25% and the 75% quantiles, with a line marking the median, and error bars showing minimum and maximum values.
Validation parameters.
The 50% limit of detection (LOD50), relative accuracy (AC), relative sensitivity (SE), relative specificity (SP), and Cohen's kappa were calculated and interpreted according to the NordVal guidelines (41).
ACKNOWLEDGMENTS
This publication is related to the International patent application number PCT/EP2015/077234.
We thank Lizzie Larsen, Helen Ludvigsen, Stine Skotte Bjerregaard, and Camilla Mejnertsen for providing excellent technical assistance.
This work was financially supported by the Ministry of Environment and Food of Denmark, the GUDP project ULTRASAL (grant 3405-11-0349), and the Danish Pig Levy Fund. The study was not sponsored by any enzyme or kit producer, and neither were they involved in the experimental design or data treatment.
REFERENCES
- 1.Gould LH, Mungai EA, Johnson SD, Richardson LTC, Williams IT, Griffin PM, Cole DJ, Hall AJ. 2013. Surveillance for foodborne disease outbreaks—United States, 2009–2010. Morb Mortal Wkly Rep 62:41–47. [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA, ECDC. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J 13:3991. doi: 10.2903/j.efsa.2015.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NMKL. 2007. NMKL no. 187. Salmonella. Detection in foods, animal faeces and environmental materials from primary animal production using MSRV. Nordic Committee on Food Analysis, Søborg, Denmark. [Google Scholar]
- 4.International Organisation for Standardization. 2002. ISO 6579:2002. Microbiology of food and animal feeding stuffs–horizontal method for the detection of Salmonella spp. International Organisation for Standardization, Geneva, Switzerland. [Google Scholar]
- 5.Eijkelkamp JM, Aarts HJM, van der Fels-Klerx HJ. 2008. Suitability of rapid detection methods for Salmonella in poultry slaughterhouses. Food Anal Methods 2:1–13. [Google Scholar]
- 6.Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D. 2011. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol 28:848–861. doi: 10.1016/j.fm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Löfström C, Krause M, Josefsen MH, Hansen F, Hoorfar J. 2009. Validation of a same-day real-time PCR method for screening of meat and carcass swabs for Salmonella. BMC Microbiol 9:85. doi: 10.1186/1471-2180-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushima H, Katsube K, Hata Y, Kishi R, Fujiwara S. 2007. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl Environ Microbiol 73:92–100. doi: 10.1128/AEM.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Ximenes E, Amalaradjou MAR, Vibbert HB, Foster K, Jones J, Liu X, Bhunia AK, Ladisch MR. 2013. Rapid sample processing for detection of food-borne pathogens via cross-flow microfiltration. Appl Environ Microbiol 79:7048–7054. doi: 10.1128/AEM.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Löfström C, Hansen F, Mansdal S, Hoorfar J. 2012. Detection of Salmonella in meat: comparative and interlaboratory validation of a noncomplex and cost-effective pre-PCR protocol. J AOAC Int 95:100–104. doi: 10.5740/jaoacint.11-093. [DOI] [PubMed] [Google Scholar]
- 11.Malorny B, Löfström C, Wagner M, Krämer N, Hoorfar J. 2008. Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl Environ Microbiol 74:1299–1304. doi: 10.1128/AEM.02489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hoek AHAM, de Jonge R, van Overbeek WM, Bouw E, Pielaat A, Smid JH, Malorny B, Junker E, Löfström C, Pedersen K, Aarts HJM, Heres L. 2012. A quantitative approach towards a better understanding of the dynamics of Salmonella spp. in a pork slaughter-line. Int J Food Microbiol 153:45–52. doi: 10.1016/j.ijfoodmicro.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Hoorfar J, Mortensen AV. 2000. Improved culture methods for isolation of Salmonella organisms from swine feces. Am J Vet Res 61:1426–1429. doi: 10.2460/ajvr.2000.61.1426. [DOI] [PubMed] [Google Scholar]
- 14.Sharma VK, Carlson SA. 2000. Simultaneous detection of Salmonella strains and Escherichia coli O157:H7 with fluorogenic PCR and single-enrichment broth culture. Appl Environ Microbiol 66:5472–5476. doi: 10.1128/AEM.66.12.5472-5476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josefsen MH, Krause M, Hansen F, Hoorfar J. 2007. Optimization of a 12-hour TaqMan PCR-based method for detection of Salmonella bacteria in meat. Appl Environ Microbiol 73:3040–3048. doi: 10.1128/AEM.02823-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krämer N, Löfström C, Vigre H, Hoorfar J, Bunge C, Malorny B. 2011. A novel strategy to obtain quantitative data for modelling: combined enrichment and real-time PCR for enumeration of salmonellae from pig carcasses. Int J Food Microbiol 145:S86–S95. doi: 10.1016/j.ijfoodmicro.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Bej AK, Mahbubani MH, Boyce MJ, Atlas RM. 1994. Detection of Salmonella spp. in oysters by PCR. Appl Environ Microbiol 60:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löfström C, Schelin J, Norling B, Vigre H, Hoorfar J, Rådström P. 2011. Culture-independent quantification of Salmonella enterica in carcass gauze swabs by flotation prior to real-time PCR. Int J Food Microbiol 145:S103–S109. doi: 10.1016/j.ijfoodmicro.2010.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Opet NJ, Levin RE. 2014. Use of β-cyclodextrin and activated carbon for quantification of Salmonella enterica ser. Enteritidis from ground beef by conventional PCR without enrichment. Food Microbiol 38:75–79. [DOI] [PubMed] [Google Scholar]
- 20.Wolffs PFG, Glencross K, Thibaudeau R, Griffiths MW. 2006. Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl Environ Microbiol 72:3896–3900. doi: 10.1128/AEM.02112-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Parliament. 2003. Regulation EC no. 2160/2003 of the European Parliament and of the Council of 17 November 2003 on the control of Salmonella and other specified food-borne zoonotic agents. European Union, Brussels, Belgium: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32003R2160. [Google Scholar]
- 22.Rådström P, Knutsson R, Wolffs P, Lövenklev M, Löfström C. 2004. Pre-PCR processing. Mol Biotechnol 26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 23.Al-Soud WA, Rådström P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol 39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bélec L, Authier J, Eliezer-Vanerot MC, Piedouillet C, Mohamed AS, Gherardi RK. 1998. Myoglobin as a polymerase chain reaction (PCR) inhibitor: a limitation for PCR from skeletal muscle tissue avoided by the use of Thermus thermophilus polymerase. Muscle Nerve 21:1064–1067. doi:. [DOI] [PubMed] [Google Scholar]
- 25.Rossen L, Norskov P, Hoimstrom K, Rasmussen OF. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol 17:37–45. doi: 10.1016/0168-1605(92)90017-W. [DOI] [PubMed] [Google Scholar]
- 26.Fachmann MSR, Josefsen MH, Hoorfar J, Nielsen MT, Löfström C. 2015. Cost-effective optimization of real-time PCR-based detection of Campylobacter and Salmonella with inhibitor tolerant DNA polymerases. J Appl Microbiol 119:1391–1402. doi: 10.1111/jam.12937. [DOI] [PubMed] [Google Scholar]
- 27.Al-Soud WA, Rådström P. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol 38:4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen WLJ, Bruggeman CA, Wolffs PFG. 2009. Evaluation of new preanalysis sample treatment tools and DNA isolation protocols to improve bacterial pathogen detection in whole blood. J Clin Microbiol 47:2629–2631. doi: 10.1128/JCM.00821-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolffs P, Knutsson R, Norling B, Radstrom P. 2004. Rapid quantification of Yersinia enterocolitica in pork samples by a novel sample preparation method, flotation, prior to real-time PCR. J Clin Microbiol 42:1042–1047. doi: 10.1128/JCM.42.3.1042-1047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanki M, Sakata J, Taguchi M, Kumeda Y, Ishibashi M, Kawai T, Kawatsu K, Yamasaki W, Inoue K, Miyahara M. 2009. Effect of sample preparation and bacterial concentration on Salmonella enterica detection in poultry meat using culture methods and PCR assaying of preenrichment broths. Food Microbiol 26:1–3. doi: 10.1016/j.fm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Pollard AJ. 2012. A novel method of selective removal of human DNA improves PCR sensitivity for detection of Salmonella Typhi in blood samples. BMC Infect Dis 12:164. doi: 10.1186/1471-2334-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horz H-P, Scheer S, Huenger F, Vianna ME, Conrads G. 2008. Selective isolation of bacterial DNA from human clinical specimens. J Microbiol Methods 72:98–102. doi: 10.1016/j.mimet.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Mayrl E, Roeder B, Mester P, Wagner M, Rossmanith P. 2009. Broad range evaluation of the matrix solubilization (matrix lysis) strategy for direct enumeration of foodborne pathogens by nucleic acids technologies. J Food Prot 72:1225–1233. doi: 10.4315/0362-028X-72.6.1225. [DOI] [PubMed] [Google Scholar]
- 34.Søndergaard MSR, Josefsen MH, Löfström C, Christensen LS, Wieczorek K, Osek J, Hoorfar J. 2014. Low-cost monitoring of Campylobacter in poultry houses by air sampling and quantitative PCR. J Food Prot 77:325–330. doi: 10.4315/0362-028X.JFP-13-268. [DOI] [PubMed] [Google Scholar]
- 35.Jensen AN, Sørensen G, Baggesen DL, Bødker R, Hoorfar J. 2003. Addition of novobiocin in pre-enrichment step can improve Salmonella culture protocol of modified semisolid Rappaport-Vassiliadis. J Microbiol Methods 55:249–255. doi: 10.1016/S0167-7012(03)00158-1. [DOI] [PubMed] [Google Scholar]
- 36.International Organisation for Standardization. 2016. ISO 16140-2:2016. Microbiology of food and animal feed–method validation–part 2: protocol for the validation of alternative (proprietary) methods against a reference method. International Organisation for Standardization, Geneva, Switzerland: http://www.iso.org/iso/catalogue_detail?csnumber=54870. [Google Scholar]
- 37.Corry JEL, Jarvis B, Passmore S, Hedges A. 2007. A critical review of measurement uncertainty in the enumeration of food micro-organisms. Food Microbiol 24:230–253. doi: 10.1016/j.fm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Jarvis B. 2007. On the compositing of samples for qualitative microbiological testing. Lett Appl Microbiol 45:592–598. doi: 10.1111/j.1472-765X.2007.02237.x. [DOI] [PubMed] [Google Scholar]
- 39.Malorny B, Paccassoni E, Fach P, Martin A, Helmuth R, Bunge C. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl Environ Microbiol 70:7046–7052. doi: 10.1128/AEM.70.12.7046-7052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Organisation for Standardization. 2007. ISO 6579/A1:2007. Microbiology of food and animal feeding stuffs–horizontal method for the detection of Salmonella spp.–amendment 1: annex D: detection of Salmonella spp. in animal faeces and in environmental samples from the primary production stage. International Organisation for Standardization, Geneva, Switzerland. [Google Scholar]
- 41.NordVal/NMKL. 2009. Protocol for the validation of alternative microbiological methods. NordVal, Oslo, Norway: http://www.nmkl.org/dokumenter/nordval/NordValprotocolmarch2009.pdf. [Google Scholar]