Abstract
Staphylococcus epidermidis and Staphylococcus aureus are opportunistic pathogens that cause nosocomial and chronic biofilm-associated infections. Indwelling medical devices and contact lenses are ideal ecological niches for formation of staphylococcal biofilms. Bacteria within biofilms are known to display reduced susceptibilities to antimicrobials and are protected from the host immune system. High rates of acquired antibiotic resistances in staphylococci and other biofilm-forming bacteria further hamper treatment options and highlight the need for new anti-biofilm strategies. Here, we aimed to evaluate the potential of marine sponge-derived actinomycetes in inhibiting biofilm formation of several strains of S. epidermidis, S. aureus, and Pseudomonas aeruginosa. Results from in vitro biofilm-formation assays, as well as scanning electron and confocal microscopy, revealed that an organic extract derived from the marine sponge-associated bacterium Streptomyces sp. SBT343 significantly inhibited staphylococcal biofilm formation on polystyrene, glass and contact lens surfaces, without affecting bacterial growth. The extract also displayed similar antagonistic effects towards the biofilm formation of other S. epidermidis and S. aureus strains tested but had no inhibitory effects towards Pseudomonas biofilms. Interestingly the extract, at lower effective concentrations, did not exhibit cytotoxic effects on mouse fibroblast, macrophage and human corneal epithelial cell lines. Chemical analysis by High Resolution Fourier Transform Mass Spectrometry (HRMS) of the Streptomyces sp. SBT343 extract proportion revealed its chemical richness and complexity. Preliminary physico-chemical characterization of the extract highlighted the heat-stable and non-proteinaceous nature of the active component(s). The combined data suggest that the Streptomyces sp. SBT343 extract selectively inhibits staphylococcal biofilm formation without interfering with bacterial cell viability. Due to absence of cell toxicity, the extract might represent a good starting material to develop a future remedy to block staphylococcal biofilm formation on contact lenses and thereby to prevent intractable contact lens-mediated ocular infections.
Keywords: marine sponges, actinomycetes, Streptomyces, staphylococci, biofilms, contact lens
Introduction
Ocular devices such as intraocular lenses, posterior contact lenses, conjunctival plugs and orbital implants have aided in restoring and improving human vision. However, contamination of these devices with bacterial biofilms can lead to device-related ocular infections such as endophthalmitis, crystalline keratopathy, corneal ulceration, keratitis, lacrimal system, and periorbital infections (Bispo et al., 2015; Cho et al., 2015). The National Institute of Health (NIH) estimates that biofilms contribute to about 75% of the human microbial infections. Biofilms are surface-associated sessile microbial communities that are enmeshed in a self-produced extracellular matrix composed of polysaccharides, proteins, lipids and DNA (Flemming and Wingender, 2010; Oja et al., 2015). Compared to the free-living planktonic counterparts, bacteria in biofilms are 1000-fold more resistant to conventional antibiotic therapies and host immune responses (Donlan, 2001; Otto, 2009; Burmolle et al., 2010). The highly persistent and detrimental nature of biofilm-associated infections and rapid emergence of multidrug resistant strains (Barros et al., 2014; Sakimura et al., 2015) has imposed a major burden on health-care and medical settings. The current inexistence of effective biofilm-based therapeutics (Bjarnsholt et al., 2013) has necessitated the need for development of novel antibiofilm strategies for prophylaxis and/or treatment of the multitude of biofilm-associated ocular infections.
Staphylococci, particularly S. epidermidis and S. aureus are the most common causative agents of device-related infections. Infections caused by other staphylococci are far less frequent (Otto, 2009). S. epidermidis and S. aureus are commensal Gram positive bacteria found on human skin and nares, causing a wide range of indwelling medical device-related infections (Rogers et al., 2009; Uribe-Alvarez et al., 2016). The biofilm-based lifestyle of S. epidermidis and S. aureus on medical devices is a hallmark of the sub-acute and chronic recalcitrant infections caused by them (Reiter et al., 2014). Biofilm formation in S. epidermidis and S. aureus is facilitated predominantly by the synthesis of the homopolymer polysaccharide intercellular adhesion (PIA) by the enzymes coded by the ica locus. PIA-independent biofilm formation, mediated by surface proteins such as biofilm associated protein (Bap/Bhp) and accumulation associated protein (Aap), eDNA release, autolysins and cell sortase-anchored proteins have also been reported in several staphylococcal strains (Otto, 2009; Archer et al., 2011; McCarthy et al., 2015).
Marine actinomycetes represent an untapped reservoir of a broad range of biologically active compounds of pharmaceutical importance (Subramani and Aalbersberg, 2012; Zotchev, 2012; Lee et al., 2014; Azman et al., 2015). Particularly, the marine sponge-associated actinomycetes are well documented for their intrinsic chemical repertoire (Abdelmohsen et al., 2014a; Santos-Gandelman et al., 2014; Reimer et al., 2015). Novel secondary metabolites with discrete biological activities have been reported from sponge-associated actinomycetes (Abdelmohsen et al., 2014a; Grkovic et al., 2014; Cheng et al., 2015). These include antimicrobial (Hentschel et al., 2001; Eltamany et al., 2014), antiparasitic (Dashti et al., 2014; Viegelmann et al., 2014), immunomodulatory (Tabares et al., 2011), antichlamydial (Reimer et al., 2015), antioxidant (Abdelmohsen et al., 2012; Grkovic et al., 2014), anticancer (Vicente et al., 2015) and anti-biofilm (Oja et al., 2015; Park et al., 2016) activities. The extreme and dynamic conditions offered by the oceans (differences in temperature, pH, pressure, light intensities etc.) are the potential reasons often linked to the production of secondary metabolites by marine actinomycetes (Abdelmohsen et al., 2017). The frequent rediscovery of bioactive compounds and redundancy of sample strains from terrestrial environment has further made the marine actinomycetes as hotspots for discovery of new compounds (Dalisay et al., 2013). Among actinomycetes, the genus Streptomyces are considered to be the most prolific producers of secondary metabolites for medical, agriculture and veterinary usage (Tan et al., 2016). Over two-thirds of natural products isolated to date are from Streptomyces which indicates their huge biosynthetic potential and chemistry profiles. The rich genetic and metabolic diversity, and the ability to catabolize a wide range of compounds, has made the genus Streptomyces to be probed for discovery of novel compounds that could be translated to clinical applications (Hassan et al., 2016; Ser et al., 2016; Yang and Sun, 2016; Perez et al., 2016; Zhao et al., 2016).
In our continuing effort for discovery of anti-biofilm agents, we employed a crystal violet-based screening method to identify anti-biofilm activity of organic extracts generated from marine sponge-derived actinomycetes. The biofilm forming reference strain S. epidermidis RP62A was employed as a model for screening. Herein we report the inhibitory effects of an organic extract from marine sponge-associated Streptomyces sp. SBT343 against the biofilm formation of S. epidermidis RP62A on polystyrene, glass and contact lens surfaces. The potential anti-biofilm activity was tested on two other strains of S. epidermidis, four different S. aureus strains and two different P. aeruginosa strains. The results obtained highlighted that the extract exhibited potent anti-biofilm effects on all the staphylococcal strains tested but did not exert any effect on the Pseudomonas strains. Preliminary evaluations on the physico-chemical characterization of active component(s) in the extract suggested their heat stable and non-proteinaceous nature.
Materials and Methods
Pathogenic Strains and Growth Conditions
Bacterial strains used in the study are listed in Table 1. All strains for the biofilm study were propagated in Tryptic Soy Broth (TSB; Becton Dickinson) (17.0 g/l pancreatic digest of casein, 3.0 g/l papaic digest of soybean meal, 5.0 g/l sodium chloride, 2.5 g/l dipotassium hydrogen phosphate, 2.5 g/l glucose) and incubated at 37°C.
Table 1.
Strains used in this study.
| Strain | Description | Reference and/or source |
|---|---|---|
| S. epidermidis ATCC 12228 | Non-infection associated strain | ATCC collection |
| S. carnosus TM300 | Meat starter culture | Rosenstein et al., 2009 |
| S. epidermidis RP62A | Reference strain isolated from intra-vascular catheter associated sepsis | ATCC collection |
| S. epidermidis O-47 | Clinical isolate from septic arthritis | Heilmann et al., 1996 |
| S. epidermidis 1457 | Clinical isolate from a patient with infected central venous catheter | Mack et al., 1992 |
| S. aureus SH1000 | MSSA; rsbU derivative of 8325-4 rsbU+ | Horsburgh et al., 2002 |
| S. aureus RN4220 | Restriction-deficient transformation recipient; originally derived from NCTC 8325-4 | Kreiswirth et al., 1983 |
| S. aureus Newman | MSSA isolate from osteomyelitis patient | Lipinski et al., 1967 |
| S. aureus USA300 | CA-MRSA isolate from a wrist abscess | McDougal et al., 2003 |
| P. aeruginosa PAO1 | Clinical isolate from wound | Dr. Vinay Pawar, Braunschweig, Germany |
| P. aeruginosa PA14 | Clinical isolate from burn wound | Dr. Vinay Pawar, Braunschweig, Germany |
Fermentation Conditions and Extract Preperation
Streptomyces sp. SBT343 was cultivated from the Mediterranean sponge Petrosia ficiformis that was collected offshore Pollonia, Milos, Greece (N36.76612°; E24.51530°) in May 2013 (Cheng et al., 2015). Briefly, 1% (v/v) inoculum of a well grown culture of Streptomyces sp. SBT343 was inoculated in 150 mL modified ISP2 medium (2.5 g/l malt extract, 1.0 g/l yeast extract, D-mannitol 25 mM, in artificial sea water) in a 250 mL conical flask and the strain was subjected to batch fermentation (incubation at 30°C with shaking at 150 rpm) for 10 days. After harvesting, the filtrate of the fermented culture was extracted twice with 250 mL ethyl acetate for each time. The extract was generated by evaporating the solvent in a rotational evaporator (Heidolph Laborota, 4001, Büchi Vacuum Controller V-805). The modified ISP2 broth medium control ensured the purity of the fermentation and was also extracted separately in a similar manner and this served as the medium control for the bioactivity testing. Extracts were dissolved in DMSO (final concentration 3.75% on the cells) and used for anti-biofilm assays.
Quantification of Biofilm Formation
Quantification of biofilm formation was performed according to Weisser et al. (Weisser et al., 2010). Briefly, overnight culture of bacterial strains were diluted with TSB medium to OD600 of 0.05, and incubated statically in a 96-well flat bottom polystyrene plate (Greiner bio-one, GmbH, Germany) with or without the varying concentrations of the Streptomyces sp. SBT343 extract (0, 31.25, 62.5, 125, and 250 μg/ml) at 37°C (for S. epidermidis and P. aeruginosa) or 30°C (for S. aureus) for 24 h. Extract from the modified ISP2 medium control (250 μg/ml) served as the control for the experiment. For biofilm quantification, planktonic bacteria were discarded, and the wells were rinsed carefully with sterile 1X phosphate buffered saline (PBS) twice, and the adherent cells in the plate were heat-fixed at 65°C for 1 h. This was followed by staining with 0.3% crystal violet for 5 min. The stained biofilm was washed with sterile water thrice. Plates were dried in an inverted position and OD492 readings were measured to compare the extent of biofilm inhibition in the extract treated sets vs. the modified ISP2 medium control treated set. The biofilm-negative S. epidermidis (ATCC12228) and S. carnosus TM300 served as the negative controls in the experiment.
Effect of the Extract on Existing Biofilms
To study the effect of the Streptomyces sp. SBT343 extract on the existing biofilm, biofilms were established by incubating the bacteria (inoculation OD600 0.05) in TSB medium in a 96-well flat bottom polystyrene plate at 37°C or 30°C for 24 h. Planktonic bacteria were discarded by inverting the plate on paper stacks. This was followed by addition of fresh TSB medium with varying concentration of the Streptomyces sp. SBT343 extract and the plate was again incubated statically at 37°C or 30°C for 24 h. The effect of the extract in eradication of preformed biofilm by the extract was measured by the crystal violet assay as explained above. Extract from the modified ISP2 medium control (250 μg/ml) and sodium metaperiodate (40 mM) served as the negative and positive controls in the experiment.
Growth Measurements
The effect of Streptomyces sp. SBT343 extract on the growth of staphylococcal strains was evaluated (Nithya et al., 2010) by growth curve analysis. The growth curves were determined up to 24 h. The extract at 50% biofilm inhibitiory concentration (BIC50) and the highest tested concentration (125 μg/ml) (non-toxic to cell lines), was added to a conical flask containing bacteria at an OD600 of 0.1. The flasks were incubated 37°C under shaking conditions at 200 rpm. TSB medium without the bacteria served as negative control. Growth medium with the modified ISP2 medium extract (125 μg/ml) and bacteria served as the control. The reading was observed continuously up to 24 h at 2 h intervals. The experiment was carried out with three independent cultures.
Investigation on the Appearance of Biofilm-Switches Upon Extract Treatment
In order to study the appearance of biofilm-switches [PIA-dependent to PIA-independent (protein-mediated) biofilms] in the presence of the extract, the residual biofilms after the extract treatments were challenged with metaperiodate (40 mM NaIO4) for PIA-dependent and proteinase K (1 mg/ml in 100 mM Tris-Cl) for PIA-independent (protein-mediated) modes of biofilm formation with the procedure described elsewhere (Wang et al., 2004).
Scanning Electron Microscopy
In order to evaluate the effect of the extract on the biofilm formation on soft contact lens (Proclear®, CooperVision® Lensbest, Germany), round pieces (diameter of 6 mm) were punched out of each contact lens using a sterile puncher. Each piece was washed twice with sterile 1XPBS and placed in a 24 well plate (Greiner bio-one, GmbH, Germany) containing 1 mL of S. epidermidis RP62A (OD600 0.05) in TSB with the extract (at concentrations of 62.5, 125, and 250 μg/ml). For testing the effect of the extract on the biofilm formation on glass surface, sterile glass cover slips (diameter of 12 mm) were placed in a 24 well plate containing 1 mL S. epidermidis RP62A (OD600 0.05) in TSB with the extract (at concentrations of 62.5, 125, and 250 μg/ml). In each case, bacteria treated with extract (250 μg/ml) from modified ISP2 medium served as the control, while wells with the TSB and the contact lens or cover slips alone served as sterile controls, respectively. Plates containing the contact lens and cover slips sets were then incubated statically at 37°C for 24 h. Samples were then washed carefully with sterile 1XPBS twice, and fixed overnight with 6.25% gluteraldehyde (in 50 mM phosphate buffer pH 7.4). After overnight fixation, samples were washed 5 times with Sörenson buffer (100 mM KH2PO4 and 100 mM Na2HPO4) and transferred to the electron microscopy unit, where they were dehydrated and then coated with gold by a low vacuum sputter coating, and scanned by scanning electron microscopy.
Confocal Microscopy
Samples on the contact lens and the cover slips were prepared and treated as above. After overnight incubation in 24 well plates, samples were subjected to a rapid epifluoroscence staining method employing the bacterial viability kit Live/Dead Bac Light, Invitrogen Ltd., Paisley, UK. The kit employs Syto 9 green and propidium iodide red fluorescent nucleic acid stains for distinguishing live and dead cells, respectively. The dye was prepared and added to the samples according to the manufacturer’s specifications. Samples with the dye were incubated in the dark for 15 minutes and then assessed by a confocal microscope. The final step comprised of acquiring photographs of the samples with a Leica Microystems (Leica TCS SP5, Leica Microsystems, Germany) at excitation levels of 488 nm for Syto 9 and 543 nm for propidium iodide. Images were acquired at 1.5 μm intervals. Further, images representing two dimensional [compressed z series (x–y sides) and compressed (x–z) views] and three dimensional views of the biofilm were acquired with IMARIS v 8.1.2. The thickness of the biofilms was also calculated with IMARIS v 8.1.2.
Cytotoxicity Studies
Human Epithelial Corneal Cells (HCEC) were cultivated in DMEM/Ham’s F12 (Invitrogen Life Technologies, USA), supplemented with 5% (v/v) FBS, 1% (w/v) L-glutamine, 0.4% (w/v) antibiotics (50 U/ml penicillin and 50 mg/ml streptomycin), insulin (5 mg/l) and EGF (10 μg/l) (PAA Laboratories GmbH, Austria). To assess the cytotoxicity on HCEC, vitality assay was performed with slight modifications (Schmitt et al., 2002). Briefly, vitality staining was performed with different concentrations of SBT343 extract for 24 h. 3.5 × 105 cells were seeded in 6-well plates for 24 h in a control medium. After treatment, cells were collected, and 70 μl of the cell suspension was stained with 30 μl staining solution [Gel RedBiotrend (Köln, Germany) and fluorescein diacetate (Kreiswirth et al., 1983)]. Twenty microliter of this mixture was applied to the slide, and the fractions of green and red cells in a total of 200 cells were counted at a 500-fold magnification with a fluorescence microscope. Macrophage (J774.1) and mouse fibroblast (NIH/3T3) cell lines were cultured in RPMI 1640 (1X)+GlutamaxTM-I and DMEM (1X)+GlutamaxTM-I (Life TechnologiesTM,USA) supplemented with 10%FCS, without antibiotics. Cytotoxicity on J774.1 and NIH/3T3 was assesssed employing alamar blue assay (Huber and Koella, 1993). 1 × 105 cells/ml were seeded in 96-well plates containing the extract at different concentrations (ranging from 31.25 to 500 μg/ml) and were incubated for 24 h at 37°C with 5% CO2. After incubation, 20 μL alamar blue (Thermofischer scientific, USA) was added to each well and the plates were further incubated for 24 h at 37°C with 5% CO2. Finally, the OD550 values of the plates were measured and normalized with OD630 values. The extent of cytoxicity was determined by comparing the extract treated sets with the control. The final DMSO concentration on the cells was 1%.
Physico-chemical Characterization of Anti-biofilm Component(s)
To understand the nature of the active component(s), the extract was subjected to thermal and enzymatic (proteinase K and trypsin) treatments. Briefly, the extract was subjected to heat treatments at 50, 75, and 100°C for 1 h and cooled on ice. For the enzymatic treatment, proteinase K or trypsin (at a final concentration of 1 mg/ml) was added to the extract (at a final concentration of 0.125 mg/ml) and the reactions were incubated for 1 h at 37°C. As controls, extracts were incubated for 1 h at 37°C without the enzymes, a treatment that did not impair the anti-biofilm effect. For each of the above tests, the biofilm inhibitory effects of treated and untreated extracts were compared using the microtitre plate assay (for biofilm formation) against all the staphylococcal strains tested. Each data point is composed of three independent experiments performed in quadruplicate.
In parallel, the activity of proteinase K and trypsin (1 mg/ml each) in the presence of DMSO were independently assessed employing the quantitative azocasein assay (Hasegawa et al., 2008).
Quantitative RT-PCR
After complementation of S. epidermidis RP62A with SBT343 extract at 62.5 (BIC50) and 250 μg/ml for 24 h at 37°C, total RNA was isolated from planktonic bacteria and those in biofilm employing Trizol reagent (Invitrogen, Paisley, UK) and FastPrep® disrupter (Thermo Savant, Qbiogene, Inc., Cedex, France). Firstly, 1 ml of cells (from planktonic and biofilm states; the biofilm was gently removed from 24 well plate using a sterile scraper and re-suspended in 1 ml of fresh TSB medium) were centrifuged at 13000 rpm for 10 min at 4°C and pellets were re-suspended in 1 ml Trizol reagent. This suspension was briefly homogenized in a Lysing Matrix E tube (MP Biomedicals Germany, GmbH, Eschwege, Germany) in the FastPrep® cell disrupter and subjected to chloroform-based RNA extraction method. Purity and concentration of the extracted total RNA was spectrophotometrically assessed using a NanoDrop ND-1000 (peqLab Biotechnologie, GmbH). The A260/A280 values of (range 1.8–2.0) indicated the purity of the RNA samples and the mean RNA yield obtained was 182.63 μg/ml. All the RNA samples were digested with DNase I (Thermo Scientific). Briefly, 1 μg of RNA was digested with 1 μl of DNase by incubation at 37°C for 30 min. This was followed by addition of 1.5 μl of DNase stop solution (50 mM EDTA) and incubation at 70°C for 10 min. After quality assessment of RNA after DNase treatment, about 50 ng/μl of RNA was used as template for qPCR experiment.
cDNA synthesis and qPCR amplification was performed simultaneously by employing the power SYBR® Green RNA-to-CTTM 1-step kit (Applied Biosystems, GmbH, Germany). Primers used in the study were designed according to the literature (Reiter et al., 2014), and were commercially produced (Eurofins MWG Synthesis GmbH, Germany). These primers were chosen based on their thermodynamic and sequence parameters. The reaction mixture for qPCR contained 1 μl of RNA template, 5 μl of power SYBR® Green RTPCR mix, 1 μl each of forward and reverse primers (10 μM), 0.08 μl of reverse transcriptase provided by the manufacturer and 1.92 μl of RNase free water. qPCR was performed using the Bio-Rad C1000 TouchTM thermal cycler with the following cycle parameters: holding stage of 48°C for 30 min and 95° C for 10 min, followed by 50 cycles of 95°C for 15 s and then 55°C for 1 min, with a final melting curve determination. Experiment was peformed with three technical and biological replicates each. A difference of ≥7 Ct (cycle threshold) between the cDNA sample and no-template PCR control was considered negligible for relative quantification analysis. Finally, the relative expression of the target genes (IcaA and IcaR) in the presence of SBT343 extract in relation to the modified ISP2 extract treated control was determined using the comparison with the expression level of the reference gene, DHFR (the expression of DHFR gene stayed constant in the conditions tested). The significance of the relative quantification was assessed by Student’s t-test (GraphPad Prism® version 6.01).
Statistical Analysis
Experiments were repeated at least three times in quadruplicates and the data were expressed as mean ± standard error mean. The Student’s t-test was used and p < 0.05 was considered as statistically significant. GraphPad Prism® version 6.01 was used for statistical analysis of the experimental data.
LC-MS Analysis
Analytical grade reagents, Methanol (MeOH), dichloromethane (DCM), acetonitrile (MeCN), and formic acid were purchased (Fisher Scientific, Hemel Hempstead, UK). In-house HPLC grade water was used from a direct Q-3 water purification system (Millipore, Watford, UK). Samples and medium control samples were prepared at a concentration of 1 mg/mL in 80:20 MeOH: DCM with a solvent blank. Experiments were performed with an Exactive mass spectrometer with an electrospray ionization source attached to an Accela 600 HPLC pump with Accela autosampler and UV/Vis detector (Thermo Scientific, Bremen, Germany). The mass accuracy was set to less than 3.0 ppm. The Orbitrap mass analyzer can limit the mass error within ±3.0 ppm. The instrument was calibrated to maintain a mass accuracy of ±1.0 ppm by applying the lock mass function. The instrument was externally calibrated per the manufacturer’s instructions before the run and was internally calibrated during the run using lock masses. Mass spectrometry was carried out over a mass range of 100–2000 m/z in positive and negative ionization modes with spray voltage of 4.5 kV and capillary temperature at 270°C. Ten microliter were injected from each vial, at a flow rate of 300 μL/min. The column used was an ACE5 C18 column (5 μm × 75 mm × 3 mm) (Hichrom Limited, Reading, UK). A binary gradient method was utilized. The two solvents were A (water and 0.1% formic acid) and B (MeCN and 0.1% formic acid). The gradient was carried out for 10 min and the program followed; at zero minutes A = 90% and B = 10%, at 30 min A = 0% and B = 100% at 36 min A = 90% and B = 10% until end at 45 min. The UV absorption wavelength was set at 254 nm, the sample tray temperature was maintained at 4°C and the column maintained at 20°C. The samples were run sequentially, with solvent and media blanks analyzed first. LC-MS data was acquired using Xcalibur version 2.2 (Thermo Scientific, Bremen, Germany). LC-MS chromatograms were subsequently obtained using MassLynx v 4.10. This was followed by dereplication strategy. Since, the modified ISP2 medium is a complex mixture of constituents and could interfere with the identification of secondary metabolites in the SBT343 extract, a medium blank was analyzed together with the bacterial extract and obtained features were regarded as interference and subtracted for detection of true bacterial secondary metabolites. Finally, the m/z values were searched for possible hits in the MarinLit® database (Abdelmohsen et al., 2014b; Macintyre et al., 2014).
Results
Anti-biofilm Effect of the Streptomyces sp. SBT343 Organic Extract
Our continuing effort for discovery of anti-biofilm agents from marine sponge-derived actinomycetes against the model isolate S. epidermidis RP62A, led to identification of the anti-biofilm Streptomyces sp. SBT343 extract. The presence of the extract at 31.25 μg/ml during the bacterial growth caused a significant (p < 0.0001) reduction in the biofilm formation after 24 h of growth. At 62.5 μg/ml about 50% of the biofilm formation was inhibited and this was designated as the Biofilm Inhibitory Concentration (BIC50). The anti-biofilm activity of the extract was dose-dependent, leading to 71.35% inhibition of biofilm formation at the maximum concentration (250 μg/ml) tested (Figure 1A). Even after the addition of extract at BIC50 and highest tested concentration, the growth of S. epidermidis RP62A was at the same level as that of the control. These results confirm that the biofilm inhibition by the extract is not due to growth effect (Figure 1B). SBT343 extract showed no effect on detaching/dispersing the bacteria from preformed biofilm at any of the tested concentrations (data not shown).
FIGURE 1.
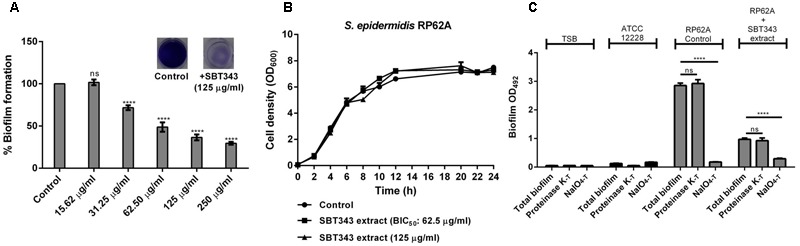
(A) Dose dependent inhibition of Streptomyces sp. SBT343 extract on biofilm formation of S. epidermidis RP62A on polystyrene flat-bottom 96-well plates. S. epidermidis RP62A treated with the extract from modified ISP2 medium (250 μg/ml) served as the appropriate control in the experiment. (B) Growth curve of S. epidermidis RP62A in the presence of BIC50 (62.5 μg/ml) and highest tested concentration (125 μg/ml) of the SBT343 extract. (C) Investigation of the Streptomyces sp. SBT343 for induction of PIA-independent biofilm formation in S. epidermidis RP62A. Control and the residual biofilms after the extract treatment (250 μg/ml) were subjected to two parallel treatments (i) with metaperiodate (NaIO4-T), which is suitable to dissolve the PIA-matrix proportion of the biofilm, and (ii) with proteinase K (proteinase K-T) which will reduce proteins engaged in the biofilm matrix. Biofilm measurements were done shortly after the respective treatments, and the total biofilm levels were compared with the untreated control and residual biofilm sets. The biofilm-negative S. epidermidis ATCC12228 served as the negative control. S. epidermidis RP62A treated with the extract from modified ISP2 medium (250 μg/ml) served as the appropriate control. Graphs represent mean values ± SEM from three independent repetitions of the experiment done in quadruplicate. ns, not significant, ∗∗∗∗p < 0.0001.
As the strain S. epidermidis RP62A is known as a strong PIA matrix biofilm producer, SBT343-mediated biofilm inhibition strongly suggests interference with PIA-mediated biofilm formation. Since, staphylococci are known to switch from PIA-dependent to PIA-independent biofilm formation under different conditions; the presence of these spontaneous switches in the presence of the extract was assessed. The ineffectiveness of proteinase K treatment on residual biofilms highlight that there is no spontaneous switch from PIA-dependent to PIA-independent (protein-mediated) biofilm formation in the presence of SBT343 extract (Figure 1C).
SEM Analysis
The biofilm inhibition potential of the SBT343 extract was studied on glass and contact lens surfaces using microscopic techniques. Electron microscopy of biofilm formation on glass cover slips and contact lenses in the presence of 62.5, 125, and 250 μg/ml SBT343 extract confirmed the results obtained by the in vitro crystal violet assay (Figure 2). In the control glass cover slips, mushroom shaped, three-dimensional biofilm was observed, whereas, multi-layered biofilm was observed on control contact lenses. In the glass cover slips and contact lenses incubated with the SBT 343 extract (62.5, 125, and 250 μg/ml), a dose-dependent reduction in the biofilm was clearly seen (Figure 2A). The reduction of biofilm with the extract was even pronounced in the contact lens model suggesting its possible application in contact lens solution and storage systems. Surfaces were distinctly seen between sporadic microcolonies in the glass cover slip and the contact lens incubated with higher concentrations of the extract. Representative SEM images at higher magnification indicated that the extract did not affect the morphology of the staphylococcal cells. Further, the presence of fibrous, net-like structures in the biofilm matrix was greatly reduced in both the glass cover slips and contact lenses incubated with SBT343 extract (Figure 2B). These findings suggest that the extract possibly works by altering the biofilm matrix composition or interferes with the production of extracellular matrix.
FIGURE 2.
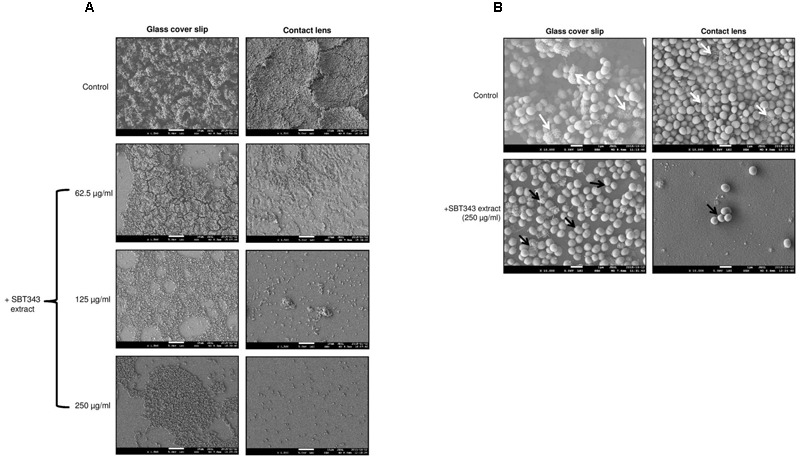
Representative SEM images of staphylococcal biofilm on (A) Glass cover slip and contact lens at X1500 magnification. (B) Glass cover slip and contact lens at X10000 magnification. The white arrows indicate the presence of fibrous, net like structures which are typical features of PIA-dependent biofilms and the black arrows indicate the absence of the same. S. epidermidis RP62A treated with the extract from modified ISP2 medium (250 μg/ml) served as the appropriate control in the microscopy experiments.
Confocal Microscopy Analysis
For the confocal microscopy studies, effect of the SBT343 extract on the biofilm formation was also examined with two systems, first in the cover slips, where the control sample showed compact and condensed biofilm, while the treatment with the extract attenuated the formation of the biofilm in a dose dependent matter. The same observations were obtained in the contact lens as a second system for examination. The ability of the extract to inhibit the biofilm formation was stronger in the coverslips experiments in comparison to the contact lens, but in both systems the inhibition was significant in comparison to the control (Figure 3). Noteworthy, very few non-viable bacteria (stained red) were spotted in the experiment both in the treated and the untreated sets. This further confirms that the extract does not interfere with bacterial cell viability. As a control, cells in biofilm were exposed to 75% ethanol for 5 mins and stained with propidium iodide and SYTO green. In this case a large population of dead cells were spotted in the microscope (data not shown). Images representing compressed x–y and x–z (side) views (Figure 3) indicated that the number of the bacteria in the biofilm and the total biofilm thickness in the extract treated cover slips and contact lens sets were greatly reduced in comparison with the compact and condensed biofilm control. These images further corroborated the results that the reduction in fluorescence in the presence of the extract, was primarily due to the repression of biofilm formation, and has no negative effects on bacterial cell viability.
FIGURE 3.
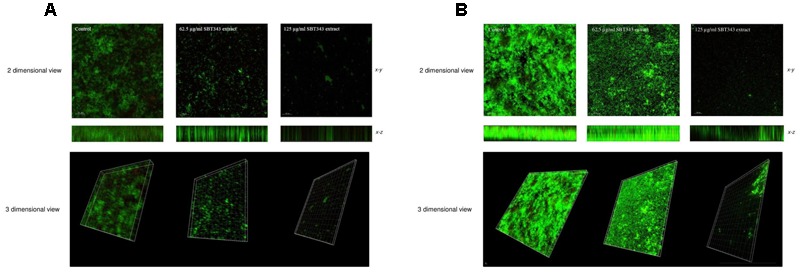
Confocal laser scanning microscopy analyses of staphylococcal biofilm in the presence of the SBT343 extract, visualized by fluorescence vital dye. Images were acquired by Leica TCS SP5 with a 20X objective lens (scale bar 30 μm). (A) Biofilm images on the glass cover slip. (B) Biofilm images on contact lens. In (A,B), compressed z series (top), where multiple x–y planes from top to bottom of the biofilm are combined and the smaller image (bottom) represents the compressed x–z (side) of the biofilm. The thickness of the biofilms in control was 28.7 μm, whereas after extract treatment it was only 15.1 μm (62.5 μg/ml) and 4.53 μm (125 μg/ml) on the glass cover slips. Similarly, the thickness of the biofilms in the extract treated contact lens samples was only 13.6 μm (62.5 μg/ml) and 7.55 μm (125 μg/ml).
Anti-biofilm Effect of SBT343 Extract on Other Pathogens
The anti-biofilm activity of the SBT343 extract was investigated with other biofilm-forming S. epidermidis, S. aureus, and P. aeruginosa strains (Table 2). SBT343 extract (125 μg/ml) significantly reduced the biofilm formation of all the S. epidermidis, MSSA and MRSA strains tested, while the biofilm formation of P. aeruginosa was unaffected (Figure 4A). A dose dependent reduction in biofilm formation of the staphylococcal strains was observed upon extract treatment (Figure 4B), while the growth of the strains were unaltered in the presence of the extract (Figure 4C). Further, the extract had no effects on the preformed biofilms of all the strains tested at the highest concentration tested (data not shown). This indicates selectivity of the extract in inhibiting staphylococcal biofilms.
Table 2.
Biofilm formation of the investigated bacterial strains employing crystal violet assay.
| Strain | Biofilm (OD 492 nm) |
|---|---|
| S. epidermidis ATCC 12228 | 0.124 ± 0.010 |
| S. carnosus TM300 | 0.204 ± 0.013 |
| S. epidermidis RP62A | 2.861 ± .0.143 |
| S. epidermidis O-47 | 1.387 ± 0.044 |
| S. epidermidis 1457 | 2.165 ± 0.069 |
| S. aureus SH1000 | 0.994 ± 0.112 |
| S. aureus RN4220 | 1.177 ± 0.092 |
| S. aureus Newman | 0.691 ± 0.149 |
| S. aureus USA300 | 0.659 ± 0.031 |
| P. aeruginosa PAO1 | 1.337 ± 0.101 |
| P. aeruginosa PA14 | 1.331 ± 0.067 |
Each data point is composed of three independent experiments performed in quadruplicate. Standard errors are reported.
FIGURE 4.
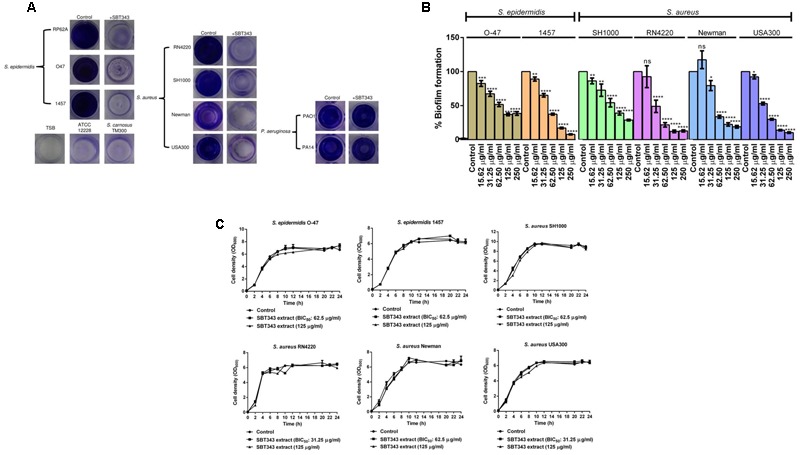
Anti-biofilm effect of SBT343 extract on other staphylococcal strains. (A) Photographs of wells of 96-well plate showing biofilms in the absence and presence of SBT343 extract after crystal violet staining. Control (modified ISP2 medium extract: 125 μg/ml) and SBT343 extract treated (125 μg/ml). (B) Dose dependent inhibitory effect of Streptomyces sp. SBT343 extract on biofilm formation of other staphylococcal strains on polystyrene flat-bottom 96-well plates. Staphylococcal strains treated with the extract from modified ISP2 medium (250 μg/ml) served as the appropriate control in the experiment; (C) Growth curves of staphylococcal strains in the presence of BIC50 and highest tested concentration (125 μg/ml) of the SBT343 extract. Graphs represent mean values ± SEM from three independent repetitions of the experiment done in quadruplicate. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Cytotoxicity Analysis
We further investigated the cytotoxicity profile of the SBT343 extract against mouse fibroblast (NIH/3T3), macrophage (J774.1) and human corneal epithelial cells (HCEC). Results from vitality test and alamar blue assay demonstrated that cells did not suffer from significant toxicity after 24 h with effective concentrations of the extract (in the range of 31.25–125 μg/ml). The highest concentration 500 μg/ml displayed moderate to high cytotoxic effects on the cell lines tested (Table 3).
Table 3.
Cytotoxic evaluation for SBT343 extract.
| Cell line | % reduction in cell viability |
||
|---|---|---|---|
| 500 μg/ml | 250 μg/ml | 31.25–125 μg/ml | |
| HCEC | 90.75 ± 1.23∗∗∗∗ | 20.66 ± 5.10∗ | NC |
| NIH/3T3 | 21.56 ± 2.43∗∗∗∗ | NC | NC |
| J774.1 | 33.83 ± 2.27∗∗∗∗ | NC | NC |
Each data point is composed of three independent experiments performed in quadruplicate. Standard errors are reported. Differences in mean were compared to untreated control and considered statistically significant when p (∗p < 0.05, ∗∗∗∗p < 0.0001) as per Student’s t-test. NC, not cytotoxic.
Physico-chemical Characterization of Anti-biofilm Component(s)
Preliminary physical and chemical characterization of the biofilm inhibiting component(s) in the extract was assessed by subjecting the extract to physical (heat) and chemical (proteinase K or trypsin) treatments. Both the physical and chemical treatments, did not reduce the activity of the extract against all the staphylococcal strains (Figure 5). However, a slight increase in the activity was observed upon heat treatment, in S. epidermidis RP62A and 1457, and S. aureus SH1000 and RN4220 strains. Similarly, a slight increase in the anti-biofilm activity was observed upon enzyme treatment in S. aureus SH1000 and RN4220 strains (Figure 5). This suggests that the active component(s) in the extract is thermo-stable and non-proteinaceous in nature, and the extract contains compound(s) with anti-biofilm activity that could work similarly on different staphylococcal strains. DMSO at the tested concentrations did not influence proteinase K and trypsin activities (Supplementary Figure 1).
FIGURE 5.
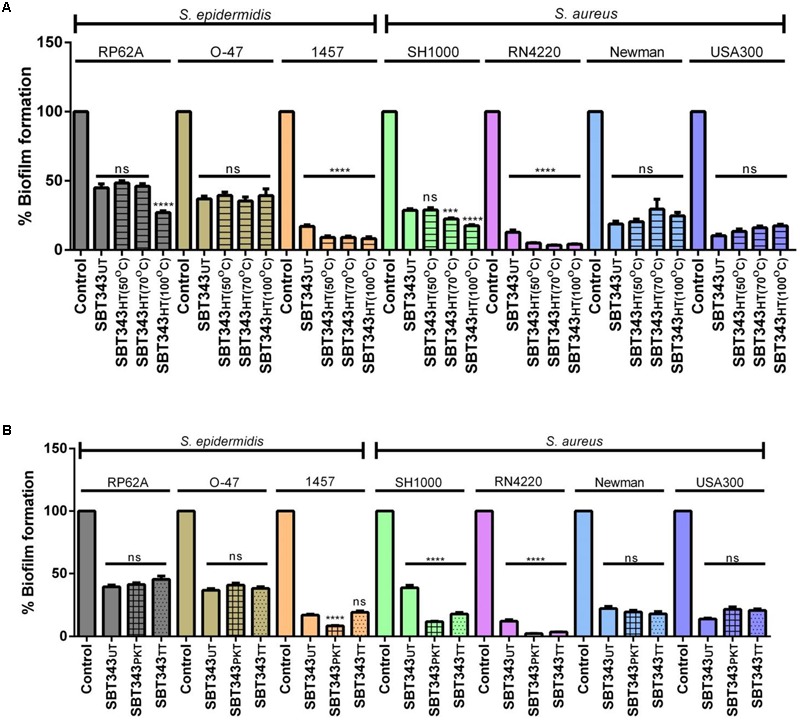
Physico-chemical characterization of anti-biofilm component(s). (A) Influence of heat treatment on anti-biofilm activity. UT, untreated; HT, heat treated. (B) Influence of enzyme treatment on anti-biofilm activity. UT, untreated; PKT, proteinase K treated; TT, trypsin treated. ns, not significant; ∗∗∗∗p < 0.0001.
LC-MS Analysis
To identify the putative bioactive secondary metabolites in the SBT343 extract, the LC-UV/MS signature of the extract generated by high resolution Fourier Transform mass spectrometry was compared with the MarinLit database (a database for marine natural products). The dereplication of the chemical profile of SBT343 extract by comparison of HRMS data with MarinLit, led to identification of several known and unknown metabolites which were previously isolated from the genus Streptomyces. Both the positive and negative modes of electrospray ionization spectral data were used for dereplication purposes. The total ion chromatogram of the SBT343 extract showing the distribution of known and unknown compounds is depicted in Figure 6A. Further, the known compounds detected from the dereplication strategy are mentioned in Table 4 and their structures are depicted in Figure 6B.
FIGURE 6.
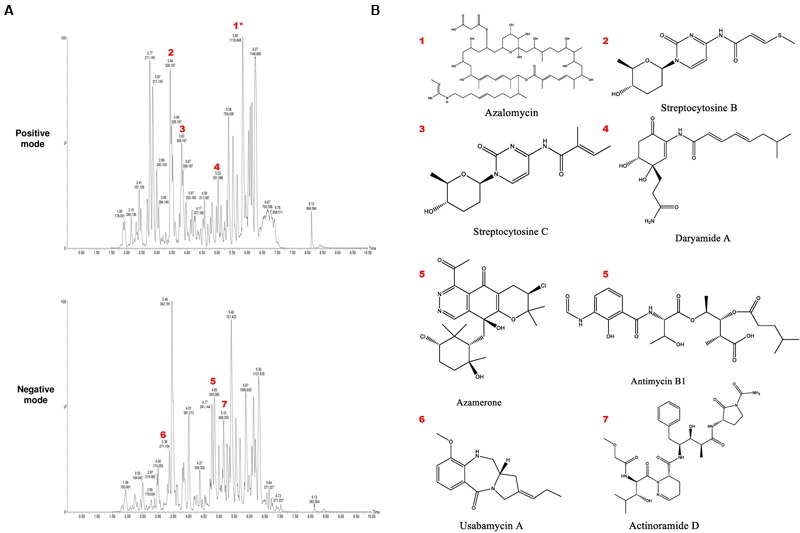
(A) Total ion chromatogram of crude extract of Streptomyces sp. SBT343 in both positive and negative modes, annotated to indicate metabolites identified in Table 4. Annotation and dereplication was done by eliminating the metabolites from the extract of modified ISP2 medium. Those annotated with an asterisk ∗ are sodium ion adducts. (B) Structures of the putatively identified and dereplicated compounds.
Table 4.
Putatively identified and dereplicated compounds from the high-resolution mass spectral data sets of the crude ethyl acetate extract of Streptomyces sp. SBT343 using MarinLit® database with a precision of ±0.1–1.0.
| Peak ID | ESI Mode | m/z∗ | Rt (min) | Hits (m/z∗∗) |
|---|---|---|---|---|
| 1 | P | 1095.624 | 5.85 | Azalomycin (1095.682) |
| 2 | P | 325.197 | 3.44 | Streptcytosine B (325.109) |
| 3 | P | 307.187 | 3.83 | Streptcytosine C (307.153) |
| 4 | P | 350.098 | 5.03 | Daryamide A (350.184) |
| 5 | N | 510.265 | 4.85 | Azamerone (510.168) |
| Antimycin B1 (510.221) | ||||
| 6 | N | 272.154 | 3.38 | Usabamycin A (272.152) |
| 7 | N | 659.355 | 5.15 | Actinoramide D (660.738) |
P, positive ESI mode; N, negative ESI mode; Rt, retention time; ∗ indicates the neutral m/z values of peaks found in the crude ethyl acetate extract of Streptomyces sp. SBT343; ∗∗ indicates the m/z values of the corresponding hits identified with the MarinLit® database.
Discussion
Biomaterials used in clinical and medical settings are ideal niches for formation of microbial biofilms (Shanmughapriya et al., 2012). Even though a number of natural and synthetic anti-biofilm agents have already been discovered (Richards and Melander, 2009; Gupta et al., 2016; Miquel et al., 2016; Tran et al., 2016), none of them has entered the market, owing to obstacles in translational research and lack of interest by pharmaceutical and biomedical companies (Romero and Kolter, 2011). Hence, there is a large unmet need for development of anti-biofilm formulations to tackle the problem of biofilms.
Various culture dependent and independent techniques have revealed the bacterial biofilm diversity in contact lens-related corneal diseases (Hall and Jones, 2010; Wiley et al., 2012). Staphylococci, particularly S. epidermidis, are frequent contaminants of a range of ocular devices. It was estimated by Hou et al. (2012) that many ophthalmic isolates of S. epidermidis could form biofilms in vitro. Strong biofilm formation by S. epidermidis has been observed on various intraocular and contact lens (IOLs) materials (Okajima et al., 2006). Bacteria in the biofilms of contact lenses are frequently resistant to antimicrobials in the soft contact lens care products (Szczotka-Flynn et al., 2009). This has sharpened the need to develop new anti-biofilm-based contact lens products for combating ocular infections. The anti-biofilm potential of natural product based preparations, extracts, and compounds have increasingly been reported in in vitro biomaterial-based models (Pacheco da Rosa et al., 2013; Nair et al., 2016). Several researchers have tried to assess the anti-biofilm aspects of marine bacteria (Nithya et al., 2011; Gowrishankar et al., 2012; Papa et al., 2015; Wu et al., 2016), particularly, marine actinomycetes (Oja et al., 2015; Saleem et al., 2015; Park et al., 2016).
Leshem et al. (2011) and Cho et al. (2015) have previously reported the inhibitory effect of natural product based extract and several compounds on staphylococcal biofilm formation on contact lens and contact lens cases. We have shown that the organic extract from marine Streptomyces sp. SBT343 exhibits similar anti-biofilm effects at much lower concentrations (62.5–250 μg/ml). The extract had no adverse effects on the contact lens material at the highest concentrations tested (data not shown). At the same time, the lowest effective concentration of the extract did not show apparent cytotoxic effects on the three different cell lines tested which indicates the possibility of using the extract for the human subjects. The selective anti-biofilm effect of the SBT343 extract against different staphylococcal strains without interference with the bacterial cell growth suggests the less possible appearance of resistant mutants with the usage of this extract. However, further experiments are needed to prove this hypothesis. The basis of employing TSB medium as the growth medium in the study is to provide optimal growth conditions at which the effects of SBT343 extract could be determined in short periods.
Under certain conditions, S. epidermidis is known to switch between the PIA-dependent and independent modes of biofilm lifestyle (Rohde et al., 2005; Hennig et al., 2007). Our findings suggest that there are no switches in the biofilm lifestyle of the organism in the presence of the extract (Figure 1C). For a better understanding of the mechanism of biofilm inhibition by the extract, the relative mRNA expression of icaA and icaR in S. epidermidis RP62A (after 12 and 24 h) was determined in the planktonic and the biofilm cells in the presence of the extract. However, no significant changes in the expression levels of icaA and icaR were noted in the extract treated planktonic and biofilm cells (data not shown). These results indicate that the extract possibly works by an alternate mechanism and a global gene expression analysis would assist in deciphering the exact mode of action of the extract. The universal anti-biofilm activity against different staphylococcal strains with no effects on preformed biofilm and low cytotoxicity of this extract suggests its potential usage in contact lenses storage cases to prevent contact lens-associated ocular infections. Further studies are necessary to determine the anti-biofilm effect of SBT343 extract on different contact lens and storage cases materials.
The preliminary physical and chemical characterization of the anti-biofilm component(s) in the extract indicates that the active component(s) is of thermo-stable and non-proteinaceous nature and could act similarly on different staphylococcal strains tested. Dereplication strategies are often used in the natural products-based research for rapid identification of secondary metabolites in the crude bacterial extracts (Abdelmohsen et al., 2014b; Cheng et al., 2015). Currently, several analytical methods and tools are available for dereplication of metabolites in complex mixtures. Comparison of the HRMS data at positive and negative modes with the MarinLit database resulted in identification of several putative compounds in the Streptomyces sp. SBT343 extract. It is clear from the chromatogram that several peaks were not identified by comparison to the database, including some of the major components that showed strong peak intensity and good resolution. The high number of unidentified compounds highlights the chemical potential of this strain as a source of new natural products. Most of the compounds identified in the extract were previously isolated from marine Streptomyces. An extensive literature search on the biological activities of these compounds revealed that none of them have been tested/reported to have anti-biofilm effects, while several of the putative compounds identified are known to possess anti-fungal, anti-cancer, anti-mycobacterial and anti-malarial effects. This further highlights the novelty in discovery of compound(s) with specific anti-biofilm effects from the SBT343 extract. Up-scaling of the fermentation process and consequent bio-assay guided fractionation would help in isolation and identification of active compound(s) in the extract. In conclusion, our results show that the chemically rich Streptomyces sp. SBT343 extract has the potential to prevent the staphylococcal biofilm formation on polystyrene, glass, and contact lens surface without exhibiting toxic effects on bacterial and mammalian cells. Future characterization of lead compounds in this extract may yield novel anti-biofilm compound(s) of pharmaceutical interest.
Author Contributions
Conceived and designed the experiments: UA, TÖ, UH, WZ. Performed the experiments: SB, EO. Analyzed the data: SB, EO, TÖ, UA. Manuscript preperation: SB, EO, DK, UA, HS, UH, WZ, TÖ. Manuscript revision: SB, EO, HS, UA, UH, WZ, TÖ. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LC and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Cheng Cheng for providing the actinomycete strains and technical assistance, Hilde Merkert and Daniela Bunsen for assisting the confocal and electron microscopy studies (all University of Würzburg). We acknowledge Mrs. Janina Dix from the Toxicology Department, Ms. Manonmani Soundararajan and Dr. Sonja Schönfelder from the IMIB for the technical assistance. We are thankful to Dr. Vinay Pawar (Helmholtz Centre for Infection Research, Braunschweig, Germany) for providing the biofilm forming P. aeruginosa strains PAO1 and PA14 for the study.
Footnotes
Funding. Financial support to TÖ and UH was provided by the Deutsche Forschungsgemeinschaft (SFB 630 TP A5 and Z1) and by the European Commission within its FP7 Programme to UH, under the thematic area KBBE.2012.3.2-01 with Grant Number 311932 (SeaBioTech). SB was supported by a fellowship of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00236/full#supplementary-material
References
- Abdelmohsen U. R., Balasubramanian S., Oelschlaeger T. A., Grkovic T., Pham N. B., Quinn R. J., et al. (2017). Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 17 e30–e41. 10.1016/S1473-3099(16)30323-1 [DOI] [PubMed] [Google Scholar]
- Abdelmohsen U. R., Bayer K., Hentschel U. (2014a). Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 31 381–399. 10.1039/c3np70111e [DOI] [PubMed] [Google Scholar]
- Abdelmohsen U. R., Cheng C., Viegelmann C., Zhang T., Grkovic T., Ahmed S., et al. (2014b). Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar. Drugs 12 1220–1244. 10.3390/md12031220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen U. R., Szesny M., Othman E. M., Schirmeister T., Grond S., Stopper H., et al. (2012). Antioxidant and anti-protease activities of diazepinomicin from the sponge-associated Micromonospora strain RV115. Mar. Drugs 10 2208–2221. 10.3390/md10102208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer N. K., Mazaitis M. J., Costerton J. W., Leid J. G., Powers M. E., Shirtliff M. E. (2011). Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2 445–459. 10.4161/viru.2.5.17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azman A. S., Othman I., Velu S. S., Chan K. G., Lee L. H. (2015). Mangrove rare actinobacteria: taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol. 6:856 10.3389/fmicb.2015.00856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros M., Branquinho R., Grosso F., Peixe L., Novais C. (2014). Linezolid-resistant Staphylococcus epidermidis, Portugal, 2012. Emerg. Infect. Dis. 20 903–905. 10.3201/eid2005.130783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bispo P. J., Haas W., Gilmore M. S. (2015). Biofilms in infections of the eye. Pathogens 4 111–136. 10.3390/pathogens4010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T., Ciofu O., Molin S., Givskov M., Hoiby N. (2013). Applying insights from biofilm biology to drug development – can a new approach be developed? Nat. Rev. Drug Discov. 12 791–808. 10.1038/nrd4000 [DOI] [PubMed] [Google Scholar]
- Burmolle M., Thomsen T. R., Fazli M., Dige I., Christensen L., Homoe P., et al. (2010). Biofilms in chronic infections – a matter of opportunity – monospecies biofilms in multispecies infections. FEMS Immunol. Med. Microbiol. 59 324–336. 10.1111/j.1574-695X.2010.00714.x [DOI] [PubMed] [Google Scholar]
- Cheng C., Macintyre L., Abdelmohsen U. R., Horn H., Polymenakou P. N., Edrada-Ebel R., et al. (2015). Biodiversity, anti-trypanosomal activity screening, and metabolomic profiling of actinomycetes isolated from mediterranean sponges. PLoS ONE 10:e0138528 10.1371/journal.pone.0138528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho P., Shi G. S., Boost M. (2015). Inhibitory effects of 2,2’-dipyridyl and 1,2,3,4,6-penta-O-galloyl-b-D-glucopyranose on biofilm formation in contact lens cases. Invest. Ophthalmol. Vis. Sci. 56 7053–7057. 10.1167/iovs.15-17723 [DOI] [PubMed] [Google Scholar]
- Dalisay D. S., Williams D. E., Wang X. L., Centko R., Chen J., Andersen R. J. (2013). Marine sediment-derived Streptomyces bacteria from British Columbia, Canada are a promising microbiota resource for the discovery of antimicrobial natural products. PLoS ONE 8:e77078 10.1371/journal.pone.0077078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti Y., Grkovic T., Abdelmohsen U. R., Hentschel U., Quinn R. J. (2014). Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 12 3046–3059. 10.3390/md12053046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M. (2001). Biofilms and device-associated infections. Emerg. Infect. Dis. 7 277–281. 10.3201/eid0702.010226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltamany E. E., Abdelmohsen U. R., Ibrahim A. K., Hassanean H. A., Hentschel U., Ahmed S. A. (2014). New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 24 4939–4942. 10.1016/j.bmcl.2014.09.040 [DOI] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Gowrishankar S., Duncun Mosioma N., Karutha Pandian S. (2012). Coral-associated bacteria as a promising antibiofilm agent against methicillin-resistant and -susceptible Staphylococcus aureus biofilms. Evid. Based Complement. Alternat. Med. 2012:862374 10.1155/2012/862374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic T., Abdelmohsen U. R., Othman E. M., Stopper H., Edrada-Ebel R., Hentschel U., et al. (2014). Two new antioxidant actinosporin analogues from the calcium alginate beads culture of sponge-associated Actinokineospora sp. strain EG49. Bioorg. Med. Chem. Lett. 24 5089–5092. 10.1016/j.bmcl.2014.08.068 [DOI] [PubMed] [Google Scholar]
- Gupta P., Sarkar S., Das B., Bhattacharjee S., Tribedi P. (2016). Biofilm, pathogenesis and prevention–a journey to break the wall: a review. Arch. Microbiol. 198 1–15. 10.1007/s00203-015-1148-6 [DOI] [PubMed] [Google Scholar]
- Hall B. J., Jones L. (2010). Contact lens cases: the missing link in contact lens safety? Eye Contact Lens 36 101–105. 10.1097/ICL.0b013e3181d05555 [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Lind E. J., Boin M. A., Hase C. C. (2008). The extracellular metalloprotease of Vibrio tubiashii is a major virulence factor for pacific oyster (Crassostrea gigas) larvae. Appl. Environ. Microbiol. 74 4101–4110. 10.1128/AEM.00061-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan R., Shaaban M. I., Abdel Bar F. M., El-Mahdy A. M., Shokralla S. (2016). Quorum sensing inhibiting activity of Streptomyces coelicoflavus isolated from soil. Front. Microbiol. 7:659 10.3389/fmicb.2016.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C., Gerke C., Perdreau-Remington F., Gotz F. (1996). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64 277–282. 10.1016/j.ijmm.2006.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Nyunt Wai S., Ziebuhr W. (2007). Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297 117–122. [DOI] [PubMed] [Google Scholar]
- Hentschel U., Schmid M., Wagner M., Fieseler L., Gernert C., Hacker J. (2001). Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35 305–312. 10.1111/j.1574-6941.2001.tb00816.x [DOI] [PubMed] [Google Scholar]
- Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. (2002). SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184 5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W., Sun X., Wang Z., Zhang Y. (2012). Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Invest. Ophthalmol. Vis. Sci. 53 5624–5631. 10.1167/iovs.11-9114 [DOI] [PubMed] [Google Scholar]
- Huber W., Koella J. C. (1993). A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop. 55 257–261. 10.1016/0001-706X(93)90083-N [DOI] [PubMed] [Google Scholar]
- Kreiswirth B. N., Lofdahl S., Betley M. J., O’reilly M., Schlievert P. M., Bergdoll M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305 709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- Lee L. H., Zainal N., Azman A. S., Eng S. K., Goh B. H., Yin W. F., et al. (2014). Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014:698178 10.1155/2014/698178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem R., Maharshak I., Ben Jacob E., Ofek I., Kremer I. (2011). The effect of nondialyzable material (NDM) cranberry extract on formation of contact lens biofilm by Staphylococcus epidermidis. Invest. Ophthalmol. Vis. Sci. 52 4929–4934. 10.1167/iovs.10-5335 [DOI] [PubMed] [Google Scholar]
- Lipinski B., Hawiger J., Jeljaszewicz J. (1967). Staphylococcal clumping with soluble fibrin monomer complexes. J. Exp. Med. 126 979–988. 10.1084/jem.126.5.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre L., Zhang T., Viegelmann C., Martinez I. J., Cheng C., Dowdells C., et al. (2014). Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 12 3416–3448. 10.3390/md12063416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D., Siemssen N., Laufs R. (1992). Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy H., Rudkin J. K., Black N. S., Gallagher L., O’neill E., O’gara J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell Infect. Microbiol. 5:1 10.3389/fcimb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal L. K., Steward C. D., Killgore G. E., Chaitram J. M., Mcallister S. K., Tenover F. C. (2003). Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41 5113–5120. 10.1128/JCM.41.11.5113-5120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S., Lagrafeuille R., Souweine B., Forestier C. (2016). Anti-biofilm activity as a health issue. Front. Microbiol. 7:592 10.3389/fmicb.2016.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S. V., Baranwal G., Chatterjee M., Sachu A., Vasudevan A. K., Bose C., et al. (2016). Antimicrobial activity of plumbagin, a naturally occurring naphthoquinone from Plumbago rosea, against Staphylococcus aureus and Candida albicans. Int. J. Med. Microbiol. 306 237–248. 10.1016/j.ijmm.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Nithya C., Begum M. F., Pandian S. K. (2010). Marine bacterial isolates inhibit biofilm formation and disrupt mature biofilms of Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 88 341–358. 10.1007/s00253-010-2777-y [DOI] [PubMed] [Google Scholar]
- Nithya C., Devi M. G., Karutha Pandian S. (2011). A novel compound from the marine bacterium Bacillus pumilus S6-15 inhibits biofilm formation in gram-positive and gram-negative species. Biofouling 27 519–528. 10.1080/08927014.2011.586127 [DOI] [PubMed] [Google Scholar]
- Oja T., San Martin Galindo P., Taguchi T., Manner S., Vuorela P. M., Ichinose K., et al. (2015). Effective antibiofilm polyketides against Staphylococcus aureus from the pyranonaphthoquinone biosynthetic pathways of Streptomyces Species. Antimicrob. Agents Chemother. 59 6046–6052. 10.1128/AAC.00991-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima Y., Kobayakawa S., Tsuji A., Tochikubo T. (2006). Biofilm formation by Staphylococcus epidermidis on intraocular lens material. Invest. Ophthalmol. Vis. Sci. 47 2971–2975. 10.1167/iovs.05-1172 [DOI] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco da Rosa J., Korenblum E., Franco-Cirigliano M. N., Abreu F., Lins U., Soares R. M., et al. (2013). Streptomyces lunalinharesii strain 235 shows the potential to inhibit bacteria involved in biocorrosion processes. Biomed Res. Int. 2013:309769 10.1155/2013/309769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R., Selan L., Parrilli E., Tilotta M., Sannino F., Feller G., et al. (2015). Anti-biofilm activities from marine cold adapted bacteria against Staphylococci and Pseudomonas aeruginosa. Front. Microbiol. 6:1333 10.3389/fmicb.2015.01333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. R., Tripathi A., Wu J., Schultz P. J., Yim I., Mcquade T. J., et al. (2016). Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nat. Commun. 7:10710 10.1038/ncomms10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M., Schleissner C., Fernandez R., Rodriguez P., Reyes F., Zuniga P., et al. (2016). PM100117 and PM100118, new antitumor macrolides produced by a marine Streptomyces caniferus GUA-06-05-006A. J. Antibiot. 69 388–394. 10.1038/ja.2015.121 [DOI] [PubMed] [Google Scholar]
- Reimer A., Blohm A., Quack T., Grevelding C. G., Kozjak-Pavlovic V., Rudel T., et al. (2015). Inhibitory activities of the marine streptomycete-derived compound SF2446A2 against Chlamydia trachomatis and Schistosoma mansoni. J. Antibiot. 68 674–679. 10.1038/ja.2015.54 [DOI] [PubMed] [Google Scholar]
- Reiter K. C., Sant’anna F. H., D’azevedo P. A. (2014). Upregulation of icaA, atlE and aap genes by linezolid but not vancomycin in Staphylococcus epidermidis RP62A biofilms. Int. J. Antimicrob. Agents 43 248–253. 10.1016/j.ijantimicag.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Richards J. J., Melander C. (2009). Controlling bacterial biofilms. Chembiochem 10 2287–2294. 10.1002/cbic.200900317 [DOI] [PubMed] [Google Scholar]
- Rogers K. L., Fey P. D., Rupp M. E. (2009). Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23 73–98. 10.1016/j.idc.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Rohde H., Burdelski C., Bartscht K., Hussain M., Buck F., Horstkotte M. A., et al. (2005). Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55 1883–1895. 10.1111/j.1365-2958.2005.04515.x [DOI] [PubMed] [Google Scholar]
- Romero D., Kolter R. (2011). Will biofilm disassembly agents make it to market? Trends Microbiol. 19 304–306. 10.1016/j.tim.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein R., Nerz C., Biswas L., Resch A., Raddatz G., Schuster S. C., et al. (2009). Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75 811–822. 10.1128/AEM.01982-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura T., Kajiyama S., Adachi S., Chiba K., Yonekura A., Tomita M., et al. (2015). Biofilm-forming Staphylococcus epidermidis expressing vancomycin resistance early after adhesion to a metal surface. Biomed Res. Int. 2015:943056 10.1155/2015/943056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem H. G., Aftab U., Sajid I., Abbas Z., Sabri A. N. (2015). Effect of crude extracts of selected actinomycetes on biofilm formation of A. schindleri, M. aci, and B. cereus. J. Basic Microbiol. 55 645–651. 10.1002/jobm.201400358 [DOI] [PubMed] [Google Scholar]
- Santos-Gandelman J. F., Giambiagi-Demarval M., Oelemann W. M., Laport M. S. (2014). Biotechnological potential of sponge-associated bacteria. Curr. Pharm. Biotechnol. 15 143–155. 10.2174/1389201015666140711115033 [DOI] [PubMed] [Google Scholar]
- Schmitt E., Lehmann L., Metzler M., Stopper H. (2002). Hormonal and genotoxic activity of resveratrol. Toxicol. Lett. 136 133–142. 10.1016/S0378-4274(02)00290-4 [DOI] [PubMed] [Google Scholar]
- Ser H. L., Tan L. T., Palanisamy U. D., Abd Malek S. N., Yin W. F., Chan K. G., et al. (2016). Streptomyces antioxidans sp. nov., a novel mangrove soil actinobacterium with antioxidative and neuroprotective potentials. Front. Microbiol. 7:899 10.3389/fmicb.2016.00899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmughapriya S., Francis A. L., Kavitha S., Natarajaseenivasan K. (2012). In vitro actinomycete biofilm development and inhibition by the polyene antibiotic, nystatin, on IUD copper surfaces. Biofouling 28 929–935. 10.1080/08927014.2012.717616 [DOI] [PubMed] [Google Scholar]
- Subramani R., Aalbersberg W. (2012). Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol. Res. 167 571–580. 10.1016/j.micres.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Szczotka-Flynn L. B., Imamura Y., Chandra J., Yu C., Mukherjee P. K., Pearlman E., et al. (2009). Increased resistance of contact lens-related bacterial biofilms to antimicrobial activity of soft contact lens care solutions. Cornea 28 918–926. 10.1097/ICO.0b013e3181a81835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabares P., Pimentel-Elardo S. M., Schirmeister T., Hunig T., Hentschel U. (2011). Anti-protease and immunomodulatory activities of bacteria associated with Caribbean sponges. Mar. Biotechnol. 13 883–892. 10.1007/s10126-010-9349-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. T., Chan K. G., Lee L. H., Goh B. H. (2016). Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol. 7:79 10.3389/fmicb.2016.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. L., Huynh E., Pham P., Lacky B., Jarvis C., Mosley T., et al. (2016). Organoselenium polymer inhibits biofilm formation in polypropylene contact lens case material. Eye Contact Lens 10.1097/ICL.0000000000000239 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Uribe-Alvarez C., Chiquete-Felix N., Contreras-Zentella M., Guerrero-Castillo S., Pena A., Uribe-Carvajal S. (2016). Staphylococcus epidermidis: metabolic adaptation and biofilm formation in response to different oxygen concentrations. Pathog. Dis. 74:ftv111 10.1093/femspd/ftv111 [DOI] [PubMed] [Google Scholar]
- Vicente J., Stewart A. K., Van Wagoner R. M., Elliott E., Bourdelais A. J., Wright J. L. (2015). Monacyclinones, new angucyclinone metabolites isolated from Streptomyces sp. M7_15 associated with the Puerto Rican sponge Scopalina ruetzleri. Mar. Drugs 13 4682–4700. 10.3390/md13084682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegelmann C., Parker J., Ooi T., Clements C., Abbott G., Young L., et al. (2014). Isolation and identification of antitrypanosomal and antimycobacterial active steroids from the sponge Haliclona simulans. Mar. Drugs 12 2937–2952. 10.3390/md12052937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Preston J. F., III, Romeo T. (2004). The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186 2724–2734. 10.1128/JB.186.9.2724-2734.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M., Schoenfelder S. M., Orasch C., Arber C., Gratwohl A., Frei R., et al. (2010). Hypervariability of biofilm formation and oxacillin resistance in a Staphylococcus epidermidis strain causing persistent severe infection in an immunocompromised patient. J. Clin. Microbiol. 48 2407–2412. 10.1128/JCM.00492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley L., Bridge D. R., Wiley L. A., Odom J. V., Elliott T., Olson J. C. (2012). Bacterial biofilm diversity in contact lens-related disease: emerging role of Achromobacter, Stenotrophomonas, and Delftia. Invest. Ophthalmol. Vis. Sci. 53 3896–3905. 10.1167/iovs.11-8762 [DOI] [PubMed] [Google Scholar]
- Wu S., Liu G., Jin W., Xiu P., Sun C. (2016). Antibiofilm and anti-infection of a marine bacterial exopolysaccharide against Pseudomonas aeruginosa. Front. Microbiol. 7:102 10.3389/fmicb.2016.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Sun C. (2016). The inhibition and resistance mechanisms of actinonin, isolated from marine Streptomyces sp. NHF165, against Vibrio anguillarum. Front. Microbiol. 7:1467 10.3389/fmicb.2016.01467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Qin Y. H., Zheng X., Zhao H. W., Chai D. Y., Li W., et al. (2016). Biogeography and adaptive evolution of Streptomyces strains from saline environments. Sci. Rep. 6:32718 10.1038/srep32718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotchev S. B. (2012). Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 158 168–175. 10.1016/j.jbiotec.2011.06.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


