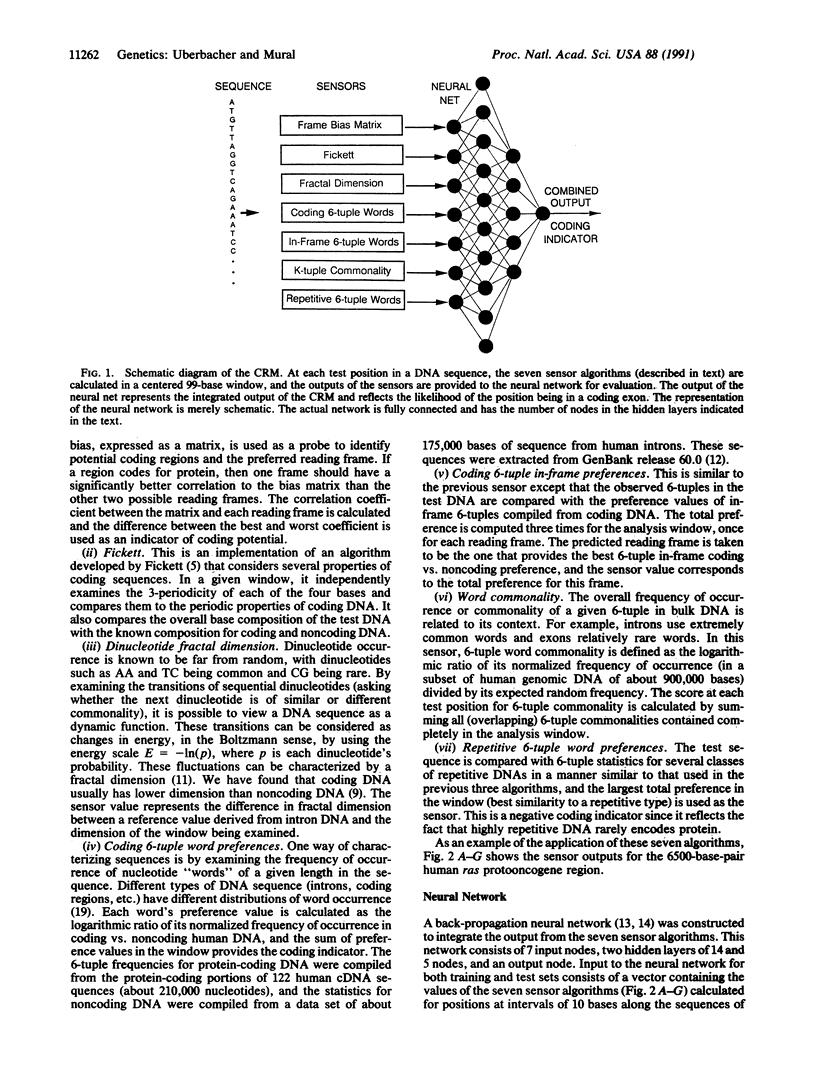
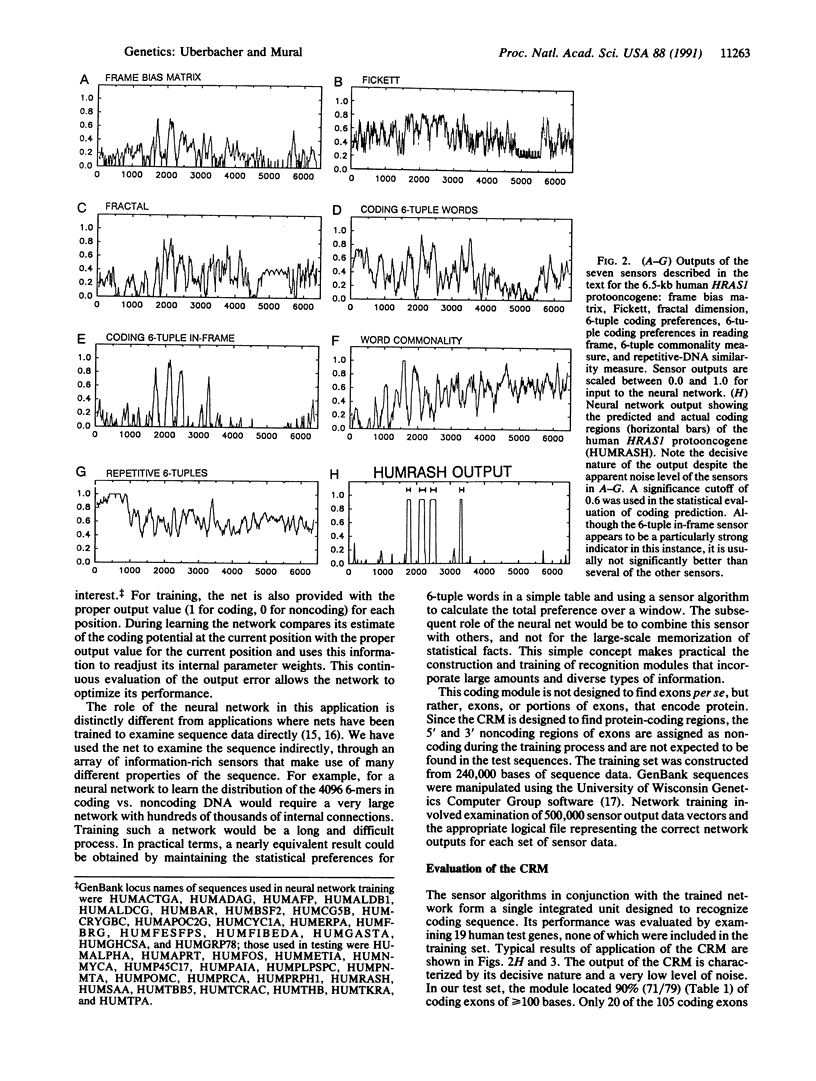
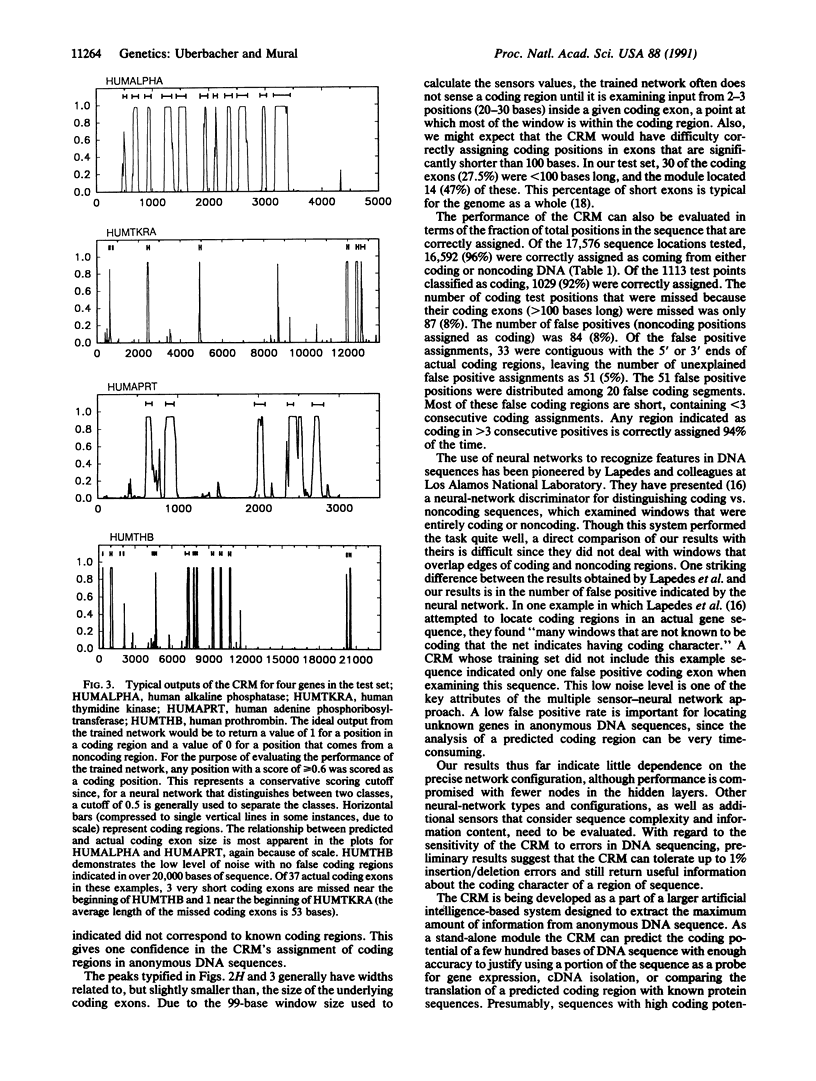
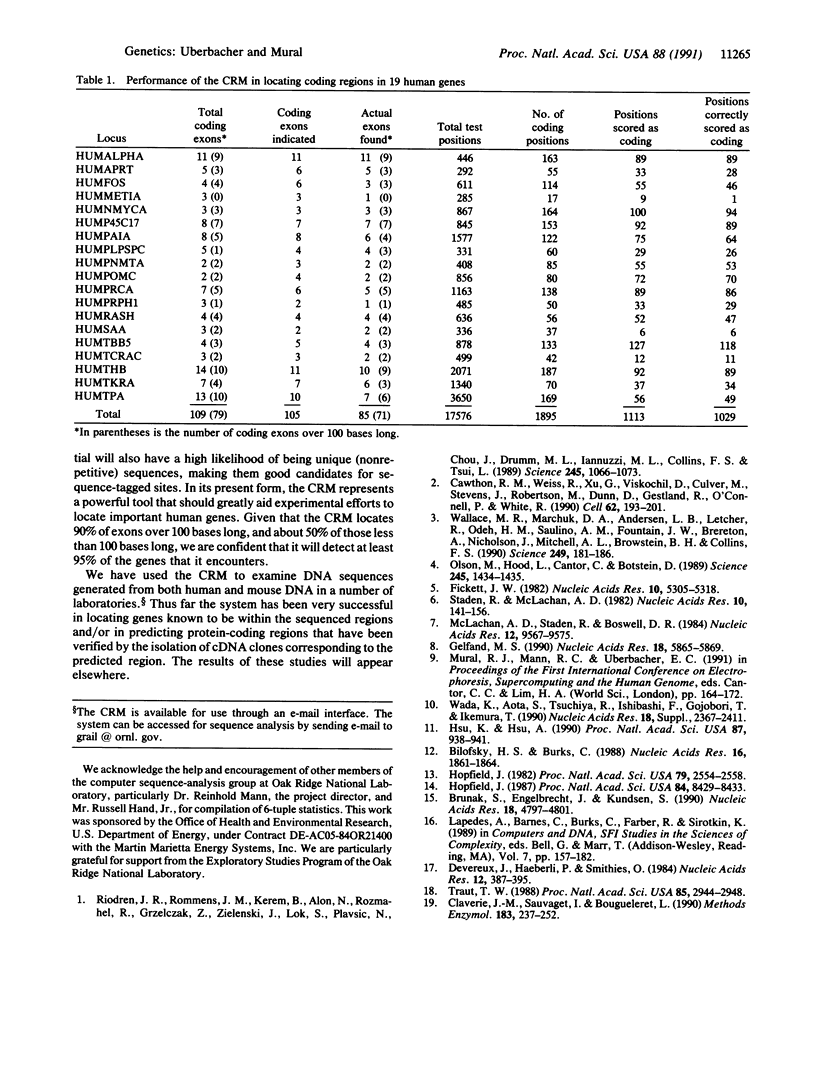
Abstract
Genes in higher eukaryotes may span tens or hundreds of kilobases with the protein-coding regions accounting for only a few percent of the total sequence. Identifying genes within large regions of uncharacterized DNA is a difficult undertaking and is currently the focus of many research efforts. We describe a reliable computational approach for locating protein-coding portions of genes in anonymous DNA sequence. Using a concept suggested by robotic environmental sensing, our method combines a set of sensor algorithms and a neural network to localize the coding regions. Several algorithms that report local characteristics of the DNA sequence, and therefore act as sensors, are also described. In its current configuration the "coding recognition module" identifies 90% of coding exons of length 100 bases or greater with less than one false positive coding exon indicated per five coding exons indicated. This is a significantly lower false positive rate than any method of which we are aware. This module demonstrates a method with general applicability to sequence-pattern recognition problems and is available for current research efforts.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. Neural network detects errors in the assignment of mRNA splice sites. Nucleic Acids Res. 1990 Aug 25;18(16):4797–4801. doi: 10.1093/nar/18.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. M., Weiss R., Xu G. F., Viskochil D., Culver M., Stevens J., Robertson M., Dunn D., Gesteland R., O'Connell P. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990 Jul 13;62(1):193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- Claverie J. M., Sauvaget I., Bougueleret L. K-tuple frequency analysis: from intron/exon discrimination to T-cell epitope mapping. Methods Enzymol. 1990;183:237–252. doi: 10.1016/0076-6879(90)83017-4. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand M. S. Computer prediction of the exon-intron structure of mammalian pre-mRNAs. Nucleic Acids Res. 1990 Oct 11;18(19):5865–5869. doi: 10.1093/nar/18.19.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Learning algorithms and probability distributions in feed-forward and feed-back networks. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8429–8433. doi: 10.1073/pnas.84.23.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2554–2558. doi: 10.1073/pnas.79.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsü K. J., Hsü A. J. Fractal geometry of music. Proc Natl Acad Sci U S A. 1990 Feb 1;87(3):938–941. doi: 10.1073/pnas.87.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Staden R., Boswell D. R. A method for measuring the non-random bias of a codon usage table. Nucleic Acids Res. 1984 Dec 21;12(24):9567–9575. doi: 10.1093/nar/12.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C., Botstein D. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989 Sep 8;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut T. W. Do exons code for structural or functional units in proteins? Proc Natl Acad Sci U S A. 1988 May;85(9):2944–2948. doi: 10.1073/pnas.85.9.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Aota S., Tsuchiya R., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2367–2411. doi: 10.1093/nar/18.suppl.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. R., Marchuk D. A., Andersen L. B., Letcher R., Odeh H. M., Saulino A. M., Fountain J. W., Brereton A., Nicholson J., Mitchell A. L. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990 Jul 13;249(4965):181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]



