Abstract
Post-transcriptional regulatory strategies that involve coupling between terminal uridyltransferase (TUTase) and exoribonuclease enzymes have recently been characterized in diverse species. Of note, the 3′ exoribonuclease Dis3L2 has received substantial attention as a factor that metabolizes uridylated substrates in contexts such as general mRNA degradation, turnover of specific miRNAs, and quality control of noncoding RNAs. To date, most studies of Dis3L2 have focused on fungi and mammalian cells. Here, we introduce Drosophila as a system that permits analysis of molecular mechanisms as well as the ability to interrogate organismal phenotypes. We started with a structure–function analysis of the Drosophila TUTase Tailor, which we recently identified to inhibit biogenesis of splicing-derived miRNA hairpins. Next, we show that Tailor/Dis3L2 form a complex via N-terminal domains in the respective proteins that are distinct from their catalytic domains. In vitro, Dis3L2 has nuclease activity, but substrate oligouridylation by Tailor stimulates their degradation by Dis3L2, especially for structured substrates. We analyzed mutants of Tailor and Dis3L2, which are viable and lack overt morphological defects. Instead, these mutants exhibit defects in female and male fertility, implying specific requirements in the germline. Dis3L2 defects are more severe than Tailor, and their requirements appear stronger in males than in females. In particular, loss of Dis3L2 completely blocks productive spermatogenesis, causing male sterility. RNA-seq analysis from single- and double-mutant testes reveals aberrant gene expression programs and suggests that noncoding RNAs may be preferentially affected by Dis3L2. Overall, our studies of a new tailing/trimming complex reveal unexpectedly specific requirements during gametogenesis.
Keywords: Drosophila, TUTase, exoribonuclease, spermatogenesis
INTRODUCTION
The steady-state levels of transcripts are determined not only by their transcription, but also by their degradation. The contribution of catabolic processes to the RNA landscape is often overlooked, but they play diverse and crucial roles (Houseley and Tollervey 2009). General RNA decay mechanisms have been studied, the most well-known of which in eukaryotes involve sequence-independent degradation of mRNAs that have lost terminal protections, namely their 5′ cap and/or 3′ poly(A) tail. Loss of the former renders transcripts amenable to 5′–3′ decay (e.g., by XRN1/2), while loss of the latter facilitates degradation by 3′–5′ decay (e.g., by the exosome). Such general pathways for RNA catabolism are a generic strategy to recognize and remove broken or otherwise damaged transcripts from the cellular pool, thereby also recycling ribonucleotides. In addition, there are many regulated pathways that feed into the general RNA decay pathway. For example, decapping enzymes can actively promote turnover, while deadenylases can be recruited to particular substrates via diverse types of RNA binding proteins (RBPs) and regulatory RNAs such as miRNAs.
In recent years, several lines of research on diverse populations of transcripts revealed a theme for the involvement of untemplated nucleotide addition in regulated RNA decay (Scheer et al. 2016). For example, oligouridylation of mammalian histone mRNAs (which are not normally adenylated) elicits decapping and degradation from both ends (Mullen and Marzluff 2008; Slevin et al. 2014). In S. pombe, the Cid1 terminal uridyltransferase (TUTase) acts upon polyadenylated mRNAs to stimulate decapping and substrate turnover (Rissland and Norbury 2009). Several studies extended this concept to the regulation of miRNA intermediates or mature species. For instance, studies in mammalian cells showed that the specificity factor Lin-28 recruits TUTases (TUT4 and TUT7) to modify pre-let-7 hairpins and mediate their turnover (Hagan et al. 2009; Heo et al. 2009; Thornton et al. 2012). Moreover, the exposure of Drosophila and mammalian mature miRNAs to perfectly complementary targets was found to stimulate their tailing and degradation (Ameres et al. 2010).
These findings implied that untemplated nucleotide addition, and in particular uridylation, serves as a signal to recruit exoribonucleases that can execute substrate trimming. Strikingly, for many unrelated types of transcripts, polyuridylation mediates recognition and degradation by the 3′–5′ exoribonuclease Dis3L2. Of note, Dis3L2 mediates the turnover of polyuridylated pre-let-7 (Chang et al. 2013; Ustianenko et al. 2013) and of polyuridylated mRNAs (Lubas et al. 2013; Malecki et al. 2013). This relatively recently studied cytoplasmic RNase is a homolog of Dis3, a core component of the multisubunit exosome complex that is a major 3′–5′ degrading machinery. However, Dis3L2 does not associate with the exosome and differs in that it features in a signal-regulated turnover pathway, whereas the exosome is commonly thought to act upon all deadenylated transcripts. Indeed, yeast and mammalian Dis3L2 were shown to prefer uridylated substrates in vitro (Chang et al. 2013; Malecki et al. 2013; Ustianenko et al. 2013), and the structural basis of mammalian Dis3L2 binding specificity to a polyuridine tract was elucidated (Faehnle et al. 2014). Among metazoan TUTases, TUT4/7 belong to a vertebrate-specific subclade that has a distinctive domain structure that includes a duplicate, inactive, nucleotidyltransferase domain. Beyond small RNAs, TUT4/7 was also found to mediate uridylation of mRNA, which marks them for degradation (Lim et al. 2014) possibly via Dis3L2 (Thomas et al. 2015). Interestingly, human Dis3L2 was recently found to mediate target-directed tailing and degradation of miRNAs via TUT1 (Haas et al. 2016), which is a more typical TUTase with only a single nucleotidyltransferase domain. Thus, multiple TUTases may partner with Dis3L2 during regulated RNA turnover.
Recently, we and others defined the Drosophila TUTase Tailor as a pre-miRNA modifying factor that prefers noncanonical substrates from the splicing-mediated “mirtron” pathway (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015). In the course of further studies, we found that Tailor forms a complex with CG16940, the Drosophila ortholog of Dis3L2. We characterize the functional properties of these enzymes, the nature of the complex, and how tailing by Tailor stimulates substrate degradation by Dis3L2. As metazoan Dis3L2 mutants have not been studied in the organismal context, we generated Dis3L2 mutant flies. Alongside analysis of Tailor mutants, we find that germline is particularly sensitive to the action of this regulatory pathway. In particular, spermatogenesis is abolished in Dis3L2 mutants, and both Tailor and Dis3L2 testis exhibit altered gene expression, including of noncoding RNAs. Thus, a seemingly ubiquitous TUTase/nuclease complex is dispensable for viability, but indispensable for shaping the transcriptome to produce functional gametes.
RESULTS AND DISCUSSION
Structure–function analysis of Tailor in hairpin tailing
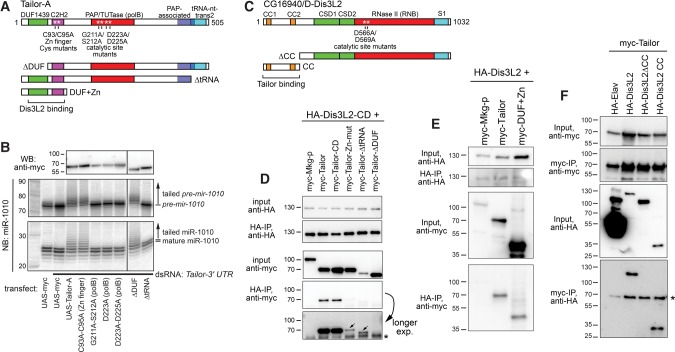
We recently identified the Drosophila TUTase Tailor as a factor that suppresses the biogenesis of noncanonical splicing-mediated miRNAs (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015). We investigated which Tailor protein domains mediate mirtron hairpin tailing. Besides its central polB-type PAP/TUTase domain, both Tailor isoforms (A and B) bear an N-terminal DUF1439 domain followed by a C2H2 zinc finger, and a C-terminal PAP-associated domain and tRNA nucleotidyltransferase 2 domain (these are predicted as overlapping). We generated additional polB mutants beyond our earlier study (Bortolamiol-Becet et al. 2015), as well as point mutants in the zinc finger, deletions of the DUF1439 or tRNA-nt-trans2 domains (Fig. 1A). We assayed these constructs by cotransfecting them into S2 cells with a mir-1010 expression construct, and analyzing its modification and maturation by Northern blotting. We evaluated the accumulation of Tailor proteins by Western blotting, and as preliminary tests showed that some mutants were less stable, we adjusted conditions to achieve more comparable levels (Fig. 1B).
FIGURE 1.
Structure–function analysis of Tailor and its association with Dis3L2. (A) Tailor domains and variants evaluated. (B) UAS-myc-Tailor variants were cotransfected into S2 cells along with ub-Gal4 and UAS-DsRed-mir-1010 mirtron vector. (WB) Western blotting of myc-Tailor variants. (NB) Northern blotting of pre-mir-1010 hairpins (middle blot) or mature miR-1010 species (bottom blot). Except for the first Northern lane, cells were depleted of endogenous Tailor by pretreatment with dsRNA against its 3′ UTR. Ectopic Tailor induces hairpin and mature miRNA tailing. This activity requires the polB and tRNA-transferase domains, but not the Zn finger or DUF1439 domain. (C) Dis3L2 domains and variants evaluated. (D) Coimmunoprecipitation (co-IP) assays of transfected HA-Dis3L2-CD and myc-Tailor variants. Mkg-p is another TUTase used as a control. Tailor and Tailor-CD exhibit comparable and specific association with Dis3L2-CD. Input blot shows that Tailor-ΔtRNA does not accumulate as well, but it can co-IP with Dis3L2-CD. In contrast, the Zn finger mutant and ΔDUF1439 variants exhibited substantially reduced association with Dis3L2-CD. Longer exposure highlights that the Zn finger mutant showed more co-IP signal than the ΔDUF1439 mutant; asterisk marks a nonspecific band. Similar overall binding profiles were seen between wild-type HA-Dis3L2 with Tailor variants (Supplemental Fig. S2). (E) The Tailor DUF1439 + Zn finger domains are sufficient to associate with Dis3L2. (F) The Dis3L2 N-terminal coiled-coil domains are necessary and sufficient to associate with Tailor. HA-Elav is used as an RNA binding protein control; it accumulates to a very high level but does not bind Tailor. Asterisk marks a nonspecific band.
We assayed these constructs in normal S2 cells as well as in cells depleted of endogenous Tailor, with similar results. The data from knockdown cells (achieved using a 3′ UTR dsRNA) are shown in Figure 1B. As expected, ectopic expression of Tailor stimulated strong tailing of the pre-mir-1010 substrate, and tailing was also observed in the mature miR-1010 pool. We earlier showed this activity was abolished in the D223A catalytic mutant, and show that additional polB point mutants as well as deletion of the tRNA-nt-trans domain are similarly inactive. On the other hand, the N-terminal domains were not necessary for tailing activity, since deletion of DUF1439 or point mutations of the Zn finger domain still induced strong mir-1010 tailing (Fig. 1B).
Tailor binds the Drosophila ortholog of Dis3L2
To understand Tailor's regulatory activities more fully, we investigated potential partner proteins. Genome-scale mass spectrometry analyses from the Drosophila Protein Interaction Map (Guruharsha et al. 2011) indicated that Tailor associated with the ribonuclease II CG16940. This seemed provocative, in light of recent literature on tailing-and-trimming pathways. CG16940 belongs to the Dis3 superfamily of CSD-RNB ribonucleases, within which Dis3 and Dis3L proteins (but not Dis3L2 proteins) also contain PIN and CR3 domains. As CG16940 lacks PIN and CR3 (Fig. 1C), it belongs to the Dis3L2 subclade (Malecki et al. 2013), a classification bolstered by the functional analogy that mammalian Dis3L2 associates with TUTases (Scheer et al. 2016). We also note that cold shock domains (CSD) were not predicted for CG16940 by several domain annotation servers, but modeling based on the murine Dis3L2 structure (Faehnle et al. 2014) indicates that CG16940 has comparable organization of α helices and β sheets in the CSD1/2 domains (Supplemental Fig. S1). CG16940 also bears N-terminal coiled-coil regions that are present within several Dipteran orthologs (e.g., other Drosophilids and mosquitos), but not in other arthropod, fungal or vertebrate Dis3L2 orthologs (Supplemental Fig. S1).
We verified this complex by showing that transfected Myc-Tailor could coimmunoprecipitate (co-IP) HA-Dis3L2 (Supplemental Fig. S2). This interaction was specific because HA-CG16940 did not co-IP with the Drosophila TUTase Mkg-p (Supplemental Fig. S2). To further map the Dis3L2-Tailor interaction, we performed structure–function tests using the Tailor variant panel (Fig. 1A). Catalytically dead Tailor could co-IP Dis3L2 as effectively as wild-type Tailor (Supplemental Fig. S2). In these tests, Tailor-ΔtRNA accumulated to a much lower level than full-length, but detectably co-IPed with Dis3L2 suggesting that this region is not essential for complex formation. The ΔDUF and Zn-mut constructs were more informative, since they accumulated robustly but did not form a complex with Dis3L2 effectively (Supplemental Fig. S2). Longer exposure of blots indicated that Tailor-Zn-mut exhibited very mild association with Dis3L2 compared to ΔDUF. We repeated these tests using catalytically inactive Dis3L2 (HA-Dis3L2-CD), and obtained similar binding profiles (Fig. 1D). If anything, binding was more robust in tests with Dis3L2-CD. The co-IP experiments with Dis3L2-CD more clearly showed that both Zn finger and DUF1439 were required for association with Tailor, but that the latter domain is more critical for the interaction. Sufficiency tests showed that an N-terminal fragment of Tailor comprising DUF1439 and the Zn finger could coimmunoprecipitate wild-type Dis3L2 (Fig. 1E).
We refined the region of Dis3L2 that associates with Tailor using deletion constructs of HA-Dis3L2 (Fig. 1C). We used HA-Elav as a control, and showed that despite its extremely robust accumulation, it does not associate with myc-Tailor (Fig. 1F). On the other hand, full-length HA-Dis3L2 could bind Tailor, but a variant lacking the N-terminal ∼200 amino acids bearing the CC domains could not. Reciprocally, a construct composed of only the N-terminal CC domains was sufficient to coimmunoprecipitate myc-Tailor (Fig. 1F). Thus, the Tailor–Dis3L2 complex is mediated by domains within their respective N termini (Fig. 1A,C).
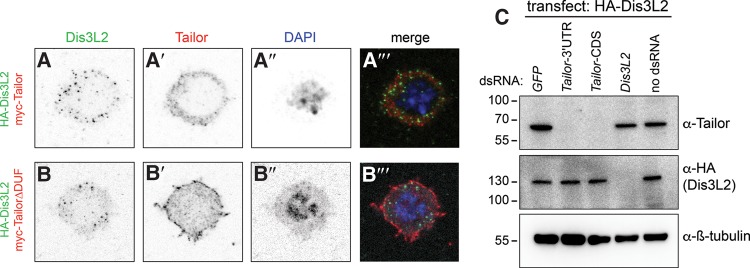
We assessed the subcellular localization of HA-Dis3L2 in transfected S2 cells. As in other species, Drosophila Dis3 is nuclear (Mamolen et al. 2010), while vertebrate Dis3L2 proteins are cytoplasmic. We observed that HA-Dis3L2 was almost completely cytoplasmic (Fig. 2A), confirming its classification. In addition to general cytoplasmic signals, we observed its accumulation in speckles. As components of protein complexes are often subcellularly colocalized, we tested for potential spatial overlap between transfected Myc-Tailor and HA-Dis3L2. Both proteins exhibited predominantly cytoplasmic signals whether transfected individually or together, and both accumulated in particles (Fig. 2A). However, closer examination showed that Tailor and Dis3L2 particles were mostly independent (Fig. 2A′′′). The results from transfection studies need to be interpreted cautiously, but these data suggest that Tailor and Dis3L2 might not exclusively reside in a complex. We performed further costaining of Tailor and Dis3L2 variants but these all showed similar patterns with no combination that was more notably colocalized (data not shown). However, we note that TailorΔDUF exhibited a more cortical pattern, which may relate to its poor association with Dis3L2 (Fig. 2B).
FIGURE 2.
Localization and codependence of transfected Tailor and Dis3L2. (A,B) Immunostaining of S2 cells transfected with HA-Dis3L2 and myc-Tailor constructs. Single channel images are shown as inverted signals. (A) Both Dis3L2 and Tailor accumulate nearly exclusively in the cytoplasm, but their punctate foci are largely nonoverlapping. (B) TailorΔDUF1439 exhibits altered localization that is more cortical than full-length Tailor. (C) Accumulation of endogenous Tailor and transfected HA-Dis3L2 proteins in reciprocal knockdown conditions. Treatment of S2 cells with Tailor-CDS or Tailor-3′ UTR dsRNAs suppresses endogenous Tailor, while Dis3L2 dsRNA suppresses protein produced from the HA-Dis3L2 construct. Similar levels of Tailor protein and HA-Dis3L2 are present in reciprocal knockdown cells.
We assessed the accumulation of Tailor and Dis3L2 in cells depleted of the reciprocal factor. For these tests, we analyzed endogenous Tailor but used transfected Dis3L2 owing to lack of availability of Dis3L2 antibodies. Western blotting showed that either Tailor-CDS or 3′ UTR dsRNAs could effectively deplete endogenous Tailor, and that Dis3L2 dsRNA effectively suppressed the accumulation of transfected HA-Dis3L2 (Fig. 2C). Under these conditions, loss of Dis3L2 did not substantially affect Tailor accumulation, while loss of Tailor did not affect the accumulation of transfected Dis3L2. These data are consistent with the apparent separable localization of these factors, as inferred from ectopically expressed proteins. These tests do not exclude the possibility that endogenous Dis3L2 may have distinct behavior, but they provide a foundation for interpreting our subsequent analyses that utilize immunoprecipitated proteins recovered from transfected cells.
Oligouridylation stimulates substrate degradation by Dis3L2
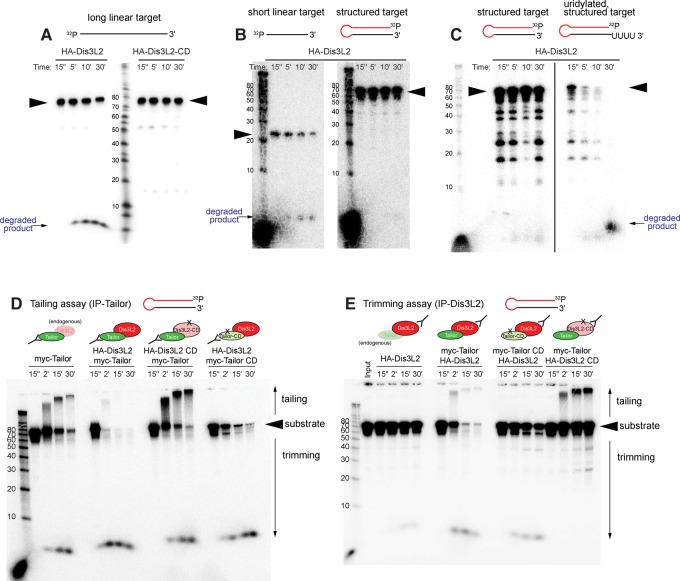
Mammalian Dis3L2 exhibits in vitro preference for uridylated substrates (Malecki et al. 2013; Faehnle et al. 2014), thus rationalizing why enzymes of the TUT4/7 subclade stimulate the action of Dis3L2 on oligouridylated pre-let-7 hairpins (Chang et al. 2013; Ustianenko et al. 2013). We therefore investigated the impact of substrate uridylation on Dis3L2. We immunoprecipitated HA-Dis3L2 from transfected S2 cells, and observed 3′–5′ degradation of a 5′-radiolabeled, unstructured, 77 nt substrate. This was a specific activity, since a catalytically inactive variant (Dis3L2-CD) did not degrade this substrate within this timecourse (Fig. 3A).
FIGURE 3.
Tailor-mediated uridylation enhances Dis3L2-mediated degradation of structured substrates. The in vitro tests shown here assess the capacity of immunoprecipitated factors to modify different 5′ radiolabeled substrates, as assayed on a reaction timecourse. (A) Parallel assays of HA-Dis3L2-IP and HA-Dis3L2-CD-IP material on an artificial linear target demonstrate exoribonuclease activity of Dis3L2. The input material (arrowhead) is decreased upon incubation with HA-Dis3L2, concomitant with accumulation of shorter species, but not with HA-Dis3L2-CD. (B) Parallel assays of HA-Dis3L2 on the model substrates’ mature miR-1010 (short linear target) and pre-mir-1010 (structured target); the miR-1010 sequence comprises the 3′ terminal 24 nt of the pre-mir-1010 hairpin. Dis3L2 is less effective at degrading the structured target. (C) Parallel assays of HA-Dis3L2 on normal and uridylated pre-mir-1010 shows that degradation of this hairpin is facilitated by a short uridine tail. (D) Tailing assays (i.e., interrogating Tailor-IP material) from cells cotransfected with combinations of wild-type and catalytically dead Tailor or Dis3L2 assayed on pre-mir-1010. Tailor-IP causes substrate tailing, while Tailor-IP from cells cotransfected with Tailor and Dis3L2 mediates robust substrate degradation. This is dependent on Dis3L2 catalytic status, and partially dependent on Tailor catalytic status. Substrate decay in the presence of Dis3L2-CD may potentially be due to endogenous Dis3L2. (E) Trimming assays (i.e., interrogating Dis3L2-IP material) from cells cotransfected with combinations of wild-type and catalytically dead Tailor or Dis3L2 assayed on pre-mir-1010. Substrate degradation by Dis3L2-IP is very strongly potentiated by cotransfection with Tailor. This is dependent on the catalytic status of Tailor and Dis3L2.
The Dis3/Rrp6-containing exosome is also capable of degrading substrates from their 3′ termini, but it has difficulty when encountering secondary structures. This is utilized, for example, during the biogenesis of Drosophila 3′ tailed mirtrons, where the unstructured 3′ tail is removed by the exosome, leaving the hairpin to proceed for further biogenesis (Flynt et al. 2010). We tested a model hairpin substrate (pre-mir-1010), and observed it was less efficiently degraded by Dis3L2 than mature miR-1010, even though they share the same 3′ terminal 24 nt (Fig. 3B). These data suggested that Dis3L2 activity is inhibited by secondary structure. We therefore tested the effects of uridylation using synthetic pre-mir-1010 bearing two or four 3′ uridines. We previously showed that these substrates were less efficiently diced in vitro than the unmodified hairpin (Bortolamiol-Becet et al. 2015). In contrast, we observe here that uridylation promotes the capacity of these substrates to be degraded by Dis3L2 (Fig. 3C and data not shown). We conclude that Dis3L2 is a 3′–5′ exonuclease whose activity is stimulated by substrate oligouridylation.
Distinct activities of Tailor, Dis3L2, and coimmunoprecipitated factors
We have shown that Tailor and Dis3L2 form a complex, and that Dis3L2 can degrade substrates. Given this, it was somewhat perplexing why we and others earlier observed efficient tailing by Tailor-IP material, which should contain Dis3L2 (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015). We tested this further by comparing tailing reactions (i.e., when assaying Tailor-IP material) and trimming reactions (i.e., when assaying Dis3L2-IP material) in cells that were singly or cotransfected with these factors.
We reproduced our earlier studies showing that incubating pre-mir-1010 with Tailor-IP across a timecourse resulted in progressively longer tailed products (Fig. 3D). These assays also yielded some degradation products, which may potentially be due to copurified endogenous Dis3L2. Notably, when preparing Tailor-IP from cells cotransfected with both Tailor and Dis3L2, we instead observed strong substrate degradation. A mild shift of tailed substrate was seen in the early timepoint, but this was accompanied by strong and rapid loss of the input substrate band indicating degradation (Fig. 3D). We infer from these results that much of Tailor isolated from singly transfected cells is not associated with Dis3L2, but that the recovery of the complex with RNA decay activity is enhanced by their coexpression.
We further tested tailing assays using Tailor-IP material in the presence of Dis3L2-CD, which resulted in robust tailing with stronger accumulation of tailed species than was generated with Tailor-IP from cells transfected only with Tailor construct (Fig. 3D). Therefore, Dis3L2 is the active nuclease in Tailor immunoprecipitates. Conversely, IP material of Tailor-CD in the presence of Dis3L2 yielded only substrate degradation, although not as efficiently as with Tailor/Dis3L2. Thus, optimal activity of the complex for substrate degradation involves both enzymatic activities, but Tailor may play some recruitment role to bring Dis3L2 to substrates.
In reciprocal trimming assays, we recapitulated that Dis3L2-IP was relatively poorly able to degrade pre-mir-1010, such that most of the input substrate was neither extended nor trimmed at the end of the timecourse. However, immunoprecipitates of Dis3L2 from cells cotransfected with Tailor yielded a subtle tailing profile, accompanied by extremely efficient substrate degradation (Fig. 3E). This enhancement of Dis3L2-mediated decay was due to Tailor enzymatic activity, since reactions of Dis3L2-IP from cells cotransfected with Tailor-CD resulted in a substantial substrate remaining intact (Fig. 3E). Nevertheless, there was more substrate turnover in this condition than with Dis3L2-IP alone, suggesting a facilitatory effect of Tailor-CD. In contrast, trimming assays with Dis3L2-CD-IP in the presence of cotransfected Tailor yielded mostly unmodified substrate but included a population of robustly tailed species, reflecting uridylation activity of the Dis3L2-CD-IP material.
Overall, these data strongly support the notion that Tailor-mediated substrate uridylation creates the preferred substrate of its partner Dis3L2 for degradation. This suggests that the rationale of the TUTase/nuclease complex is to permit direct handoff of Tailor-modified substrate for Dis3L2 to act upon. However, the data also support the possibility that Tailor may also serve to recruit Dis3L2 to substrates, a scenario consistent with recent evidence that Tailor is a unique TUTase with intrinsic substrate preference (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015), in contrast to other characterized TUTases that are themselves recruited to targets (e.g., the RBP Lin28 is a specificity factor that recruits TUT4/7 to pre-let-7 hairpins). Finally, there might be independent functions of these enzymes. This scenario is perhaps more relevant to Tailor, given that its activity in substrate tailing is clearly observed in vivo (Fig. 1B) and vitro (Fig. 3D,E). At least for certain substrates (e.g., pre-miRNA hairpins), tailing itself is sufficient to impede further biogenesis (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015), and so substrate turnover does not necessarily have to occur for Tailor to exert a regulatory effect.
Dis3L2 is required for fertility in both sexes and essential for spermatogenesis
Most studies of metazoan TUTase/nuclease complexes, particularly in mammalian systems, have focused on cell systems. Molecular analyses suggest that these tailing and trimming factors might operate fairly broadly in the animal. Consistent with this, Tailor and Dis3L2 are expressed ubiquitously across modENCODE developmental and tissue RNA-seq data sets (Brown et al. 2014). Both factors are expressed at somewhat higher levels in ovaries and in maternal deposits into embryos, but not more than a few-fold higher than in most other locations (Supplemental Fig. S3).
Nevertheless, despite its broad expression, Tailor knockout animals are viable, but exhibit subfertility in both males and females (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015). We note the severity of fertility defects differed between these studies, which might be due to the utilization of different alleles and/or to the different assay regimens; we also cannot discount the impact of different food or other environmental conditions. Therefore, we reanalyzed several trans-heterozygous combinations of the Tailor pBac allele, two Tailor-CRISPR alleles. Under the conditions assayed, the Tailor loss-of-function genotypes exhibited some variability, but both combinations tested were substantially (∼50%–70%) subfertile in both females and males relative to controls (Fig. 4A,B).
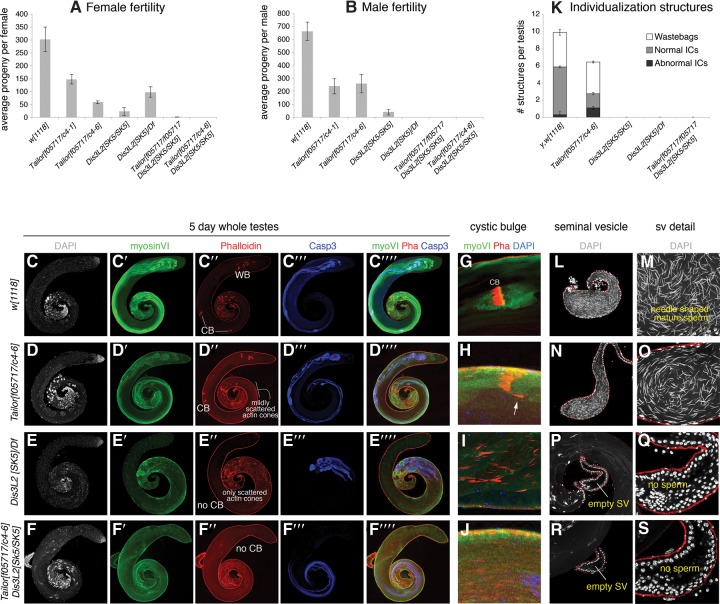
FIGURE 4.
Tailor and Dis3L2 are required for fertility and gametogenesis, particularly in males. We tested the phenotypic characteristics of Tailor, Dis3l2, and double mutant animals. We analyzed two trans-heterozygous combinations of Tailor, homozygous and hemizygous combinations of Dis3l2, and double mutants. (A) Quantification of individual female fertility. Tailor and Dis3L2 mutants are subfertile, but the double mutants are nearly fully sterile. (B) Quantification of individual male fertility. Tailor mutants are subfertile, while Dis3L2 single or double mutants are nearly completely sterile. (C–J) Immunostaining of 5-d-old testes, using DAPI to label DNA (in gray), anti-myosin VI (in green), phalloidin to label F-actin (in red), and anti-activated-caspase-3 (in blue). (C) Control w[1118] testis. The 64 haploid meiotic products remain together in a cyst and their cytoplasmic contents are extruded synchronously and form a cystic bulge (CB), they detach from the distal sperm tails to form a wastebag (WB). (D) Tailor trans-heterozygous mutant exhibits cystic bulges, but these exhibit mildly scattered actin cones. (E) Dis3L2 hemizygous mutant completely lacks cystic bulges; all actin cones are separate and only reach the middle portion of the testis. (F) Tailor, Dis3l2 double mutant is similar to the Dis3l2 single mutant. (G–J) Close-ups of individualization complexes in which actin cones are normally well organized within a cystic bulge (G, control) and mildly scattered in Tailor mutants (H). Dis3L2 single (I) or Tailor, Dis3L2 double (J) mutants lack cystic bulges and present only scattered actin cones in the testis basal region. (K) Quantification of ICs and wastebags in Tailor and Dis3L2 single and double mutants. (L–S) Seminal vesicles stained for DAPI; the sperm bear needle-shaped nuclei. Compared to control (L,M), Tailor mutant has a reduced seminal vesicle (N,O), while Dis3L2 (P,Q) mutant, and Tailor, Dis3L2 (R,S) double mutants bear empty seminal vesicles lacking sperm.
As specific Dis3L2 mutants have not yet been reported, we used the transgenic gRNA system (Kondo and Ueda 2013) to generate a frameshift allele upstream of all its annotated functional domains (Supplemental Fig. S4). Similar to Tailor mutants, we found that homozygous Dis3L2 mutants are viable, but exhibit infertility in both sexes. This is consistent with the notion that these physically interacting partners participate in similar genetic processes. However, Dis3L2 homozygous mutants notably exhibited stronger defects, since Dis3L2 females were less fertile than Tailor females, while Dis3L2 males were nearly completely sterile (Fig. 4A,B). We retested the Dis3L2 mutant over a deficiency, and observed that the hemizygotes recapitulated a stronger sterility defect in males (Fig. 4A,B). We also generated different combinations of Tailor; Dis3L2 double mutant animals and found that they were still viable, but now completely sterile in both sexes (Fig. 4A,B). Therefore, even though molecular processes mediated by Tailor/Dis3L2 might operate throughout development, the genetics points to gonads as settings of paramount biological importance.
We studied the phenotypic impact of Tailor and Dis3L2, and focused on the male germline due to the complete sterility of the nuclease mutants. In the testis, the germline stem cells (GSCs) are arranged as a characteristic ring surrounding the somatic hub. Following the meiotic divisions of the GSC, the sister spermatid nuclei remain bundled together in the basal region of the testis. An actin-rich cone or individualization complex (IC) forms around each spermatid nucleus, with the group of 64 ICs forming a cystic bulge (CB). The ICs separate from the nuclear region and move synchronously down the flagella to the testis apical region, extruding the spermatid cytoplasm to form wastebags (WB). Finally, the individualized sperm are deposited in the seminal vesicle.
We used the nuclear marker DAPI, the germline marker Vasa, and Fas III to examine the testis GSC niche. However, the somatic hub was contacted by similar numbers of GSC in both Tailor and Dis3L2 single mutants, as well as double mutants (Supplemental Fig. S5). Instead, we observed defective spermatid differentiation. We stained for DAPI to label DNA, phalloidin to label actin cones, anti-Myosin VI to label cystic bulges, and anti-cleaved Caspase 3 to read out activation of the nonapoptotic differentiation program in sperm. Compared to control testes (Fig. 4C), trans-heterozygous Tailor mutants initially bundled their spermatid nuclei appropriately, and activated Caspase-3 appropriately during terminal differentiation. However, Tailor mutants exhibited mild IC scattering during the individualization process (Fig. 4D). The defects in hemizygous Dis3L2 mutants were more profound. Spermatid nuclei were poorly bundled in the basal region (Fig. 4E). Actin cones were assembled and activated Caspase-3 reactivity was initiated. However, no CBs were formed, and the randomly moving ICs did not generate any wastebags. These data indicate the failure of differentiation of the meiotic products. We observed similar defects in Dis3L2 homozygous mutants (Supplemental Fig. S6), indicating that the allele selected for study is likely functionally null. We also generated Tailor, Dis3L2 double mutants and found the Dis3L2 phenotype to be largely epistatic, as the double mutants also lacked CBs and WBs (Fig. 4F).
Figure 4G–J are close-ups that depict examples of actin cone organization within individualization complexes (ICs). In wild type (Fig. 4G), each cystic bulge contains 64 ICs that are closely apposed in synchrony, whereas in Tailor mutants there are frequently some lagging cones or scattered cones not associated with a CB (Fig. 4H). In contrast, no CBs can be found in Dis3L2 or Tailor, Dis3L2 double mutants (Fig. 4I,J). We quantified normal and abnormal ICs in different genotypes, along with wastebags (Fig. 4K).
These defects were mirrored in the ultimate contents of the seminal vesicle (SV). That is, compared to control testis (Fig. 4L,M), Tailor mutants exhibited smaller SV, reflecting a reduction in sperm production (Fig. 4N,O). Meanwhile, Dis3L2 mutants and Tailor, Dis3L2 double mutants bore rudimentary SVs that were essentially devoid of mature sperm (Fig. 4P–S; Supplemental Fig. S6), thus explaining the complete sterility of males of these genotypes.
Gene expression defects in Tailor and Dis3L2 mutant gonads
To gain insight into the molecular defects associated with loss of tailing and trimming pathways, we performed total RNA sequencing of Tailor trans-heterozygous and Dis3L2 hemizygous mutants, as well as double mutants. In light of the stronger defects of Dis3L2 mutants on the male reproductive system, we dissected biologically replicate testis RNA samples for each genotype and analyzed them using total RNA sequencing, typically obtaining 20–30 million reads per sample (Supplemental Table S1).
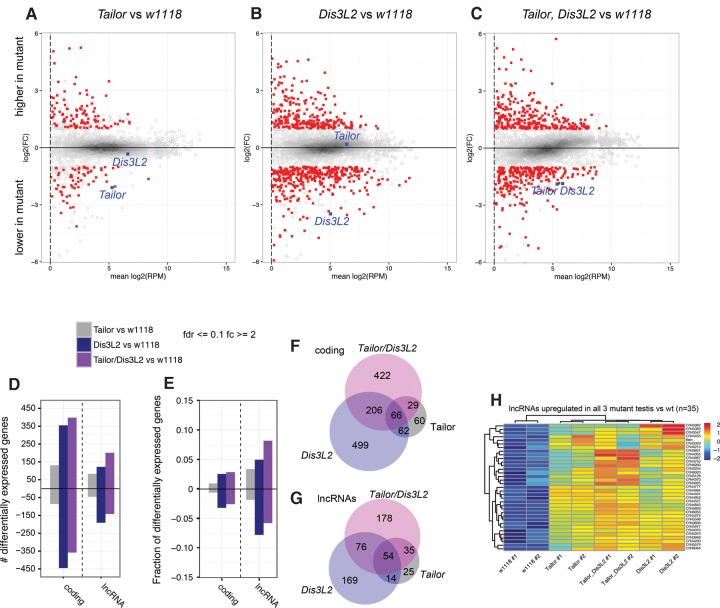
We identified differentially expressed genes in these different genotypes, using the replicates to define a fold change of ≥2 at a false discovery rate of ≤0.1. Principal component analysis on the differentially expressed genes showed that the biological replicates were reasonably well clustered, indicating that the data were reliable (Supplemental Fig. S7). We represented the data in MA scatter plots (Fig. 5A–C). While it was not a given that the Tailor and Dis3L2 indel alleles would affect RNA accumulation, we observed that among well-expressed genes, Tailor and Dis3L2 were among the most down-regulated transcripts in their respective RNA-seq data sets (Fig. 5A–C, circled loci). This confirms the genotypes of these data sets and suggests that the indel alleles are subject to nonsense mediated decay.
FIGURE 5.
Gene expression changes in Tailor and Dis3L2 single and double mutants. (A–C) MA plots comparing Tailor (A), Dis3L2 (B), and double mutants (C) to w[1118] control testis. These plots show protein-coding genes with FC ≥ 2 and FDR ≤ 0.1 in red; loci that do not meet these cutoffs are in gray. Genes above the x-axis are higher in mutants and ones below the y-axis are lower in mutants relative to control. For references, Tailor and Dis3L2 are shown as blue boxes, and they are specifically lower in cognate mutant data sets. (D,E) Barplots of numbers (D) and fractions (E) of differentially expressed protein-coding genes and lncRNAs in the different mutants. (F,G) Venn diagrams depicting the degree of overlap in differentially expressed protein-coding genes (F) and lncRNAs (G) among the different mutants. These plots include all loci; Venn diagrams that separate up-regulated and down-regulated loci are shown in Supplemental Figure S8. (H) Heatmap depicting a cluster of lncRNAs that is coordinately up-regulated in all three mutant conditions, relative to control.
We note that overall mRNA expression was less altered in Tailor mutants than in either Dis3L2 or double mutant testes (Fig. 5A–C). This is summarized in barplots in Figure 5D,E. These profiles are consistent with the interpretation that Tailor-mediated modifications are not always coupled to changes in gene expression, whereas removal of the nuclease might be inferred to be more directly causal to changing target levels. However, these comparisons also showed that relatively similar numbers of transcripts were called as elevated or down-regulated in each mutant, indicating that they are likely to reflect a mixture of direct and indirect effects. Analysis of overlapping gene expression changes between mutants indicates that a substantial fraction of genes deregulated in Dis3L2 and Tailor, Dis3L2 mutants are shared (Fig. 5F), consistent with the overall similarity in their morphological defects (Fig. 4); nevertheless, a large set of genes are individually deregulated in these conditions (Fig. 5F). Venn diagrams that separate the up-regulated genes and down-regulated genes are shown in Supplemental Figure S8.
The Drosophila testis is known to specifically express a large population of long noncoding RNAs (lncRNAs) (Brown et al. 2014). We therefore plotted annotated lncRNAs separately from protein-coding genes in gene expression analyses (Supplemental Fig. S9). We observed that lncRNA changes exhibited a similar profile of changes in the mutants as the mRNAs, in that Tailor showed fewer alterations than Dis3L2 or double mutant testis (Fig. 5D,G). However, as the number of lncRNAs is much smaller than protein-coding genes, the fraction of affected lncRNAs was greater than for mRNAs (Fig. 5D,E). This is consistent with the notion that lncRNAs are proportionally more sensitive to these post-transcriptional regulatory pathways.
A limitation of these data is that RNA-seq data are not suitable for identifying uridylated transcripts, as this would require TAIL-sequencing (Lim et al. 2014), and we also lack data on direct Dis3L2 targets. Still, these data sets provide a foundation for future studies to dissect how tailing and trimming enzymes are required for gametogenesis. We present clusters of loci that are altered in a co-regulated fashion between one or more mutant conditions in Supplemental Figure S10. Perhaps particularly notable is the set of lncRNAs that are up-regulated in the testes of all three mutant conditions relative to control (Fig. 5H). These sets might be enriched for candidate loci that might be directly suppressed by a shared tailing-and-trimming pathway.
Concluding remarks: specialized biological requirements for RNA tailing and trimming pathways
As mentioned in the Introduction, a slew of recent studies have analyzed coupled uridyltransferase/exonuclease systems that extend and trim 3′ ends of diverse transcripts (including small, intermediate, and large RNA species) in a variety of eukaryotic systems (including fungi, plants, and animals). Recent studies linked mammalian Dis3L2 to the concept of quality control of noncoding RNAs (Labno et al. 2016; Pirouz et al. 2016; Ustianenko et al. 2016). These studies provided important molecular evidence on direct Dis3L2 substrates, by immunoprecipitating RNAs bound to catalytically inactive Dis3L2 proteins and subjecting these to sequencing. These studies converged upon the finding that many large and small noncoding RNAs, as well as some mRNAs, are targeted by Dis3L2, with a common theme being that Dis3L2 is needed to degrade aberrant RNAs bearing secondary structures. Moreover, while this work was in revision, an independent study of the Drosophila Tailor/Dis3L2 complex and its direct role in noncoding RNA quality control was reported (Reimao-Pinto et al. 2016). That study provides important direct evidence for direct Drosophila Dis3L2 substrates in cultured cells; otherwise, our work on the nature and mechanisms of the Tailor/Dis3L2 complex reinforce each other. Conceptually, cytoplasmic TUTase/Dis3L2 complexes may have parallels with the nuclear quality control pathway in which substrate adenylation by the TRAMP complex enhances their degradation by the nuclear exosome bearing the exonucleases Rrp6 and Dis3/Rrp46 (LaCava et al. 2005; Schmidt and Butler 2013).
We note that the bulk of studies on metazoan RNA tailing and trimming factors have been conducted in cultured cells. This work has yielded many insights into molecular regulatory strategies, but has largely left open the question of how important these pathways are in the intact animal. In mammals, Zcchc11/TUT4 knockout mice lack obvious embryonic or fetal defects, but exhibit general postnatal fitness defects and growth retardation that may be due in part to insufficient IGF-1 expression (Jones et al. 2012). On the other hand, loss-of-function Dis3l2 mutations were found to cause Perlman syndrome of overgrowth, and were also detected in some sporadic Wilms tumor patients (Astuti et al. 2012). The finding of mitotic defects and growth enhancement in Dis3L2 knockdown cells suggested roles for this factor in cell and organismal growth, but might appear to be opposite in phenotypes of TUT4 mutants. Thus, while TUT4 (and TUT7) are molecularly and mechanistically linked to Dis3L2, how these factors and regulatory strategies link to organismal phenotypes remains to be understood.
Given that Tailor and Dis3L2 are broadly expressed in Drosophila, are amenable to functional study in cultured cells, it might have been reasonable to wonder if mutants might be lethal and/or exhibit developmental defects. While we do not exclude the possibility of developmental or physiological issues in Tailor and Dis3L2 mutants, the fact is both are viable and relatively normally patterned but exhibit overt defects in the germline of both males and females, especially in the testis. These genetic observations support the notion that germline cells are particularly sensitive to the proper function of these seemingly ubiquitous regulatory mechanisms. In this regard, we note that it was recently proposed that during apoptosis in mammalian cells, the TUT4/7-Dis3L2 system for mRNA decay is globally activated (Thomas et al. 2015). Perhaps it is relevant that caspase activation is required in a nonapoptotic manner in the Drosophila testis to mediate spermatid individualization (Arama et al. 2003; Huh et al. 2004), and thus may represent a naturally sensitized setting for the role of tailing and trimming pathways.
An appealing model is that the complex of Tailor/Dis3L2 facilitates a direct handoff, in which substrate tagging by the uridyltransferase permits their subsequent degradation. Evidence to support this notion comes from in vitro enzymatic assays, which show that substrate degradation by Drosophila Dis3L2 is stimulated by oligouridylation, similar to what was shown for mammalian Dis3L2 (Scheer et al. 2016) and recently independently shown in Drosophila (Reimao-Pinto et al. 2016). We observe overlaps in the gene expression profiles of these mutants, as well as similarities in gonadal defects in Tailor and Dis3L2 mutants. Nevertheless, Tailor and Dis3L2 mutants do not phenocopy each other, and the double mutants appear to have enhanced defects (e.g., in females). It is conceivable that Tailor and Dis3L2 do not function exclusively as a complex, but instead have some unique substrates and functions. Alternatively, Dis3L2 might be able to take advantage of uridylation action of some other TUTase(s). These possibilities might be addressed in the future by comparing wt or catalytically inactive Tailor and Dis3L2 proteins in vivo using CLIP-sequencing strategies, or by testing whether Tailor mutant phenotypes can be enhanced by removing other TUTase(s) in combination.
MATERIALS AND METHODS
Expression constructs
Tailor variant constructs. We cloned Tailor-A mutant constructs into pENTR-d-TOPO from Tailor-A expression constructs (Bortolamiol-Becet et al. 2015). For point mutants, using primer sets in Supplemental Table S2, we performed initial amplifications using primer pairs 1 + 2 and 3 + 4 (see scheme, Supplemental Table S2) and the wt clone as template. A second PCR was performed using the gel-purified first-round products as template for primers 1 and 4 only. For deletion mutants, a single PCR was performed with primer 4 and -DUF_fwd for ΔDUF, 1 and -tRNANucl_rev for ΔtRNA. We then transferred them into ptMW, a pUAS-6xMyc vector.
Dis3L2 constructs. To make 3xHA tagged Dis3L2, we used primers in Supplemental Table S2 to amplify from Dis3L2 cDNA as template and cloned the product into pUAST using the indicated restriction sites. Truncation versions of Dis3L2 were also cloned by PCR. Catalytic inactive pUAS-3xHA-Dis3L2 was generated by site-directed mutagenesis PCR using primers described in Supplemental Table S2.
Northern blotting
Northern blotting for small RNAs followed previously described probes and procedures (Okamura et al. 2007). To obtain single-nucleotide resolution of 55–80 nt pre-miRNAs, 8–15 mg of total RNA was loaded on 12% polyacrylamide–urea sequencing gels (40 cm long) as previously described (Bortolamiol-Becet et al. 2015). Briefly, gels were pre-run in 1× TBE for 30 min at 50 mA, and samples were run at 20 mA for 20 min, then at 50 mA for 5–6 h. We transferred them to GeneScreen Plus Hybridization Transfer Membranes (PerkinElmer) in 0.5× TBE for an hour at 300 mA on a semi-dry transfer system and then prehybridized and hybridized them using standard procedures (Okamura et al. 2007).
Coimmunoprecipitation assay
For each group of test, one well of a six-well S2 cell is transfected with 1 µg of each indicated plasmid and 500 ng ubi-Gal4 using Effectene (Qiagen). Cells are washed with PBS and lysed with co-IP lysis buffer (10 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× Protease inhibitor [Roche]), incubated on ice for 30 min, and followed by two centrifugations at 14,000 rpm, 10 min in cold room. The cleared cell lysates are incubated with HA antibody (Santa Cruz) or Myc antibody (Santa Cruz) conjugated beads for 2 h. Beads are washed with co-IP wash buffer (10 mM Tris–HCl, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100) five times and then resuspended in 3× sample buffer for Western blot analysis.
Preparation of RNA substrates for in vitro assays
Radiolabeled mature miR-1010, pre-mir-1010, and pre-mir-1010 + 2U or +4U substrates were prepared as previously described (Bortolamiol-Becet et al. 2015); the unstructured linear target was a synthetic let-7/hpCG4068 target RNA whose preparation was also described in Okamura et al. (2008).
Tailing and trimming assays
We used our previously described method to assay substrate tailing in vitro using immunoprecipitated proteins (Bortolamiol-Becet et al. 2015). After 3 d of transfection with myc-tagged Tailor and/or HA-tagged Dis3L2, either in wild-type or catalytically inactive form, cells were washed with PBS and resuspended in 5 volumes of lysis buffer (500 mM NaCl, 1 mM EDTA, 10 mM Tris–HCl, pH 7.5, 1% Triton-X and 1× protease inhibitor [Roche]). Cell lysate was obtained by 20 min incubation on ice followed by five passages through a 25G needle. We cleared the lysate by two 10 min centrifugations at 10,000g. The immunopurified proteins were obtained by incubating cleared lysate with beads coated with anti-Myc and rotated for 2 h at 4°C. Beads were washed three times with lysis buffer and four times with wash buffer (200 mM KCl, 10 mM Tris pH 8.0, 0.1 mM EDTA), then resuspended in wash buffer. Fifteen microliters of beads were mixed with 15 µL of 2× reaction mix (6.4 mM MgCl2, 2 mM DTT, 0.5 mM rNTP, 1000 cpm of radiolabeled RNA substrate) and incubated at 25°C for the indicated time.
For trimming assays, we incubated cleared cell lysate with beads coated with anti-HA and rotated for 3 h at 4°C. Beads were washed five times with lysis buffer and five times with reaction buffer (50 mM KCl, 10 mM Tris, pH 7.5, 5 mM MgCl2 and 10 mM DTT), then resuspended in reaction buffer. Of note, 1000 cpm of radiolabeled RNA substrate is then added to the IP material and incubated at 25°C for the indicated time.
Reactions were stopped by the same volume of 2× RNA loading buffer. RNAs were run on polyacrylamide–urea gels. The gel is incubated with fixing buffer (40% EtOH, 10% acetic acid and 5% glycerol) for 1 h and vacuum-dried at 80°C for 3 h.
Cell staining
To coat transfected S2 cells onto cover slides, cover slides are treated with 1% poly-lysine for 30 min, washed with distilled water, 70% EtOH and then air-dried. Cells are loaded onto the treated slides for 30 min incubation, then fixed for 15 min with 4% paraformaldehyde and washed four times for 5 min with PBS. Cells are permeabilized with three 10 min incubations with 0.1% Triton in PBS (PBST) and followed by incubation with blocking buffer (1% Goat serum in PBST) for 1 h. Primary antibody (1:200 of anti-Myc [A14] from Santa Cruz and 1:50 of anti-HA [F7] from Santa Cruz) are added to the blocking buffer and incubated with cells at 4°C overnight. Cells are washed with four 15 min PBST incubation, treated with the secondary antibody (488 anti-mouse, and 568 anti-rabbit) for 1 h, and then washed again with four 15 min PBST incubations. Cells are finally mounted with Vectorshield mounting buffer with DAPI.
Drosophila genetics
We used published alleles of tailor[f05717], [c4-1], and [c4-6] (Bortolamiol-Becet et al. 2015; Reimao-Pinto et al. 2015), and Df(3R)ED201 that uncovers Dis3L2. To generate specific alleles of Dis3L2, we used the transgenic gRNA system and the published crossing scheme (Kondo and Ueda 2013). In brief, we annealed a gRNA pair (Supplemental Table S2) and ligated it into the BbsI-digested pBFv-U6.2. The gRNA transgene was crossed to nos-Cas9 flies, and from their progeny, animals bearing a candidate mutagenized chromosome and that had segregated away the gRNA and Cas9 transgenes. Four out of seven candidates tested had passed Dis3L2 indels through the germline, of which two were in frame and two were out of frame (Supplemental Fig. S4). We characterized a [-4] deletion allele in further detail.
Testis immunostaining
Dissected tissues were fixed for 25 min in fresh 4% paraformaldehyde, washed extensively with 0.5% Triton in PBS (PBST), and blocked for 1 h in 2% bovine serum albumin BSA in PBST. We used the following primary antibodies in overnight incubations at 4°C: rabbit anti-cleaved Caspase-3 (1:50; Cell Signaling), mouse anti-Myosin-6 (1:20), Alexa Fluor 568-conjugated Phalloidin (1:1000; Thermo Fisher). We used Alexa Fluor-conjugated secondary antibodies (1:500, Molecular Probes) and DAPI to stain DNA (1:1000).
Fertility assay
We tested fertility of males and females from indicated genetic backgrounds by crossing them with w[1118] strain flies. We crossed 3-d-old flies with 5 w[1118] virgin females for the tested males and three virgin males for tested females. We flipped every 2 d during 2 wk and counted the progeny during the third instar larvae stage.
Transcriptome analysis
We isolated total RNA from 5-d-old flies using TRIzol. We made two independent dissections to generate biologically replicate RNA samples, whose quality was assessed by a Bioanalyzer. We used the Illumina Truseq Total RNA library Prep Kit LT to make RNA-seq libraries from 650 ng of total RNA. Manufacturer's protocol was followed except for using eight cycles of PCR to amplify the final library instead of the recommended 15 cycles, to minimize artifacts caused by PCR amplification. All samples were pooled together, using the barcoded adapters provided by the manufacturer, over two flow cells of a HiSeq2500 and sequenced using PE50 at the New York Genome Center. The raw sequencing data have been deposited to the NCBI Gene Expression Omnibus (GEO) under accession GSE92305.
We mapped total RNA-seq reads from four data sets to the UCSC Drosophila melanogaster (dm6) genome assembly. We normalized RNA-seq by the trimmed mean of M-values (TMM) normalization method in the edgeR/Limma Bioconductor library (Oshlack et al. 2010). We used the voom method of Limma (Smyth 2005) to correct for the Poisson noise due to the discrete counts of RNA-seq. The gene annotation (R6.12) was from the flybase FB2016_04 release. Differentially expressed mRNA and IncRNA loci between Tailor/Dis3L2 mutants versus wild type were identified by a moderated t-test, and FDR (Benjamini–Hochberg) was estimated, using the Limma library (Smyth 2005) in Bioconductor. The genes with FDR ≤0.1 and FC greater than or equal to twofold were considered to be differentially expressed. Transcript per million (TPM) was calculated to give gene expression per gene.
Principal component analyses (PCA) were used to give an overview of RNA-seq expression across replicated samples. Hierarchical clustering was used for clustering of mRNA and IncRNA expression. The input of these analyses was normalized total RNA-seq expression (log2 TPM). GO enrichment analysis for targets of highly and lowly expressed miRNAs was performed using the topGO Bioconductor library (Alexa and Rahnenfuhrer 2010).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stefan Ameres for Tailor mutants and antibody, and the Bloomington Stock Center, Vienna Drosophila RNAi Center, Drosophila Genomics Resource Center, and the Developmental Studies Hybridoma Bank for other fly stocks, cells, and antibodies. Work in E.C.L.’s group was supported by the National Institutes of Health (R01-GM083300) and Memorial Sloan Kettering Core Grant P30-CA008748 (National Cancer Institute).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059527.116.
REFERENCES
- Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for gene ontology. http://www.bioconductor.org/packages/2.12/bioc/html/topGO.html.
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. 2003. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell 4: 687–697. [DOI] [PubMed] [Google Scholar]
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, et al. 2012. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284. [DOI] [PubMed] [Google Scholar]
- Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin CJ, Ameres SL, Lai EC. 2015. Selective suppression of the splicing-mediated microRNA pathway by the terminal uridyltransferase tailor. Mol Cell 59: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May G, Stoiber M, Duff M, Booth B, Park S, Suzuki A, Wan K, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. 2014. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC. 2010. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, Pfeffer S. 2016. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res 44: 2873–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. [DOI] [PubMed] [Google Scholar]
- Huh JR, Vernooy SY, Yu H, Yan N, Shi Y, Guo M, Hay BA. 2004. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol 2: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, Powers JT, Daley GQ, Quinton LJ, Mizgerd JP. 2012. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS Genet 8: e1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno A, Warkocki Z, Kulinski T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. 2016. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res 44: 10437–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J 32: 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32: 1842–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamolen M, Smith A, Andrulis ED. 2010. Drosophila melanogaster Dis3 N-terminal domains are required for ribonuclease activities, nuclear localization and exosome interactions. Nucleic Acids Res 38: 5507–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC. 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, Young MD. 2010. From RNA-seq reads to differential expression results. Genome Biol 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M, Du P, Munafo M, Gregory RI. 2016. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep 16: 1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimao-Pinto MM, Ignatova V, Burkard TR, Hung JH, Manzenreither RA, Sowemimo I, Herzog VA, Reichholf B, Farina-Lopez S, Ameres SL. 2015. Uridylation of RNA hairpins by tailor confines the emergence of microRNAs in Drosophila. Mol Cell 59: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimao-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, Ameres SL. 2016. Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J 35: 2417–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer H, Zuber H, De Almeida C, Gagliardi D. 2016. Uridylation earmarks mRNAs for degradation…and more. Trends Genet 32: 607–619. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Butler JS. 2013. Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip Rev RNA 4: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF. 2014. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol Cell 53: 1020–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (ed. Gentleman R, et al. ), pp. 397–420. Springer, New York. [Google Scholar]
- Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, Walch M, Lieberman J. 2015. Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Rep 11: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. 2012. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. 2013. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19: 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, Vanacova S. 2016. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J 35: 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.