ABSTRACT
Although minerals represent important soil constituents, their impact on the diversity and structure of soil microbial communities remains poorly documented. In this study, pure mineral particles with various chemistries (i.e., obsidian, apatite, and calcite) were considered. Each mineral type was conditioned in mesh bags and incubated in soil below different tree stands (beech, coppice with standards, and Corsican pine) for 2.5 years to determine the relative impacts of mineralogy and mineral weatherability on the taxonomic and functional diversities of mineral-associated bacterial communities. After this incubation period, the minerals and the surrounding bulk soil were collected to determine mass loss and to perform soil analyses, enzymatic assays, and cultivation-dependent and -independent analyses. Notably, our 16S rRNA gene pyrosequencing analyses revealed that after the 2.5-year incubation period, the mineral-associated bacterial communities strongly differed from those of the surrounding bulk soil for all tree stands considered. When focusing only on minerals, our analyses showed that the bacterial communities associated with calcite, the less recalcitrant mineral type, significantly differed from those that colonized obsidian and apatite minerals. The cultivation-dependent analysis revealed significantly higher abundances of effective mineral-weathering bacteria on the most recalcitrant minerals (i.e., apatite and obsidian). Together, our data showed an enrichment of Betaproteobacteria and effective mineral-weathering bacteria related to the Burkholderia and Collimonas genera on the minerals, suggesting a key role for these taxa in mineral weathering and nutrient cycling in nutrient-poor forest ecosystems.
IMPORTANCE Forests are usually developed on nutrient-poor and rocky soils, while nutrient-rich soils have been dedicated to agriculture. In this context, nutrient recycling and nutrient access are key processes in such environments. Deciphering how soil mineralogy influences the diversity, structure, and function of soil bacterial communities in relation to the soil conditions is crucial to better understanding the relative role of the soil bacterial communities in nutrient cycling and plant nutrition in nutrient-poor environments. The present study determined in detail the diversity and structure of bacterial communities associated with different mineral types incubated for 2.5 years in the soil under different tree species using cultivation-dependent and -independent analyses. Our data showed an enrichment of specific bacterial taxa on the minerals, specifically on the most weathered minerals, suggesting that they play key roles in mineral weathering and nutrient cycling in nutrient-poor forest ecosystems.
KEYWORDS: mineral chemistry, bacterial communities, forest soil, mesh bags, mineral weatherability
INTRODUCTION
In temperate regions, most forest ecosystems are developed on acidic and nutrient-poor soils, making access to and recycling of nutrients key processes for the long-lasting function of these ecosystems (1). In contrast to the case with agricultural soils, this aspect is important in forestry, for which fertilization remains a very rare practice. Most of the nutritive cations required for tree growth and for the function of the forest biosphere come from atmospheric deposits, the recycling of the nutrients contained in dead organic matter or from the soil rocks and minerals. Although part of these nutritive elements comes in a soluble form and are directly bioaccessible, another large portion remains entrapped in the crystal structure of the soil rocks and mineral particles, which are not directly accessible to tree roots. In this context, mineral weathering is essential to provide nutritive elements, especially for the most recalcitrant minerals. Indeed, the rocks and minerals present in soil vary in chemistry, weatherability and surface area, making certain minerals such as calcite quickly weathered under acidic conditions, while phyllosilicates remain comparatively resistant (2).
In addition to the action of abiotic processes related to water circulation, the repeated freeze-thaw events, or acidification, plants, lichens, and microorganisms have been reported to contribute to mineral and rock dissolution (3–7). Plant roots and fungal hyphae have been shown to physically disrupt mineral particles through the pressure applied to mineral surfaces and cracks and to form pores and tunnels at the surface of these minerals (8, 9). Bacteria and lichens also produce acidifying and chelating metabolites such as protons, organic acids or siderophores, which are known to increase mineral dissolution (4, 5, 7, 10–12). Several studies have reported that mineral weathering in soil varies according to the plant species (3) and is intensified in the rhizosphere compared to the surrounding bulk soil (BS) (13–16). Notably, accumulating evidence indicates that this intensification is partly related to the activity of root-associated microorganisms (1, 10, 17–22). In this sense, several microcosm experiments comparing tree seedlings inoculated or not with effective mineral weathering bacterial and/or fungal strains demonstrated that microorganisms significantly increase mineral dissolution, providing more nutritive cations available for plant nutrition (18, 20, 23).
Although mineral weathering is intensified in the plant root vicinity, less information is available concerning the processes that occur in the surrounding bulk soil, especially on the surfaces of rocks and minerals. However, rocks and minerals can be viewed as nutritive reserves and biogeochemical interfaces (24, 25), where biotic mineral weathering represents a key process in nutrient cycling. Mineral colonization by macro- and microorganisms has been largely reported in many terrestrial and aquatic environments, where minerals are directly accessible and in contact with air or water (4, 26–29). In contrast, few studies have investigated bacterial communities that colonize soil minerals and their role in nutrient cycling (30–34). This discrepancy is possibly due to the fact that soil is composed of a complex mosaic of rocks and minerals that interact to various degrees with organic matter, making their analysis difficult. Based on in vitro experiments and in situ analyses primarily performed on outcrop rocks or mineral particles incubated in aquifers, it has been suggested that, apart from a passive accumulation of microorganisms in cracks and pores, the surface charge and the chemistry of rocks and mineral particles as well as local physico-chemical conditions are the main parameters that determine microbial colonization of mineral surfaces (27, 34–41). Certini et al. (30) revealed that under soil conditions, the metabolic potential of sandstone-associated microorganisms differed from the surrounding bulk soil and that higher metabolic potentials were observed for smaller mineral particles. More recently, Lepleux et al. (33) showed that after a 4-year incubation time in a nutrient-poor and acidic forest, apatite particles were significantly enriched in effective mineral-weathering bacteria compared to the surrounding bulk soil. These results suggest that bacterial colonization of minerals may not be a coincidental event but rather an adaptive strategy to access limiting nutritive elements. Based on all these observations, the concept of a “mineralosphere,” which defines the microorganisms preferentially selected on the surface and around mineral particles, was proposed (25).
Deciphering how soil mineralogy influences the diversity, structure, and function of soil bacterial communities in relation to soil conditions is crucial for better understanding the relative role of soil bacterial communities in nutrient cycling and plant nutrition in nutrient-poor environments. In this context, our study aimed to investigate the diversity, structure, and functional potential of the bacterial communities colonizing different mineral types in the soil of three different tree stands (beech, coppice with standard, and Corsican pine [Bc, CwS, and Cp, respectively]). Pure mineral particles with various chemistries (calcite, apatite, and obsidian) were conditioned in mesh bags and incubated in the soil of the long-term forest observatory (LTO) of Breuil-Chenue. After 2.5 years of incubation, which is a very short period for such minerals, we combined soil chemical analyses and 16S rRNA gene amplicon pyrosequencing to determine how bacterial communities were affected in the early stages of interactions between bacteria and mineral surfaces, by mineral chemistry and by the soil conditions occurring under the different tree stands. In parallel, Biolog microplate assays were used to determine the carbon source utilization patterns of the bacterial communities in the bulk soil and those associated with mineral particles. Finally, a cultivation-dependent analysis was performed to determine the mineral weathering potential of each bacterial strain through different bioassays allowing measurement of the ability to solubilize inorganic phosphorus and calcite and to mobilize iron. The weatherability of the different mineral types used was determined in abiotic conditions to determine if this parameter was a potential driving factor of the bacterial communities. The present study allowed us to better characterize the distribution and the weathering potential of bacterial communities in forest soil and to further corroborate their role in mineral weathering and nutrient cycling.
RESULTS
Soil chemical properties and mineral mass loss.
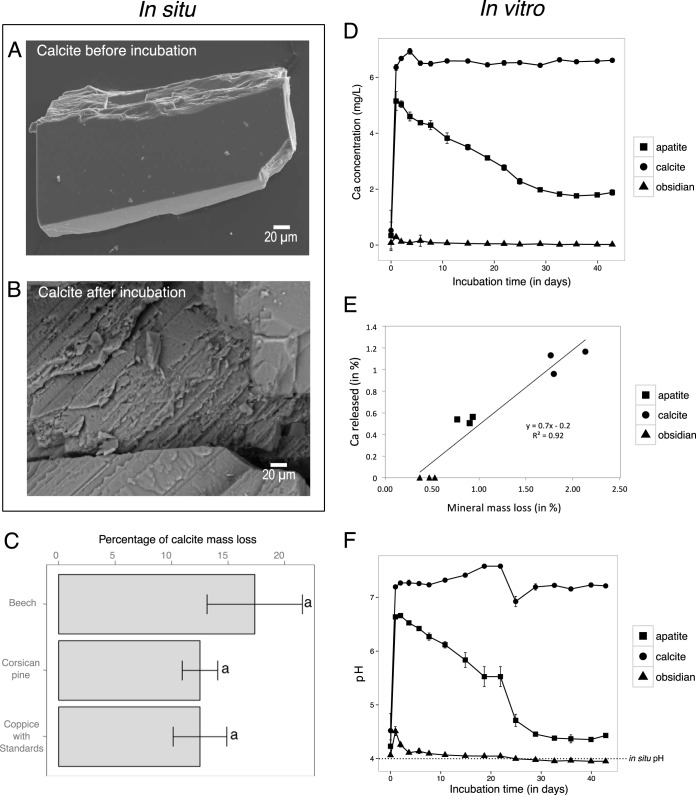
Soil chemical analyses revealed that the soil samples collected under the beech (Bc) stand were significantly more acidic than under the coppice with standards (CwS) and Corsican pine (Cp) stands (Bc < CwS < Cp; P < 0.05) (Table 1). In contrast, the organic matter content and the total carbon and total nitrogen contents were significantly higher under the Bc stand than under the CwS and Cp stands (Bc < CwS < Cp; P < 0.05). Similarly, the concentrations of most of the exchangeable nutritive cations were significantly higher under the Bc stand than under the other two tree stands (P < 0.05). After the 2.5-year incubation, the mass of calcite particles collected in the mesh bags was significantly reduced compared to initial mass (P < 0.05) regardless of the tree stand. In contrast, no visible mass loss was observed for the apatite and obsidian minerals. Calcite particles appeared significantly weathered for all tree stands considered compared to the control. Visible dissolution marks were observed for calcite particles as revealed by electron microscopy imaging (Fig. 1A and B) but not for apatite and obsidian particles (data not shown). While there was no significant difference among the different tree stands (P > 0.05), the most intensively weathered calcite particles were recovered from the Bc stand (percentage of mass loss, 17.3 ± 8.4%). The least weathered particles were recovered from the CwS and Cp stands (percentage of mass loss, 12.5% ± 4.7% and 12.5% ± 3.1%, respectively) (Fig. 1C).
TABLE 1.
Chemical properties of the bulk soil collected below each tree standa
| Parameter | Value for stand type |
||
|---|---|---|---|
| Beech | Coppice with standards | Corsican pine | |
| pH | 3.81 ± 0.01 C | 4.16 ± 0.06 B | 4.32 ± 0.02 A |
| C tot (g · kg−1) | 67.33 ± 0.83 A | 57.27 ± 1.79 B | 36.53 ± 1.61 C |
| N tot (g · kg−1) | 3.29 ± 0.07 A | 3.01 ± 0.06 B | 1.85 ± 0.04 C |
| C/N | 20.47 ± 0.27 A | 19.03 ± 0.19 B | 19.80 ± 0.46 AB |
| OM (g · kg−1) | 116.33 ± 1.45 A | 99.03 ± 3.03 B | 63.23 ± 2.78 C |
| P Duch. (g · kg−1) | 0.109 ± 0.001 B | 0.126 ± 0.002 A | 0.102 ± 0.001 B |
| P Ols. (g · kg−1) | 0.015 ± 0.001 A | 0.011 ± 0.000 B | 0.007 ± 0.000 C |
| CEC (cmol+ · kg−1) | 8.07 ± 0.19 A | 6.18 ± 0.3 B | 4.60 ± 0.39 C |
| H+ (cmol+ · kg−1) | 0.913 ± 0.009 A | 0.303 ± 0.033 B | 0.187 ± 0.023 C |
| Al (cmol+ · kg−1) | 6.09 ± 0.13 A | 5.70 ± 0.18 A | 4.42 ± 0.28 B |
| K (cmol+ · kg−1) | 0.223 ± 0.008 A | 0.209 ± 0.017 A | 0.096 ± 0.004 B |
| Ca (cmol+ · kg−1) | 0.537 ± 0.042 A | 0.198 ± 0.034 B | 0.101 ± 0.005 B |
| Mg (cmol+ · kg−1) | 0.197 ± 0.009 A | 0.139 ± 0.016 B | 0.078 ± 0.006 C |
| Na (cmol+ · kg−1) | 0.021 ± 0.001 A | 0.020 ± 0.002 A | 0.016 ± 0.000 A |
| Fe (cmol+ · kg−1) | 0.242 ± 0.006 A | 0.101 ± 0.015 B | 0.044 ± 0.006 C |
Each value is the mean of three replicates ± the standard error of the mean. One-factor ANOVA was carried out to compare the sampling sites. Different letters indicate significant differences (P < 0.05). Abbreviations: N tot, total nitrogen; C tot, total carbon; OM, organic matter; CEC, cationic exchange capacity; H+, protons; P. Duch., phosphorus extracted according to the Duchaufour method; P. Ols, phosphorus extracted according to the Olsen method.
FIG 1.
Mineral weatherability under soil conditions and in a microcosm experiment. (A and B) Representative scanning electron micrographs from calcite particle observed before (A) and after (B) a 2.5-year incubation in the soil under a beech stand. As no surface alteration was observed for apatite and obsidian, scanning electron micrographs are not presented. (C) Estimation of the calcite mass loss after a 2.5-year incubation under beech stands (Bc), coppice with standards (CwS), and Corsican pine (Cp) stands. Each value is the mean of three (bulk soil) or four (minerals) replicates with standard error of the mean. As no mass loss was measured for apatite and obsidian, data are not presented. (D) Concentration of the Ca released from obsidian, apatite, and calcite particles over time in the microcosm experiment. (E) Relationship between the relative mineral mass loss and the percentage of Ca released for each mineral. This relationship was determined using the data generated in the mineral weatherability experiment. Three independent replicate reactors were done for each mineral type and are presented. (F) Evolution of the pH of the output solution (initial pH = 4) during the abiotic-dissolution assay performed with obsidian, apatite, and calcite. The dotted line represents the initial pH and corresponds to the pH measured in situ at the Breuil-Chenue experimental site.
Mineral weatherability and acid-buffering capacity.
The abiotic mineral dissolutions of obsidian, apatite, and calcite were determined under controlled conditions mimicking the soil pH occurring in the Breuil-Chenue LTO using a microcosm setup. After 42 days of incubation, the mass loss measured for calcite (1.9% of mass loss) was significantly higher than for apatite and obsidian (0.85 and 0.45%, respectively; P < 0.05) (data not shown). For calcite, the concentration of Ca in the output solution appeared almost constant over time for calcite particles, with a value close to 6.5 mg · liter−1 (Fig. 1D). In contrast, apatite exhibited nonstoichiometric dissolution during the first 25 days. After this period, the Ca concentration measured in the output solution reached 1.7 mg · liter−1, and the release rates for Ca and P became stoichiometric (Ca/P = 2.1). Similarly, for obsidian particles, the Ca concentration exhibited an increase during the first step, followed by a decrease until steady state was reached (0.03 mg · liter−1) at approximately 25 days, when the rates of release of Ca and Si became stoichiometric (Ca/Si = 0.5). A comparison of the data obtained for the three mineral types revealed that the relative percentage of released Ca in the output solution was positively correlated with the total mineral mass loss (R2 = 0.92; P < 0.0001 [Fig. 1E]). In addition to the variations in the concentrations of nutrients in the output solution, our measurements also revealed significant changes in the pH. These modifications are due to the buffering capacity of each mineral type (the solid phase of each mineral consumes protons during the dissolution process). This was particularly the case for calcite, for which the pH measured in the output solution of calcite appeared significantly increased, from 4 in the initial solution to ca. 7 at the end of the experiment (Fig. 1F). Similarly, the pH measured in the output solution of apatite increased to 6.6 during the first 20 days of the experiment and then decreased until it stabilized at 4.4 at the end of the experiment. Such an acid-buffering effect was not reported for obsidian, for which the pH remained at 4.
Metabolic assay.
To test the impact of the compartment (bulk soil [BS] versus minerals), soil conditions, and mineral type on the substrate utilization potential of the microbial communities, a Biolog-based approach was utilized. Our analysis revealed that the most metabolized carbon sources were identical for bulk soil and mineral-associated bacterial communities and corresponded to l-asparagine, N-acetylglucosamine, and β-methyl-d-glucoside. However, microbial communities colonizing the three types of minerals metabolized a significantly broader range of carbon sources than those from the surrounding bulk soil for all tree stands considered (P < 0.05) (Table 2). Indeed, mineral-associated microbial communities exhibited the highest average well color development (AWCD) and Shannon-Weaver indices. Focusing exclusively on mineral samples, no significant difference was observed for the AWCD and Shannon-Weaver indices among the three types of minerals, except under the Cp stand, where apatite-associated microbial communities were characterized by significantly lower AWCD and Shannon-Weaver indices than those occurring on obsidian and calcite (P < 0.05) (Table 2).
TABLE 2.
Estimates of the functional and taxonomic diversity in bulk soil and mineral samples according to Biolog EcoPlates and 16S rRNA gene pyrosequencing data analysesa
| Tree stand | Compartment | Functional diversity |
Taxonomic diversity |
||||
|---|---|---|---|---|---|---|---|
| Metabolized substrates | Shannon | AWCD | Sobs | Shannon | Coverage | ||
| Beech | Bulk soil | 19.67 ± 1.86 b | 2.70 ± 0.08 b | 0.45 ± 0.03 b | 714 ± 9 a | 4.99 ± 0.01 ab | 0.96 ± 0.00 |
| Obsidian | 25.75 ± 0.48 a | 3.05 ± 0.02 a | 0.84 ± 0.03 a | 768 ± 64 a | 4.45 ± 0.47 b | 0.95 ± 0.00 | |
| Apatite | 25.25 ± 1.11 a | 3.01 ± 0.04 a | 0.77 ± 0.05 a | 1059 ± 250 a | 5.43 ± 0.31 ab | 0.93 ± 0.02 | |
| Calcite | 26.00 ± 0.00 a | 3.04 ± 0.02 a | 0.92 ± 0.06 a | 931 ± 27 a | 5.63 ± 0.04 a | 0.95 ± 0.00 | |
| Coppice with standards | Bulk soil | 14.00 ± 1.53 b | 2.41 ± 0.08 c | 0.21 ± 0.04 b | 748 ± 36 a | 4.99 ± 0.04 ab | 0.95 ± 0.00 |
| Obsidian | 24.50 ± 0.87 a | 2.99 ± 0.05 a | 0.79 ± 0.05 a | 816 ± 37 a | 5.00 ± 0.11 ab | 0.95 ± 0.00 | |
| Apatite | 28.25 ± 1.38 a | 3.11 ± 0.04 a | 0.80 ± 0.07 a | 678 ± 80 a | 4.62 ± 0.30 ab | 0.96 ± 0.01 | |
| Calcite | 25.00 ± 0.41 a | 3.05 ± 0.02 a | 0.84 ± 0.03 a | 731 ± 63 a | 4.90 ± 0.29 ab | 0.95 ± 0.00 | |
| Corsican pine | Bulk soil | 11.00 ± 1.00 c | 2.21 ± 0.04 c | 0.19 ± 0.05 b | 688 ± 12 a | 4.39 ± 0.08 b | 0.95 ± 0.00 |
| Obsidian | 25.75 ± 0.63 a | 3.05 ± 0.02 a | 0.85 ± 0.06 a | 973 ± 36 a | 5.40 ± 0.09 ab | 0.94 ± 0.00 | |
| Apatite | 20.00 ± 1.68 b | 2.76 ± 0.06 b | 0.45 ± 0.11 b | 793 ± 29 a | 5.08 ± 0.09 ab | 0.95 ± 0.00 | |
| Calcite | 24.00 ± 0.41 ab | 3.00 ± 0.01 a | 0.80 ± 0.04 a | 820 ± 64 a | 5.21 ± 0.17 ab | 0.95 ± 0.01 | |
Each value is the mean ± the standard error of the mean of three or four biological replicates. Different letters indicate significant differences (P < 0.05).
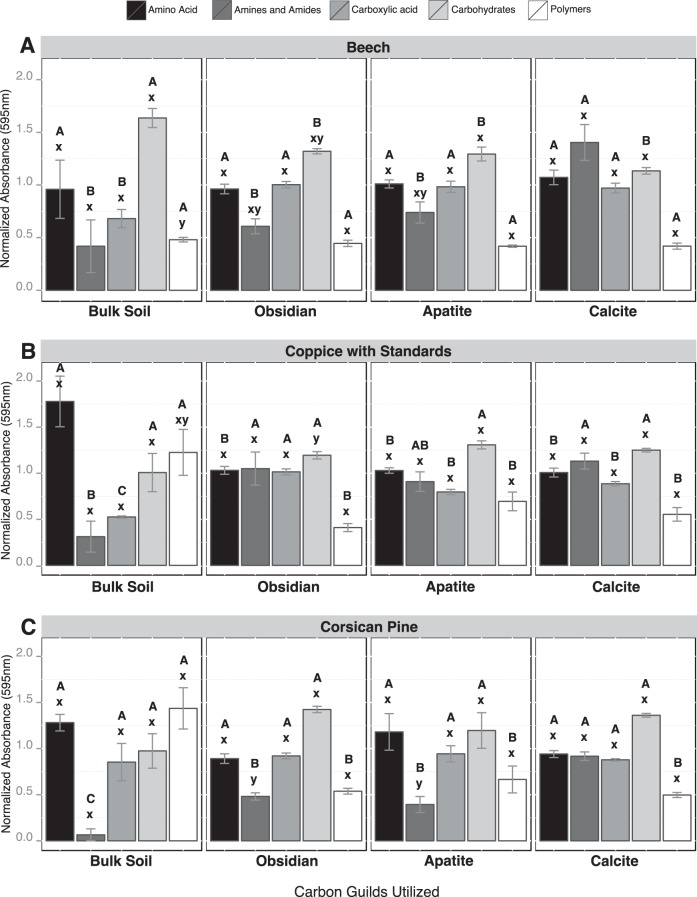
When the Biolog data were considered based on substrate guilds, significant differences were observed between bulk soil and mineral samples for amines/amides (AM), carboxylic and acetic acids (CA) and polymers (PL) (Fig. 2). Under the CwS and Cp stands, AM were significantly more metabolized by mineral-associated microbial communities than those from the bulk soil (P < 0.05), while PL were more metabolized by bulk soil microbial communities (P < 0.05) (Fig. 2B and C). In addition, under the Bc and CwS stands, CA were more metabolized by mineral-associated bacterial communities than those from the bulk soil (Fig. 2A and B). A detailed analysis of the substrates that were metabolized revealed that d-galacturonic acid, 4-hydroxybenzoic acid, glycogen, l-serine, and d-xylose were significantly more metabolized by mineral-associated microbial communities than by bulk soil microbial communities for all tree stands considered (P < 0.05). Soil conditions did not impact significantly the substrate utilization potential of the microbial communities, except for the BS samples, for which PL were significantly more metabolized under the Cp stand than under the Bc stand. Similarly, the Biolog profiles obtained for each mineral type were relatively similar across the different tree stands, even if AM were significantly more metabolized by obsidian and apatite-associated bacterial communities under the CwS stand than under the Cp stand.
FIG 2.
Metabolic potentials of microbial communities based on Biolog EcoPlate analysis. The relative use of carbon-substrate guilds by microbial communities (amino acids, amines and amides, carboxylic acid, carbohydrates, and polymers) was estimated under beech stands (A), coppice with standards (B), and Corsican pine stands (C) and for each compartment considered (bulk soil, obsidian, apatite, and calcite). For the different substrate guilds, different capital letters (A, B, or C) above the bars indicate significant differences in the same tree stand according to a one-factor ANOVA and Tukey's multiple pairwise comparisons test, while different lowercase letters (x or y) indicate significant differences between the tree stands. Each value is the mean of three (bulk soil) or four (minerals) replicates, and the error bars indicate the standard errors of the means.
Molecular diversity, richness, and structure of the bulk soil and mineral-associated bacterial communities.
For the same number of 16S rRNA gene pyrosequences, the bacterial richness occurring in the bulk soil and mineral samples did not vary significantly, although it ranged from 714 operational taxonomic units (OTUs) in bulk soil under the Bc stand to 1,059 OTUs on apatite under the Bc stand (Table 2). These observations were confirmed by the rarefaction curves and Chao1 analyses (Table 2; see also Fig. S2 in the supplemental material). Good's coverage was ≥93% for all conditions, indicating that the 16S rRNA gene sequences identified in these samples represent the majority of the bacterial sequences present in situ (Table 2). The Shannon-Weaver index also had low variations between samples, with values ranging from 4.39 in bulk soil under the Cp stand to 5.63 on calcite under the Bc stand. No significant differences were observed for this index among the compartments (bulk soil versus minerals), the different mineral types, or the different tree stands, except under the Bc stand. In this tree stand, the Shannon index was significantly higher for calcite than for obsidian samples (P < 0.05) (Table 2). Under each tree stand, the numbers of OTUs shared by the different sample types were investigated considering the total and the top 500 OTUs and are summarized in Fig. S3A and B. When each tree stand was considered independently, the proportion of OTUs common between BS and at least one mineral type ranged between 19.2 and 23% (Fig. S3A) when all the OTUs were considered, while it ranged from 67.6 and 74.8% (Fig. S3B) for the top 500 OTUs. Furthermore, a large fraction of the OTUs was exclusively detected on mineral particles, especially when all the OTUs have been taken into account, representing 65.5 to 75.1% of the total bacterial diversity (Fig. S3A). The same analysis done on the top 500 OTUs showed that the proportion of OTUs specifically associated with minerals ranged from 25.2 to 32.0% (Fig. S3B) of the bacterial diversity. Notably, whatever the threshold considered (total number of OTUs versus top 500 OTUs), calcite was the mineral type characterized by the highest number of specific OTUs for all tree stands considered. When the same sample type was compared across the different tree stands (i.e., mineral type or soil samples under Bc, Cp, and CwS), 71.0 to 86.2% of the top 500 OTUs were common to the tree stands considered, while this value reached only ca. 20% when all the OTUs were considered (Fig. S3A and B). To disentangle the effects of soil conditions (beech, coppice with standards, and Corsican pine) and compartment type (BS versus mineral) on bacterial community composition, we conducted variance partitioning analyses. Although 83.7% of the variance remained unexplained, the compartment effect explained 10.0% of the variance, while the soil conditions explained 6.3% (P < 0.001). A second variance partitioning analysis was performed considering only the mineral samples and not the BS compartment. This analysis revealed that the mineral type (obsidian, apatite, and calcite) explained 12.3% of the inertia in the bacteria community, while the soil conditions explained only 5.7% (P < 0.001). Again, the larger portion of the variance remained unexplained (82%).
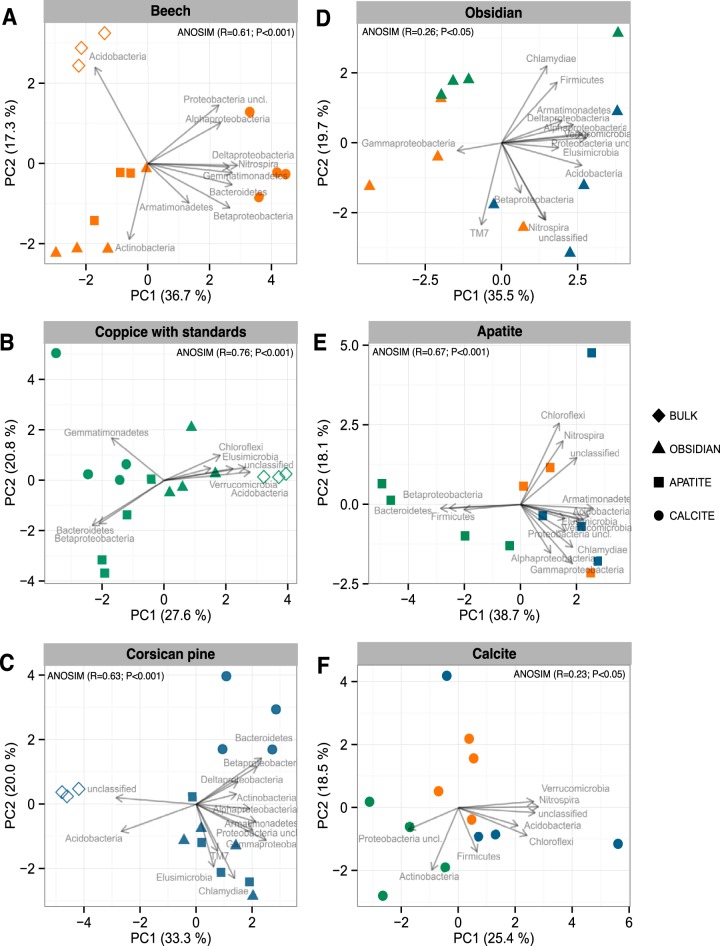
Based on taxonomic affiliation of the 16S rRNA gene sequences, the dominant phyla across all the samples were Proteobacteria (37.9%), Actinobacteria (18.2%), Acidobacteria (16.0%), and Bacteroidetes (10.8%) (Fig. 3). Multivariate analyses demonstrated that the bacterial phyla were not distributed uniformly among the samples (Fig. 4). At the phylum level, BS bacterial communities appeared significantly different from the colonizing minerals, as confirmed by an analysis of similarity (ANOSIM) for each tree stand (P < 0.001). Similar results were obtained when the multivariate analysis was conducted at the OTU level (Fig. S4A to F). Notably, BS bacterial communities appeared separated according to the tree stand (Fig. S4G). At the phylum level, BS samples harbored a significantly higher relative abundance of sequences related to Acidobacteria (34.8 to 46.1% of total reads) than did mineral samples (3.1 to 16.4%; P < 0.05) for all tree stands considered (Fig. 3 and 4A to C). Notably, under the CwS and Cp stands, 16S rRNA gene sequences of nonassigned bacteria were significantly more abundant in the BS samples (22.7 to 37.4%) than in the mineral samples (<19.1%; P < 0.05) (Fig. 3 and 4B and C). In contrast, mineral-associated bacterial communities were significantly enriched in representatives of Betaproteobacteria than the communities from the BS samples for all the tree stands considered (P > 0.05) (Fig. 3). Moreover, under the CwS and Cp stands, representatives of Bacteroidetes were significantly enriched on minerals (4.6 to 29.0%) compared to bulk soil samples (<2.09%, P > 0.05) (Fig. 3). Last, under the Cp stand, representatives of Gammaproteobacteria were significantly enriched on minerals (9.6 to 11.8%) compared to those of the surrounding BS (1.9%) (P < 0.05) (Fig. 3).
FIG 3.
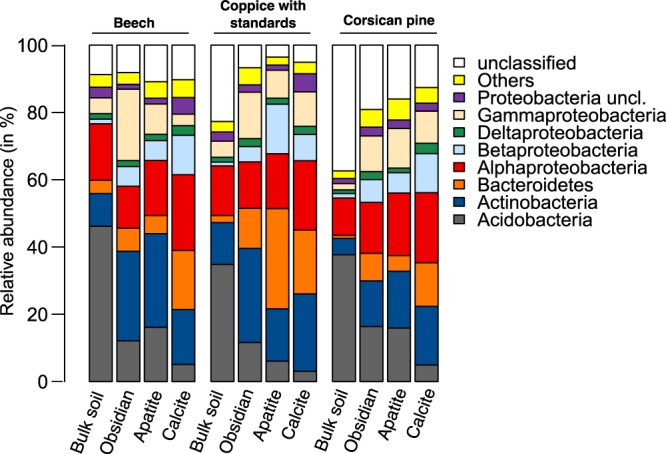
Relative distributions of the major bacterial taxa. The relative abundances of the major phyla and class were calculated as the percentage of sequences belonging to a particular lineage of all 16S rRNA gene sequences under beech and Corsican pine stands and under coppice with standards and for each compartment considered (bulk soil, obsidian, apatite, and calcite).
FIG 4.
Multivariate analysis of the bacterial communities based on the relative abundances of bacterial phyla and proteobacterial classes estimated by 16S rRNA gene pyrosequencing analysis. Multivariate analysis was conducted separately for each type of tree stand: beech, coppice with standards, and Corsican pine. The same analysis was conducted for each mineral type, excluding the bulk soil samples: obsidian, apatite, and calcite. For legibility, the samples are presented as follows: orange, stand of beech; green, coppice with standards; blue, Corsican pine stand; circles, calcite samples; squares, apatite samples; triangles, obsidian samples; open diamonds, bulk soil samples. Vectors show the direction of maximum change for variables and longer arrows indicate a greater change in relative abundance. The percentages of the total variance explained by the first two axes, PC1 and PC2, are presented on each graph.
When focusing on both mineral and BS samples, calcite-associated bacterial communities were found to differ significantly from bacterial communities colonizing apatite and obsidian particles for all tree stands considered (Fig. 4A to C). At the phylum level, significantly higher abundances of sequences assigned to Bacteroidetes (>13.0%) and Betaproteobacteria (>11.6%) were observed on calcite than on obsidian and apatite (<8.2% and <6.8%, respectively) under the Bc and Cp stands (P < 0.05) (Fig. 3). At the genus level, sequences related to Ferruginibacter were significantly more abundant on calcite (>2.41% of total reads) than on apatite and obsidian (<0.21%) for all tree stands considered (P < 0.05) (Table S1). To a lower extent, several other genera affiliated with Alphaproteobacteria (Afipia, Bosea, Devosia, Hyphomicrobium, Novosphingobium, and Sphingomonas) and Gammaproteobacteria (Rhizobacter and Arenimonas) or with ammonia-oxidizing related genera (Nitrospira and Nitrosospira) appeared significantly more abundant on calcite surfaces than on other minerals (P < 0.05) (Table S1). Apatite- and obsidian-associated bacterial communities exhibited similar patterns at different levels of classification (phylum and genus). Notably, these two mineral types were significantly enriched in sequences related to the genera Mucilaginibacter and Burkholderia (>2.4% and >1.3%, respectively) compared to calcite and BS samples (<0.8% and <0.6%, respectively) in all tree stands (P < 0.05) (Table S1). In addition to the mineral type effect, significant differences were observed for some bacterial genera depending on the tree stand. Under the CwS stand, apatite-associated bacterial communities were significantly enriched in sequences related to the genera Streptomyces (7.8%), Chitinophaga (7.3%), and Pedobacter (2.2%) compared to the bulk soil, calcite, and obsidian samples (<0.1%, <0.5%, and < 0.2%, respectively) (P < 0.05) (Table S1). In contrast, the genera Nevskia and Dyella were significantly enriched on obsidian and apatite (>1.4% and >1.1%, respectively) compared to bulk soil and calcite samples (>0.1% and >0.7%, respectively) (P < 0.05) (Table S1). Last, under the Cp stand, the genus Mycobacterium was significantly more abundant on mineral surfaces than in the bulk soil samples (P < 0.05).
Considering only the minerals, multivariate analyses demonstrated that for the same mineral type, mineral-associated bacterial communities were significantly structured according to the tree stand considered (Fig. 4D to F). These results were confirmed for all mineral types by ANOSIM (P < 0.05). According to the ordination plots, the mineral-associated bacterial communities that developed under the CwS stand tended to differ from those under the Bc and Cp stands (Fig. 4D to F). This differentiation according to the tree stand was particularly evident for apatite (Fig. 4E). A detailed analysis of the taxa present revealed that under the CwS stand, bacterial communities were significantly enriched in sequences related to Bacteroidetes and Betaproteobacteria compared to those occurring under the Bc and Cp stands (Fig. 3 and 4E). At the genus level, the relative abundances of representatives of the Mucilaginibacter, Pedobacter, Sphingomonas, Burkholderia, and Dokdonella genera were significantly enriched on apatite incubated under the CwS stand compared to apatite particles incubated under the other two tree stands (Table S2).
Quantification of the culturable bacterial communities.
Total culturable bacterial communities from the bulk soil and mineral samples collected in each tree stand were recovered on 1/10 tryptic soy agar (TSA) plates at densities ranging from 5.7 log CFU · g−1 (dry weight of soil/mineral) for apatite particles incubated under the Cp stand to 7.3 log CFU · g−1 for apatite particles incubated under the CwS stand (Fig. S5). Based on a one-factor (tree species or mineral type) analysis of variance (ANOVA), significant differences in bacterial density were observed. Under the Bc stand, obsidian and calcite samples displayed a higher culturable bacterial density than apatite samples. Moreover, under the CwS stand, the culturable bacterial density was significantly higher on apatite than on other samples, while the bulk soil and calcite samples harbored a significantly higher density than obsidian. Last, under the Cp stand, the culturable bacterial density was significantly higher on calcite than on the other samples.
Mineral weathering assays. (i) Phosphorus solubilization and medium acidification.
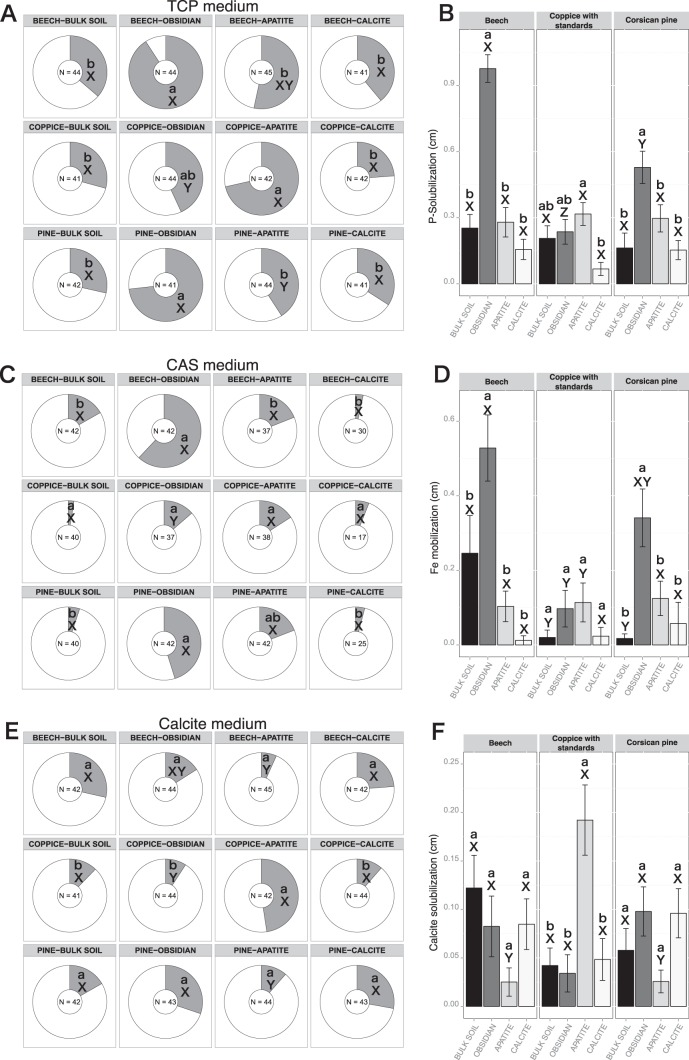
The screening of the bacterial strains for the ability to solubilize inorganic P revealed that the BS and the calcite samples from the three tree stands harbored 23.8 to 39.0% effective P-solubilizing bacteria. No significant differences were reported for frequency and efficacy between the different tree stands for these two conditions (BS and calcite). In contrast, the frequency of effective P-solubilizing bacteria was significantly higher on obsidian than on the bulk soil samples under the Bc (90.9% of effective P-solubilizing bacteria; n = 44) and Cp (73.2%; n = 41) (χ2Bc = 28.3 and P < 0.001; χ2CP = 16.5 and P < 0.001) stands (Fig. 5A). Under the CwS stand, the frequency of effective P-solubilizing bacteria was significantly higher on apatite particles (71.4%; n = 41) than on the bulk soil samples (Fig. 5A). Moreover, a one-factor ANOVA demonstrated that the obsidian-associated bacteria from the Bc and Cp stands had the highest P-solubilizing efficacy off all the samples (P < 0.05) (Fig. 5B).
FIG 5.
Relative distributions and efficacies of bacterial strains capable of mobilizing phosphorus, iron, and calcite. Pie charts represent the distribution of bacterial isolates recovered from bulk soil, obsidian, apatite, and calcite samples under beech stands, coppice with standards, and Corsican pine stands and tested on TCP medium (A), CAS medium (C), and CAL medium (E). Effective bacterial strains for each bioassay are represented in gray, and noneffective strains are represented in white. For each bioassay, the number of strains considered is presented in the center of the pie chart. For the same compartment, this number may vary between the different bioassays, due to the ability of the bacterial strains to grow on the different culture media. Significant differences in frequencies were evaluated by χ2 test. Bar plots represent the averaged efficacy of the bacterial strains recovered from bulk soil, obsidian, apatite, and calcite samples under beech stands, coppice with standards, and Corsican pine stands and tested on TCP medium (B), CAS medium (D), and CAL medium (F). Efficacies were obtained by measuring the discoloration zone (in centimeters) around bacterial colonies on the different media. Significant differences in efficacy were evaluated according to a one-factor ANOVA and Tukey's multiple pairwise comparisons test. Each value is the mean of three (bulk soil) or four (minerals) replicates and the error bars indicate the standard error of the mean. For both pie charts and bar plots, different lowercase letters (a or b) indicate significant differences (P < 0.05) in the same tree stand, while different capital letters (X or Y) indicate significant differences (P < 0.05) between the tree stands.
As P solubilization by bacteria is often related to the production of acidifying metabolites, acidification of the modified liquid TCP medium was determined. Our analyses revealed that after a 5-day incubation period, the pH measured in the culture medium for the different bacterial strains tested ranged from 3.4 to 6.2. Under the same tree stand, no significant differences were observed between bacterial strains from the different mineral types, except under the Bc stand, where obsidian-associated bacteria acidified significantly more (average pH = 4.2 ± 0.18) than those associated with calcite (average pH = 4.9 ± 0.11) (P < 0.05). For obsidian and calcite minerals, our results revealed that bacterial strains originating from the Bc and the Cp stands induced significantly more acidification of the culture medium (average pH = 4.7 ± 0.08) than those collected from the CwS stand (average pH = 5.27 ± 0.08) (P < 0.05). Notably, a linear regression analysis (y = −0.94x + 5.09; R2 = 0.3; P < 0.001) demonstrated that the P solubilization efficacy and the pH data were significantly correlated (Fig. S6). This relationship was reinforced for the bacterial strains isolated from obsidian surfaces under the Bc and the Cp stands, which were characterized for most of them by the highest P solubilizing efficacy and the strongest acidifying ability (pH 4.5 to 3.4).
(ii) Iron mobilization.
The frequency of Fe-mobilizing bacteria in the BS compartment from the Bc stand (16%; n = 42) was slightly, but not significantly, higher than that in the BS samples from the CwS (2.5%; n = 40) and Cp (5%; n = 40) stands. This frequency did not significantly vary between bulk soil, calcite, and apatite samples for all tree stands considered (Fig. 5C). However, the frequency of Fe-mobilizing bacteria was significantly higher on obsidian incubated under the Bc (61.9%; n = 42) and Cp (45.2%; n = 42) stands than on the bulk soil samples (χ2Bc = 18.0 and P < 0.0001; χ2CP = 17.4 and P < 0.0001) (Fig. 5C). Under the Bc and Cp stands, the relative efficacy of Fe-mobilizing bacteria associated with obsidian was significantly higher than in the other samples (P < 0.05) (Fig. 5D), while no differences were observed under the CwS stand.
(iii) Calcite solubilization.
Our analyses revealed that the frequencies of calcite-solubilizing bacteria were relatively similar among most of the samples. Indeed, ca. 15% of the total bacterial strains was capable of solubilizing calcite (Fig. 5E). In the CwS stand, the highest frequency of calcite-solubilizing bacteria (47.6%; n = 42) was detected for the bacterial isolated from apatite. In terms of efficacy, our analyses showed that in the CwS stand, apatite-associated bacteria were significantly more effective at solubilizing calcite than bacteria associated with the other mineral types (P < 0.05) (Fig. 5F).
Taxonomic affiliation of the bacterial strains from the mineral and bulk soil samples.
Based on their 16S rRNA gene sequence (ca. 800 bp), the bacterial strains from the BS or mineral samples were primarily assigned to Proteobacteria (51.5%), Actinobacteria (29.1%), Firmicutes (16.6%), and Bacteroidetes (2.8%). Among Proteobacteria, bacterial strains belonged to the beta (25.5%), alpha (19.6%), and gamma (6.4%) subclasses. These 16S rRNA gene sequences exhibited high similarity (95 to 100%) with the 16S rRNA gene sequences of environmental bacterial strains. In our collection, the most represented genera were Burkholderia (n = 75 strains), Microbacterium (n = 64), Rhizobium (n = 45), Paenibacillus (n = 39), Collimonas (n = 30), Arthrobacter (n = 25), Bacillus (n = 24), Variovorax (n = 16), Mesorhizobium (n = 15), and Sphingomonas (n = 15) (Fig. S7). Notably, the 16S rRNA gene sequences assigned to the Burkholderia and Collimonas genera presented high sequence homology with 16S rRNA gene sequences of strains isolated from the same experimental site. A detailed analysis of the relative distributions of the different genera detected according to their ecological origin revealed that for all tree stands considered, the BS samples were characterized by higher proportions of Bacillus and Paenibacillus strains than in the mineral samples (Fig. S7). Notably, calcite samples harbored a higher proportion of Microbacterium strains (>39%) than did the other mineral types or the BS samples in the three tree stands. Several bacterial genera, such as Arthrobacter, Rhizobium, Mesorhizobium, Bosea, Leifsonia, Microbacterium, Mycobacterium, and Sphingomonas, were exclusively isolated from the mineral samples (Fig. S7).
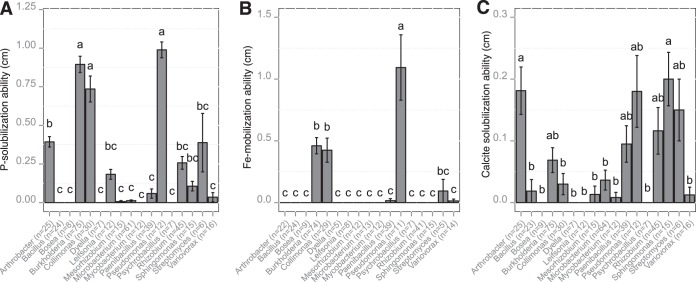
For each bacterial genus containing a minimum of five strains, we estimated the relative efficacies at solubilizing inorganic P and calcite and at mobilizing Fe (Fig. 6). Among these genera, representatives of Bosea, Dyella, and Psychrobacillus were not effective in any of the bioassays used (tricalcium phosphate [TCP], chrome azurol S [CAS] medium, or calcite medium [CAL medium]). In contrast, representatives of Burkholderia, Collimonas, and Pseudomonas harbored the highest efficacy at solubilizing inorganic P and at mobilizing Fe (Fig. 6A and B). Representatives of Arthrobacter solubilized significantly more P than several other genera (Variovorax, Paenibacillus, and Microbacterium) but were unable to mobilize Fe under our experimental conditions. For calcite, the most effective mineral-weathering bacteria were related to the Arthrobacter, Pseudomonas, and Sphingomonas genera (Fig. 6C). Interestingly, among the bacterial strains belonging to the Burkholderia genus, the most effective P-solubilizing strains were also those that were the most effective at mobilizing iron (Fig. S8). This trend was not observed for the Collimonas strains. Moreover, based on a one-factor ANOVA, we highlighted that the P and Fe weathering efficacies of the Burkholderia strains differed according to their origin. Indeed, Burkholderia strains coming from obsidian particles were significantly more effective at mineral weathering than those from the bulk soil (P < 0.05) (Fig. S9).
FIG 6.
Relationship between taxonomic belonging and functional efficacy. The averaged P, Fe, and calcite solubilization (in centimeters) were estimated for the major bacterial genera on TCP (A), CAS (B), and CAL (C) media, respectively. The number of strains tested for each bacterial genus and in each biotest is presented in parentheses. For the same genus, this number may vary between the different bioassays, due to the ability of the bacterial strains to grow on the different culture media. For each bioassay, different letters (a, b, and c) above the bars indicate significant differences between the bacterial genera (P < 0.05), according to a one-factor ANOVA and Tukey's multiple pairwise comparisons test (P < 0.05). The error bars indicate the standard errors of the means.
Comparison of taxonomic data obtained from the culture-dependent and -independent approaches revealed relatively good overlap. Indeed, 11 of the 30 most abundant bacterial genera identified through 16S rRNA gene pyrosequencing (10.4% of total reads) were also detected by the culture-dependent approach (Table S4) and were related to bacterial genera such as Burkholderia, Mycobacterium, Serratia, Phenylobacterium, and Streptomyces. In addition, the most abundant OTU obtained from the 16S rRNA gene-based pyrosequencing analysis and related to the genus Burkholderia shared strong sequence homologies (99 to 100%) with sequences recovered from the bacterial strains using Sanger sequencing technology (overlapping portion of 120 bases). This result suggests that major taxa identified in situ were successfully cultured and tested in this work for their mineral weathering abilities.
DISCUSSION
Mineral-associated bacterial communities and compartment (bulk soil versus mineral) effect.
Our 16S rRNA gene pyrosequencing analyses indicated that after a 2.5-year incubation period under different tree stands, obsidian, apatite, and calcite mineral particles harbor a high bacterial diversity on their surface, comparable to that of adjacent soils. Nevertheless, beta-diversity data revealed significant differences between the two compartments (BS versus mineral). At the OTU level, our results suggest that 25.2 to 32.0% of the top 500 OTUs were exclusively recovered from mineral surfaces. The nondetection of these OTUs in the BS compartment might be explained by insufficient sequence coverage and the fact that they probably represent rare species in the bulk soil. However, between 67.6 and 74.8% of the dominant OTUs (i.e., top 500) were shared between bulk soil and mineral-associated bacterial communities for all tree stands considered. These results contrast with those obtained by Hutchens and coworkers (37), who have demonstrated using an automated method of ribosomal intergenic spacer analysis (ARISA) that only 9% of the bacterial ribotypes were common to both minerals and soil compartments on pegmatite granite outcrops. In our study, the differentiation of mineral-associated bacterial communities from those of the surrounding bulk soil is one of the most significant differences among the samples. Notably, although the BS bacterial communities differed between the different tree stands, the separation of the BS and mineral-associated bacterial communities was conserved, highlighting an important mineral effect. The selective effect of minerals the soil bacterial communities is supported by other studies that were carried out on managed pasture and forest soils (30, 32–34). However, most of these previous studies relied exclusively on fingerprinting techniques (phospholipid-derived fatty acids [PLFA] and ARISA), cloning-sequencing that does not allow for conclusive taxonomic identification of individual phylotypes. Lepleux et al. (33) were the first to use 16S rRNA gene pyrosequencing on apatite particles incubated in forest soil but without biological replicates. In our study, we applied 16S rRNA gene pyrosequencing on replicate samples from the bulk soil and from different mineral types incubated under the same conditions. Our analyses revealed that Acidobacteria were the overall most abundant division detected in the bulk soil samples of each tree stand and accounted for 34 to 46% of the total reads. Although few representatives of this division are culturable (42), the ubiquity of Acidobacteria in soil, their abundant presence in acidic soils, and the recent results obtained by the 13C-based method (43) suggest that they are key players in soil biogeochemical cycles (44). Notably, while Acidobacteria are very abundant in the bulk soil compartment, the relative abundance of sequences assigned to Acidobacteria strongly decreases on mineral surfaces. Their low abundance was previously emphasized on rock substrate of the Matsch Valley, but no driving factor was identified (45). At the experimental site of Breuil-Chenue, the abundance of Acidobacteria was also shown to be lower in the rhizosphere or on the surfaces of minerals than in the surrounding bulk soil. Such results suggest that Acidobacteria are less competitive than other taxa or that the physicochemical conditions (e.g., increased pH) are not favorable to them. In contrast, representatives of Bacteroidetes, Actinobacteria, and Proteobacteria were significantly enriched in our study for all mineral types considered. Notably, these taxa have been repeatedly detected on mineral or rock surfaces such as granite, volcanic basaltic glass, marine basalts, or pure particles of apatite (33, 46–48), indicating that they may be adapted to colonize and inhabit rock and mineral surfaces in terrestrial and aquatic ecosystems.
Physicochemical properties of minerals affect soil bacterial communities.
The compositional differences of the bacterial communities observed between bulk soil and minerals can be explained by different factors. First, bacteria may accumulate on mineral surfaces due to passive diffusion and preferential water circulation. Another explanation is that the pores, fissures, cracks, concave surfaces, and little depressions present on the mineral surfaces ensure physical protection and selective advantages to bacteria and can be considered bacterial sanctuaries (49). In addition, bacterial adhesion on mineral surfaces was formerly shown to be determined by ionic strength and pH (41), suggesting that environmental conditions and geochemistry may influence the composition and attachment of bacterial communities. Despite these effects, our work demonstrated that the chemical composition and the intrinsic properties of the mineral substrates impact mineral-associated bacterial communities. Under each tree stand, calcite-associated bacterial communities significantly differed from the communities colonizing apatite and obsidian. Shifts in bacterial community composition according to mineralogy have been previously described for mica basalt and rock phosphate in managed pasture soil conditioned in microcosms (32); hematite, saprolite, and quartz in aquifers (50); quartz, plagioclase, potassium feldspar, and muscovite on an exposed pegmatite (51); or in artificial soils (52). Together, these results suggest that minerals may provide key nutritive or toxic elements that structure bacterial communities and thus represent bacterial microhabitats. This is particularly the case when minerals contain limiting nutrients that are absent or in very low concentrations in the surrounding environment. In this sense, Rogers and Bennett (39) found that bacteria preferentially colonized manufactured glasses containing apatite (contains P) or goethite (contains Fe) rather than glasses without these chemical elements. In contrast, Roberts (53) found that silicates containing more than 1.2% aluminum (Al) had less bacterial biomass than Al-poor silicates, indicating a possible Al toxicity or repellent effect on bacterial communities. In our study, we showed that 16S rRNA gene sequences assigned to Proteobacteria (Aquabacterium, Sphingomonas, Novosphingobium, Hyphomicrobium, Bosea, Afipia, Nitrosomonas, and Brevundimonas), Bacteroidetes (Ferruginibacter), and Nitrospira were notably enriched on calcite compared to apatite and obsidian. Notably, several of the bacterial phylotypes related to these taxa showed significant and positive correlations with the manganese contained in the mineral particles (Table S3). Manganese is known to affect a variety of biological processes, such as photosynthesis, carbon fixation, and scavenging of reactive oxygen species (ROS) (54–56). The potential benefits of Mn(II) oxidation are numerous, and a wide variety of microorganisms have been reported to catalyze this reaction (57). However, regarding whether the oxidation of Mn(II) to either Mn(III) or Mn(IV) is thermodynamically favorable, no direct evidence demonstrating that Mn oxidation supplies energy to microorganisms has been reported thus far. Apart from these phylotypes, ammonia-oxidizing bacteria (AOB) appeared also enriched on calcite surface. Notably, Nitrospira organisms were significantly enriched on calcite surfaces only below the beech and pine stands, while Nitrosospira organisms were significantly enriched only below the coppice stand. These data suggest a potential role of AOB in calcite dissolution and/or a better adaptation to the physicochemical conditions occurring on calcite surfaces. Notably, nitrifying bacteria have been proposed to play an important role in weathering of minerals (58) in other acid forest soils.
The weatherability of the different minerals considered in our study and their buffering capacity may also determine the structure and composition of the bacterial communities that develop on mineral surfaces and in the surrounding bulk soil. Indeed, minerals according to their physicochemical properties are more or less recalcitrant to environmental conditions (2). The abiotic experiments conducted in our study to highlight such differences in weatherability revealed that at pH values similar to those measured under field conditions of Breuil-Chenue (pH 4), calcite was the most weatherable mineral, followed by apatite and obsidian. In addition to the differences in weatherability, this study also highlighted that the carbonates released during calcite dissolution neutralized the protons of the acid solution provided, raising the pH of the outflow solution to neutral values (pH 7). The consumption of protons during the mineral weathering process is a well-known phenomenon which corresponds to the relative buffering capacity of each mineral type. Under our experimental conditions, such buffering capacity was not observed for obsidian and was observed for only the first days of incubation for apatite. These results suggest that calcite may locally impact its surrounding environment not only by releasing calcium but also by modifying local soil pH. Notably, large-scale studies have highlighted that soil pH is one of the main driving factors of belowground bacterial communities (59–61). As a consequence, the calcite dissolution occurring in the Breuil-Chenue forest soil likely modified the local pH, thereby indirectly promoting the development of bacterial communities adapted to neutral pH. This hypothesis fits very well with the results obtained for subsurface microbial communities incubated in laboratory reactors, for which neutrophilic bacterial populations have been shown to preferentially colonize highly buffering carbonates contained in mineral and rock surfaces (62).
Functional adaptation of mineral-associated bacteria.
Measuring the global functional diversity revealed that two monosaccharides derivatives of glucose (N-acetylglucosamine and β-methyl-d-glucoside) and one amino-acid (l-asparagine) were the most metabolized carbon sources for both mineral- and bulk soil-associated microbial communities. However, regarding the taxonomic differentiation that was observed for the bacterial communities according to their ecological origin, a functional differentiation was observed for the microbial communities occurring on minerals and in the surrounding bulk soil. Notably, mineral-associated microorganisms were found to metabolize a larger spectrum of carbon sources than those from the surrounding bulk soil. Such versatility in the use of carbon sources suggests a higher ecological fitness for the mineral-associated microbial communities than for the bulk soil, allowing them to adapt to the relatively oligotrophic conditions that occur on mineral surfaces compared to the surrounding BS. To our knowledge, functional analyses of soil mineral-associated bacterial communities carried out on pure and size-calibrated mineral particles are very rare (33). Nevertheless, at the field scale, the effect of mineral fertilizers on the microbial functional diversity has already been investigated on different soil types and revealed contrasting results of functional diversity. Hu et al. (63) demonstrated that mineral fertilization (NPK) treatment increased microbial functional diversity in soils. In contrast, Lepleux et al. (64) suggested that dolomite [CaMg(CO3)2] amendment of soil did not induce changes in functional diversity but rather induced a quantitative difference in the metabolism of l-asparagine, which was more metabolized in the amended soil treatment.
In addition to the use of a global bioassay (i.e., Biolog), the screening of the bacterial strains for their ability to mobilize inorganic nutrients revealed strong differences between the different mineral types and/or the surrounding bulk soil. Notably, effective P-solubilizing bacteria strains also appeared to be effective in producing siderophores for Fe scavenging. Among the mineral types, obsidian particles displayed the highest proportion of effective mineral weathering bacteria on their surface. Interestingly, this volcanic glass, mainly composed of SiO2, is also the mineral type characterized by the higher content of iron. This is also the most recalcitrant to weathering agents, and our abiotic experiment confirmed this point. As a consequence, our results suggest that the mineral-weathering ability represents a prerequisite for bacterial communities to colonize minerals in order to access the nutrients entrapped in their structure. Unlike the Bc and Cp stands, obsidian buried under the CwS stand did not harbor a higher proportion of effective weathering bacteria than did the surrounding bulk soil. This result indicates that the mineral weathering potential of bacterial communities may be influenced not only by mineral chemistry but also by the soil conditions (i.e., land cover effect). Vegetation cover is known to influence belowground bacterial communities through leaf litter degradation and root exudates (65, 66) and to modify the soil physicochemical parameters (67). However, our results suggest that this land cover effect did occur on all the mineral types. Indeed, for calcite particles, no significant enrichment in effective weathering bacteria was reported compared to the surrounding bulk soil. Such an absence of structure may be explained by the high weatherability of calcite and the potential variation of pH, which promoted the development of opportunistic bacteria in the vicinity of the weathered calcite particles. Of course, these results have to be moderated, as the cultivation-dependent approach gave only a partial view of the bacterial communities.
Relationships between mineral weathering potential and taxonomic identity.
A detailed analysis of the functional and taxonomic distribution of bacterial communities revealed a relationship between taxonomy and function. Indeed, our cultivation-dependent analysis revealed that representatives of the Burkholderia, Collimonas, and Pseudomonas genera were among the most effective at solubilizing P and mobilizing iron. Bacterial strains affiliated with these genera were essentially isolated from obsidian and apatite but not from calcite. In addition, our cultivation-independent approach highlighted a significant enrichment of 16S rRNA gene sequences related to Burkholderia and Collimonas on obsidian and apatite. Moreover, several effective mineral-weathering bacterial strains isolated in this study presented a strong homology of their 16S rRNA gene sequences with those of effective mineral-weathering bacterial strains previously isolated from the same experimental site in the rhizosphere or on mineral surfaces (33, 68, 69). Interestingly, significant positive correlations between the level of mineral dissolution and the relative abundance of 16S rRNA gene sequences assigned to the Betaproteobacteria class or to the Burkholderiales order were found for apatite particles incubated for 4 years in forest soil (33, 34). However, in our study, the shorter incubation time (2.5 years) did not allow us to measure mineral dissolution for apatite and obsidian, on which sequences related to the Burkholderiales and Burkholderia strains were enriched. As these bacteria are effective at weathering minerals, the facts that they are regularly found on minerals and are enriched on the most weathered minerals or under nutrient-poor conditions suggest that these bacteria are specially adapted to such environments and may be considered good functional markers for mineral weathering in situ. Such analyses should be extended to other soil microorganisms, such as archaea and fungi, to determine if specific patterns also exist for them.
Conclusions.
This study demonstrated that after a 2.5-year incubation, minerals incubated in nutrient-poor forest soils enrich specific bacterial communities on their surfaces compared to the surrounding bulk soil. The combination of culture-dependent and -independent analyses highlighted that this enrichment occurred at both taxonomical and functional levels, supporting the mineralosphere concept (25). Moreover, our results show that the characteristics of the mineral tested (i.e., chemical composition, weatherability, and capacity to modify their surrounding environment) should also be considered important drivers of bacterial communities. Although it is difficult to disentangle the relative effect of each characteristic on natural minerals, it is clear that the nutrients released during mineral dissolution and the modification of pH or ionic strength both determine the structure of bacterial communities, as evidenced in the case of calcite. The combination of culture-dependent and -independent analyses also highlighted that representatives of the Burkholderia and Collimonas genera, also found in the tree rhizosphere, were significantly enriched on apatite and obsidian surfaces and were very effective at weathering minerals, confirming their important role in nutrient cycling in acidic forest soils. Together, the new results obtained in this study show that minerals need to be considered reactive interfaces at both biological and chemical levels. These results also suggest that besides the chemistry and weatherability of minerals, their heterogeneous distribution in the soil matrix may partly explain the spatial variation of bacterial communities in soil.
MATERIALS AND METHODS
Site description and minerals.
The study was carried out in the forest LTO of Breuil-Chenue located in the Morvan region (47°18′N, 4°5′E, France). The native forest is a 150-year-old stand of coppice with standards (CwS) dominated by beech (Fagus sylvatica L.) and oak (Quercus sessiliflora Smith). The native forest was partially clear-felled and replaced in 1976 by monospecific plantations distributed in plots of 0.1 ha, such as beech (Fagus sylvatica L.) and Corsican pine (Pinus nigra Arn. subsp. laricio Poiret var. Corsicana). As the different plots are separated from the others by only few hundred meters, they share the same climatic conditions. The soil is classified as a Typic Dystrochrept (70) developed on the “Pierre qui Vire” granite. The bulk soil is sandy-loam textured (60% sands and <20% clays) and acidic (pHH2O ca. 4) (67).
To investigate the taxonomic and functional diversity of the bacterial communities colonizing the minerals, we selected three types of minerals with various chemistries: calcite, apatite, and obsidian. Pure minerals were obtained from Krantz (Bonn, Germany) and from the Compagnie Générale de Madagascar (Paris, France). The total chemical characteristics of each mineral type were determined by inductively coupled plasma spectrometry-atomic emission spectrometry (ICP-AES) for the major elements (SiO2 total ± 1%, Al2O3 total ± 1%, Fe2O3 total ± 2%, MgO total ± 5%, K2O total ± 5%, etc.) and by inductively coupled plasma spectrometry (ICP-MS) for trace elements (Zr total ± 8%, etc.) after using LiBO2 and dissolution by HNO3. Their compositions are presented in Table 3. The mineral types used are frequently found in soils, although apatite is encountered in the Breuil-Chenue granite as an accessory mineral, while obsidian and calcite are not naturally present. Pure crystals of apatite, calcite, and obsidian (initial size, ca. 2 cm) were ground, sonicated, washed, and calibrated (size, 100 to 200 μm). Three grams was introduced into nylon bags (mesh size, 50 μm; 4 by 10 cm) to prevent root colonization. On 13 June 2012, four replicate mesh bags of each mineral type were buried within the organomineral layer (5 to 15 cm) of the soil under coppice with standards (i.e., native forest, CwS) and under planted beech (Bc) and Corsican pine (Cp) stands. The different mesh bags were placed (approximately 10 cm apart one another) in an alternating manner (apatite/calcite/obsidian) along a 3-m transect.
TABLE 3.
Composition of the mineral particles testeda
| Mineral | % of indicated compound |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SiO2 | Al2O3 | Fe2O3 | MnO | MgO | CaO | Na2O | K2O | TiO2 | P2O5 | |
| Obsidian | 76.38 | 12.60 | 1.19 | 0.06 | 0.08 | 0.62 | 3.95 | 4.82 | 0.13 | <LD |
| Apatite | 0.81 | 0.10 | 0.10 | 0.03 | 0.02 | 52.64 | 0.23 | <LD | <LD | 41.26 |
| Calcite | 0.07 | 0.04 | <LD | 0.11 | <LD | 55.47 | <LD | <LD | <LD | 0.11 |
The total chemical characteristics of each mineral type were determined by ICP-AES for the major elements. The data present the relative composition of each mineral type. <LD, value was below the detection limit.
Mineral weatherability.
The weatherability of obsidian, apatite, and calcite was determined in flowthrough reactors for 42 days at 20°C. This exposure time was necessary to obtain a stoichiometric dissolution of the three minerals tested. For each mineral type, 3 g of 100-μm mineral particles was placed in a reactor and maintained by using 0.5-μm nylon filters placed at each extremity of the reactor. The reactors were placed on a rack that was continuously shaken (180° swing; frequency = 2 s) to homogenize mineral particles in the column (Fig. S1). Abiotic mineral weathering was performed with hydrochloric acid (HCl) at pHH2O 4, which is representative of the pH values measured in the soil of the forest LTO of Breuil-Chenue. The flow (Q) of HCl solution supplied in each reactor was fixed at 2 ml · h−1 using a peristaltic pump (ISM444; Ismatec) with Tygon R3607 0.13-mm-inner-diameter tubing (catalog no. 070535-00–/.AME00-SC0189; Ismatec). The volume of the column reactors was ca. 15 ml and led to a residence time of the HCl solution in the reactor of 7.5 h. Prior to starting the experiment, the entire experimental setup was cleaned by a continuous flow of HCl solution for 3 days. Three independent replicate reactors were used for each mineral type. During the experiment, the outflowing solutions were collected at days 0 to 42 at regular intervals, and samples were stored at 4°C in previously acid-washed polypropylene bottles. The concentrations of the major elements were determined by ICP-AES, and pHH2O values of the outflowing solutions were estimated. The amount (in milligrams) of each major element accumulated during the experiment was calculated. For each mineral type and each reactor, mineral particles were dried at 60°C; the mass of particles remaining after the dissolution experiment was determined by weighing (dry weight), and the relative mass loss was calculated (final weight/initial weight × 100).
Sampling and soil analyses.
On 4 November 2014 (2 years and 5 months later), all the mesh bags and the biological replicates of the surrounding bulk soil (BS) were sampled. Briefly, the litter layer was removed, and the mesh bags and the soil found in the soil horizon were collected (5 to 15 cm). Three spatially distant samples of the surrounding BS samples were collected per tree stand. These BS samples were collected at 10 cm of the mesh bags to avoid an impact of mineral dissolution on the soil properties and microbial communities. All the samples were then transported to the laboratory. Mesh bags (n = 36) were opened, and the mass of particles of each mesh bag was determined by weighing (wet weight). In addition, the relative mass loss was calculated (final weight/initial weight × 100). All the BS samples (n = 3) were sieved (mesh size 2 mm) and homogenized. A subsample of each mineral sample was used to perform electron microscopy imaging. For each sample (BS or mineral particles), 0.25 g was suspended in 1 ml of sterile ultrapure water mixed with 40% glycerol, vortexed for 5 min, and cryopreserved at −80°C for cultivation-dependent analyses. Bulk soil samples were chemically characterized at the Laboratoire d'Analyze des Sols (Arras, France). The cation exchange capacity (CEC) was determined using the cobaltihexamine method. Titration of the cobaltihexamine chloride soil extract was performed at 472 nm and compared to a reference of 0.05 N cobaltihexamine chloride extract. The pH was determined by the water method using a soil/water ratio of 1:5 (wt/vol). Total carbon (C) and total nitrogen (N) contents (both obtained after combustion at 1,000°C) and phosphorus (P) content were determined according to methods published by Duchaufour and Bonneau (71), Duval (72), and Olsen (73). Exchangeable cations (Ca, Mg, Na, K, Fe, Mn, and Al) and H+ were extracted using cobaltihexamine, and their concentrations were determined by ICP-AES for cations and by potentiometric measurement using 0.05 M KOH for protons.
DNA extraction, 16S rRNA gene PCR, and pyrosequencing.
Genomic DNA was extracted from 0.25 g of bulk soil and mineral particles using a PowerMax soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) as recommended by the manufacturer. DNA purity and concentration were assessed using a NanoDrop 1000 spectrometer (NanoDrop Technologies, Wilmington, DE). The 16S rRNA gene amplicon libraries were generated in one step, using primers 799F (5′-AACMGGATTAGATACCCKG-3′) and 1115R (3′-AGGGTTGCGCTCGTTG-3′) (74, 75) containing the specific Roche 454-pyrosequencing adaptors and 5-base barcodes. Primer 799F was associated with the adaptor sequence A (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′) and primer 1115R to the adaptor sequence B (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG). PCR mixtures contained 1× PCR master mix (5 PRIME), 500 nM 799f primer, 500 nM 1115r primer, and 8 ng of DNA in a final volume of 50 μl. For each sample, 3 independent PCRs were performed. Amplifications were performed using the following cycle parameters: 95°C for 5 min (initial denaturation), followed by 30 cycles of 95°C for 30 s, 57°C for 35 s, and 72°C for 30 s with a final extension step at 72°C for 10 min. The PCR products were checked by gel electrophoresis and were purified by using the QIAquick purification kit (Qiagen, Valencia, CA) as recommended by the manufacturer. The concentration of each purified PCR product was measured using a NanoDrop 1000 spectrometer (NanoDrop Technologies), and an equimolar mix of all 45 amplicon libraries was used for pyrosequencing. Pyrosequencing was performed by Beckman Coulter genomics (Danvers, MA) on an FLX 454 Titanium System genome sequencer (GS) (Roche). After pyrosequencing, a total of 537,776 reads of 16S rRNA gene sequences were obtained from the 45 samples. Reads were filtered for length (>300 bp), quality score (mean, ≥25), number of ambiguous bases (0), and length of homopolymer runs (<8) using the trim.seqs script in mothur v.1.30.2 (76). High-quality sequences were aligned, and the operational taxonomic units (OTUs) were defined with a 3% dissimilarity level. Chimeric sequences were detected using the chimera.uchime command and were removed from further analysis. To avoid any biases associated with different numbers of sequences in each of the samples, we randomly subsampled a total of 7,560 sequences (corresponding to the smaller set of sequences after mothur processing) from each sample, which was used for all downstream analysis. Taxonomy was assigned to each OTU by aligning sequences against the RDP alignment database with a bootstrap value of 80 for taxonomic assignment.
Substrate utilization assays.
To compare the substrate utilization potentials of soil microbial communities inhabiting the surface of mineral particles and those of the surrounding bulk soil, we used Biolog EcoPlates (31 different carbon substrates; Biolog Inc., Hayward, CA) on all samples (n = 45). Briefly, 0.25 g of mineral particles or bulk soil was vortexed for 5 min in 5 ml of sterile ultrapure water. These suspensions were used to inoculate 150 μl in each well of the microplates. Optical density (OD) was measured after 48 h of incubation at 25°C at 595 nm with an automatic microplate reader (Bio-Rad; model iMark). The OD at 595 nm (OD595) of the control well (containing no carbon source) was subtracted from the OD595 of each of the other wells. These corrected values were used for the subsequent analysis. The average well color development (AWCD) reflecting the total ability of microorganisms to use carbon substrates was determined according to the formula AWCD = Σ(Ci − R)/n, where Ci corresponds to the OD measured for each substrate, R corresponds to OD of the control well, and n is the total number of the sole carbon substrates (EcoPlates; n = 31) (77). In addition, substrates were subdivided into five groups: carbohydrates (CH), carboxylic and ketonic acids (CA), amines and amides (AM), amino acids, and polymers (PL) (78). For each group, the values of all substrates were averaged and standard errors of the means (SEMs) were calculated.
Isolation of bacterial strains and phylogenetic assignment.
For each tree stand, a bacterial collection was generated from the BS and mineral particle samples collected under the three tree stands considered. Briefly, 0.25 g of soil or mineral particles stored at −80°C in 40% glycerol was suspended in 20 ml of sterile Milli-Q water and homogenized by vortexing. These suspensions were then used to perform serial dilutions. Dilutions were spread onto a 1/10 tryptic soy agar (TSA) medium (tryptic soil broth from Difco, 3 g · liter−1, and agar, 15 g · liter−1). Plates were incubated for 5 days at 25°C, and bacterial concentration was estimated by plate counting. For each sample, a total of 15 bacterial isolates were picked up from the same dilution (10−3), giving a total of 45 isolates per sample type and tree stand. Each bacterial isolate was purified by three successive plating on 1/10 TSA and then stored at −80°C in LB medium supplemented with 20% glycerol. Phylogenetic assignment of bacterial isolates was carried out by sequence analysis of the 16S rRNA gene. A fragment of ca. 900 bp was PCR amplified using the universal set of primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and 907r (5′-CCGTCAATTCMTTTGAGTTT-3′) (79, 80). The reactions were performed using the following cycling parameters: one step of 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 54°C for 1 min, and 72°C for 1 min 30 s, and a final extension step of 10 min at 72°C. PCR products were then purified by using a QIAquick purification kit (Qiagen, Valencia, CA) as recommended by the manufacturer. PCR products were checked by gel electrophoresis, and their concentration was estimated using a NanoDrop 1000 spectrometer (NanoDrop Technologies). Last, PCR products were sequenced using the Sanger method at Eurofins MWG (Ebersberg, Germany).
Mineral weathering assays.
The mineral weathering potential of bacterial isolates was assessed using two bioassays measuring the abilities (i) to mobilize iron and (ii) to solubilize tricalcium orthophosphate. The ability to mobilize iron was tested by following the protocol of Frey-Klett et al. (81) using chrome azurol S (CAS) medium. This bioassay is used to highlight siderophore production (82). This medium contains, per liter, 800 ml of solution 1 {34.36 g of PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] Na2 buffer [Sigma], 0.3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 0.246 g of MgSO4·7H2O, 0.0147 g of CaCl2·2H2O, and 15 g of agar in 800 ml of H2O; pH 6.8}, 100 ml of solution 2 (4 g of glucose in 100 ml of H2O), and 100 ml of solution 3 (mixture of 0.0905 g of chrome azurol S in 75 ml, 0.0024 g of FeCl3 in 15 ml, and 0.1640 g of hexa-decyl trimethyl ammonium bromide in 60 ml). Solutions 2 and 3 are added after autoclaving, at a temperature of ca. 70°C. The ability to solubilize inorganic phosphorous was tested using a bioassay with 1/10 diluted tricalcium phosphate (TCP), according to the method of Lepleux et al. (64). This bioassay allows the detection of bacterial strains producing organic acids and protons. This medium contains, per liter of distilled water, 0.5 g of NH4Cl, 0.1 g of NaCl, 0.1 g of MgSO4·7H2O, 1 g of glucose, 0.4 g of Ca3(PO4)2, and 15 g of agar. These two bioassays are commonly used to determine the mineral weathering potential of bacterial strains (64, 83, 84). As the mineral-associated bacterial strains were isolated from different mineral types, adaptation of the TCP bioassay was done using pure mineral particles of apatite, calcite, or obsidian instead of Ca3(PO4)2. For each mineral type (obsidian, apatite, and calcite), size fractions of <50 μm and <100 μm have been tested, introducing, per liter of medium, 0.4, 0.8, and 1.2 g of these mineral fractions. Under our experimental conditions, only calcite allowed to the development of measurable dissolution haloes. The CAL medium contains, per liter of distilled water, 1.2 g of calcite, 1 g of glucose, 0.1 g of MgSO4, 0.1 g of NaCl, 0.5 g of NH4Cl, and 15 g of agar.
For each bioassay, bacterial inoculum preparation was the same. Briefly, bacterial isolates were cultivated for 48 h at 25°C in liquid LB medium. Two milliliters of each culture was then centrifuged and washed three times with sterile ultrapure water and suspended in 2 ml of sterile ultrapure water. The absorbance at 595 nm of each resulting suspension was adjusted at an OD595 of 0.8 (ca. 108 cell·ml−1). Five microliters of inoculum was dropped in triplicates on the surface of agar medium of each bioassay. After incubation at 25°C for 7 days for TCP and CAS media and 14 days for CAL medium, clear haloes surrounding bacterial colonies were measured and averaged. Based on the colony diameter (Cdiam) and the halo diameter (Hdiam), a solubilization index was calculated (SI = Hdiam − Cdiam). As mineral solubilization may be related to the production of acidifying metabolites by the bacterial isolates, we tested a subsample of the bacterial collection under the same culture conditions as in the TCP bioassay, with some modifications. Briefly, acidification was assessed in liquid TCP medium lacking tricalcium orthophosphate (TCPm). Microplates containing 180 μl of liquid TCPm medium were inoculated with 20 μl of bacterial inoculum in triplicates. After incubation for 5 days at 25°C, 180 μl of culture supernatant was recovered in a new microplate containing 20 μl of bromocresol green (1 g·liter−1; Sigma). The pH of the culture medium was determined at 595 nm with an automatic microplate reader (Bio-Rad; model iMark) according to the method of Uroz et al. (85). The average values of the 3 replicates for pH measurements were taken as the acidification potential. For each series, the bacterial strain M2R3 (Collimonas fungivorans) was used as a positive control.
Statistical analyses.
The effects of the compartment type (mineral versus BS), mineral type (apatite, calcite, or obsidian), or soil conditions (beech stands, CwS, and Corsican pine stands) were determined for the Biolog and 16S rRNA pyrosequence data by one-factor ANOVA followed by Tukey's post hoc tests. For the 16S rRNA gene pyrosequence data, the relative abundances of taxa (from OTU to class) were transformed using the arcsine square root to achieve a normal distribution and to allow ANOVA. Moreover, alpha diversity estimates were calculated for the taxonomic and functional data set using the R package vegan (86). The effect of the tree species on the soil chemical parameters was also determined by one-factor ANOVA followed by Tukey's post hoc tests. Multivariate analyses were conducted on the 16S rRNA gene pyrosequence, and analyses of similarity (ANOSIM) were performed to test for differences within groups, based on Bray-Curtis distances. Both analyses were done at class to OTU levels. The FactoMineR package (87) was used to perform all the multivariate analyses. Mantel tests (1,000 permutations, Pearson correlations) were performed to test correlations between mineral composition and pyrosequencing data based on Bray-Curtis dissimilarity matrices, using the ade4 package in R (88). The relationship between the relative amount of calcium released during the mineral weatherability assays and the relative mass loss was determined using Pearson correlation. Last, variance partitioning was performed to determine the proportion of variation in 16S rRNA gene pyrosequences, explained by each set of explanatory variables, namely, (i) compartment type (bulk soil and mineral), (ii) mineral type (obsidian, apatite, and calcite), (iii) soil conditions (beech stands, coppice with standards, and Corsican pine stands), and (iv) unexplained variance. For mineral weathering assays achieved on TCP, calcite medium, and CAS media, frequencies of distribution of effective weathering bacteria were compared by a χ2 test (P < 0.05) using the NCStats package in R, while the relative efficacies of bacterial isolates to weather minerals were compared by one-factor ANOVA followed by Tukey's post hoc tests.
Accession number(s).
The 454 pyrosequencing data generated for this study were submitted to the Sequence Read Archive (SRA) and are available under Bioproject no. PRJNA326237. The partial 16S rRNA gene sequences of the strains isolates determined in this study have been deposited in the GenBank database under accession numbers KX418770 to KX419241.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Laboratory of Excellence Arbre (ANR-11-LABX-0002-01) in the frame of the multidisciplinary project INABACT and the ANR JC BACTOWEATHER (ANR-11-JSV7-0001). Y. Colin is a postdoctoral scientist supported by grants from ANR, ANDRA, and Lorraine Region. O. Nicolitch is a PhD student supported by a fellowship from the Laboratory of Excellence Arbre (ANR-11-LABX-0002-01).
We thank A. Legout for helpful comments and for giving us access to the site of Breuil-Chenue. We thank C. Rose from the certified facility in Functional Ecology (PTEF OC 081) from UMR 1137 EEF and UR 1138 BEF for imaging mineral particles. The UMR1136 and UR1138 as well as the PTEF platform are supported by the French Agency through the Laboratory of Excellence Arbre (ANR-11-LABX-0002-01). We thank C. Calvaruso and C. Cochet, C. Pantigny and M. Beuret for helpful discussion and for technical assistance. The English language was reviewed by American Journal Experts (certificate A626-4389-7F05-178A-F8FC). We thank the four reviewers for their helpful comments to improve and clarify our manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02684-16.
REFERENCES
- 1.Uroz S, Oger P, Lepleux C, Collignon C, Frey-Klett P, Turpault M-P. 2011. Bacterial weathering and its contribution to nutrient cycling in temperate forest ecosystems. Res Microbiol 162:820–831. doi: 10.1016/j.resmic.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Brantley SL, Kubicki JD, White AF. 2008. Kinetics of water-rock interaction. Springer New York, New York, NY. [Google Scholar]
- 3.Augusto L, Turpault M-P, Ranger J. 2000. Impact of forest tree species on feldspar weathering rates. Geoderma 96:215–237. doi: 10.1016/S0016-7061(00)00021-5. [DOI] [Google Scholar]
- 4.Banfield JF, Barker WW, Welch SA, Taunton A. 1999. Biological impact on mineral dissolution: application of the lichen model to understanding mineral weathering in the rhizosphere. Proc Natl Acad Sci U S A 96:3404–3411. doi: 10.1073/pnas.96.7.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landeweert R, Hoffland E, Finlay RD, Kuyper TW, van Breemen N. 2001. Linking plants to rocks: ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol Evol 16:248–254. doi: 10.1016/S0169-5347(01)02122-X. [DOI] [PubMed] [Google Scholar]
- 6.Smits MM, Bonneville S, Benning LG, Banwart SA, Leake JR. 2012. Plant-driven weathering of apatite—the role of an ectomycorrhizal fungus. Geobiology 10:445–456. doi: 10.1111/j.1472-4669.2012.00331.x. [DOI] [PubMed] [Google Scholar]
- 7.Uroz S, Calvaruso C, Turpault M-P, Frey-Klett P. 2009. Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol 17:378–387. doi: 10.1016/j.tim.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Bonneville S, Smits MM, Brown A, Harrington J, Leake JR, Brydson R, Benning LG. 2009. Plant-driven fungal weathering: early stages of mineral alteration at the nanometer scale. Geology 37:615–618. doi: 10.1130/G25699A.1. [DOI] [Google Scholar]
- 9.Jongmans AG, Van Breemen N, Lundström U, Van Hees PAW, Finlay RD, Srinivasan M, Unestam T, Giesler R, Melkerud P-A, Olsson M. 1997. Rock-eating fungi. Nature 389:682–683. doi: 10.1038/39493. [DOI] [Google Scholar]
- 10.Barker WW, Welch SA, Banfield JF. 1997. Biogeochemical weathering of silicate minerals. Rev Mineral Geochem 35:391–428. [Google Scholar]
- 11.Hinsinger P. 2001. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237:173–195. doi: 10.1023/A:1013351617532. [DOI] [Google Scholar]
- 12.Liermann LJ, Kalinowski BE, Brantley SL, Ferry JG. 2000. Role of bacterial siderophores in dissolution of hornblende. Geochim Cosmochim Acta 64:587–602. doi: 10.1016/S0016-7037(99)00288-4. [DOI] [Google Scholar]
- 13.Drever JI, Stillings LL. 1997. The role of organic acids in mineral weathering. Colloids Surf A Physicochem Eng Asp 120:167–181. [Google Scholar]
- 14.Calvaruso C, Mareschal L, Turpault M-P, Leclerc E. 2009. Rapid clay weathering in the rhizosphere of Norway spruce and oak in an acid forest ecosystem. Soil Sci Soc Am J 73:331–338. doi: 10.2136/sssaj2007.0400. [DOI] [Google Scholar]
- 15.Rengel Z, Marschner P. 2005. Nutrient availability and management in the rhizosphere: exploiting genotypic differences. New Phytol 168:305–312. doi: 10.1111/j.1469-8137.2005.01558.x. [DOI] [PubMed] [Google Scholar]
- 16.Turpault M-P, Claude N, Christophe C. 2009. Rhizosphere impact on the dissolution of test minerals in a forest ecosystem. Geoderma 153:147–154. doi: 10.1016/j.geoderma.2009.07.023. [DOI] [Google Scholar]
- 17.Robert M, Berthelin J. 1986. Role of biological and biochemical factors in soil mineral weathering, p 453–495. In Huang PM, Schnitzer M (ed), Interactions of soil minerals with natural organics and microbes. Soil Science Society of America, Madison, WI. [Google Scholar]
- 18.Calvaruso C, Turpault M-P, Frey-Klett P. 2006. Root-associated bacteria contribute to mineral weathering and to mineral nutrition in trees: a budgeting analysis. Appl Environ Microbiol 72:1258–1266. doi: 10.1128/AEM.72.2.1258-1266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gadd GM. 2010. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 20.Koele N, Turpault M-P, Hildebrand EE, Uroz S, Frey-Klett P. 2009. Interactions between mycorrhizal fungi and mycorrhizosphere bacteria during mineral weathering: budget analysis and bacterial quantification. Soil Biol Biochem 41:1935–1942. doi: 10.1016/j.soilbio.2009.06.017. [DOI] [Google Scholar]
- 21.Toro M, Azcon R, Barea J. 1997. Improvement of arbuscular mycorrhiza development by inoculation of soil with phosphate-solubilizing rhizobacteria to improve rock phosphate bioavailability (32P) and nutrient cycling. Appl Environ Microbiol 63:4408–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallander H. 2000. Uptake of P from apatite by Pinus sylvestris seedlings colonised by different ectomycorrhizal fungi. Plant Soil 218:249–256. [Google Scholar]
- 23.Leyval C, Berthelin J. 1991. Weathering of a mica by roots and rhizospheric microorganisms of pine. Soil Sci Soc Am J 55:1009–1016. doi: 10.2136/sssaj1991.03615995005500040020x. [DOI] [Google Scholar]
- 24.Totsche KU, Rennert T, Gerzabek MH, Kögel-Knabner I, Smalla K, Spiteller M, Vogel H-J. 2010. Biogeochemical interfaces in soil: the interdisciplinary challenge for soil science. J Plant Nutr Soil Sc 173:88–99. doi: 10.1002/jpln.200900105. [DOI] [Google Scholar]
- 25.Uroz S, Kelly LC, Turpault M-P, Lepleux C, Frey-Klett P. 2015. The mineralosphere concept: mineralogical control of the distribution and function of mineral-associated bacterial communities. Trends Microbiol 23:751–762. doi: 10.1016/j.tim.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Barton H, Taylor N, Kreate M, Springer A, Oehrle S, Bertog J. 2007. The impact of host rock geochemistry on bacterial community structure in oligotrophic cave environments. Int J Speleol 36:93–104. doi: 10.5038/1827-806X.36.2.5. [DOI] [Google Scholar]
- 27.Bennett PC, Hiebert FK, Choi WJ. 1996. Microbial colonization and weathering of silicates in a petroleum-contaminated groundwater. Chem Geol 132:45–53. doi: 10.1016/S0009-2541(96)00040-X. [DOI] [Google Scholar]
- 28.Gorbushina AA. 2007. Life on the rocks. Environ Microbiol 9:1613–1631. doi: 10.1111/j.1462-2920.2007.01301.x. [DOI] [PubMed] [Google Scholar]
- 29.Mauck BS, Roberts JA. 2007. Mineralogic control on abundance and diversity of surface-adherent microbial communities. Geomicrobiol J 24:167–177. doi: 10.1080/01490450701457162. [DOI] [Google Scholar]
- 30.Certini G, Campbell CD, Edwards AC. 2004. Rock fragments in soil support a different microbial community from the fine earth. Soil Biol Biochem 36:1119–1128. doi: 10.1016/j.soilbio.2004.02.022. [DOI] [Google Scholar]
- 31.Carson JK, Rooney D, Gleeson DB, Clipson N. 2007. Altering the mineral composition of soil causes a shift in microbial community structure. FEMS Microbiol Ecol 61:414–423. doi: 10.1111/j.1574-6941.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- 32.Carson JK, Campbell L, Rooney D, Clipson N, Gleeson DB. 2009. Minerals in soil select distinct bacterial communities in their microhabitats. FEMS Microbiol Ecol 67:381–388. doi: 10.1111/j.1574-6941.2008.00645.x. [DOI] [PubMed] [Google Scholar]
- 33.Lepleux C, Turpault MP, Oger P, Frey-Klett P, Uroz S. 2012. Correlation of the abundance of betaproteobacteria on mineral surfaces with mineral weathering in forest soils. Appl Environ Microbiol 78:7114–7119. doi: 10.1128/AEM.00996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uroz S, Turpault MP, Delaruelle C, Mareschal L, Pierrat J-C, Frey-Klett P. 2012. Minerals affect the specific diversity of forest soil bacterial communities. Geomicrobiol J 29:88–98. doi: 10.1080/01490451.2010.523764. [DOI] [Google Scholar]
- 35.Bennett PC, Rogers JR, Choi WJ, Hiebert FK. 2001. Silicates, silicate weathering, and microbial ecology. Geomicrobiol J 18:3–19. doi: 10.1080/01490450151079734. [DOI] [Google Scholar]
- 36.Gleeson DB, Clipson N, Melville K, Gadd GM, McDermott FP. 2005. Characterization of fungal community structure on a weathered pegmatitic granite. Microb Ecol 50:360–368. doi: 10.1007/s00248-005-0198-8. [DOI] [PubMed] [Google Scholar]
- 37.Hutchens E, Gleeson D, McDermott F, Miranda-CasoLuengo R, Clipson N. 2010. Meter-scale diversity of microbial communities on a weathered pegmatite granite outcrop in the Wicklow Mountains, Ireland; evidence for mineral induced selection? Geomicrobiol J 27:1–14. doi: 10.1080/01490450903232157. [DOI] [Google Scholar]
- 38.Kelly LC, Colin Y, Turpault MP, Uroz S. 2016. Mineral type and solution chemistry affect the structure and composition of actively growing bacterial communities as revealed by bromodeoxyuridine immunocapture and 16S rRNA pyrosequencing. Microb Ecol 72:428–442. doi: 10.1007/s00248-016-0774-0. [DOI] [PubMed] [Google Scholar]
- 39.Rogers JR, Bennett PC. 2004. Mineral stimulation of subsurface microorganisms: release of limiting nutrients from silicates. Chem Geol 203:91–108. doi: 10.1016/j.chemgeo.2003.09.001. [DOI] [Google Scholar]
- 40.Rogers JR, Bennett PC, Choi WJ. 1998. Feldspars as a source of nutrients for microorganisms. Am Mineral 83:1532–1540. doi: 10.2138/am-1998-11-1241. [DOI] [Google Scholar]
- 41.Scholl MA, Mills AL, Herman JS, Hornberger GM. 1990. The influence of mineralogy and solution chemistry on the attachment of bacteria to representative aquifer materials. J Contam Hydrol 6:321–336. doi: 10.1016/0169-7722(90)90032-C. [DOI] [Google Scholar]
- 42.Lladó S, Žifčáková L, Větrovsky̌ Eichlerová TI, Baldrian P. 2016. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol Fert Soils 52:251–260. doi: 10.1007/s00374-015-1072-6. [DOI] [Google Scholar]
- 43.Eichorst SA, Kuske CR. 2012. Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl Environ Microbiol 78:2316–2327. doi: 10.1128/AEM.07313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catão EC, Lopes FA, Araújo JF, de Castro AP, Barreto CC, Bustamante M, Quirino BF, Krüger RH. 2014. Soil acidobacterial 16S rRNA gene sequences reveal subgroup level differences between savanna-like cerrado and Atlantic forest Brazilian biomes. Int J Microbiol 2014:56341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito A, Ahmed E, Ciccazzo S, Sikorski J, Overmann J, Holmström SJ, Brusetti L. 2015. Comparison of rock varnish bacterial communities with surrounding non-varnished rock surfaces: taxon-specific analysis and morphological description. Microb Ecol 70:741–750. doi: 10.1007/s00248-015-0617-4. [DOI] [PubMed] [Google Scholar]
- 46.Lapanje A, Wimmersberger C, Furrer G, Brunner I, Frey B. 2012. Pattern of elemental release during the granite dissolution can be changed by aerobic heterotrophic bacterial strains isolated from Damma glacier (Central Alps) deglaciated granite sand. Microb Ecol 63:865–882. doi: 10.1007/s00248-011-9976-7. [DOI] [PubMed] [Google Scholar]
- 47.Cockell CS, Olsson K, Knowles F, Kelly L, Herrera A, Thorsteinsson T, Marteinsson V. 2009. Bacteria in weathered basaltic glass, Iceland. Geomicrobiol J 26:491–507. doi: 10.1080/01490450903061101. [DOI] [Google Scholar]
- 48.Mason OU, Di Meo-Savoie CA, Van Nostrand JD, Zhou J, Fisk MR, Giovannoni SJ. 2009. Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts. ISME J 3:231–242. doi: 10.1038/ismej.2008.92. [DOI] [PubMed] [Google Scholar]
- 49.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 50.Boyd ES, Cummings DE, Geesey GG. 2007. Mineralogy influences structure and diversity of bacterial communities associated with geological substrata in a pristine aquifer. Microb Ecol 54:170–182. doi: 10.1007/s00248-006-9187-9. [DOI] [PubMed] [Google Scholar]
- 51.Gleeson D, McDermott F, Clipson N. 2006. Structural diversity of bacterial communities in a heavy metal mineralized granite outcrop. Environ Microbiol 8:383–393. doi: 10.1111/j.1462-2920.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 52.Hemkemeyer M, Pronk GJ, Heister K, Kögel-Knabner I, Martens R, Tebbe CC. 2014. Artificial soil studies reveal domain-specific preferences of microorganisms for the colonisation of different soil minerals and particle size fractions. FEMS Microbiol Ecol 90:770–782. doi: 10.1111/1574-6941.12436. [DOI] [PubMed] [Google Scholar]
- 53.Roberts JA. 2004. Inhibition and enhancement of microbial surface colonization: the role of silicate composition. Chem Geol 212:313–327. doi: 10.1016/j.chemgeo.2004.08.021. [DOI] [Google Scholar]
- 54.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, Foster SJ. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol 44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 55.Kehres DG, Maguire ME. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Ecol 27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 56.Tebo BM, Johnson HA, McCarthy JK, Templeton AS. 2005. Geomicrobiology of manganese (II) oxidation. Trends Microbiol 13:421–428. doi: 10.1016/j.tim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 57.Das AP, Sukla LB, Pradhan N, Nayak S. 2011. Manganese biomining: a review. Bioresour Technol 102:7381–7387. doi: 10.1016/j.biortech.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 58.Berthelin J, Bonne M, Belgy G, Wedraogo FX. 1985. A major role for nitrification in the weathering of minerals of brown acid forest soils. Geomicrobiol J 4:175–190. doi: 10.1080/01490458509385930. [DOI] [Google Scholar]
- 59.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 61.Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 62.Jones AA, Bennett PC. 2014. Mineral microniches control the diversity of subsurface microbial populations. Geomicrobiol J 31:246–261. doi: 10.1080/01490451.2013.809174. [DOI] [Google Scholar]
- 63.Hu J, Lin X, Wang J, Dai J, Chen R, Zhang J, Wong MH. 2011. Microbial functional diversity, metabolic quotient, and invertase activity of a sandy loam soil as affected by long-term application of organic amendment and mineral fertilizer. J Soils Sediments 11:271–280. doi: 10.1007/s11368-010-0308-1. [DOI] [Google Scholar]
- 64.Lepleux C, Uroz S, Collignon C, Churin J-L, Turpault M-P, Frey-Klett P. 2013. A short-term mineral amendment impacts the mineral weathering bacterial communities in an acidic forest soil. Res Microbiol 164:729–739. doi: 10.1016/j.resmic.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 65.Prescott CE, Grayston SJ. 2013. Tree species influence on microbial communities in litter and soil: current knowledge and research needs. Forest Ecol Manag 309:19–27. doi: 10.1016/j.foreco.2013.02.034. [DOI] [Google Scholar]
- 66.Xu W, Shi L, Chan O, Li J, Casper P, Zou X. 2013. Assessing the effect of litter species on the dynamic of bacterial and fungal communities during leaf decomposition in microcosm by molecular techniques. PLoS One 8:e84613. doi: 10.1371/journal.pone.0084613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mareschal L, Bonnaud P, Turpault MP, Ranger J. 2010. Impact of common European tree species on the chemical and physicochemical properties of fine earth: an unusual pattern. Eur J Soil Sci 61:14–23. doi: 10.1111/j.1365-2389.2009.01206.x. [DOI] [Google Scholar]
- 68.Collignon C, Uroz S, Turpault MP, Frey-Klett P. 2011. Seasons differently impact the structure of mineral weathering bacterial communities in beech and spruce stands. Soil Biol Biochem 43:2012–2022. doi: 10.1016/j.soilbio.2011.05.008. [DOI] [Google Scholar]
- 69.Calvaruso C, Turpault M-P, Leclerc E, Ranger J, Garbaye J, Uroz S, Frey-Klett P. 2010. Influence of forest trees on the distribution of mineral weathering-associated bacterial communities of the Scleroderma citrinum mycorrhizosphere. Appl Environ Microbiol 76:4780–4787. doi: 10.1128/AEM.03040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.USDA. 1999. Soil taxonomy: a basic system of soil classification for making and interpreting soil surveys, 2nd ed US Government Printing Office, Washington, DC. [Google Scholar]
- 71.Duchaufour P, Bonneau M. 1959. Une nouvelle méthode de dosage du phosphore assimilable dans les sols forestiers. Bull Afes 4:193–198. [Google Scholar]
- 72.Duval L. 1963. Etude des conditions de validité du dosage céruléomolybdique de l'acide phosphorique. Conséquences pratiques. Chim Anal 45:237–250. [Google Scholar]
- 73.Olsen SR. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. US Department of Agriculture, Washington, DC. [Google Scholar]
- 74.Chelius MK, Triplett EW. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L Microb Ecol 41:252–263. [DOI] [PubMed] [Google Scholar]
- 75.Reysenbach AL, Pace NR. 1995. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction, p 101–107. In Robb FT, Place AL (ed), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 76.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garland JL, Mills AL. 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber KP, Legge RL. 2009. One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J Microbiol Methods 79:55–61. doi: 10.1016/j.mimet.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 79.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–176. In Stackebrandt E, Goodfellow H (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom. [Google Scholar]
- 81.Frey-Klett P, Chavatte M, Clausse M-L, Courrier S, Roux CL, Raaijmakers J, Martinotti MG, Pierrat J-C, Garbaye J. 2005. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol 165:317–328. [DOI] [PubMed] [Google Scholar]
- 82.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 83.Calvaruso C, Turpault M-P, Leclerc E, Frey-Klett P. 2007. Impact of ectomycorrhizosphere on the functional diversity of soil bacterial and fungal communities from a forest stand in relation to nutrient mobilization processes. Microb Ecol 54:567–577. doi: 10.1007/s00248-007-9260-z. [DOI] [PubMed] [Google Scholar]
- 84.Uroz S, Calvaruso C, Turpault MP, Sarniguet A, De Boer W, Leveau JHJ, Frey-Klett P. 2009. Efficient mineral weathering is a distinctive functional trait of the bacterial genus Collimonas. Soil Biol Biochem 41:2178–2186. doi: 10.1016/j.soilbio.2009.07.031. [DOI] [Google Scholar]
- 85.Uroz S, Calvaruso C, Turpault M-P, Pierrat J-C, Mustin C, Frey-Klett P. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microbiol 73:3019–3027. doi: 10.1128/AEM.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. [Google Scholar]
- 87.Oksanen J. 2015. Vegan: an introduction to ordination. http://cran.r-project.org/web/packages/vegan/vignettes/intro-vegan.pdf.
- 88.Dray S, Dufour A-B. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.