Abstract
Hepatoblastoma is the most common pediatric liver malignancy, typically striking children within the first 3 years of their young lives. While advances in chemotherapy and newer surgical techniques have improved survival in patients with localized disease, unfortunately, for the 25% of patients with metastasis, the overall survival remains poor. These tumors, which are thought to arise from hepatic progenitors or hepatoblasts, hence the name hepatoblastoma, can be categorized by histological subtyping based on their level of cell differentiation. Genomic and histological analysis of human tumor samples has shown exon-3 deletions or missense mutations in gene coding for β-catenin, a downstream effector of the Wnt signaling pathway, in up to 90% of hepatoblastoma cases. The current article will review key aberrations in molecular pathways that are implicated in various subtypes of hepatoblastoma with an emphasis on Wnt signaling. It will also discuss cooperation among components of pathways such as β-catenin and Yes-associated protein in cancer development. Understanding the complex network of molecular signaling in oncogenesis will undoubtedly aid in the discovery of new therapeutics to help combat hepatoblastoma.
Key words: Wnt signaling, Liver development, Liver tumor, Pediatric, Yap
HEPATOBLASTOMA: BACKGROUND
Incidence and Risk Factors
Hepatoblastoma is the most common pediatric liver cancer accounting for 80% of malignant liver tumors in childhood. The incidence is estimated at 1.2–1.5/million children per year, comprising about 1% of all pediatric cancers1,2. The rarity of this tumor poses challenges to therapeutic advances, as limited resources make investigation into disease pathology more difficult. Fewer patients transcend into fewer biological samples for study and lower levels of enrollment for clinical trials. Just recently, the four major pediatric liver cancer treatment groups worldwide—The International Childhood Liver Tumour Strategy Group (SIOPEL), The Children’s Oncology Group (COG) [formerly the Pediatric Oncology Group (POG) and the Children’s Cancer Group (CCG)], the German Society of Pediatric Oncology and Haematology (GPOH), and the Japanese Study Group for Pediatric Liver Tumors (JPLT)—have come together to share data and form a consensus group called the Children’s Hepatic Tumors International Collaboration (CHIC) in order to pool data and identify risk factors that would otherwise be difficult to identify due to the rarity of this tumor3. This large cohort of more than 1,000 hepatoblastoma patients was able to confirm suspected clinical risk factors as well as identify new groups that would require more aggressive therapies. While children with low-risk hepatoblastoma have survival rates of >90%, children in the high-risk category, historically, have a dismal overall survival of only 25%–40%.
While the causes of hepatoblastoma are unknown, a few risk factors have been identified. A leading factor associated with the high incidence of hepatoblastoma is low birth weight4. In fact, very low birth weight may be the cause of doubling of the incidence of hepatoblastoma from 1975 to 1999 due to improved survival of the very low birth weight infants over this period5. While additional validation is essential, other suspected risk factors for this tumor include younger maternal age, use of infertility treatment, maternal smoking, and higher maternal prepregnancy body mass index2.
Patient Stratification and Management of Hepatoblastoma
Treatment of hepatoblastoma varies depending on risk stratification, which is based on a combination of histology, radiographic staging, and distant metastatic disease. Because all patients require local control with surgical resection, the extent of liver lobes unaffected determines the presurgical stage of tumor. Most treatment centers in the US and Europe are now using the presurgical extent of disease (PRETEXT) staging system developed by SIOPEL, which is determined by radiographic imaging. PRETEXT stage I indicates tumor involvement in only one liver segment, leaving three contiguous areas of liver segments that are disease free. Conversely, PRETEXT stage IV indicates multifocal involvement of all liver lobes or, rarely, one large diffuse tumor encompassing all lobes. The amount and type of adjuvant chemotherapy vary depending on the risk category. Fortunately, most hepatoblastoma tumors are chemosensitive, and more than 80% of tumors that were previously unresectable do eventually become amenable to surgery after initiation of chemotherapy6. In cases of PRETEXT stage I–III in which hepatoblastoma remains in close proximity to essential vascular structures, or in all cases of multifocal PRETEXT IV, liver transplantation becomes the only hope for cure. These patients rely on the availability of donor organs for survival, which is a limited resource. Timing of transplant can become critical as availability of organs cannot always be planned with the timing of chemotherapy response and recovery from treatment. Early referral for transplant for these select patients is now strongly recommended7. Although advances in chemotherapeutics, such as the discovery of cisplatin, have greatly increased survival for some, we must still continue to work to optimize therapies for these children at highest risk8,9.
There are a group of tumors that are categorized as high-risk status and poor prognosis. These include hepatoblastoma with small cell undifferentiated (SCU) histology, those with distant metastatic disease, and hepatoblastomas associated with serum α-fetoprotein (AFP) levels <100 ng/ml3,10. While there have been some incidence reports of decreased serum AFP levels in up to 11% of patients, a recent large study of 833 hepatoblastoma patients showed just 21 of those patients, or 2.7%, had initial levels less than 100 ng/ml9,11,12. Initial incidence was closer to 10%; however, on repeated testing, false negatives were identified (which can occur with very high AFP values), and the true incidence was much lower. Interestingly, a little over half of these samples had SCU histology, although all histological subsets were present, including two patients with pure fetal histology, a histological subset with better prognosis. Of these high-risk hepatoblastomas, 17%–22% will have distant metastasis to the lungs at the time of initial diagnosis3,13,14. In fact, metastatic spread appears to be the most common contributor to the high-risk status. Understanding the genetic and molecular origins of hepatoblastoma subtypes may lead to the development of novel ways to manage these cases and also pave the way to molecular therapies, with the hope of improving survival of these children.
CELLULAR AND MOLECULAR BASIS OF HEPATOBLASTOMA
Cellular Basis of Hepatoblastoma
The origin of hepatoblastoma is complex as it arises from primary hepatoblasts, and likely from even less differentiated hepatic stem cells or human fetal liver multipotent progenitor cells (hFLMPCs). These small blast-like cells when isolated and cultured from normal fetal livers are highly proliferative and able to differentiate into a variety of tissues including hepatocytes, bile ducts, cartilage, bone, fat, and endothelium15. The potential origin of hepatoblastomas from hFLMPCs could explain not only why there are histological differences between the various subgroups of hepatoblastoma but also why 40% of tumor samples have a mixed histological picture including both epithelial and mesenchymal elements. A possible explanation is that as these multipotential stem cells get exposed to local spatiotemporal cues, either molecules or other cell types, they differentiate and proliferate into various lineages that may eventually dictate both the histology and composition of these tumors. Further, it is likely that the stage of hFLMPCs that get inflicted with specific mutations may also determine the eventual histological and differentiation status of a hepatoblastoma.
Molecular Basis of Hepatoblastoma
For decades we have known that β-catenin plays a critical role in the development of various human organs, including skeletal muscle and brain16. Over the last several years, its role in various processes during liver growth and regeneration has also been discovered17. This is well demonstrated in a murine mouse model where conditional knockout of CTNNB1 in hepatic epithelial cells resulted in decreased basal liver weight and also affected hepatocyte proliferation after partial hepatectomy18. Importantly, its role in hepatoblast proliferation and hepatocyte differentiation and maturation during normal hepatic development makes it an intriguing target of transformation in hepatoblastoma pathogenesis19,20. Indeed, hepatoblastoma is the tumor with the most interstitial deletions or missense mutations in CTNNB1 gene reported in more than 60%–70% of all hepatoblastoma cases.
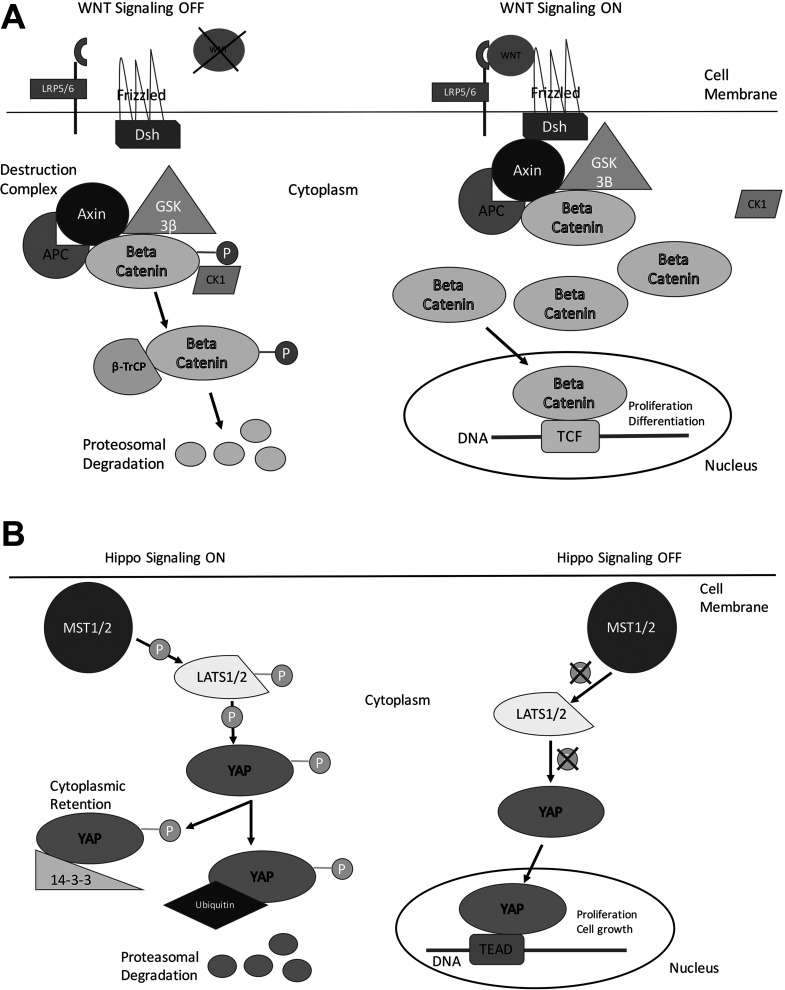
β-Catenin activation in physiological context is most commonly regulated by the canonical Wnt pathway21 (Fig. 1A). Secreted Wnt binds to its receptor Frizzled and coreceptors LDL-related protein-5/6 (LRP5/6) to activate a protein called dishevelled, which then inactivates the destruction complex. The hypophosphorylated β-catenin cannot be degraded and accumulates in the cytoplasm and eventually translocates to the nucleus where it binds to the T-cell factor (TCF) family of transcription factors to induce expression of target genes in a context- and tissue-specific manner. When Wnt is absent, β-catenin is phosphorylated by a cytoplasmic destruction complex consisting of glycogen synthase kinase 3β (GSK3β), casein kinase, axin, and adenomatosis polyposis coli gene product (APC). The phosphorylation occurs at specific serine and threonine residues located in exon 3 of the β-catenin protein, which makes it amenable to detection by βTrCP1, a component of the ubiquitin ligase complex and eventual ubiquitin-mediated proteosomal degradation. If phosphorylation of β-catenin is affected due to mutations or deletions in exon 3 of CTNNB1 or mutations affecting the degradation machinery of β-catenin, the pathway becomes active as is evident in many cancers.
Figure 1.
Simplified overview of the Wnt/β-catenin signaling and the Hippo signaling pathways. (A) In “off” state, β-catenin complexes with the members of its degradation complex where it is phosphorylated at specific serine and threonine residues for recognition by proteasome for degradation. In its “on” state, Wnt binds to receptor Frizzled and coreceptor LRP5/6 to recruit dishevelled, allowing for the β-catenin degradation complex to be inactivated. This leads to hypophosphorylation of β-catenin, its stabilization in the cytoplasm followed by its nuclear translocation. In the nucleus, β-catenin acts as a cofactor for TCF family of transcription factors to induce target gene expression. (B) In its “on” state, the Hippo signaling kinases MST1/2 are activated to in turn activate the LATS1/2 kinases. The activated LATS1/2 in turn phosphorylates YAP (or its paralog TAZ), the major effector of the Hippo cascade. Once phosphorylated, YAP (or TAZ) is either retained in the cytoplasm or degraded by proteasome. In its “off” state, the kinase complexes in the Hippo signaling are inactive, and YAP (or TAZ) can translocate to the nucleus to act as cofactor for the TEAD family of transcription factors and regulate expression of key target genes.
Most mutations in hepatoblastoma are interstitial deletions or missense point mutations in exon 3 of CTNNB1, which either directly alter the serine/threonine phosphorylation sites of β-catenin or alter a residue in the immediate vicinity of the phosphorylation sites, making the β-catenin protein unrecognizable for degradation22. It has been shown that larger deletions affecting exon 3 of CTNNB1 are more frequent in hepatoblastoma with fetal histology, while point mutations are relatively more abundant in embryonal tumors and SCU hepatoblastomas23. However, mutations in other components of the β-catenin degradation complex including those affecting APC and AXIN2 have also been reported24,25. Overall, Wnt pathway abnormalities collectively account for up to 90% of genetic defects in hepatoblastoma tumors23–26. In another study of 23 patient hepatoblastoma samples, all 19 available samples expressed positive nuclear and cytoplasmic immunohistochemistry (IHC) staining of β-catenin. Furthermore, mRNA expression of known inducible inhibitory Wnt pathway genes NKD-1, βTrCP1, DKK-1, and Axin2 was upregulated in all samples compared to normal adjacent liver tissue. Disruptions of the negative feedback loop suggest that aberrations within the Wnt inhibitory pathway are occurring downstream of these overexpressed Wnt targets proteins, since despite their overexpression, they fail to inhibit β-catenin nuclear translocation27. This study did not supply data for histological subtypes of tumor samples or other clinical factors to determine if Wnt inhibition is related to the degree of hepatoblast differentiation.
More recently, data are beginning to be reported using more timely technologies including whole-genome sequencing (WGS) and whole-exome sequencing (WES). A study utilized WES to identify mutations in exon 3 of CTNNB1 in 12 of 15 cases28. In another study using six paired hepatoblastomas and lymphocytes subjected to WES, the authors identified 24 somatic nonsynonymous mutations in 21 genes, many of which were novel, including 3 novel mutations targeting the CTNNB1 (G512V) and CAPRIN2 (R968H/S969C) genes in the Wnt pathway, and genes previously shown to be involved in the ubiquitin ligase complex (SPOP, KLHL22, TRPC4AP, and RNF169)29. The CTNNB1 (G512V) and CAPRIN2 (R968H/S969C) were gain-of-functional mutations, and CAPRIN2 (R968H/S969C) also activated the Wnt pathway in hepatoblastoma cells. This study also points to the diverse mechanistic basis of β-catenin activation evident in major subsets of hepatoblastomas. But overall, these types of studies have consistently shown a low rate of mutations in hepatoblastomas compared to adult liver tumors28.
Exploring the Second Hit Hypothesis
In the 1970s Knudson and Strong, by observing the incidence of retinoblastoma within affected families, astutely hypothesized that certain germline mutations, or one inherited genetically, created an increased risk of acquiring cancer30. They predicted that two separate mutations were needed for tumor formation, and since these patients were already born with one genetic aberration, these patients needed an additional mutation or a “second hit” to develop cancer. Since then, the Rb gene has been validated as a tumor suppressor gene and has paved the way for discoveries of other important genes in familial cancer syndromes such as Li–Fraumeni syndrome and familial adenomatous polyposis (FAP)30,31. Furthermore, it has led to the discoveries of important oncogenes and tumor suppressor genes necessary for oncogenesis applying the second hit theory to even noninherited forms of cancer.
Likewise, in hepatoblastoma, certain genetic aberrations are well-known predisposing risk factors. One example is children with the Beckwith–Weiderman syndrome (BWS), an overgrowth syndrome characterized by hemihypertrophy and organomegaly, in which 1.7% of patients develop hepatoblastoma. Risk is especially increased in cases of BWS with uniparental disomy on chromosome 11p15.5, in which incidence is increased to 4.7%. For this reason, routine screening with ultrasound and serum AFP is recommended in order to detect the disease at earlier stages32. Another example is that of FAP, where children have a 750- to 7,500-fold increased risk of hepatoblastoma development supporting the utility for routine ultrasound screening26. In fact, historically, reports of β-catenin mutations in adult colon cancer, as well as the increased risk of hepatoblastoma in children with FAP, are what led to the investigation of the role β-catenin could be playing in hepatoblastoma tumorigenesis when, in 1999, Koch et al. discovered that nearly half of a group of hepatoblastoma samples possessed such mutations, paving the way for discoveries into hepatoblastoma molecular and genetic pathogenesis.
The YAP–β-Catenin Connection
The Yes-associated protein (Yap) is a well-conserved downstream effector of the Hippo tumor suppressor signaling cascade that plays key roles in organ growth and stem cell differentiation. Activation of Yap increases liver size in transgenic mice due to enhanced cell proliferation, without any effect on cell size33. Yap activation was also found to expand populations of undifferentiated progenitor cells in the intestinal cells of mice, indicating its potential role in cancer development33. Yap is a component of the Hippo signaling pathway. When the pathway is active, Yap largely remains cytoplasmic and is phosphorylated by the large tumor suppressors 1 and 2 (LATS1/2), which target it for proteasome-mediated degradation (Fig. 1B). However, when the pathway is turned off, the kinases are inactive, and Yap is not phosphorylated. In its unphosphorylated form, it is able to translocate into the nucleus, where it binds and activates TEA domain family member or TEAD family of transcription factors, functioning as a transcription cofactor that initiates target gene expression, with eventual effect on growth and regeneration34 (Fig. 1B). Liver-specific knockout mice of LATS1/2 show an upregulation of Yap target genes35. More recently, forced activation of Yap in adult hepatocytes was shown to dedifferentiate these cells into early liver progenitor cells and eventually reprogram them into biliary epithelial cells36.
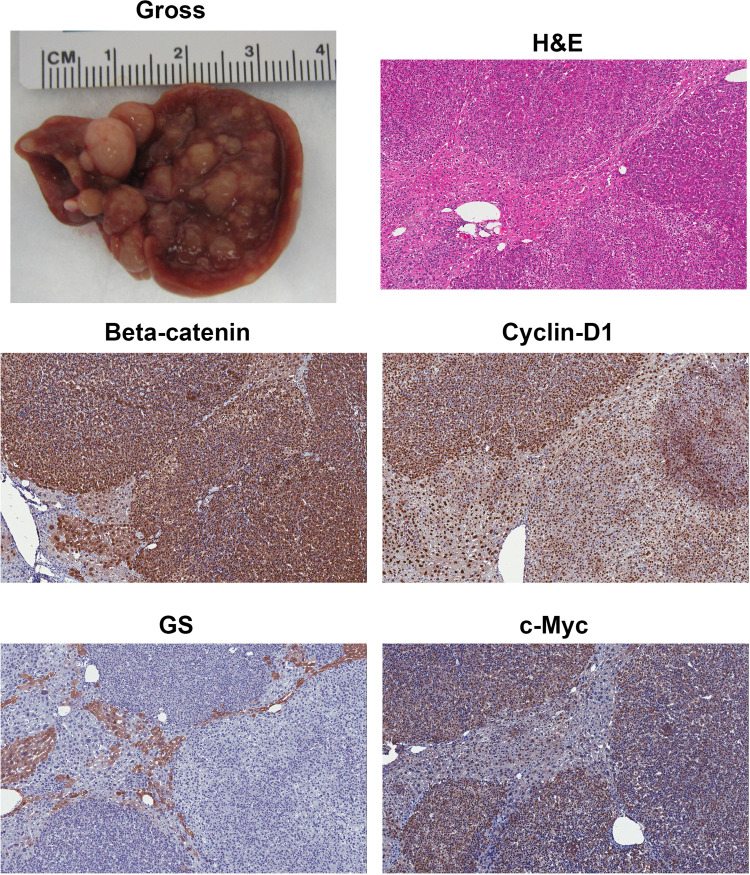
The discovery of the Hippo pathway is still in its infancy, having only been implicated in oncogenesis within the past 5 years. However, signaling connections between the Yap and the Wnt/β-catenin pathway are already being described, suggesting a cooperation in tumor development37. Recently dysfunctional Hippo signaling was demonstrated in liver tumor including hepatoblastomas38. This study used IHC to show nuclear staining of Yap in 73% of all hepatoblastoma samples (n = 22). More recently, we showed concurrent nuclear staining of Yap and β-catenin in 79% of the 94 human hepatoblastoma samples39. A functional synergism was identified by studies in several hepatoblastoma cell lines where silencing of β-catenin and Yap showed decreased cell proliferation and increased apoptosis, and this effect was amplified upon dual silencing of both genes. In the most convincing evidence shown, coexpression of active Yap and β-catenin via hydrodynamic tail vein injection of sleeping beauty transposons containing S127A–Yap and Δ90–β-catenin led to hepatoblastoma development (Fig. 2). There was no tumor development when the mutant β-catenin or mutant Yap was expressed individually, supporting the idea that a second hit is required for hepatoblastoma to occur39. Grossly, the tumors in this model were pale, multinodular, and abundant, covering almost the entire surface of the liver especially in advanced stages (Fig. 2). This was due to rapid clonal expansion of the hepatocytes that stably expressed mutant β-catenin and mutant Yap. Histologically, the tumors appeared to display fetal or intermediate differentiation (histological categories discussed in the forthcoming section) (Fig. 2). However, these tumors showed absence of GS staining, which is traditionally a target of the Wnt signaling pathway reflective of a more differentiated hepatocyte state (Fig. 2)22. While these tumors started early on as GS+ tumors, they lose GS staining rapidly as tumors expand and grow. GS− hepatoblastomas despite the presence of nuclear β-catenin is reflective of a more embryonal hepatoblastoma22. This is likely due to the fact that β-catenin activation during normal liver development is biphasic19,20. While it controls hepatoblast proliferation during early liver development through regulation of targets such as cyclin D1, at later stages of hepatic development it controls hepatocyte maturation by regulating targets such as GS. Thus, embryonal hepatoblastomas are GS−, while fetal hepatoblastomas are GS+, although both show nuclear/cytoplasmic β-catenin22. These observations show that in the Yap–β-catenin model, the tumors may be starting with more fetal morphology, but as the tumors progress, they may dedifferentiate more and at least at a molecular level resemble embryonal hepatoblastomas39. In addition to being positive for β-catenin and negative for GS, the hepatoblastoma in the Yap–β-catenin model was positive for c-Myc and cyclin D1 (Fig. 2).
Figure 2.
Yap and β-catenin coexpression in liver leads to hepatoblastomas in mice. Gross image of tumor-bearing liver at 9 weeks after hydrodynamic tail vein injection of sleeping beauty plasmids carrying mutant Yap and β-catenin. Histology from a representative 9-week postinjection liver reveals the tumors to be hepatoblastomas. Tumors were notably positive for nuclear and cytoplasmic β-catenin and strongly positive for the β-catenin target cyclin D1. Tumors were negative for another β-catenin target, glutamine synthetase (GS). Tumors were strongly positive for β-catenin target cyclin D1.
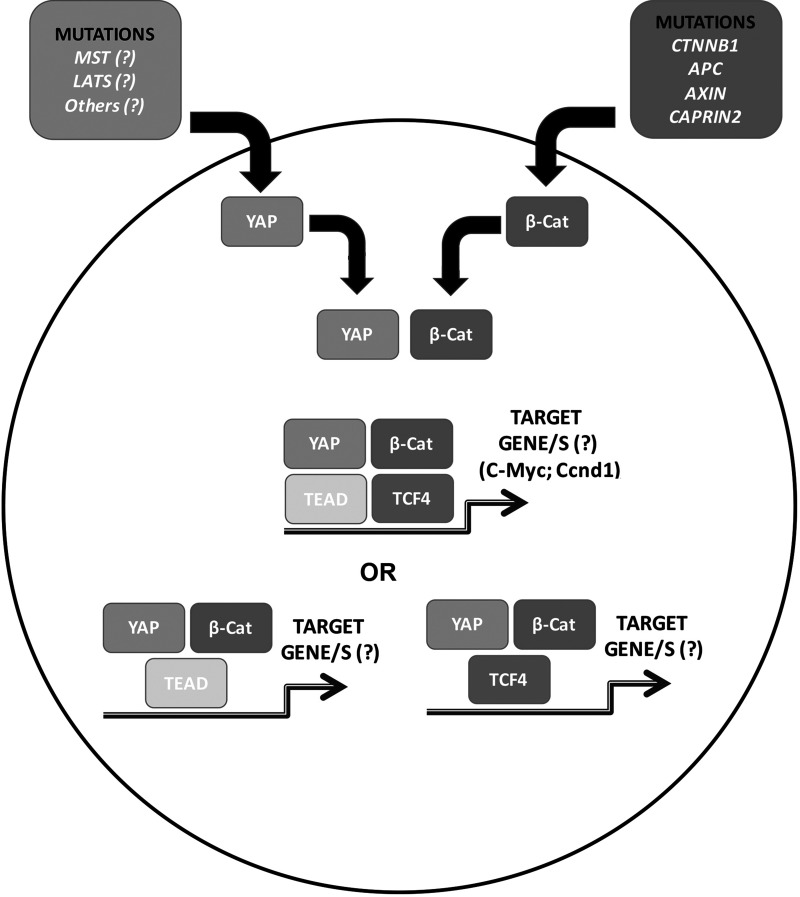
In this model, there was also a clear evidence of an association of β-catenin and Yap1 such as by coprecipitation studies39. Further, introduction of dominant-negative TCF4 or a mutant of Yap, which could not bind TEAD at the time of injection of S127A–Yap and Δ90–β-catenin, completely prevented hepatoblastoma development. This demonstrates the relevance of respective transcription factors for both Yap and β-catenin in disease pathogenesis. However, whether all four proteins are in one complex or whether Yap and β-catenin bind to TCF4 or TEAD distinctly to drive specific target genes essential for hepatoblastoma initiation and progression remains unknown (Fig. 3).
Figure 3.
Model depicting Yap and β-catenin cooperation in hepatoblastoma. Nuclear translocation of β-catenin in the majority of hepatoblastomas occurs due to various mechanisms such as mutations in CTNNB1 or due to mutations affecting genes encoding for proteins like Axin1 and APC, which are required for β-catenin degradation. Yap nuclear translocation also is evident in a majority of these tumors; however, the mechanism remains elusive. In the nucleus, the two proteins interact with each other and either interact their respective transcription factors together in a complex or in two separate complexes as indicated to eventually dictate target genes such as cyclin D1 and c-Myc or others that are not yet identified, to lead to hepatoblastoma development and growth.
Last, while activation of β-catenin in hepatoblastoma is due to mutations or deletions affecting exon 3 of CTNNB1 or loss-of-function mutations in APC and AXIN1 or gain-of-function mutations in CAPRIN2, the mechanisms leading to Yap activation remain obscure (Fig. 3). Suffice to say the interaction between the two pathways seems critical, and further characterization should have novel biological and therapeutic implications.
GENOTYPE–PHENOTYPE RELATIONSHIP IN HEPATOBLASTOMA
Histological or Cellular Correlates of Tumor Prognosis
Broadly, hepatoblastomas can be separated into two main histological groups: epithelial and epithelial mixed with mesenchymal elements, including bone, cartilage, or skeletal muscle tissue40. The latter group comprises about 20%–56% of tumors9,10,41,42. The epithelial group can be further classified into well-differentiated fetal (WDF), crowded fetal (fetal with mitoses), embryonal, macrotrabecular (MT) (either fetal or embryonal), and SCU histology (Figs. 4 and 5). The majority of subtypes are combined fetal and embryonal, both of which are present in 30%–60% of cases, with the minority of samples being either pure WDF, MT histology, or SCU subtype (again usually mixed in with other epithelial elements), accounting for <10% of specimens. It is important to remember however that most initial histological diagnosis is performed with a small biopsy sample that may or may not be representative of the much larger tumor it was obtained from.
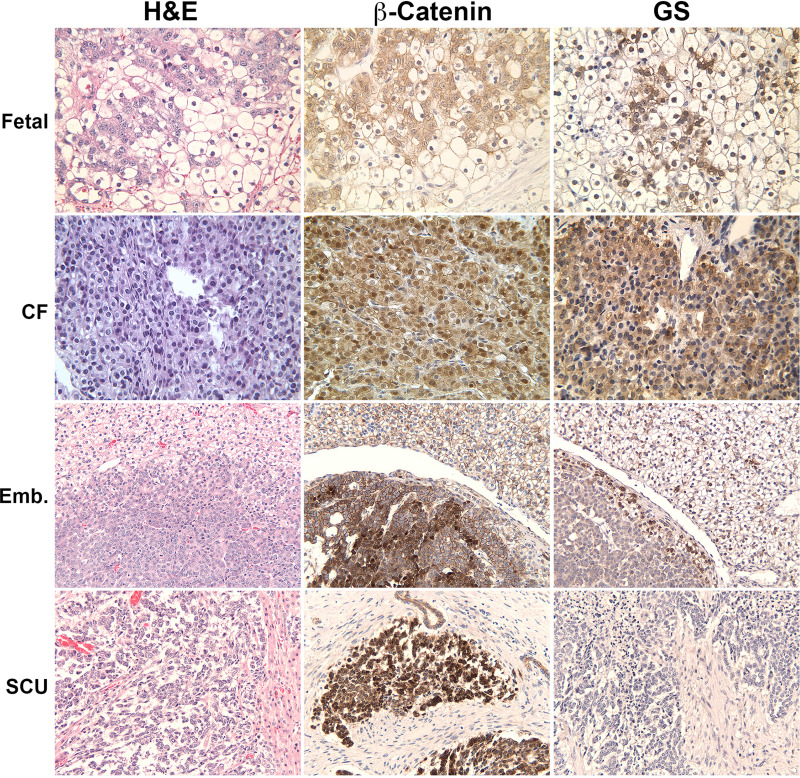
Figure 4.
Wnt signaling status in various histologic subtypes of hepatoblastoma in patients. A representative fetal hepatoblastoma shows the presence of nuclear and cytoplasmic β-catenin along with patchy positivity for glutamine synthetase (GS). A crowded fetal (CF) hepatoblastoma is positive for β-catenin as well as GS, which is more homogeneous. Embryonal hepatoblastoma is strongly positive for nuclear and cytoplasmic β-catenin but negative for GS. Occasional area may show some staining for GS. Small cell undifferentiated (SCU) hepatoblastoma is again strongly positive for β-catenin but completely lacks any GS staining.
Figure 5.
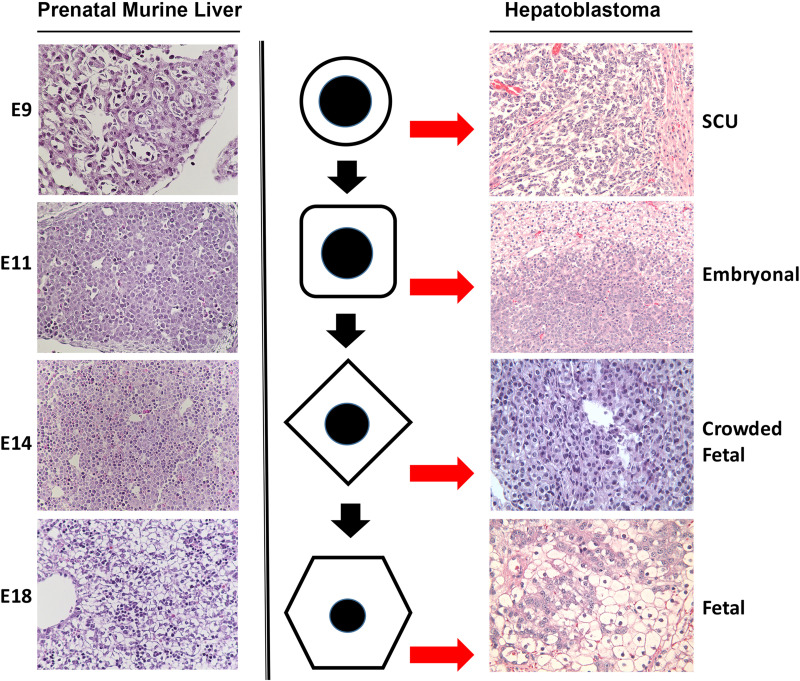
Comparative histology of murine prenatal hepatic developmental stages and histologic subtypes of human hepatoblastoma. Hepatoblastomas are classified based on their cell composition. There is resemblance in histology of various stages of hepatic development to various subtypes of hepatoblastoma, which is dictated by the predominant cell type that arrests and expands within the tumor. Shown to the left are representative images of developing mouse liver from embryonic day 9 (E9), E11, E14, and E18. Shown to the right are the sections from patient hepatoblastomas of various subtypes including small cell undifferentiated (SCU), embryonal (Emb), crowded fetal (CF), and fetal hepatoblastoma. The middle row is a representation of the predominant cell type that exists at specific stages during normal liver development, but an arrest at specific stages of maturation leads to their expansion and enrichment to eventually contribute to a specific histology and eventually a histological subtype of hepatoblastoma.
The histology of hepatoblastoma has a significant impact on patient outcomes. In looking at possible risk factors that could predict outcome for these children, Haas et al. showed a clear difference in overall survival depending on these subgroup classifications with 24-month survival rates being 92% for pure fetal, 63% for embryonal, 50% for MT, and a dismal 0% for SCU10. This group also found a significant impact of number of mitotic figures in a sample on overall patient survival. The most mature, differentiated subtype referred to as pure fetal histology has the lowest mitotic index. This histology has the best prognosis of all subtypes as disease presentation is local, usually confined to only a liver lobe that is amenable to surgical resection. In the most recent COG study (AHEP0731), patients with pure fetal histology are classified as very low risk, as their survival rate is close to 100% with surgical resection alone43. Adjuvant chemotherapy is usually omitted in these cases. On the other hand, embryonal histology poses a greater risk for invasion into multiple liver segments and blood vessels, and lymphatics, as well as greater risk of metastasis as it is less differentiated and can contain anaplastic and teratoid elements. Mitotic index is usually high in these cases.
Molecular Correlates of Tumor Prognosis
The morphology of hepatoblastoma can be distinctively correlated with different stages of prenatal liver development (Figs. 4 and 5). IHC has demonstrated several differences between pure fetal and embryonal histology that correlate with markers indicative of later or earlier stages of embryonic maturation, respectively. The WDF histology appears to show patchy β-catenin nuclear stain and strong staining for GS, a marker for differentiation, and β-catenin activation (Fig. 4). Histologically, fetal hepatoblastomas are composed of cells that resemble hepatocytes present in the livers of late prenatal stages of mouse embryonic development such as embryonic days 17–18 (Fig. 5). The tumor is composed of sheets of uniformed cuboidal cells with clear cytoplasm. The CF shows intense and strong nuclear β-catenin as well as strong GS staining (Fig. 4). Histologically, the cells contained in CF hepatoblastoma look similar to cells contained in embryonic day 14–16 murine livers minus the ongoing hematopoiesis (Fig. 5). The embryonal hepatoblastoma shows diffuse nuclear β-catenin staining and variable but mostly negative GS (Fig. 4). This type of tumor is composed of cells that resemble hepatoblasts in mouse embryonic days 11–13 (Fig. 5). The SCU hepatoblastoma or the SCU component of a hepatoblastoma is also positive for nuclear and cytoplasmic β-catenin, while being negative for GS (Fig. 4). The SCU hepatoblastoma histology may represent an even earlier phase of embryonic liver development in mice. We have included an area from embryonic day 9 of mouse showing cardiac mesenchyme and hepatoblasts at the time of hepatic bud formation (Fig. 5).
Based on histological comparison between stages of hepatic development and histologic subtypes of hepatoblastoma, one can hypothesize on the origin of hepatoblastomas. It is likely that resident hepatic cells at specific stages of normal liver development undergo molecular perturbations due to genetic or environmental factors that lead to their clonal expansion to yield hepatoblastoma with a corresponding histological subtype (Fig. 5).
Since β-catenin localization differs in histological subtypes of hepatoblastoma, it may have prognostic significance. As mentioned previously, poorly differentiated hepatoblasts (embryonal and SCU) have intense nuclear staining of β-catenin, while pure fetal hepatocytes (WDF) have predominantly membranous staining of this protein with only focal nuclear staining in many cases. The nuclear visualization of β-catenin by IHC has been correlated to more aggressive tumor growth, worse patient prognosis, and advanced disease23,24,44,45. Similarly, there are more recent data available to suggest that more stem cell marker expression (e.g., SALL4 or LIN28B) may be associated with worse outcome, possibly related to staining of more primitive hepatoblastoma cell types (personal observation by S.R.).
MODELS TO STUDY HEPATOBLASTOMA
Cell Lines
While there are innate limitations to the use of cancer cell lines, they do remain an important tool to study molecular aberrations and cell behavior. Several human hepatoblastoma cell lines exist, although some are more characterized than others. In Table 1, we list all known human hepatoblastoma cells along with the corresponding mutations in CTNNB1 in the form of either a missense mutation or an interstitial deletion. The most well-known and readily available cells include the HepG2 cells, which have been erroneously used as human hepatocellular carcinoma (HCC) cells in many previous studies. However, human HCC cells do not exhibit large interstitial deletion in CTNNB1 as is evident in HepG2 cells, but instead show mostly point mutations, although hepatoblastomas can have either point mutations or deletions in exon 3 of CTNNB1 (Table 1). Also, while HC-AFW1 cells were initially isolated from a pediatric HCC case, based on, again, a large interstitial deletion in exon 3 of CTNNB1, it may exhibit more hepatoblastoma-like characteristics and hence has also been included in Table 1 46.
Table 1.
Human Cell Lines With β-Catenin Mutations
| Human Cell Lines | CTNNB1 Mutation |
|---|---|
| HepG2 | Δ116 amino acids* |
| HepT1 | Δ76 amino acids* |
| HC-AFW1 | Δ49 amino acids* † |
| HepT5 | Δ75 amino acids* |
| Hep2933TT | Δ117 amino acids* |
| HepT3 | T41A |
| HuH6 | T41A‡ |
Exon 3 deletions.
HC-AFW1 cells were originally derived by a 4-year-old patient with hepatocellular carcinoma, not hepatoblastoma; however, these cell were found to have a typical truncated mutation as seen in hepatoblastoma and, thus, behavior in a similar manner to other hepatoblastoma cell lines with similar mutations46.
Threonine-to-alanine substitution at codon 41.
Animal Models
Having an animal model of a human disease is highly relevant to study mechanism, biology, and therapies. Animal models for hepatoblastomas are outlined in Table 2. As can be appreciated, there is an array of models available. There are transgenic mice models like c-Myc transgenic mice and Lin28b transgenic mice that display mixed tumors throughout the liver. These mice display HCCs as well as hepatoblastomas47,48. As discussed previously, we have generated a novel mouse model where coexpression of mutant Yap and mutant β-catenin leads to hepatoblastoma development in mice39. Traditionally, using several hepatoblastoma cell lines outlined in Table 1, various groups have generated tumor xenograft models for hepatoblastoma. The sites for injections are either subcutaneous or through the spleen, which allow for some cells to escape through circulation to eventually form tumor nodules in the liver. A list of these models, which have been widely used to test therapies and tumor behavior, is presented in Table 2. Last, hepatoblastoma cells derived from explanted livers postsurgical resection have been used to derive xenograft models. Patient-derived xenografts are indeed an innovative tool to study cancer and can have implications in personalized medicine. Intriguingly, a patient-derived xenograft study involving hepatoblastoma showed not only development of tumors in the liver but also pulmonary metastasis49.
Table 2.
Summary of Hepatoblastoma Mouse Models
| Mouse Models | Tumor Location | Metastasis | References |
|---|---|---|---|
| MYC transgenic mouse | Liver | No | 48 |
| Lin28b transgenic mouse | Liver | No | 47 |
| β-Catenin–YAP dual plasmid injections using SB and hydrodynamic tail vein injection | Liver | No | 39 |
| Subcutaneous injection of human hepatoblastoma cell lines | Subcutaneous tumors at the site of injection (flank, thigh, and paravertebral) | No | 54–59 |
| HuH6 splenic injection in immunocompromised mice | Largely splenic, but 82% developed liver tumors after immediate splenectomy postinjection | No | 60,61 |
| Patient-derived xenograft transplanted into mouse liver | Liver | Yes, 50% with pulmonary nodules | 49 |
SUMMARY AND FUTURE PERSPECTIVES OF Wnt SIGNALING IN HEPATOBLASTOMA
Hepatoblastoma is a classical pediatric tumor where only limited numbers of mutations are found per case. In fact, recent WES on these tumor types has revealed 1–7 mutations/case (average of 2.9 mutations/case)28. Many of these mutations affect the Wnt pathway such that almost 75% of all hepatoblastomas fall into this category due to mutations/deletions in CTNNB1, APC, or AXIN1. Clearly, hepatoblastoma has β-catenin activation, but is that sufficient to cause this tumor if activated appropriately during development? This remains an unanswered question, although APC deletion during hepatic development led to embryonic lethality50. In this model, APC deletion occurred in hepatoblasts, but due to this untimely and excessive activation, where correct partners of β-catenin may not be expressed, the liver became hypoplastic, and hepatocyte differentiation failed. β-Catenin activation in this model did promote bile duct development50. Indeed, the role of β-catenin in bile duct development has been intriguing and requires further analysis51,52. However, future studies will be critical to determine if β-catenin activation in immature or mature hepatocytes during hepatic development may be sufficient to cause hepatoblastoma. Equally relevant will be to determine why hepatoblastomas occur more frequently in low birth weight babies.
Yap signaling pathway cooperates with β-catenin in hepatoblastoma pathogenesis39. However, the mechanism of Yap activation in hepatoblastoma cases remains unknown, and hence more studies are needed. The mechanism of how Yap and β-catenin contribute to hepatoblastoma development and growth remains to be discovered. Recently, we have identified c-Myc to be one downstream target of Yap–β-catenin in the hepatoblastoma model. When sleeping beauty transposons with mutant Yap and mutant β-catenin were injected in hepatocyte-specific c-Myc knockout mice, there was a significant decrease in hepatoblastoma burden, which was attributable to a compromise in optimum tumor metabolism53. Further, this model will be useful to test novel therapies for hepatoblastoma treatment.
WGS and WES studies are already revealing new molecular aberrations in hepatoblastoma. A recent study showed mutations in NFE2L2 (encoding for Nrf2) in a small group of hepatoblastoma cases29. Nrf2 plays an important role in regulating redox state of a cell specifically transcribing target genes involved in antioxidant response. Continued analysis is expected to yield additional new aberrations and may eventually help understand the biology of a subset of tumors that are more aggressive and have poor prognosis. Since molecular therapies are lacking for hepatoblastoma, such studies may also yield druggable targets that may improve.
ACKNOWLEDGMENTS
This study was funded by NIH grants 1R01DK62277, 1R01DK100287, and R01CA204586 and Endowed Chair for Experimental Pathology to S.P.S.M. S.P.S.M. is a consultant for Abbvie Pharmaceutical and Dicerna Pharmaceuticals and has corporate research agreements with the two companies.
REFERENCES
- 1. Darbari A, Sabin KM, Shapiro CN, Schwarz KB. Epidemiology of primary hepatic malignancies in U.S. children. Hepatology 2003;38(3):560–6. [DOI] [PubMed] [Google Scholar]
- 2. McLaughlin CC, Baptiste MS, Schymura MJ, Nasca PC, Zdeb MS. Maternal and infant birth characteristics and hepatoblastoma. Am J Epidemiol. 2006;163(9):818–28. [DOI] [PubMed] [Google Scholar]
- 3. Czauderna P, Haeberle B, Hiyama E, Rangaswami A, Krailo M, Maibach R, Rinaldi E, Feng Y, Aronson D, Malogolowkin M, Yoshimura K, Leuschner I, Lopez-Terrada D, Hishiki T, Perilong G, von Schewinitz D, Schmid I, Watanabe K, Derosa M, Meyers R. The Children’s Hepatic tumors International Collaboration (CHIC): Novel global rare tumor database yields new prognostic factors in hepatoblastoma and becomes a research model. Eur J Cancer 2016;52:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tanimura M, Matsui I, Abe J, Ikeda H, Kobayashi N, Ohira M, Yokoyama M, Kaneko M. Increased risk of hepatoblastoma among immature children with a lower birth weight. Cancer Res. 1998;58(14):3032–5. [PubMed] [Google Scholar]
- 5. Spector LG, Johnson KJ, Soler JT, Puumala SE. Perinatal risk factors for hepatoblastoma. Br J Cancer 2008;98(9):1570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schnater JM, Aronson DC, Plaschkes J, Perilongo G, Brown J, Otte JB, Brugieres L, Czauderna P, MacKinlay G, Vos A. Surgical view of the treatment of patients with hepatoblastoma: Results from the first prospective trial of the International Society of Pediatric Oncology Liver Tumor Study Group. Cancer 2002;94(4):1111–20. [PubMed] [Google Scholar]
- 7. Czauderna P, Lopez-Terrada D, Hiyama E, Haberle B, Malogolowkin MH, Meyers RL. Hepatoblastoma state of the art: Pathology, genetics, risk stratification, and chemotherapy. Curr Opin Pediatr. 2014;26(1):19–28. [DOI] [PubMed] [Google Scholar]
- 8. Davies JQ, de la Hall PM, Kaschula RO, Sinclair-Smith CC, Hartley P, Rode H, Millar AJ. Hepatoblastoma—Evolution of management and outcome and significance of histology of the resected tumor. A 31-year experience with 40 cases. J Pediatr Surg. 2004;39(9):1321–7. [DOI] [PubMed] [Google Scholar]
- 9. Fuchs J, Rydzynski J, Von Schweinitz D, Bode U, Hecker H, Weinel P, Burger D, Harms D, Erttmann R, Oldhafer K, Mildenberger H, Study Committee of the Cooperative Pediatric Liver Tumor Study Hb 94 for the German Society for Pediatric Oncology and Hematology. Pretreatment prognostic factors and treatment results in children with hepatoblastoma: A report from the German Cooperative Pediatric Liver Tumor Study HB 94. Cancer 2002;95(1):172–82. [DOI] [PubMed] [Google Scholar]
- 10. Haas JE, Muczynski KA, Krailo M, Ablin A, Land V, Vietti TJ, Hammond GD. Histopathology and prognosis in childhood hepatoblastoma and hepatocarcinoma. Cancer 1989;64(5):1082–95. [DOI] [PubMed] [Google Scholar]
- 11. von Schweinitz D, Hecker H, Harms D, Bode U, Weinel P, Burger D, Erttmann R, Mildenberger H. Complete resection before development of drug resistance is essential for survival from advanced hepatoblastoma—A report from the German Cooperative Pediatric Liver Tumor Study HB-89. J Pediatr Surg. 1995;30(6):845–52. [DOI] [PubMed] [Google Scholar]
- 12. De Ioris M, Brugieres L, Zimmermann A, Keeling J, Brock P, Maibach R, Pritchard J, Shafford L, Zsiros J, Czaudzerna P, Perilongo G. Hepatoblastoma with a low serum alpha-fetoprotein level at diagnosis: The SIOPEL group experience. Eur J Cancer 2008;44(4):545–50. [DOI] [PubMed] [Google Scholar]
- 13. Meyers RL, Tiao G, de Ville de Goyet J, Superina R, Aronson DC. Hepatoblastoma state of the art: Pre-treatment extent of disease, surgical resection guidelines and the role of liver transplantation. Curr Opin Pediatr. 2014;26(1):29–36. [DOI] [PubMed] [Google Scholar]
- 14. Perilongo G, Brown J, Shafford E, Brock P, De Camargo B, Keeling JW, Vos A, Philips A, Pritchard J, Plaschkes J. Hepatoblastoma presenting with lung metastases: Treatment results of the first cooperative, prospective study of the International Society of Paediatric Oncology on childhood liver tumors. Cancer 2000;89(8):1845–53. [DOI] [PubMed] [Google Scholar]
- 15. Dan YY, Riehle KJ, Lazaro C, Teoh N, Haque J, Campbell JS, Fausto N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc Natl Acad Sci USA 2006;103(26):9912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes Dev. 1997;11(24):3286–305. [DOI] [PubMed] [Google Scholar]
- 17. Monga SP. Role and regulation of β-catenin signaling during physiological liver growth. Gene Expr. 2014;16(2):51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology 2006;131(5):1561–72. [DOI] [PubMed] [Google Scholar]
- 19. Lade AG, Monga SP. Beta-catenin signaling in hepatic development and progenitors: Which way does the WNT blow? Dev Dyn. 2011;240(3):486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan X, Yuan Y, Zeng G, Apte U, Thompson MD, Cieply B, Stolz DB, Michalopoulos GK, Kaestner KH, Monga SP. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology 2008;47(5):1667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monga SP. β-Catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology 2015;148(7):1294–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armengol C, Cairo S, Fabre M, Buendia MA. Wnt signaling and hepatocarcinogenesis: The hepatoblastoma model. Int J Biochem Cell Biol. 2011;43(2):265–70. [DOI] [PubMed] [Google Scholar]
- 23. Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, Goga A, Balakrishnan A, Semeraro M, Gresh L, Gresh L, Pontoglio M, Strick-Marchand H, Levillayer F, Nouet Y, Rickman D, Gauthier F, Branchereau S, Brugières L, Laithier V, Bouvier R, Boman F, Basso G, Michiels JF, Hofman P, Arbez-Gindre F, Jouan H, Rousselet-Chapeau MC, Berrebi D, Marcellin L, Plenat F, Zachar D, Joubert M, Selves J, Pasquier D, Bioulac-Sage P, Grotzer M, Childs M, Fabre M, Buendia MA. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell 2008;14(6):471–84. [DOI] [PubMed] [Google Scholar]
- 24. Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res. 1999;59(2):269–73. [PubMed] [Google Scholar]
- 25. Koch A, Weber N, Waha A, Hartmann W, Denkhaus D, Behrens J, Birchmeier W, von Schweinitz D, Pietsch T. Mutations and elevated transcriptional activity of conductin (AXIN2) in hepatoblastomas. J Pathol. 2004;204(5):546–54. [DOI] [PubMed] [Google Scholar]
- 26. Aretz S, Koch A, Uhlhaas S, Friedl W, Propping P, von Schweinitz D, Pietsch T. Should children at risk for familial adenomatous polyposis be screened for hepatoblastoma and children with apparently sporadic hepatoblastoma be screened for APC germline mutations? Pediatr Blood Cancer 2006;47(6):811–8. [DOI] [PubMed] [Google Scholar]
- 27. Koch A, Waha A, Hartmann W, Hrychyk A, Schuller U, Waha A, Wharton KA Jr, Fuchs SY, von Schweinitz D, Pietsch T. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res. 2005;11(12):4295–304. [DOI] [PubMed] [Google Scholar]
- 28. Eichenmuller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Haberle B, Cairo S, Leuschner I, von Schweinitz D, Strom TM, Kappler R. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61(6):1312–20. [DOI] [PubMed] [Google Scholar]
- 29. Jia D, Dong R, Jing Y, Xu D, Wang Q, Chen L, Li Q, Huang Y, Zhang Y, Zhang Z, Liu L, Zheng S, Xia Q, Wang H, Dong K, He X. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology 2014;60(5):1686–96. [DOI] [PubMed] [Google Scholar]
- 30. Knudson A. Retinoblastoma: Teacher of cancer biology and medicine. PLoS Med. 2005;2(10):e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chompret A, Brugieres L, Ronsin M, Gardes M, Dessarps-Freichey F, Abel A, Hua D, Ligot L, Dondon MG, Bressac-de Paillerets B, Frébourg T, Lemerle J, Bonaïti-Pellié C, Feunteun J. P53 germline mutations in childhood cancers and cancer risk for carrier individuals. Br J Cancer 2000;82(12):1932–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mussa A, Molinatto C, Baldassarre G, Riberi E, Russo S, Larizza L, Riccio A, Ferrero GB. Cancer risk in Beckwith-Wiedemann syndrome: A systematic review and meta-analysis outlining a novel (epi)genotype specific histotype targeted screening protocol. J Pediatr. 2016. [DOI] [PubMed] [Google Scholar]
- 33. Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17(23):2054–60. [DOI] [PubMed] [Google Scholar]
- 34. Barry ER, Camargo FD. The Hippo superhighway: Signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol. 2013;25(2):247–53. [DOI] [PubMed] [Google Scholar]
- 35. Lee DH, Park JO, Kim TS, Kim SK, Kim TH, Kim MC, Park GS, Kim JH, Kuninaka S, Olson EN, Saya H, Kim SY, Lee H, Lim DS. LATS-YAP/TAZ controls lineage specification by regulating TGFbeta signaling and Hnf4alpha expression during liver development. Nat Commun. 2016;7:11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, Camargo FD. Hippo pathway activity influences liver cell fate. Cell 2014;157(6):1324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Konsavage WM Jr, Yochum GS. Intersection of Hippo/YAP and Wnt/beta-catenin signaling pathways. Acta Biochim Biophys Sin. (Shanghai) 2013;45(2):71–9. [DOI] [PubMed] [Google Scholar]
- 38. Li H, Wolfe A, Septer S, Edwards G, Zhong X, Abdulkarim AB, Ranganathan S, Apte U. Deregulation of Hippo kinase signalling in human hepatic malignancies. Liver Int. 2012;32(1):38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, Fan B, Terracciano L, Armeanu-Ebinger S, Ribback S, Dombrowski F, Evert M, Chen X, Monga SP. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice . Gastroenterology 2014;147(3):690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopez-Terrada D, Alaggio R, de Davila MT, Czauderna P, Hiyama E, Katzenstein H, Leuschner I, Malogolowkin M, Meyers R, Ranganathan S, Tanaka Y, Tomlinson G, Fabrè M, Zimmermann A, Finegold MJ, Children’s Oncology Group Liver Tumor Committee. Towards an international pediatric liver tumor consensus classification: Proceedings of the Los Angeles COG liver tumors symposium. Mod Pathol. 2014;27(3):472–91. [DOI] [PubMed] [Google Scholar]
- 41. Lack EE, Neave C, Vawter GF. Hepatoblastoma. A clinical and pathologic study of 54 cases. Am J Surg Pathol. 1982;6(8):693–705. [PubMed] [Google Scholar]
- 42. Gonzalez-Crussi F, Upton MP, Maurer HS. Hepatoblastoma. Attempt at characterization of histologic subtypes. Am J Surg Pathol. 1982;6(7):599–612. [PubMed] [Google Scholar]
- 43. Malogolowkin MH, Katzenstein HM, Meyers RL, Krailo MD, Rowland JM, Haas J, Finegold MJ. Complete surgical resection is curative for children with hepatoblastoma with pure fetal histology: A report from the Children’s Oncology Group. J Clin Oncol. 2011;29(24):3301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lopez-Terrada D, Gunaratne PH, Adesina AM, Pulliam J, Hoang DM, Nguyen Y, Mistretta TA, Margolin J, Finegold MJ. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol. 2009;40(6):783–94. [DOI] [PubMed] [Google Scholar]
- 45. Ferrari A, De Salvo GL, Brennan B, van Noesel MM, De Paoli A, Casanova M, Francotte N, Kelsey A, Alaggio R, Oberlin O, Carli M, Ben-Arush M, Bergeron C, Merks JH, Jenny M, Stevens MC, Bisogno G, Orbach D. Synovial sarcoma in children and adolescents: The European Pediatric Soft Tissue Sarcoma Study Group prospective trial (EpSSG NRSTS 2005). Ann Oncol. 2015;26(3):567–72. [DOI] [PubMed] [Google Scholar]
- 46. Armeanu-Ebinger S, Wenz J, Seitz G, Leuschner I, Handgretinger R, Mau-Holzmann UA, Bonin M, Sipos B, Fuchs J, Warmann SW. Characterisation of the cell line HC-AFW1 derived from a pediatric hepatocellular carcinoma. PLoS One 2012;7(5):e38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, Comerford SA, Ramezani S, Sun X, Parikh MS, Yang EH, Powers JT, Shinoda G, Shah SP, Hammer RE, Daley GQ, Zhu H. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014;26(2):248–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004;431(7012):1112–7. [DOI] [PubMed] [Google Scholar]
- 49. Bissig-Choisat B, Kettlun-Leyton C, Legras XD, Zorman B, Barzi M, Chen LL, Amin MD, Huang YH, Pautler RG, Hampton OA, Prakash MM, Yang D, Borowiak M, Munzy D, Doddapaneni HV, Hu J, Shi Y, Gaber MW, Hicks MJ, Thompson PA, Lu Y, Mills GB, Finegold M, Goss JA, Parsons DW, Vasudevan SA, Sumazin P, López-Terrada D, Bissig KD. Novel patient-derived xenograft and cell line models for therapeutic testing of pediatric liver cancer. J Hepatol. 2016;65(2):325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Decaens T, Godard C, de Reynies A, Rickman DS, Tronche F, Couty JP, Perret C, Colnot S. Stabilization of beta-catenin affects mouse embryonic liver growth and hepatoblast fate. Hepatology 2008;47(1):247–58. [DOI] [PubMed] [Google Scholar]
- 51. Cordi S, Godard C, Saandi T, Jacquemin P, Monga SP, Colnot S, Lemaigre FP. Role of beta-catenin in development of bile ducts. Differentiation 2016;91(1–3):42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Monga SP, Monga HK, Tan X, Mule K, Pediaditakis P, Michalopoulos GK. Beta-catenin antisense studies in embryonic liver cultures: Role in proliferation, apoptosis, and lineage specification. Gastroenterology 2003;124(1):202–16. [DOI] [PubMed] [Google Scholar]
- 53. Wang H, Lu J, Edmunds LR, Kulkarni S, Dolezal J, Tao J, Ranganathan S, Jackson L, Fromherz M, Stolz DB, Uppala R, Bharathi S, Monga SP, Goetzman ES, Prochownik EV. Coordinated activities of multiple Myc-dependent and Myc-independent biosynthetic pathways in hepatoblastoma. J Biol Chem. 2016;291(51):26241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berger M, Neth O, Ilmer M, Garnier A, Salinas-Martin MV, de Agustin Asencio JC, von Schweinitz D, Kappler R, Munoz M. Hepatoblastoma cells express truncated neurokinin-1 receptor and can be growth inhibited by aprepitant in vitro and in vivo. J Hepatol. 2014;60(5):985–94. [DOI] [PubMed] [Google Scholar]
- 55. Eicher C, Dewerth A, Thomale J, Ellerkamp V, Hildenbrand S, Warmann SW, Fuchs J, Armeanu-Ebinger S. Effect of sorafenib combined with cytostatic agents on hepatoblastoma cell lines and xenografts. Br J Cancer 2013;108(2):334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hata Y, Uchino J, Sato K, Sasaki F, Une Y, Naito H, Manabe K, Kuwahara T, Kasai Y. Establishment of an experimental model of human hepatoblastoma. Cancer 1982;50(1):97–101. [DOI] [PubMed] [Google Scholar]
- 57. Hayashi S, Fujita K, Matsumoto S, Akita M, Satomi A. Isolation and identification of cancer stem cells from a side population of a human hepatoblastoma cell line, HuH-6 clone-5. Pediatr Surg Int. 2011;27(1):9–16. [DOI] [PubMed] [Google Scholar]
- 58. Klein JL, Nguyen TH, Laroque P, Kopher KA, Williams JR, Wessels BW, Dillehay LE, Frincke J, Order SE, Leichner PK. Yttrium-90 and iodine-131 radioimmunoglobulin therapy of an experimental human hepatoma. Cancer Res. 1989;49(22):6383–9. [PubMed] [Google Scholar]
- 59. Lieber J, Dewerth A, Wenz J, Kirchner B, Eicher C, Warmann SW, Fuchs J, Armeanu-Ebinger S. Increased efficacy of CDDP in a xenograft model of hepatoblastoma using the apoptosis sensitizer ABT-737. Oncol Rep. 2013;29(2):646–52. [DOI] [PubMed] [Google Scholar]
- 60. Ellerkamp V, Armeanu-Ebinger S, Wenz J, Warmann SW, Schafer J, Ruck P, Fuchs J. Successful establishment of an orthotopic hepatoblastoma in vivo model in NOD/LtSz-scid IL2Rgammanull mice. PLoS One 2011;6(8):e23419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schnater JM, Bruder E, Bertschin S, Woodtli T, de Theije C, Pietsch T, Aronson DC, von Schweinitz D, Lamers WH, Kohler ES. Subcutaneous and intrahepatic growth of human hepatoblastoma in immunodeficient mice. J Hepatol. 2006;45(3):377–86. [DOI] [PubMed] [Google Scholar]