ABSTRACT
Brochothrix thermosphacta is a dominant but poorly studied meat spoilage organism. It is a close relative of the foodborne pathogen Listeria monocytogenes, and Brochothrix constitutes the second genus in the Listeriaceae family. Here, the genomes of 12 B. thermosphacta strains were sequenced, assembled into draft genomes, characterized, and compared with the genomes of Brochothrix campestris and L. monocytogenes. Phenotypic properties including biogenic amine production and antibiotic and heavy metal susceptibilities were tested. Comparative genomic analyses revealed a high degree of similarity among the B. thermosphacta strains, with bacteriophage genes constituting a significant proportion of the accessory genome. Genes for the production of the malodorous compounds acetate, acetoin, butanediol, and fatty acids were found, as were stress response regulatory genes, which likely play important roles in the spoilage process. Amino acid decarboxylases were not identified in the genomes, and phenotypic testing confirmed their absence. Orthologs of Listeria virulence proteins involved in virulence regulation, intracellular survival, and surface protein anchoring were found; however, key virulence genes were absent. Analysis of antibiotic susceptibility showed that strains were sensitive to the four tested antibiotics, except for one tetracycline-resistant isolate with plasmid-mediated tetracycline resistance genes. Strains tolerated higher levels of copper and cobalt than of cadmium although not at concentrations high enough to categorize the strains as being resistant. This study provides insight into the Brochothrix genome, links previous phenotypic data and data provided here to the gene inventory, and identifies genes that may contribute to the persistence of this organism in the food chain.
IMPORTANCE Despite increasing knowledge and advances in food preservation techniques, microbial spoilage of foods causes substantial losses, with negative social and economic consequences. To better control the contamination and microbial spoilage of foods, fundamental knowledge of the biology of key spoilage bacteria is crucial. As a common meat spoilage organism, B. thermosphacta contributes substantially to spoilage-associated losses. Nonetheless, this organism and particularly its genome remain largely unstudied. This study contributes to improving our knowledge of the Brochothrix genus. Spoilage-relevant pathways and genes that may play a role in the survival of this organism in a food processing environment were identified, linking previous phenotypic data and data provided here to the gene inventory of Brochothrix and establishing parallels to and differences from the closely related foodborne pathogen L. monocytogenes.
KEYWORDS: Brochothrix, Listeria monocytogenes, meat spoilage, spoilage
INTRODUCTION
Brochothrix thermosphacta is one of the most abundant spoilage organisms of fresh and cured meats, fish, and fish products, due to its tolerance to high-salt and low-pH conditions, its ability to grow at refrigeration temperatures, and its production of organoleptically unpleasant compounds (1–3). This Gram-positive, fermentative bacterium can become the dominant spoilage species of modified atmosphere-packaged (MAP) and vacuum-packaged meats when sufficient oxygen is present (3–6). Although aerobic spoilage is dominated largely by Gram-negative pseudomonads, B. thermosphacta can also play an important role in shortening the shelf-life of aerobically stored meat, particularly when bacteriostatic agents such as sulfites are used (7–11).
B. thermosphacta is considered a nonproteolytic spoilage organism and is associated with spoilage characterized by cheesy, buttery, or sour odors rather than putrefaction (2, 12). Under anaerobic conditions or oxygen limitation, B. thermosphacta produces lactic acid and ethanol as primary end products, with smaller amounts of formate and acetate also being produced (13, 14). Under aerobic conditions, the end products of its metabolism include lactic acid, acetic acid, acetoin, diacetyl, 2,3-butanediol, ethanol, isobutyric acid, isovaleric acid, and 2-methylbutyric acid (12, 14). These fatty acids are thought to be formed from amino acids and not by lipolysis (2). Information on the lipolytic activity of B. thermosphacta strains is scarce and ambiguous, and the role of lipolysis in meat spoilage caused by B. thermosphacta remains unclear (5, 15, 16).
The Brochothrix genus comprises only two known species, B. thermosphacta and the lesser-known species B. campestris (17). Brochothrix bacteria are low-GC-content bacteria (Firmicutes) that have been placed into the Listeriaceae family, and members of the Listeria and Brochothrix genera share many common features, such as the same main fatty acids, menaquinones, and meso-diaminopimelic acid in the peptidoglycan; similar GC contents; and catalase production (18). Their 16S rRNA gene sequences share 92.8 to 96.6% sequence similarity (19).
While B. campestris has been isolated only from soil and grass (17), B. thermosphacta is thought to be ubiquitous throughout the meat production chain (2, 20, 21). As a saprophytic organism capable of growth at cold temperatures, B. thermosphacta shares its environmental niche with a member of its sister taxon, Listeria monocytogenes, the foodborne pathogen and causative agent of listeriosis. The persistence of microorganisms like B. thermosphacta and L. monocytogenes in the food chain and other ecosystems reflects their ability to adapt to numerous stresses such as osmotic, pH, and temperature stresses (22). Furthermore, the use of antibiotics in animal husbandry and the presence of heavy metals in the environment provide selective pressure for the survival and dominance of resistant bacteria (23). Even low levels of antibiotics and heavy metals are thought to be sufficient to select and enrich bacteria carrying multiresistance plasmids (24). Resistance to heavy metals appears to be one of the longest-known environmental adaptations of Listeria, and although it remains unclear whether heavy metal resistance contributes to overall fitness in food processing environments or foods, it is interesting that heavy metal-resistant Listeria strains are more common among food isolates than among those recovered sporadically (22).
While genome characterization of L. monocytogenes and other Listeria species has been extensive, until now, genomic analyses have focused only on bacteriophages of B. thermosphacta (25). A recent study also used the genome sequences of B. thermosphacta DSM 20171 and B. campestris DSM 4712 to investigate the dynamics of genome evolution of the Listeria genus but without any in-depth analysis of Brochothrix genomes (26). To better understand the physiology of this problematic spoilage organism, we have sequenced the genomes of 12 B. thermosphacta strains and conducted comparative genomic analyses of these strains with B. campestris DSM 4712. Emphasis was placed on identifying genes and pathways that may play an important role in food spoilage and the persistence of this organism throughout the food chain. Numerous virulence genes of Listeria are involved in adhesion to mammalian cells and the transition from an environmental organism to a human pathogen. As Brochothrix is a close relative of Listeria that is commonly found to be associated with meat, we conducted a search for Listeria virulence orthologs that may play a role in the adaptation of B. thermosphacta from a farm to a carcass ecosystem. To link the gene inventory identified in this study with phenotypic properties, amino acid decarboxylase activity and antibiotic and heavy metal tolerances of the strains were investigated.
RESULTS
Genomic features and intergenomic sequence similarity of strains.
Whole-genome sequencing was undertaken for 12 B. thermosphacta isolates, including the type strain B. thermosphacta DSM 20171. Table 1 contains a summary of the assembly and annotation metrics of these draft genomes. The estimated sizes of the 12 draft genomes were highly similar, varying by ∼1.7 kb, and were between 2.47 and 2.64 Mb. Their GC content was between 36.2 and 36.4%, and the genomes harbored 2,244 to 2,417 predicted protein-coding sequences.
TABLE 1.
B. thermosphacta strains
| Strain | Meat sourcea | Genome size (bp) | No. of contigs | G+C content (%) | No. of protein-coding genes | No. of tRNAs | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| DSM 20171b | Pork sausage | 2,494,302 | 41 | 36.4 | 2,290 | 84 | MDLK00000000 |
| 7803c | Beef (MAP) | 2,575,404 | 61 | 36.3 | 2,348 | 80 | MDLL00000000 |
| 7804c | Beef (MAP) | 2,613,933 | 56 | 36.3 | 2,415 | 83 | MDLU00000000 |
| 7806c | Beef (air) | 2,539,264 | 41 | 36.3 | 2,328 | 80 | MDLM00000000 |
| 7807c | Lamb (air) | 2,482,452 | 31 | 36.4 | 2,277 | 91 | MDLN00000000 |
| 7808c | Lamb (air) | 2,557,615 | 68 | 36.3 | 2,328 | 86 | MDLO00000000 |
| 7809c | Beef (air) | 2,491,703 | 22 | 36.3 | 2,273 | 80 | MDLV00000000 |
| 7810c | Beef (air) | 2,471,990 | 22 | 36.4 | 2,244 | 80 | MDLP00000000 |
| 7811d | Beef (air) | 2,548,405 | 186 | 36.3 | 2,292 | 81 | MDLT00000000 |
| 7813d | Veal mince (MAP) | 2,642,200 | 166 | 36.2 | 2,417 | 90 | MDLQ00000000 |
| 7816d | Lamb (air) | 2,584,597 | 157 | 36.3 | 2,342 | 71 | MDLR00000000 |
| 7818d | Beef (air) | 2,587,934 | 130 | 36.3 | 2,364 | 82 | MDLS00000000 |
MAP, modified atmosphere packaged. Air indicates storage under aerobic conditions.
Type strain (isolated during a study on the microflora of fresh pork sausage by Sulzbacher and Mclean [37]).
Strains were isolated in 1993 (meat sources obtained in Melbourne, Australia) during a meat spoilage study (H. M. Craven and N. Baxter-Keene, CSIRO Agriculture and Food, unpublished data).
Strains were isolated for this study from meat sources obtained in Melbourne, Australia.
A high degree of genomic sequence similarity among the B. thermosphacta strains was demonstrated by two calculated genomic indices (see Tables S1 and S2 in the supplemental material). Genomic comparisons of the B. thermosphacta isolates to B. thermosphacta DSM 20171 revealed OrthoANI (Orthologous Average Nucleotide Identity) values of between 98.99 and 99.54% (the recommended cutoff range for species delimitation is 95 to 96%), while GGDC DDH (Genome-to-Genome Distance Calculator Digital DNA:DNA Hybridization) estimates, which correlate well with wet laboratory DNA-DNA hybridization percentages, were between 90.7 and 93.1%. For B. campestris DSM 4712 and B. thermosphacta DSM 20171, OrthoANI and GGDC DDH values of 75.05 and 20.60%, respectively, were obtained.
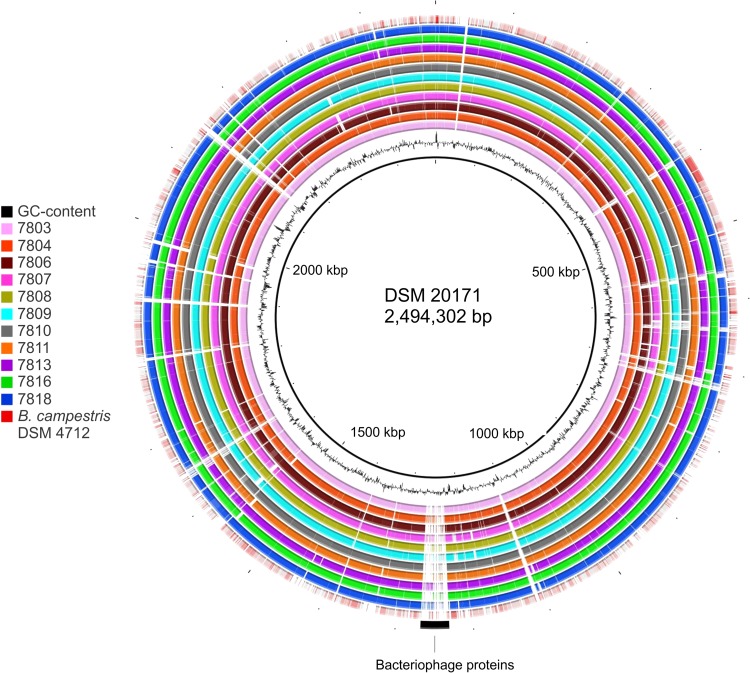
The genomic sequence similarity of B. thermosphacta isolates is also illustrated in a BLASTn ring diagram (Fig. 1), where the 11 Australian strains and B. campestris DSM 4712 were compared to the central reference genome of B. thermosphacta DSM 20171. The most prominent region absent in the 11 Australian isolates was an ∼37-kb region of the type strain genome harboring phage genes.
FIG 1.
B. thermosphacta strains share a high degree of genomic similarity. Shown is a schematic of BLAST comparisons of nucleotide sequences of the draft genomes of B. thermosphacta strains and B. campestris DSM 4712 (shown as concentric rings) to B. thermosphacta DSM 20171 (central black ring). The GC content of the genome of B. thermosphacta DSM 20171 is shown in black outside the inner ring. BLASTn matches are colored on a sliding scale from 70 to 90%, demonstrating the degree of sequence similarity between the query strains (shown in the key) and the B. thermosphacta type strain. Spaces in the rings indicate regions of the type strain genome that were absent in the query genomes. Bacteriophage proteins encoded in the type strain genome, but largely absent in the query genomes, are depicted with a black arc in the outer ring.
Pangenome investigation.
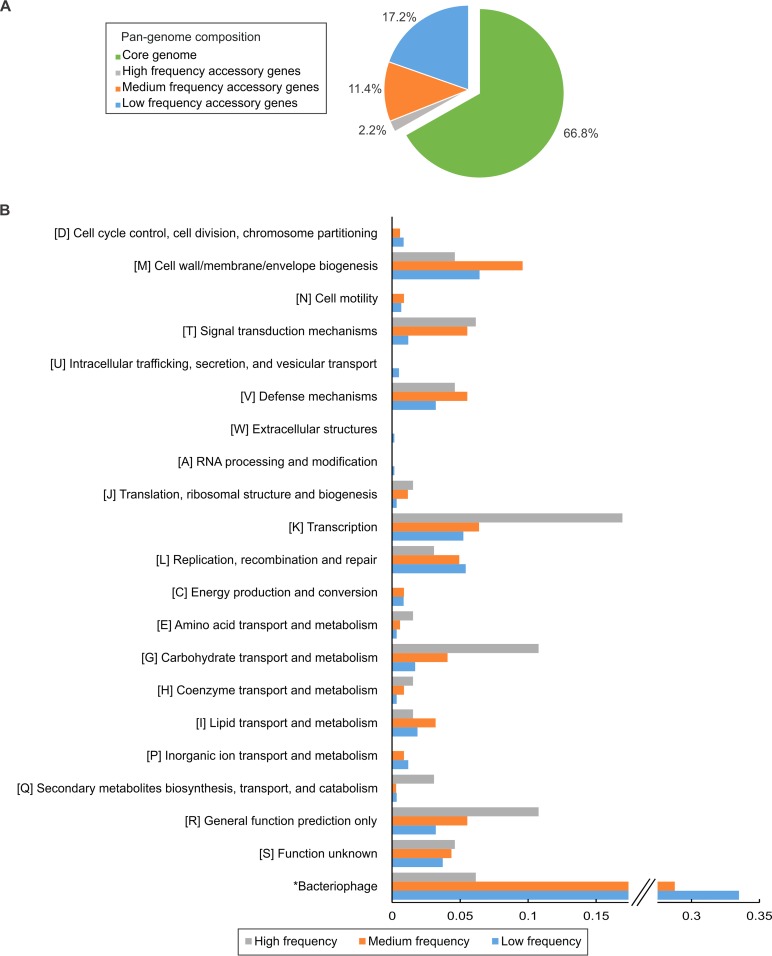
Strain diversity was investigated with a pangenome analysis. The pangenome of the 12 strains comprised 3,012 protein-coding sequences, with 2,012 core genes (66.8% of the pangenome) and 1,000 accessory genes (30.8% of the pangenome) (Fig. 2A). With between 2,244 and 2,417 predicted protein-coding genes identified in the draft genomes (Table 1), the 2,012 core genes constituted between 83 and 90% of the predicted proteome of each isolate.
FIG 2.
Pangenome analysis shows that bacteriophage genes represent a large proportion of the accessory genome. (A) The pangenome of the 12 B. thermosphacta strains comprises 3,012 genes, with the core genome constituting 66.8% of the genes, the high-frequency accessory group constituting 2.2%, the medium-frequency accessory group representing 11.4%, and the low-frequency accessory group representing 17.2%. (B) The relative abundances of COG functional categories for the high-, medium-, and low-frequency accessory groups are shown. On the vertical axis, COG functional categories are depicted with their letter association, while the horizontal axis indicates their relative abundance. The relative abundance of prophage coding sequences is included at the bottom of the graph, and the corresponding label on the vertical axis has been marked with an asterisk (*Bacteriophage) to highlight the different method by which these gene functions were determined. To aid visualization, the x axis and the low- and medium-frequency bars associated with bacteriophage coding sequences are truncated, and the breakpoints are marked with //.
To investigate possible links between genes of the accessory genome and different evolutionary processes influencing their frequency of occurrence, this group was separated into high-, medium-, and low-frequency accessory genes. With 591 genes, the low-frequency accessory group was the largest, followed by the medium-frequency group, with 344 genes, and the high-frequency group, with 65 coding sequences (Fig. 2A). Functional annotation of their gene products assigned pools of 46, 109, and 223 categories to the high-, medium-, and low-frequency accessory groups, respectively (Fig. 2B). As prophages typically contain significant numbers of hypothetical genes to which no annotation can be assigned, bacteriophage-associated coding sequences were determined by using PHAST. Of the high-frequency accessory genes, 6.2% were bacteriophage coding sequences, while 28.8% and 33.5% of the medium- and low-frequency accessory genomes were bacteriophage genes. Thus, prophage coding sequences constituted a significant proportion of the accessory genome, in particular the medium- and low-frequency groups.
In the high-frequency pool, genes involved in the regulation of gene expression were strongly represented, with this group having the highest relative number of coding sequences involved in transcription processes but also in signal transduction and carbohydrate transport and metabolism. In addition to bacteriophage genes, which were notably the most abundant within the medium- and low-frequency accessory groups, genes involved in the biogenesis of the cell envelope; defense mechanisms such as clustered regularly interspaced short palindromic repeat (CRISPR) systems; transcription processes; and DNA replication, recombination, and repair were also abundant in both groups. Furthermore, genes with functions in signal transduction and carbohydrate and lipid transport and metabolism were well represented in the medium-frequency group.
Annotation of the draft genomes involved a limited amount of manual curation. Thus, to help gauge the possible impact of miscalled or spurious open reading frames (ORFs) identified by the gene-calling algorithm, the statistics of the lengths of the proteins belonging to the accessory group were determined (see Fig. S1 in the supplemental material). The median lengths of core and accessory proteins were 273 and 202 amino acids, respectively, and individual groupings of accessory genes had median lengths of 214 residues (high frequency), 219 residues (medium frequency), and 189 residues (low frequency), indicating possible miscalled ORFs in the accessory genome.
Phylogeny of B. thermosphacta strains.
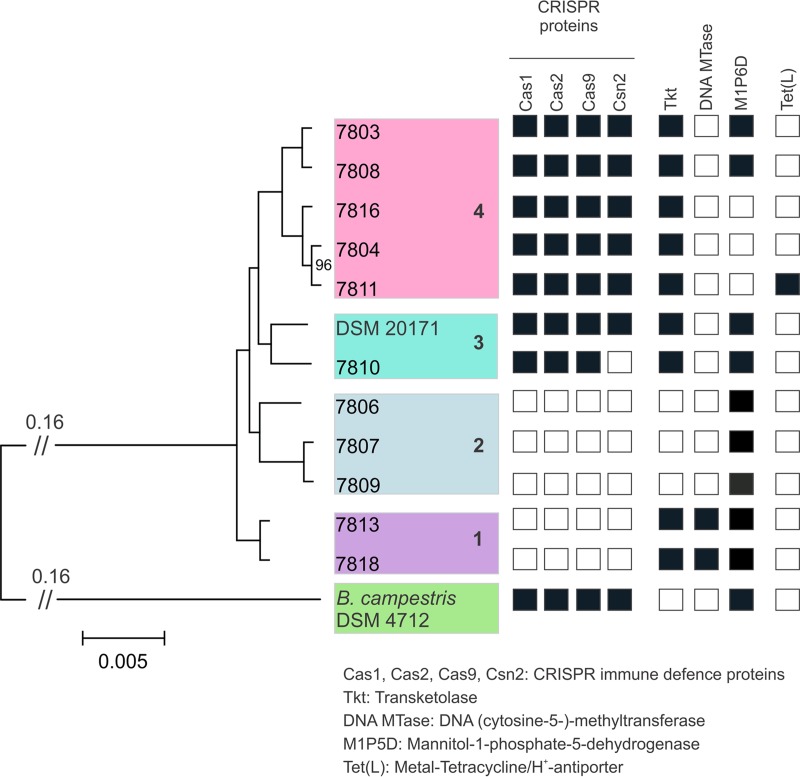
To better understand the relationship among these geographically and temporally diverse strains, a phylogenetic tree of the 12 strains and B. campestris DSM 4712 was inferred by using alignments of orthologous genomic regions of the strains and an approximated maximum likelihood approach. The resulting tree shows four distinct phylogroups (groups 1 to 4) of strains (Fig. 3). Genes specific to certain phylogroups, which were identified during the pangenome analysis, are listed to the right of this tree, and locus tags of these genes are given in Table 2.
FIG 3.
Phylogenetic tree showing the relationships of the 12 B. thermosphacta strains and accessory genes present or absent in these strains. The tree is based on alignments of orthologous regions of the genomes and was generated by using an approximated maximum likelihood method. All interior node values are 100 or otherwise as marked and represent local support values from 1,000 bootstrap replicates. The scale bar denotes the number of substitutions per site. Branches that were truncated to aid visualization are marked with //, and the original branch length is indicated above. The four phylogroups of B. thermosphacta strains are represented in colored boxes, with the phylogroup number shown in boldface type at the right of the box. Accessory genes present or absent in strains are shown to the right of the tree, and black boxes represent genes that are present, while white boxes represent the absence of genes from the genome.
TABLE 2.
Locus tags of coding sequencesa
| Coding region | Locus tag(s) (GenBank accession no.) |
|
|---|---|---|
| B. thermosphactab | B. campestris DSM 4712 | |
| Cas1 | BFR34_02990 | BCAMP_RS03535 |
| Cas2 | BFR34_02995 | BCAMP_RS03530 |
| Cas9 | BFR34_02985 | — |
| Csn2 | BFR34_03000 | BCAMP_RS03525 |
| Transketolase (N-terminal peptide) | BFR34_08475 | — |
| Transketolase (C-terminal peptide) | BFR34_08480 | — |
| DNA (cytosine-5-)methyltransferase | BFR35_12330 (MDLQ00000000) | — |
| Mannitol-1-phosphate-5-dehydrogenase | BFR34_07330 | BCAMP_RS09425 |
| Glucokinase | BFR34_10620 | BCAMP_RS05020 |
| Fructokinases | BFR34_11880, BFR34_10080 | BCAMP_RS00590 |
| Pyruvate carboxylase | BFR34_02675 | BCAMP_RS02425 |
| Lactate dehydrogenase | BFR34_12075 | BCAMP_RS09650 |
| Pyruvate decarboxylase | BFR34_04065 | — |
| Alcohol dehydrogenase | BFR34_11980 | BCAMP_10295c |
| Acetaldehyde dehydrogenase | BFR34_12025 | BCAMP_RS09610 |
| Pyruvate formate-lyase | BFR34_00380 | BCAMP_RS07680 |
| Phosphate acetyltransferase | BFR34_05080 | BCAMP_RS07720 |
| Acetate kinase | BFR34_09255 | BCAMP_RS05095 |
| Acetolactate synthase | BFR34_08185 | BCAMP_RS01540 |
| Acetolactate decarboxylase | BFR34_08180 | BCAMP_RS01545 |
| Butanediol dehydrogenase | BFR34_03815 | BCAMP_RS09100 |
| Acyl-CoA thioesterase | BFR34_05700 | BCAMP_RS02760 |
| PotA | BFR34_09935 | BCAMP_RS06135 |
| PotB | BFR34_09930 | BCAMP_RS06130 |
| PotC | BFR34_09925 | BCAMP_RS06125 |
| PotD | BFR34_09920 | BCAMP_RS06120 |
| Lipase | BFR34_09910 | BCAMP_RS06110 |
| Tet(L) | BFR45_11435 (MDLT00000000) | — |
| Alkyl mercury lyase | BFR36_11230 (MDLR00000000) | — |
| Mercury(II)-responsive transcriptional regulator | BFR36_11260 (MDLR00000000) | — |
| RNA polymerase sigma factor sigma B | BFR34_09145 | BCAMP_RS05220 |
— indicates that the gene was not identified.
Gene locus tags are from B. thermosphacta DSM 20171 (GenBank accession number MDLK00000000) unless otherwise specified in parentheses (accession numbers of other B. thermosphacta genomes).
This gene was identified in the sequence reported under GenBank accession number AODH00000000 but is missing from the RefSeq version of the B. campestris DSM 4712 draft genome under accession number NZ_AODH00000000.1.
Group 1 isolates 7813 and 7818 formed a clade separated from the other B. thermosphacta strains, and these isolates were found to encode an immune defense restriction modification protein [DNA (cytosine-5-)methyltransferase] unique to this group. Group 2 comprised the three isolates 7806, 7807, and 7809. Absent in the genomes of group 1 and group 2 isolates were genes for the two universally conserved CRISPR-prokaryotic immune defense proteins, Cas1 and Cas2, as well as genes for predicted Cas9 and Csn2 proteins. Group 2 strains also lacked two genes (N- and C-terminal peptides) for an additional transketolase (Tkt) encoded by all of the other genomes. Group 3 comprised B. thermosphacta DSM 20171 and a second isolate, 7810, and group 4 contained the largest number of strains, with five isolates (isolates 7803, 7808, 7816, 7804, and 7811). While all of the group 3 and 4 strains carried genes for the CRISPR proteins and Tkt peptide chains, a mannitol-1-phosphate-5-dehydrogenase (M1P5D) gene carried by all of the other strains was absent in the genomes of group 4 isolates 7811, 7804, and 7816. These three strains also encoded a truncated mannitol-specific phosphotransferase transporter and were missing an additional mannitol uptake gene, both of which are located upstream of the M1P5D gene, and tested negative for mannitol fermentation (Table 3).
TABLE 3.
Phenotypic properties of B. thermosphacta isolatesb
| Strain | Mannitol fermentation abilitya | Antibiotic tolerance (MIC [μg/ml])c |
Heavy metal tolerance (MTC [μg/ml])d |
|||||
|---|---|---|---|---|---|---|---|---|
| Pen G | Cip | Tet | Ery | CdCl | CoCl2 | CuSO4 | ||
| DSM 20171 | + | 0.12 | 0.12 | 0.25 | 0.02 | 10 | 200 | 200 |
| 7803 | + | 0.25 | 0.25 | 0.5 | 0.06 | 10 | 200 | 200 |
| 7804 | − | 0.5 | 0.25 | 0.5 | 0.06 | 15 | 200 | 200 |
| 7806 | + | 0.12 | 0.25 | 0.25 | 0.03 | 10 | 200 | 200 |
| 7807 | + | 0.5 | 0.25 | 0.25 | 0.06 | 10 | 200 | 200 |
| 7808 | + | 0.25 | 0.25 | 0.5 | 0.06 | 10 | 200 | 200 |
| 7809 | + | 0.25 | 0.12 | 0.25 | 0.06 | 15 | 200 | 200 |
| 7810 | + | 0.5 | 0.5 | 0.25 | 0.12 | 10 | 200 | 200 |
| 7811 | − | 0.5 | 0.25 | 32 | 0.12 | 10 | 200 | 200 |
| 7813 | + | 0.25 | 0.12 | 0.25 | 0.06 | 10 | 200 | 200 |
| 7816 | − | 0.25 | 0.25 | 0.5 | 0.06 | 10 | 200 | 200 |
| 7818 | + | 0.25 | 0.25 | 0.25 | 0.12 | 10 | 200 | 200 |
+ indicates an ability to ferment mannitol, and − indicates an inability to ferment mannitol.
Voges-Proskauer tests were positive for all strains. There were negative decarboxylase activity reactions for all strains with lysine, arginine, ornithine, histidine, tryptophan, and phenylalanine on decarboxylase medium.
Pen G, penicillin G; Cip, ciprofloxacin; Tet, tetracycline; Ery, erythromycin.
The maximum tolerance concentrations (MTCs) are shown for the three heavy metal salts.
Neither the meat environment nor the packaging type from which the strains were derived appeared to be congruent with the phylogrouping of the strains.
Cell metabolism.
Inspection of the genomes of B. thermosphacta strains and B. campestris DSM 4712 revealed a diverse repertoire of substrate-specific genes from the phosphotransferase system (PTS), including glucose-, maltose-, sucrose-, fructose-, mannose-, trehalose-, cellobiose-, β-glucoside-, mannitol-, and N-acetylglucosamine-specific EIIA, EIIB, and EIIC genes. Ribose, glycerol-3-phosphate, maltose/maltodextrin, and inositol transporter genes as well as lactate, gluconate, and malate permease genes were also identified.
For both Brochothrix species, all genes required for glycolysis were found, including genes encoding the sugar-specific kinases glucokinase and fructokinase (Table 2; see also Fig. S2 in the supplemental material). The full complement of genes required for the pentose phosphate pathway were also found (Fig. S3), while coding sequences for only four of the eight enzymes of the citrate cycle (tricarboxylic acid [TCA] cycle) were present (Fig. S4). Genes identified as being absent included α-ketoglutarate dehydrogenase, succinate dehydrogenase, succinate thiokinase, and malate dehydrogenase genes. Furthermore, both species encoded a pyruvate carboxylase (Table 2), while a fumarate reductase was not found.
The Brochothrix genomes were examined for the presence of genes involved in the conversion of pyruvate to various end products, particularly those considered malodorous compounds, as described by pathways in the KEGG database (see Fig. S2 and S5 in the supplemental material). Genes involved in the production of lactate, ethanol, acetate, and formate were present in both Brochothrix species and included genes for the enzymes lactate dehydrogenase (EC 1.1.1.27), alcohol dehydrogenase (EC 1.1.1.1), aldehyde dehydrogenase (EC 1.2.1.3), acetaldehyde dehydrogenase (EC 1.2.1.10), pyruvate formate-lyase (EC 2.3.1.54), phosphate acetyltransferase (EC 2.3.1.8), and acetate kinase (EC 2.7.2.1) (Table 2). A pyruvate decarboxylase (EC 4.1.1.1) gene was identified only in the draft genomes of the B. thermosphacta strains (Table 2). Furthermore, in both species, genes involved in butanediol fermentation were found, including α-acetolactate synthase (EC 2.2.1.6) and acetolactate decarboxylase (EC 4.1.1.5) genes with contiguous locations for the production of acetoin from pyruvate and an (S,S)-butanediol dehydrogenase gene required for the reduction of acetoin to 2,3-butanediol (EC 1.1.1.76) (Table 2 and Fig. S5). The production of acetoin, as determined by positive Voges-Proskauer reactions, was observed for all B. thermosphacta strains (Table 3).
Amino acid decarboxylase genes were not identified in the B. thermosphacta strains or B. campestris. Furthermore, lysine, arginine, ornithine, histidine, phenylalanine, and tryptophan decarboxylase activities were not detected in the 12 strains by using both API 20E strips and decarboxylase screening medium (Table 3), while the control strain Escherichia coli ATCC 25922 was clearly positive for lysine and ornithine decarboxylase reactions by using both testing procedures. However, both Brochothrix species were found to possess a homologous gene cluster with conserved synteny in L. monocytogenes comprising the spermidine/putrescine-specific uptake genes potA, potB, potC, and potD. The translated products of these genes also have significant homology to the PotABCD components of the spermidine/putrescine ABC transporter in Escherichia coli K-12 (Table 2).
As it is thought that free fatty acids result from amino acid metabolism in B. thermosphacta (2), amino acid pathways were investigated. Genes were present in both Brochothrix species for the production of isovaleryl coenzyme A (CoA) (3-methylbutanoyl-CoA), isobutyryl-CoA, and 2-methylbutanoyl-CoA from the degradation of valine, leucine, and isoleucine, respectively (see Fig. S6 in the supplemental material). These pathways involve the common activity of a branched-chain amino acid transaminase (EC 2.6.1.42), which catalyzes the reaction between the amino acids and their α-keto acids. Further steps involve the activity of a branched-chain keto acid dehydrogenase (EC 1.2.4.4), a dihydrolipoyllysine residue (2-methylpropanoyl)transferase (EC 2.3.1.168), and a dihydrolipoyl dehydrogenase (EC 1.8.1.4) for the conversion of the resulting 2-oxo acids to the respective branched-chain fatty acyl-CoAs. Furthermore, genes of the distal pathways of valine and isoleucine degradation were identified in B. thermosphacta strains for the synthesis of propanoyl-CoA (Fig. S6). An acyl-CoA thioesterase gene for the hydrolysis of the acyl-CoAs to free fatty acids and coenzyme A was also identified in both Brochothrix species (Table 2).
A search for lipase and esterase genes revealed the presence of lipases in B. thermosphacta and B. campestris with 54% and 52% identities, respectively, to lipases in Listeria species (Table 2) and multiple esterase genes in both Brochothrix species.
Resistance genes.
Numerous multidrug resistance efflux transporters were annotated in the draft genomes. As a close relative of L. monocytogenes and an organism that shares its environmental niche with this foodborne pathogen, we were interested in testing the MICs of the strains against antibiotics traditionally used to treat listeriosis, such as penicillin, erythromycin, and tetracycline, as well as the therapeutically important fluoroquinolone ciprofloxacin, to which L. monocytogenes shows decreased susceptibility (27, 28). B. thermosphacta strains were sensitive to all four tested antibiotics, with the exception of one isolate (7811) that displayed resistance to tetracycline (the MIC was 32 μg/ml) (Table 3). Inspection of the draft genome of this strain revealed the presence of a tetracycline efflux transporter gene on a contig with notably high coverage (read coverage ∼14-fold higher than the average genome read coverage) (Table 2 and Fig. 3). This gene product was found to share >80% identity with known class L Tet determinants and higher identity with determinants from the plasmid subgroup [88% identity to the Tet(L) protein from Bacillus subtilis plasmid pNS1981 (GenBank accession number BAA00005)] than with those from the chromosomal subgroup (83% identity to the Bacillus subtilis GSY908 Tet(L) protein [accession number CAA30827]) (29–31). A tetR regulatory gene was also identified on this contig in close proximity to the efflux gene.
Subsystems of SEED predicted numerous genes in both Brochothrix species to be involved in copper, arsenic, cobalt, zinc, and cadmium homeostasis/resistance. Metal resistance is commonly defined based on the metal tolerance levels of other bacteria, and until now, data on metal tolerance levels of B. thermosphacta have been missing. The heavy metal tolerance of the isolates was analyzed by determining the maximum tolerance concentrations (MTCs) of CoCl2, CdCl, and CuSO4 (Table 3). Overall, significantly higher concentrations of CuSO4 and CoCl2 than of CdCl were tolerated; however, none of the observed MTCs were at levels high enough to categorize the strains as being resistant to these metals (32–35). An MTC of 200 μg/ml of CoCl2 and CuSO4 was observed for all of the strains. Although impaired, growth of the isolates was still observed in the range of 200 to 500 μg/ml CoCl2, while 300 to 350 μg/ml was the upper limit for growth on medium containing CuSO4. For CdCl, the MTCs of the strains were between 10 and 15 μg/ml, and although impaired, growth of all isolates was still observed at 50 μg/ml CdCl. At these higher concentrations, confluent growth was replaced by the emergence of single colonies.
Examination of the draft genomes of the strains revealed differences in the numbers of predicted copper, cobalt, and cadmium resistance genes. The genomes of the strains harbored one or two genes predicted to be involved in cadmium transport, although no correlation between the number of these genes and the tolerated cadmium concentrations was observed. The strains were also found to carry two putative cobalt transporter genes and three or four genes with a predicted function in copper transport and homeostasis.
Presence of Listeria virulence genes in Brochothrix.
The genomes of Brochothrix were searched for orthologs of 44 well-characterized Listeria virulence factors with the aim of identifying gene candidates that may play a role in contamination and growth on meat.
The key virulence genes hly (listeriolysin O), actA (nucleation-promoting factor), plcA, plcB (membrane-damaging phospholipase), inlA (internalin A), and inlB (internalin B) were absent from the Brochothrix genomes. However, orthologs of other virulence factors involved in virulence regulation, intracellular survival, and surface protein anchoring were identified (Table 4). Conserved synteny in the Brochothrix genomes was observed for only 8 of the 22 identified virulence gene orthologs. These orthologs included two factors involved in adherence to mammalian cells (dltA and fbpA), one gene required for intracellular survival (prsA2), a factor with a function in iron uptake (hbp2), homologous virR and virS two-component regulatory genes, a threonine phosphatase (stpA), and the surface anchoring factor srtB (Table 4).
TABLE 4.
Orthologs of L. monocytogenes virulence factors present in Brochothrix genomes according to functional categorya
| Function | Virulence factor | Related gene | Locus tag |
|
|---|---|---|---|---|
| B. thermosphacta DSM 20171 | B. campestris DSM 4712 | |||
| Adherence | d-Alanine-phosphoribitol ligase | dltA | BFR34_01920 | BCAMP_RS06480 |
| Fibronectin-binding protein | fbpA | BFR34_07800 | BCAMP_RS01735 | |
| Bile resistance | Bile salt hydrolase | bsh | BFR34_01045 | No hit |
| Enzyme | Threonine phosphatase | stp | BFR34_07765 | BCAMP_RS01770 |
| Metalloprotease | mpl | No hit | BCAMP_RS07830 | |
| Intracellular survival | Lipoate protein ligase A1 | lplA1 | BFR34_04540 | BCAMP_RS04420 |
| Oligopeptide-binding protein | oppA | BFR34_01280 | BCAMP_RS03095 | |
| Posttranslation chaperone | prsA2 | BFR34_04575 | BCAMP_RS10760 | |
| Sugar uptake system | hpt | BFR34_09035 | BCAMP_RS05315 | |
| Iron uptake | Hemoglobin-binding protein | hbp2 | BFR34_05250 | BCAMP_RS12765 |
| Invasion | Cell wall teichoic acid glycosylation protein | gtcA | BFR34_03990 | BCAMP_RS09170 |
| Promoting-entry protein | lpeA | BFR34_01515 | BCAMP_RS03260 | |
| Peptidoglycan modification | OatA | oatA | BFR34_04360 | BCAMP_RS07115 |
| Regulation | Positive regulatory factor | prfA | BFR34_00370 | No hit |
| LisR | lisR | BFR34_11190 | BCAMP_RS07275 | |
| LisK | lisK | BFR34_11195 | BCAMP_RS07280 | |
| VirR | virR | BFR34_09230 | BCAMP_RS05130 | |
| VirS | virS | BFR34_09225 | BCAMP_RS05135 | |
| Surface protein anchoring | Diacylglyceryl transferase | lgt | BFR34_06385 | BCAMP_RS05615 |
| Specific signal peptidase II | lspA | BFR34_07940 | BCAMP_RS09260 | |
| Sortase A | srtA | BFR34_10065 | BCAMP_RS06070 | |
| Sortase B | srtB | BFR34_05235 | BCAMP_RS10940 | |
Orthologous genes with conserved synteny are highlighted in boldface type.
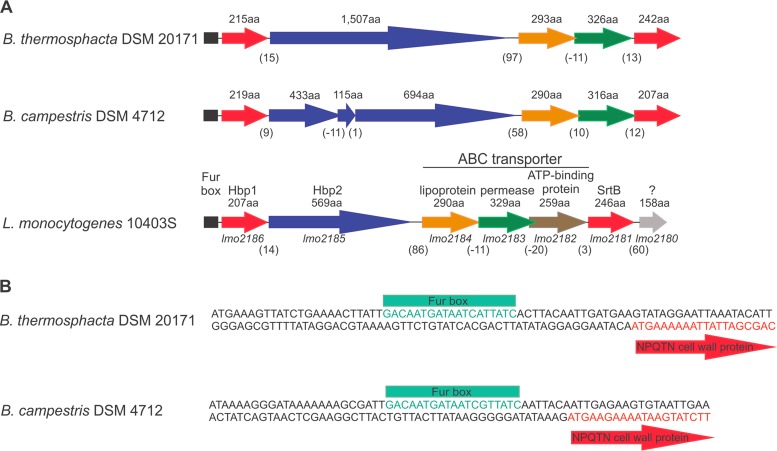
In L. monocytogenes, the srtB gene (lmo2181) is located on a Fur-regulated operon, which also encodes the heme-binding virulence protein Hbp2 (lmo2185), the cell wall anchored heme-binding protein Hbp1 (lmo2186), and components of a ferric hydroxymate ABC transporter (lmo2182, lmo2183, and lmo2184) (Fig. 4A) (36). A conserved srtB locus was found in the Brochothrix genomes (Fig. 4A), as was a well-conserved Fur box (Fig. 4B) identified upstream of the first gene of this putative operon. In B. thermosphacta, the srtB locus carried five open reading frames, while in B. campestris DSM 4712, there were seven genes. Counterparts to the L. monocytogenes genes lmo2182 and lmo2180, encoding a membrane ATPase of the ABC transporter and a soluble protein of unknown function, respectively, were missing. The first gene of this putative operon was predicted to carry an SrtB sorting signal (NPXTN) and encoded proteins in B. thermosphacta and B. campestris that shared 51 and 44% identities, respectively, with their L. monocytogenes counterpart Hbp1. This gene was followed by a homologous hbp2 gene, which showed striking variability in sequence and length among the B. thermosphacta strains and also between both species. The evolution of this gene in B. campestris appears to have led to three separate hbp2 coding sequences. Reading frames downstream of hbp2 encoded proteins with homology to Lmo2184 and Lmo2183, which are lipoprotein and permease components of a ferric hydroxymate ABC transporter. The srtB operon in Brochothrix lacked an ortholog for lmo2180 and terminated with srtB.
FIG 4.
Schematic representation of the conserved synteny of the srtB locus in Brochothrix. (A) In L. monocytogenes, the srtB operon contains genes for cell wall-associated and secreted proteins (lmo2186 and lmo2185), sortase B that ligates them to the peptidoglycan (lmo2181), and an ABC transporter (lmo2182 to lmo2184). Genes encoding an ATP-binding component of the putative ABC transporter were absent from this region in Brochothrix species. In B. thermosphacta, the length of the second gene of this locus varied considerably among the isolates, while in B. campestris, the evolution of this gene led to three separate coding regions. Numbers in parentheses indicate the number of spacer nucleotides between the genes or bases that overlap. aa, amino acids. (B) A Fur box was identified upstream of the first gene of the srtB locus in Brochothrix species.
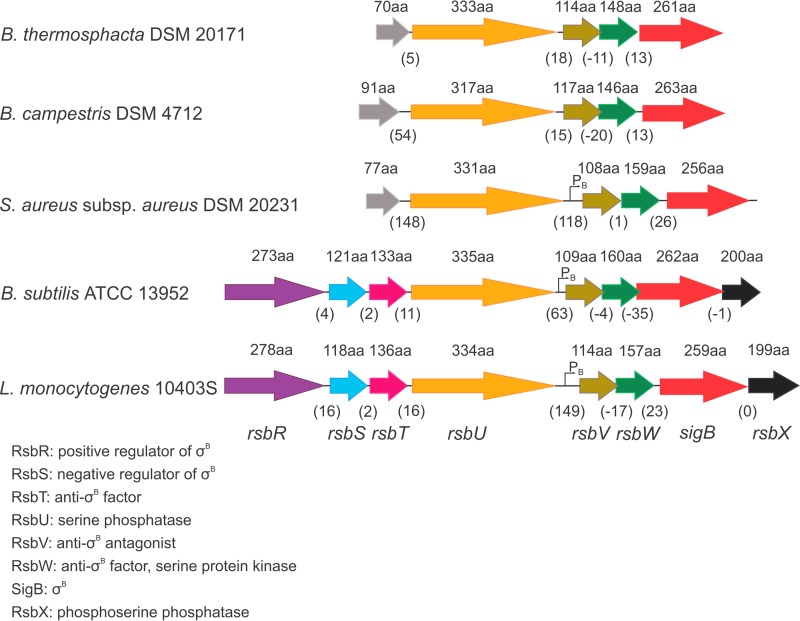
Stress response genes.
Another region of notable synteny identified in the Brochothrix genomes, which extended beyond the Listeriaceae family to other Gram-positive organisms, was a locus encoding the key stress regulator in L. monocytogenes, the alternative sigma factor σB (Fig. 5 and Table 2). Although the highest sequence similarity between amino acid sequences of σB and σB-related gene sequences from Brochothrix was shared with members of the Listeria genus, the gene components of this genomic region were most similar to those of the respective region in Staphylococcus aureus (Fig. 5). In addition to the σB gene, a gene predicted to encode an anti-sigma factor (rsbW) was found, as were anti-sigma factor antagonist (rsbV) and RsbV phosphatase (rsbU) genes. In contrast to L. monocytogenes, B. subtilis, and S. aureus, where the rsbU gene was followed by a significant spacer sequence that holds a known σB-dependent promoter, the spacer region following the rsbU gene was notably shorter in Brochothrix. The length of this spacer in B. thermosphacta was only 18 bp, and it was only 15 bp in B. campestris (Fig. 5). Consequently, a σB-dependent promoter is likely not present between these genes in Brochothrix due to the short length of the respective spacer regions. The rsbU gene was followed by the rsbV, rsbW, and sigB genes. As is the case in S. aureus, an rsbX gene was absent in Brochothrix genomes.
FIG 5.
Conserved synteny of the sigB locus in Gram-positive bacteria. The conserved synteny of the sigB locus identified in the Brochothrix genomes extends beyond the Listeriaceae family to other Gram-positive organisms. In both Brochothrix species, this region harbored predicted rsbU, rsbV, rsbW, and sigB genes. Large spacer regions between the rsbU and rsbV genes in S. aureus, B. subtilis, and L. monocytogenes contain σB-dependent promoters, which are likely not present in this intergenic region in Brochothrix species due to the short length of the respective spacer regions. Gray arrows depict a hypothetical protein-coding sequence.
DISCUSSION
Intergenomic sequence similarity of strains.
A high degree of genomic similarity was observed for the 12 B. thermosphacta strains by using genome relatedness indices and BLASTn ring comparisons. The core genome phylogeny showed that there are several intraspecies clades and that strains 7813 and 7818 from the group 1 lineage were clearly separated from the remaining 10 strains. The high degree of similarity between the type strain and the 11 Australian isolates, and also among the Australian strains, was surprising considering their geographic and temporal differences and the different meat packaging types used for the meat sources of the isolates (37). Although, overall, the temporal and, in particular, the geographic diversities of the isolates were limited, the striking genomic similarity between the Australian isolates and the type strain could indicate that a high degree of genomic similarity is a feature of this species. However, B. thermosphacta is an environmental organism and has been isolated from a variety of foods (2), and therefore, sequencing of isolates from alternative sources may shed more light on the overall intraspecies diversity.
Pangenome investigation.
The core genome of the strains contained an estimated 2,012 coding sequences, which is similar in size to the predicted core genome of the Listeria genus (1,994 core genes) (38). Determination of the pangenome from draft genomic data must be considered an estimate given the variable numbers of contig breakpoints and difficulties in assembling large repeated regions. However, it can be anticipated that the number of genes in the core genome will decrease as the numbers of available B. thermosphacta genomes increase. The accessory genome was estimated to constitute 30.8% of the pangenome of the strains. While this illustrates a relatively limited amount of strain diversity, this number was nonetheless higher than anticipated based on the BLASTn comparisons that we conducted. A notable reduction in the median sequence length of accessory proteins was observed and may be the result of incorrect ORF assignments during the automated annotation process. Hence, it is possible that the prediction of 30.8% accessory genome content of the 12 strains, and, therefore, their strain diversity, is an overestimate.
The accessory genome was categorized into high-, medium-, and low-frequency genes in the place of a typical binary analysis of core and accessory genomes, enabling a linkage of genes in these groups to different evolutionary processes, as suggested by Cordero and Polz (39). High-frequency genes are thought to be maintained primarily by vertical inheritance and homologous recombination, typically encode core metabolic functions, and are potentially involved in habitat-specific tasks (39). In line with these assumptions, the high-frequency accessory pool in B. thermosphacta contained genes involved in signal transduction, transcription regulation, and carbohydrate uptake and metabolism. All of these processes may enable the adaptation of this organism to new environments and the uptake and utilization of different carbon sources and can be explained by the saprophytic lifestyle of this bacterium. It is important to note that as the genomes are drafts, it cannot be excluded that some of the genes from this group may in fact belong to the core genome.
Bacteriophage genes represented the largest proportions of the medium- and low-frequency genes. Also identified among the medium- and low-frequency pools were bacterial defense systems, including CRISPR and restriction modification elements. Interestingly, there was no noticeable correlation between the presence of these genes and the abundance of prophages in the isolates. Low-frequency genes, which tend to be carried on mobile regions of the genome, allow a higher rate of DNA turnover (39), and congruent with this hypothesis, the low-frequency group comprised genes conferring resistance to the antibiotic tetracycline that were located on a putative plasmid.
Cell metabolism.
In line with previous observations, the draft genome sequences of the B. thermosphacta strains and B. campestris DSM 4712 indicated that these organisms should be able to utilize a variety of carbon sources, including C3 compounds such as glycerol (18). Additionally, work by Chiara et al., which identified ethanolamine utilization genes (eut genes) in Brochothrix, as well as the annotation of the draft genomes presented here suggest that this organism may also be capable of utilizing this compound (26). Evidence suggests that the utilization of ethanolamine, a major constituent of lipids in eukaryotic and bacterial cells, may play a role in the coexistence of bacteria like L. monocytogenes with other microorganisms that are incapable of using these substrates (40).
Genes encoding enzymes for the major catabolic pathways, the glycolysis and pentose phosphate pathways, were present in Brochothrix; however, four enzymes of the TCA cycle appear to be absent. Previous reports have shown that Brochothrix does not have an operational TCA cycle, and indeed, this organism must have alternate means to provide amino acid precursors such as oxaloacetate. Like in L. monocytogenes, it is possible that the pyruvate carboxylase-catalyzed carboxylation of pyruvate is the predominant reaction leading to oxaloacetate in B. thermosphacta (41). Furthermore, both the requirement for CO2 for oxaloacetate production and the absence of a decarboxylating enzyme such as malate dehydrogenase, which may be inhibited by elevated CO2 concentrations, could help explain why CO2 has only a limited inhibitory effect on B. thermosphacta.
B. thermosphacta has been shown to be capable of producing a number of organoleptically unpleasant compounds such as acetoin, butanediol, and a variety of fatty acids and alcohols, imparting cheesy, sour, and malty odors that characterize its growth on meat (12, 42, 43). One study also investigated the spoilage ability of B. campestris in vacuum-packaged lamb, demonstrating that this species has the potential to cause spoilage due to the production of off-odors and discoloration of drip in the packs (6). Despite the multitude of studies demonstrating the ability of B. thermosphacta to produce these malodorous substances, the gene inventory responsible for their production has remained unknown. Under anaerobic conditions, Brochothrix species likely engage a pyruvate formate-lyase for the decarboxylation of pyruvate to acetyl-CoA and formate, which could initiate the transformation of pyruvate to acetate via the sequential activity of phosphate acetyltransferase and acetate kinase enzymes and to ethanol via the sequential activity of acetaldehyde dehydrogenase and alcohol dehydrogenase enzymes. These enzymes, in addition to an identified lactate dehydrogenase, likely explain the production of lactate, acetate, formate, and ethanol by B. thermosphacta under anaerobic conditions, as previously described (44). Under aerobic conditions, pyruvate decarboxylase activity may initiate the production of both ethanol and acetate with the reduction of acetaldehyde to ethanol via alcohol dehydrogenase activity and the oxidation of acetaldehyde to acetate by aldehyde dehydrogenase activity. Genes were also found in both species for the production of the key spoilage products acetoin and butanediol, which have been shown to be produced under aerobic conditions when glucose is metabolized (45). Diacetyl production is also characteristic of spoilage caused by B. thermosphacta, and it can be formed via the nonenzymatic oxidative decarboxylation of acetoin (45). All 12 strains demonstrated acetoin production in a Voges-Proskauer test, further underlining the importance of these substances for spoilage caused by Brochothrix.
Previous analyses have shown conflicting results regarding the presence and activity of secreted lipases of B. thermosphacta (5, 15, 16). In both Brochothrix species, a lipase gene with significant homology to lipases in Listeria species as well as numerous esterase genes were identified. Hence, the gene inventory for the production of fatty acid end products from lipolysis may be present in Brochothrix. It is also possible that lipid metabolism in the form of uncharacterized or poorly characterized lipid oxidation pathways may play a role in spoilage caused by this organism. However, there is strong evidence for the production of fatty acid moieties from amino acids (45, 46). In line with these observations, genes for the conversion of the branched-chain amino acids leucine, valine, and isoleucine to their branched-chain acyl-CoA derivatives were present in both species. These steps, which are common for all three amino acids, involve the conversion of the amino acids to their respective α-keto acids and the subsequent conversion of the α-keto acids to acyl-CoAs. In addition, B. thermosphacta strains may be able to produce propanoyl-CoA from distal leucine and valine degradation pathways. Thus, it is likely that together with the activity of acyl-CoA thioesterases present in both species, Brochothrix produces the branched-chain fatty acids isovaleric acid, isobutyric acid, and 2-methylbutyric acid from the degradation of leucine, valine, and isoleucine, while propionic acid production by B. thermosphacta may also be the result of valine and/or isoleucine metabolism.
The secretion of biogenic amines into the environment can contribute to spoilage due to their putrid aroma and can have serious health implications (47, 48). Biogenic amines are produced primarily by the decarboxylation of amino acids via amino acid decarboxylase activity. A number of studies have demonstrated that different B. thermosphacta strains are capable of producing various biogenic amines such as histamine, tyramine, tryptamine, putrescine, and cadaverine (42, 49, 50). Nonetheless, amino acid decarboxylase genes were not found in the genomes of these strains. Moreover, lysine, arginine, and ornithine reactions were negative by using API 20E strips, and an amino acid decarboxylase screen also failed to detect decarboxylase activity for a wide range of target amino acid decarboxylases. The B. thermosphacta strains from this study and B. campestris DSM 4712 may be reliant on the uptake of polyamines such as spermidine or putrescine, via the homologous potA, potB, potC, and potD genes, which enable spermidine-preferential uptake in E. coli (51).
Considering the similarity of the biochemical profiles of B. thermosphacta and B. campestris, it is not surprising that for the metabolic reactions analyzed in this study, the gene inventories of both organisms are so similar. The presence of all genes required for acetoin and butanediol formation also provides further evidence that B. campestris may be capable of spoilage.
Resistance genes.
Although B. thermosphacta is not a pathogen, antibiotic resistance genes in spoilage bacteria increase the gene pool from which pathogens can acquire antibiotic resistance traits (23). Previously, both Brochothrix species were described as being susceptible to a wide range of antibiotics (18). In this study, a tetracycline-resistant isolate (7811) was identified and found to possess a tetL gene, which encodes transmembrane tetracycline efflux pumps in known tetL-carrying organisms such as B. subtilis (30, 31). This gene was located on a contig with ∼14-fold-higher read coverage than the average read coverage of the genome, suggesting that this contig may represent a high-copy-number plasmid.
There is growing concern about bacterial cross-resistance to antibiotics, disinfectants, and/or heavy metals in various environments (52–55). Until now, there have been no available data on the heavy metal tolerance of B. thermosphacta strains. The MTCs of the 12 isolates for the tested heavy metals were not at levels considered high enough for the isolates to be resistant to these substances. However, growth, although impaired, was still observed at much higher concentrations of the metals and could be attributed to the presence of putative copper, cobalt, and cadmium resistance genes identified in the draft genomes. Considering that heavy metal-resistant Listeria isolates are more common among food isolates than among those recovered sporadically (22), further studies on the heavy metal resistance of B. thermosphacta and other spoilage-related organisms, which could also act as potential reservoirs for resistance determinants, may be warranted.
Presence of Listeria virulence genes in Brochothrix.
Notable among the Listeria virulence orthologs detected in the Brochothrix genomes was the strong representation of factors involved in intracellular survival and surface protein anchoring, while the majority of factors with a function in adherence and invasion of mammalian host cells were not found. Homologs of virulence proteins involved in immune modulation were also missing, which is not surprising considering the nonpathogenic nature of this bacterium and the specialized functions that many of these proteins have. Speculation can be made that some of the homologous virulence genes present in the Brochothrix genomes are required for the attachment to and growth on animal carcasses. For example, FbpA (fibronectin-binding protein) facilitates the adhesion of Listeria to host fibronectin (56), while OppA (oligopeptide-binding protein) enables oligopeptide uptake with pleiotropic effects on growth at cold temperatures, and PrsA2 (posttranslocation secretion protein) contributes to the integrity of the cell wall and resistance to osmotic stress in Listeria (57, 58). In addition, sortase B (SrtB) plays an indirect role in virulence in L. monocytogenes and heme/hemoglobin uptake by anchoring Hbp1 and Hbp2 to the peptidoglycan (36, 59). Hbp2 promotes intracellular survival by facilitating bacterial escape from phagosomes in macrophages (59), and while SrtB binds Hbp2 to the cell wall, a pool of Hbp2 is also secreted into the environment, where this protein operates as a hemophore (36). Due to the conserved nature of the srtB locus and the finding of a Fur box upstream of this region in Brochothrix, it is likely that this locus is iron regulated and may contribute to the uptake of heme, facilitating the use of the tightly bound iron present in animal carcasses.
The absence of both key Listeria virulence genes and conserved synteny for the majority of virulence gene orthologs identified in the Brochothrix genomes highlights the degree of genetic divergence between these organisms that we detected. A number of studies suggest that Listeria species have evolved from a pathogenic ancestor and that nonpathogenic Listeria members arose primarily via gene loss (38, 60, 61). Hence, it is possible that Brochothrix species have evolved from a more recent nonpathogenic member of the Listeria genus; however, further phylogenetic studies would be required to answer this intriguing question.
Stress response genes.
In L. monocytogenes, σB plays a crucial role in growth and survival at cold temperatures, at low pH, and under oxidative and osmotic stress and carbon starvation (62–65). It is also involved in regulating a number of genes of virulence determinants, including genes from the internalin family (66). It is tempting to speculate that σB and σB-related genes may regulate gene expression in B. thermosphacta, facilitating survival and growth under high-salt conditions and adaptation to refrigeration temperatures. The practical importance of this key regulatory factor for the food industry was previously recognized, as the activation of σB results in increased stress resistance of the bacterial strains (65). A better fundamental knowledge of the σB activation process and the σB regulon will be crucial for the effective design of food processing steps to ensure that a stress response leading to enhanced resistance of the bacteria to inactivation and food preservation treatments is not activated.
Conclusion.
This study provides a detailed analysis of the B. thermosphacta genome, identifying spoilage-relevant pathways and establishing parallels to and differences from the closely related foodborne pathogen L. monocytogenes. New insights into the gene inventory of Brochothrix are provided, and genes involved in meat spoilage pathways, which were previously described in the literature, are discussed. The limited genetic diversity of the isolates in this study may reflect their niche specificity, and therefore, as more strains of B. thermosphacta are sequenced, it will be interesting to see if this is true of the species as a whole.
MATERIALS AND METHODS
Bacterial strains.
In total, 12 B. thermosphacta strains were used in this study (Table 1). B. campestris DSM 4712 (GenBank accession number AODH00000000.1) and L. monocytogenes 10403S (GenBank accession number CP002002) (67, 68) were included for genomic comparisons. Strains 7803, 7804, 7806, 7807, 7808, 7809, and 7810 were isolated in 1993 during a meat spoilage study (Craven and Baxter-Keene, CSIRO Agriculture and Food, unpublished data) by cutting 25-cm2 cores from the meat and placing them into sterile bags with 300 ml of maximum recovery diluent (MRD). The bags were macerated in a stomacher for 1 min and stored in ice water. Samples of the homogenate were serially diluted in MRD, and 50 μl of appropriate dilutions was plated onto nonselective plate count agar (Oxoid).
Strains 7811, 7813, 7816, and 7818 were isolated for the purpose of this study. The respective meat sources were kept at 4°C until their use-by date, and the strains were then isolated according to methods described previously by Gribble and Brightwell (6).
Strain identification.
PCR was performed prior to whole-genome sequencing to confirm that all isolates were Brochothrix strains. Primers were designed to target the glyceraldehyde-3-phosphate dehydrogenase gene in Brochothrix, following the alignment of this gene with Listeria orthologs and the determination of conserved bases present only in the Brochothrix gene. The forward primer bound to bases 611 to 632 of the gene on the plus strand. The reverse primer targeted bases 107 to 125 of the gene on the minus strand.
Template preparation was performed by immersing a small single colony in 25 μl H2O, heating the sample for 15 min at 95°C before centrifugation at 15,000 × g for 2 min, and using 2 μl of the supernatant as the template. The amplification mixture also contained 0.25 μM each primer (gapdhfwd [5′-GTAGTGTTTGGAACAATGTTAG-3′] and gapdhrev [5′-CTGATGCTTCACAATTAGC-3′]) and 1× HotStarTaq master mix (Qiagen). PCR amplification was performed as follows: an initial denaturation step at 95°C for 15 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min, and polymerization at 72°C for 1 min and a final extension step at 72°C for 4 min.
DNA isolation, whole-genome sequencing, and gene annotation.
B. thermosphacta strains from Table 1 were grown for 18 h (±2 h) in tryptone soya broth (TSB; Oxoid) at 25°C. Genomic DNA was isolated with the Qiagen DNeasy blood and tissue kit according the manufacturer's protocol for Gram-positive bacteria. Library preparation and genome sequencing were carried out at The Ramaciotti Centre at the University of NSW by using the Nextera XT DNA library 300-bp paired-end preparation kit and were performed on the Illumina MiSeq platform (Illumina USA). A public Galaxy instance supported by the CSIRO Bioinformatics Core and IM&T (69) was used to run the assembly software. FastQC:Read QC (version 0.10.1) was applied for quality control of sequence reads and adaptor sequences, and trailing bases were removed by using Trimmomatic (version 1.0.0) (70). A minimum read length of 150 bases was chosen. Assembly of the sequencing reads was performed by using the SPAdes genome assembler tool (version 3.5) with k-mer sizes of 21, 33, 55, and 77 (71). For annotation, the genomes were submitted to RAST (version 2.0; FIGfam version, release 70) (72) and the NCBI Prokaryotic Genome Automatic Annotation Pipeline (version 3.3).
Genome sequence similarity analyses.
Overall genome relatedness indices, Orthologous Average Nucleotide Identity (OrthoANI) (73) and Digital DNA:DNA Hybridization (DDH) estimates (74), were determined by using the OrthoANI tool (version 0.93; BLAST+) and Genome-to-Genome Distance Calculator (GGDC) (version 2, BLAST+) with recommended formula 2 (identities/high-scoring segment pair lengths).
BLASTn comparisons were performed with BRIG (BLAST Ring Image Generator application) (75). B. thermosphacta DSM 20171 was used as the central reference sequence, and 70 and 90% identities were chosen as the lower and upper identity thresholds, respectively. Mauve (version 2.3.1) (76) and PHAST (PHAge Search Tool) (77) were used for identifying bacteriophage genes present in B. thermosphacta DSM 20171 but absent in the query genomes.
Pangenome analysis.
Pangenome analysis was performed with usearch (version 8.1.1861), using a minimum identity of 0.6. Seed sequences were parsed into a tabular form with a custom script (https://github.com/bioinformatics-deakin/gene-matrix-from-uclust3.py), and genes less than 153 nucleotides long (protein sequences <50 amino acids long) were eliminated from the analysis. The core genome of the 12 isolates was determined as the set of genes shared by all strains, while the accessory genome was the set of genes present in a subset of strains or in a single strain.
The accessory genome was split into high-frequency (genes shared by 10 or 11 of the isolates), medium-frequency (genes common to at least 3 and up to 9 strains), and low-frequency (unique genes and genes shared by only 2 isolates) coding sequences. Functional annotation of the translated seed sequences of these accessory groups was conducted with the NCBI CD-Search tool to identify conserved domains by using a mirrored COG (Clusters of Orthologous Groups of Proteins) database with the default settings (with an E value threshold of 0.01, a maximum number of hits of 500, and composition-corrected scoring turned on). As each COG can have one or more general category letter associations, the pools of functional categories were compared for each accessory group. Due to the anticipated high number of nonassignable (i.e., hypothetical proteins) bacteriophage gene products, PHAST was used to determine the prophage regions in each of the 12 B. thermosphacta genomes. All genes within these identified prophage regions were then classified as “bacteriophage” genes.
The abundances of general categories from COG assignment and prophage genes determined by PHAST were normalized for high-, medium-, and low-frequency genes by dividing these values by the number of genes present in each frequency group. Statistical analysis of the lengths of the core and accessory proteins was conducted by using BoxPlotR as implemented in BoxPlotR (http://boxplot.tyerslab.com/).
Phylogeny of Brochothrix strains.
Progressive Mauve (78) was used to create whole-genome alignments of B. thermosphacta strains and B. campestris DSM 4712. StripSubset removed locally collinear blocks (LCBs) of <2,000 bases, and Gblocks (version 0.91b) (79, 80) was run with the default settings to remove poorly aligned positions and divergent regions of the alignments prior to phylogenetic analysis. Tree building was performed by using FastTree (version 2.1.7) (81, 82) with the general time reversible (GTR) gamma model of nucleotide evolution (determined by jModelTest [version 2.1.7] [83, 84] as the most suitable model) and 1,000 bootstrapping replicates. The tree was rooted on B. campestris DSM 4712.
Identification of genes involved in cell metabolism.
SEED Viewer was used to identify reactions for cellular processes in the B. thermosphacta strains and B. campestris DSM 4712. Pathway details were obtained by using KEGG pathway reconstructions, and comparisons were accessed via SEED Viewer (85). Reconstructed KEGG pathways for which many reactions were present and only single or a few steps were missing were manually controlled for gap filling. Genes not found within these models or gene candidates found to be missing critical protein domains are reported as being not identified in the genomes.
Virulence factor search in Brochothrix.
A Brochothrix protein database was generated by using all protein sequences from the 12 B. thermosphacta strains and B. campestris DSM 4712. Listeria virulence factors included in this analysis were obtained from the table supplied for pathogenomic comparisons of Listeria spp. from the Virulence Factors of Pathogenic Bacteria (VFPB) database (http://www.mgc.ac.cn/cgi-bin/VFs/compvfs.cgi?Genus=Listeria). This list of 44 Listeria virulence proteins was searched against the Brochothrix database by using the scoring matrix BLOSUM62 and the BLASTp algorithm. Proteins with statistically significant sequence similarity (BLASTp E value score cutoff of <10−5 and bit score of ≥50) were subjected to domain analysis by using the InterProScan program, and gene synteny was manually assessed.
Biochemical tests of strains.
API 20E strips (bioMérieux) were used to test the following reactions: arginine, lysine, and ornithine decarboxylase activity; Voges-Proskauer reactions; and mannitol fermentation. Two strips per strain were inoculated according to the manufacturer's instructions. One strip was incubated at 25°C for 24 h. The second strip was incubated at 25°C for 30 h. Reactions were read according to the product's instructions. Quality control of the strips was tested by using Escherichia coli strain ATCC 25922.
Screening for biogenic amine production.
An improved decarboxylase medium as described previously by Bover-Cid and Holzapfel (86) was used, with one adjustment: the pH of the medium was set to pH 5.6 to better mimic the pH of raw meat. Strains were grown at 25°C for 18 h (±2 h) in nutrient standard broth (Oxoid) (with 1% precursor amino acids and 0.005% pyridoxal-5-phosphate added). Strains were then streaked onto decarboxylase medium containing the amino acids l-phenylalanine and l-tryptophan and the amino acid salts l-histidine monohydrochloride (Sigma), l-lysine hydrochloride (BDH Chemicals), and dl-ornithine monohydrochloride (Sigma) and without amino acids (as controls) and were incubated for 7 days at 25°C under aerobic and anaerobic conditions in parallel. Positive reactions increase the pH of the medium, changing the color of the medium from yellow/brown to purple. E. coli ATCC 25922 was used as a positive control for testing this system due to its known lysine and ornithine decarboxylase activities.
MIC testing of antibiotics.
MICs of tetracycline, erythromycin, penicillin G, and ciprofloxacin were established by using Thermo Scientific Oxoid MICE strips according to the manufacturer's protocol for Listeria, with one exception: the plate incubation temperature was 25°C to allow the growth of Brochothrix.
MTC testing of heavy metals.
The MTCs of three heavy metal salts (CoCl2, CdCl, and CuSO4) were tested by using a modified plate method (87). B. thermosphacta strains were grown at 25°C for 18 h (±2 h) in TSB medium. Subsequently, 100 μl of each cultured strain were spread onto tryptic soya agar (TSA) plates containing no heavy metal (control plates); 50, 100, 200, 300, 400, 450, 500, and 600 μg/ml of CoCl2; 5, 10, 15, 30, 50, 70, 100, and 200 μg/ml of CdCl; and 100, 200, 300, 350, and 400 μg/ml of CuSO4. Plates were incubated at 25°C and checked for growth at daily intervals for up to 5 days. The MTC was determined as the highest metal concentration at which the growth of bacteria was not altered compared to their growth on control plates. MTCs are lower than MICs by a factor of ∼2 (88, 89).
Accession number(s).
GenBank accession numbers of the 12 B. thermosphacta strains are listed in Table 1.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02786-16.
REFERENCES
- 1.Borch E, Kant-Muermans M-L, Blixt Y. 1996. Bacterial spoilage of meat and cured meat products. Int J Food Microbiol 33:103–120. doi: 10.1016/0168-1605(96)01135-X. [DOI] [PubMed] [Google Scholar]
- 2.Holley RA. 2014. Brochothrix, p 331–334. In Tortorello CABL. (ed), Encyclopedia of food microbiology, 2nd ed Academic Press, Oxford, United Kingdom. doi: 10.1016/B978-0-12-384730-0.00048-3. [DOI] [Google Scholar]
- 3.Mamlouk K, Macé S, Guilbaud M, Jaffrès E, Ferchichi M, Prévost H, Pilet M-F, Dousset X. 2012. Quantification of viable Brochothrix thermosphacta in cooked shrimp and salmon by real-time PCR. Food Microbiol 30:173–179. doi: 10.1016/j.fm.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Ercolini D, Ferrocino I, Nasi A, Ndagijimana M, Vernocchi P, La Storia A, Laghi L, Mauriello G, Guerzoni ME, Villani F. 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl Environ Microbiol 77:7372–7381. doi: 10.1128/AEM.05521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowak A, Rygala A, Oltuszak-Walczak E, Walczak P. 2012. The prevalence and some metabolic traits of Brochothrix thermosphacta in meat and meat products packaged in different ways. J Sci Food Agric 92:1304–1310. doi: 10.1002/jsfa.4701. [DOI] [PubMed] [Google Scholar]
- 6.Gribble A, Brightwell G. 2013. Spoilage characteristics of Brochothrix thermosphacta and campestris in chilled vacuum packaged lamb, and their detection and identification by real time PCR. Meat Sci 94:361–368. doi: 10.1016/j.meatsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Prieto M, Garcialopez ML, Garciaarmesto MR, Otero A, Lopez TM, Moreno B. 1993. Factors affecting spoilage microflora succession on lamb carcasses at refrigeration temperatures. J Appl Bacteriol 74:521–525. [PubMed] [Google Scholar]
- 8.Barlow J, Kitchell AG. 1966. A note on the spoilage of prepacked lamb chops by Microbacterium thermosphactum. J Appl Bacteriol 29:185–188. doi: 10.1111/j.1365-2672.1966.tb03466.x. [DOI] [Google Scholar]
- 9.Grau FH, Eustace IJ, Bill BA. 1985. Microbial flora of lamb carcasses stored at 0°C in packs flushed with nitrogen or filled with carbon dioxide. J Food Sci 50:482–485. [Google Scholar]
- 10.Banks JG, Dalton HK, Nychas GJ, Board RG. 1985. Sulfite, an elective agent in the microbiological and chemical changes occurring in uncooked comminuted meat products. J Appl Biochem 7:161–179. [PubMed] [Google Scholar]
- 11.Stringer SC, Chaffey BJ, Dodd CER, Morgan MRA, Waites WM. 1995. Specific antibody-mediated detection of Brochothrix thermosphacta in situ in British fresh sausage. J Appl Bacteriol 78:335–340. doi: 10.1111/j.1365-2672.1995.tb03415.x. [DOI] [PubMed] [Google Scholar]
- 12.Nychas GJE, Drosinos EH. 2014. Meat and poultry—spoilage of meat, p 514–519. In Tortorello CABL. (ed), Encyclopedia of food microbiology, 2nd ed Academic Press, Oxford, United Kingdom. doi: 10.1016/B978-0-12-384730-0.00194-4. [DOI] [Google Scholar]
- 13.Labadie J. 1999. Consequences of packaging on bacterial growth. Meat is an ecological niche. Meat Sci 52:299–305. doi: 10.1016/S0309-1740(99)00006-6. [DOI] [PubMed] [Google Scholar]
- 14.Blickstad E, Molin G. 1984. Growth and end-product formation in fermenter cultures of Brochothrix thermosphacta ATCC 11509T and two psychrotrophic Lactobacillus spp. in different gaseous atmospheres. J Appl Bacteriol 57:213–220. doi: 10.1111/j.1365-2672.1984.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 15.Collins-Thompson DL, Sørhaug T, Witter LD, Ordal ZJ. 1971. Glycerol ester hydrolase activity of Microbacterium thermosphactum. Appl Microbiol 21:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papon M, Talon R. 1988. Factors affecting growth and lipase production by meat lactobacilli strains and Brochothrix thermosphacta. J Appl Bacteriol 64:107–115. doi: 10.1111/j.1365-2672.1988.tb02729.x. [DOI] [PubMed] [Google Scholar]
- 17.Talon R, Grimont PAD, Grimont F, Gasser F, Boeufgras JM. 1988. Brochothrix campestris sp. nov. Int J Syst Bacteriol 38:99–102. doi: 10.1099/00207713-38-1-99. [DOI] [Google Scholar]
- 18.Stackebrandt E, Jones D. 2006. The genus Brochothrix, p 477–491. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, 3rd ed Springer, New York, NY. doi: 10.1007/0-387-30744-3_12. [DOI] [Google Scholar]
- 19.Ludwig W, Schleifer K-H, Stackebrandt E. 1984. 16S rRNA analysis of Listeria monocytogenes and Brochothrix thermosphacta. FEMS Microbiol Lett 25:199–204. doi: 10.1111/j.1574-6968.1984.tb01456.x. [DOI] [Google Scholar]
- 20.Patterson JT, Gibbs PA. 1978. Sources and properties of some organisms isolated in two abattoirs. Meat Sci 2:263–273. doi: 10.1016/0309-1740(78)90027-X. [DOI] [PubMed] [Google Scholar]
- 21.Nychas G-JE, Skandamis PN, Tassou CC, Koutsoumanis KP. 2008. Meat spoilage during distribution. Meat Sci 78:77–89. doi: 10.1016/j.meatsci.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Ratani SS, Siletzky RM, Dutta V, Yildirim S, Osborne JA, Lin W, Hitchins AD, Ward TJ, Kathariou S. 2012. Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl Environ Microbiol 78:6938–6945. doi: 10.1128/AEM.01553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verraes C, Van Boxstael S, Van Meervenne E, Van Coillie E, Butaye P, Catry B, de Schaetzen M-A, Van Huffel X, Imberechts H, Dierick K, Daube G, Saegerman C, De Block J, Dewulf J, Herman L. 2013. Antimicrobial resistance in the food chain: a review. Int J Environ Res Public Health 10:2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5:e01918-14. doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilcher S, Loessner MJ, Klumpp J. 2010. Brochothrix thermosphacta bacteriophages feature heterogeneous and highly mosaic genomes and utilize unique prophage insertion sites. J Bacteriol 192:5441–5453. doi: 10.1128/JB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiara M, Caruso M, D'Erchia AM, Manzari C, Fraccalvieri R, Goffredo E, Latorre L, Miccolupo A, Padalino I, Santagada G, Chiocco D, Pesole G, Horner DS, Parisi A. 2015. Comparative genomics of Listeria sensu lato: genus-wide differences in evolutionary dynamics and the progressive gain of complex, potentially pathogenicity-related traits through lateral gene transfer. Genome Biol Evol 7:2154–2172. doi: 10.1093/gbe/evv131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 28.Hof H, Nichterlein T, Kretschmar M. 1997. Management of listeriosis. Clin Microbiol Rev 10:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy SB, McMurry LM, Barbosa TM, Burdett V, Courvalin P, Hillen W, Roberts MC, Rood JI, Taylor DE. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother 43:1523–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi R, Amano H, Shishido K. 1988. Nucleotide sequence homology of the tetracycline-resistance determinant naturally maintained in Bacillus subtilis Marburg 168 chromosome and the tetracycline-resistance gene of B. subtilis plasmid pNS1981. Biochim Biophys Acta 950:441–444. doi: 10.1016/0167-4781(88)90142-X. [DOI] [PubMed] [Google Scholar]
- 31.Takayuki H, Takayuki I, Noboru T, Kensuke F. 1985. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene 37:131–138. doi: 10.1016/0378-1119(85)90265-3. [DOI] [PubMed] [Google Scholar]
- 32.Mullapudi S, Siletzky RM, Kathariou S. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl Environ Microbiol 74:1464–1468. doi: 10.1128/AEM.02426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLauchlin J, Hampton MD, Shah S, Threlfall EJ, Wieneke AA, Curtis GDW. 1997. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J Appl Microbiol 83:381–388. doi: 10.1046/j.1365-2672.1997.00238.x. [DOI] [PubMed] [Google Scholar]
- 34.Najiah M, Lee SW, Wendy W, Tee LW, Nadirah M, Faizah SH. 2009. Antibiotic resistance and heavy metals tolerance in gram-negative bacteria from diseased American bullfrog (Rana catesbeiana) cultured in Malaysia. Agric Sci China 8:1270–1275. doi: 10.1016/S1671-2927(08)60338-7. [DOI] [Google Scholar]
- 35.Dekker L, Osborne TH, Santini JM. 2014. Isolation and identification of cobalt- and caesium-resistant bacteria from a nuclear fuel storage pond. FEMS Microbiol Lett 359:81–84. doi: 10.1111/1574-6968.12562. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Q, Jiang X, Moore KJ, Shao Y, Pi H, Dubail I, Charbit A, Newton SM, Klebba PE. 2011. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol 80:1581–1597. doi: 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sulzbacher WL, Mclean RA. 1951. The bacterial flora of fresh pork sausage. Food Technol 5:7–8. [Google Scholar]
- 38.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 40.Tang S, Orsi RH, den Bakker HC, Wiedmann M, Boor KJ, Bergholz TM. 2015. Transcriptomic analysis of the adaptation of Listeria monocytogenes to growth on vacuum-packed cold smoked salmon. Appl Environ Microbiol 81:6812–6824. doi: 10.1128/AEM.01752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joseph B, Goebel W. 2007. Life of Listeria monocytogenes in the host cells' cytosol. Microbes Infect 9:1188–1195. doi: 10.1016/j.micinf.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Casaburi A, De Filippis F, Villani F, Ercolini D. 2014. Activities of strains of Brochothrix thermosphacta in vitro and in meat. Food Res Int 62:366–374. doi: 10.1016/j.foodres.2014.03.019. [DOI] [Google Scholar]
- 43.Casaburi A, Piombino P, Nychas G-J, Villani F, Ercolini D. 2015. Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol 45(Part A):83–102. doi: 10.1016/j.fm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Grau FH. 1983. End products of glucose fermentation by Brochothrix thermosphacta. Appl Environ Microbiol 45:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dainty RH, Hibbard CM. 1983. Precursors of the major end products of aerobic metabolism of Brochothrix thermosphacta. J Appl Bacteriol 55:127–133. doi: 10.1111/j.1365-2672.1983.tb02656.x. [DOI] [PubMed] [Google Scholar]
- 46.Dainty RH, Hofman FJK. 1983. The influence of glucose concentration and culture incubation time on end-product formation during aerobic growth of Brochothrix thermosphacta. J Appl Bacteriol 55:233–239. doi: 10.1111/j.1365-2672.1983.tb01320.x. [DOI] [Google Scholar]
- 47.Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, Ishihama A, Igarashi K. 2004. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem 279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
- 48.Shalaby AR. 1996. Significance of biogenic amines to food safety and human health. Food Res Int 29:675–690. doi: 10.1016/S0963-9969(96)00066-X. [DOI] [Google Scholar]
- 49.Nowak A, Czyzowska A. 2011. In vitro synthesis of biogenic amines by Brochothrix thermosphacta isolates from meat and meat products and the influence of other microorganisms. Meat Sci 88:571–574. doi: 10.1016/j.meatsci.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Paleologos EK, Savvaidis IN, Kontominas MG. 2004. Biogenic amines formation and its relation to microbiological and sensory attributes in ice-stored whole, gutted and filleted Mediterranean Sea bass (Dicentrarchus labrax). Food Microbiol 21:549–557. doi: 10.1016/j.fm.2003.11.009. [DOI] [Google Scholar]
- 51.Igarashi K, Kashiwagi K. 1999. Polyamine transport in bacteria and yeast. Biochem J 344:633–642. doi: 10.1042/bj3440633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chapman JS. 2003. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegrad 51:271–276. doi: 10.1016/S0964-8305(03)00044-1. [DOI] [Google Scholar]
- 53.Langsrud S, Sidhu MS, Heir E, Holck AL. 2003. Bacterial disinfectant resistance—a challenge for the food industry. Int Biodeterior Biodegrad 51:283–290. doi: 10.1016/S0964-8305(03)00039-8. [DOI] [Google Scholar]
- 54.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Elhanafi D, Dutta V, Kathariou S. 2010. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl Environ Microbiol 76:8231–8238. doi: 10.1128/AEM.02056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osanai A, Li S-J, Asano K, Sashinami H, Hu D-L, Nakane A. 2013. Fibronectin-binding protein, FbpA, is the adhesin responsible for pathogenesis of Listeria monocytogenes infection. Microbiol Immunol 57:253–262. doi: 10.1111/1348-0421.12030. [DOI] [PubMed] [Google Scholar]
- 57.Borezee E, Pellegrini E, Berche P. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun 68:7069–7077. doi: 10.1128/IAI.68.12.7069-7077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cahoon LA, Freitag NE. 2014. Listeria monocytogenes virulence factor secretion: don't leave the cell without a chaperone. Front Cell Infect Microbiol 4:13. doi: 10.3389/fcimb.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borezée E, Pellegrini E, Beretti J-L, Berche P. 2001. SvpA, a novel surface virulence-associated protein required for intracellular survival of Listeria monocytogenes. Microbiology 147:2913–2923. doi: 10.1099/00221287-147-11-2913. [DOI] [PubMed] [Google Scholar]
- 60.Hain T, Steinweg C, Kuenne CT, Billion A, Ghai R, Chatterjee SS, Domann E, Kärst U, Goesmann A, Bekel T, Bartels D, Kaiser O, Meyer F, Pühler A, Weisshaar B, Wehland J, Liang C, Dandekar T, Lampidis R, Kreft J, Goebel W, Chakraborty T. 2006. Whole-genome sequence of Listeria welshimeri reveals common steps in genome reduction with Listeria innocua as compared to Listeria monocytogenes. J Bacteriol 188:7405–7415. doi: 10.1128/JB.00758-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmid MW, Ng EYW, Lampidis R, Emmerth M, Walcher M, Kreft J, Goebel W, Wagner M, Schleifer K-H. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst Appl Microbiol 28:1–18. doi: 10.1016/j.syapm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 62.Becker LA, Çetin MS, Hutkins RW, Benson AK. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol 180:4547–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Becker LA, Evans SN, Hutkins RW, Benson AK. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J Bacteriol 182:7083–7087. doi: 10.1128/JB.182.24.7083-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferreira A, O'Byrne CP, Boor KJ. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol 67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wemekamp-Kamphuis HH, Wouters JA, de Leeuw PPLA, Hain T, Chakraborty T, Abee T. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl Environ Microbiol 70:3457–3466. doi: 10.1128/AEM.70.6.3457-3466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edman DC, Pollock MB, Hall ER. 1968. Listeria monocytogenes L forms. I. Induction, maintenance, and biological characteristics. J Bacteriol 96:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol 139:2005–2009. [PubMed] [Google Scholar]
- 69.Goecks J, Nekrutenko A, Taylor J, Galaxy Team . 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee I, Kim YO, Park S-C, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 74.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darling AE, Mau B, Perna NT. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 80.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 81.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 84.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bover-Cid S, Holzapfel WH. 1999. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol 53:33–41. doi: 10.1016/S0168-1605(99)00152-X. [DOI] [PubMed] [Google Scholar]
- 87.Hu N, Zhao B. 2007. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol Lett 267:17–22. doi: 10.1111/j.1574-6968.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- 88.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmidt T, Stoppel R-D, Schlegel HG. 1991. High-level nickel resistance in Alcaligenes xylosoxydans 31A and Alcaligenes eutrophus KTO2. Appl Environ Microbiol 57:3301–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.