Abstract
Nuclear pore complexes (NPCs), which are composed of nucleoporins (Nups) and regulate transport between the nucleus and cytoplasm, significantly impact the replicative life span (RLS) of Saccharomyces cerevisiae. We previously reported that deletion of the nonessential gene NUP100 increases RLS, although the molecular basis for this effect was unknown. In this study, we find that nuclear tRNA accumulation contributes to increased longevity in nup100Δ cells. Fluorescence in situ hybridization (FISH) experiments demonstrate that several specific tRNAs accumulate in the nuclei of nup100Δ mutants. Protein levels of the transcription factor Gcn4 are increased when NUP100 is deleted, and GCN4 is required for the elevated life spans of nup100Δ mutants, similar to other previously described tRNA export and ribosomal mutants. Northern blots indicate that tRNA splicing and aminoacylation are not significantly affected in nup100Δ cells, suggesting that Nup100 is largely required for nuclear export of mature, processed tRNAs. Distinct tRNAs accumulate in the nuclei of nup100Δ and msn5Δ mutants, while Los1-GFP nucleocytoplasmic shuttling is unaffected by Nup100. Thus, we conclude that Nup100 regulates tRNA export in a manner distinct from Los1 or Msn5. Together, these experiments reveal a novel Nup100 role in the tRNA life cycle that impacts the S. cerevisiae life span.
Keywords: aging, nuclear pore complex, tRNA
INTRODUCTION
Increased age is a significant risk factor for a variety of human diseases including cancers, neurodegenerative disorders, diabetes, and heart attacks (López-Otín et al. 2013). Moreover, aging produces other debilitating effects that impair the well-being of otherwise healthy adults. Several model systems, including the replicative life span (RLS) of the budding yeast Saccharomyces cerevisiae, are used to study the cellular and molecular causes of aging. Importantly, many factors and signaling pathways that regulate RLS affect longevity in other metazoan model organisms (Steinkraus et al. 2008; Longo et al. 2012), suggesting the insights revealed from this system have the potential to inform our understanding of the aging process in higher eukaryotes. S. cerevisiae cells divide asymmetrically, and RLS refers to the number of daughters a single mother cell produces prior to senescence or death. Recently, we demonstrated that mutations in specific genes encoding components of nuclear pore complexes (NPCs) directly alter S. cerevisiae RLS (Lord et al. 2015).
NPCs are embedded in the nuclear envelope and regulate macromolecular transport between the nucleus and cytoplasm (Knockenhauer and Schwartz 2016; Kosinski et al. 2016; Lin et al. 2016). The ∼30 different nucleoporins (Nups) present in S. cerevisiae NPCs play specific structural and/or functional roles. In the NPC central channel as well as at the cytoplasmic and nuclear faces, the FG Nups are a class that contain domains rich in phenylalanine-glycine (FG) repeat motifs separated by polar spacers (for review, see Terry and Wente 2009). The FG repeats interact with nuclear transport receptors (NTRs) to facilitate translocation of cargo-bound NTRs through NPCs (Jovanovic-Talisman et al. 2009; Terry and Wente 2009; Labokha et al. 2012; Zahn et al. 2016). In addition, a subset of FG Nups with glycine-leucine-phenylalanine-glycine (GLFG) repeats also limits the passive diffusion of large molecules through NPCs (Laurell et al. 2011; Hülsmann et al. 2012; Lord et al. 2015). Thus, GLFG Nups contribute toward formation of a permeability barrier for nonspecific transport and provide NTR binding sites for transport of select molecules. Based on studies of mutants lacking specific FG regions, the 17 different S. cerevisiae NTRs are likely transported by preferred interactions with one or more of the 11 FG Nups (Strawn et al. 2004; Terry and Wente 2007). Since the FG domains themselves are not required for NPC structural integrity, removal of specific FG domains therefore inhibits different NTR-mediated transport events.
We previously reported connections between FG Nup function and aging in S. cerevisiae (Lord et al. 2015). Life span decreases in mutant cells lacking the GLFG domain of Nup116 (nup116ΔGLFG). Although the passive diffusion barrier of nup116ΔGLFG and other ΔGLFG mutants is significantly increased relative to wild-type cells, the passive permeability of NPCs has a negligible correlation with RLS. Instead, inhibited nuclear transport of the NTR Kap121 appears to contribute to the decreased life spans of nup116ΔGLFG mutants, which in turn disrupts mitochondrial function (Lord et al. 2015). We also found that RLS significantly increases in nup100Δ and nup100ΔGLFG mutants (Lord et al. 2015); however, the molecular basis for increased RLS in the nup100 mutants is unknown.
A recent study showed that RLS is significantly increased in los1Δ cells (McCormick et al. 2015). Los1 is an NTR required for the nuclear export of intron-containing pre-tRNAs as well as mature tRNAs (Grosshans et al. 2000; Murthi et al. 2010; Huang and Hopper 2015), and is one of several factors that influence tRNA transport in S. cerevisiae. Msn5 is a separate NTR that binds Tef1/2 and preferentially mediates the nuclear export of mature, aminoacylated tRNAs (Murthi et al. 2010; Huang and Hopper 2015). Interestingly, mature cytoplasmic tRNAs appear to be continuously imported back into the nucleus in S. cerevisiae (Takano et al. 2005); this retrograde import of tRNAs indirectly requires the NTR Mtr10 (Murthi et al. 2010; Huang and Hopper 2015) and may be important for tRNA quality control (Hopper 2013) as well as certain tRNA modifications (Ohira and Suzuki 2011). Environmental conditions play a significant role in regulating tRNA re-export, as mature tRNAs accumulate in the nuclei of wild-type yeast that are amino acid- or glucose-starved (Whitney et al. 2007; Huang and Hopper 2014; Pierce et al. 2014b). Mutation or inhibition of CEX1, SOL1/2, UTP8, or UTP9 also causes nuclear tRNA accumulation (Steiner-Mosonyi et al. 2003; Stanford et al. 2004; McGuire and Mangroo 2007; Eswara et al. 2009). Given the complexity of the tRNA maturation pathways, the connection to aging is intriguing.
Genetic analyses suggest nuclear tRNA accumulation in los1Δ cells impacts longevity similarly to dietary restriction and deletion of TOR1 (McCormick et al. 2015). Additionally, protein levels of the starvation-responsive transcription factor Gcn4 are elevated in los1Δ mutants, and GCN4 is partially required for their increased life spans (Ghavidel et al. 2007; McCormick et al. 2015). GCN4 is also necessary for the extended life spans of several ribosomal mutants including rpl20bΔ and rpl31aΔ, which display elevated Gcn4 levels (Steffen et al. 2008). Although GCN4 clearly plays an essential downstream role in life span regulation of several mutants, it is unclear which Gcn4 transcriptional targets are important in this capacity, and it is also unknown whether moderately increased Gcn4 protein levels are actually sufficient to affect RLS. Overall, these studies suggest that inhibiting tRNA export increases S. cerevisiae life span through a GCN4-dependent signaling pathway.
Since different FG Nups regulate the transport of specific nuclear import and export events (Strawn et al. 2004; Terry and Wente 2007; Adams et al. 2014) and los1Δ, nup100Δ, and nup100ΔGLFG cells display increased life spans (Lord et al. 2015; McCormick et al. 2015), we investigate here whether NUP100 is required for tRNA export. We find that the export of specific tRNAs is impaired in nup100Δ and nup100ΔGLFG cells. Moreover, protein levels of Gcn4 are increased when Nup100 is inhibited, and the increased life spans of nup100Δ cells require GCN4. Both tRNA splicing and aminoacylation are largely unaffected in nup100Δ cells, suggesting Nup100 likely regulates re-export of mature, processed tRNAs. Together, these experiments demonstrate a novel role for Nup100 in tRNA export that affects RLS and aging.
RESULTS
Nup100 is required for tRNA export
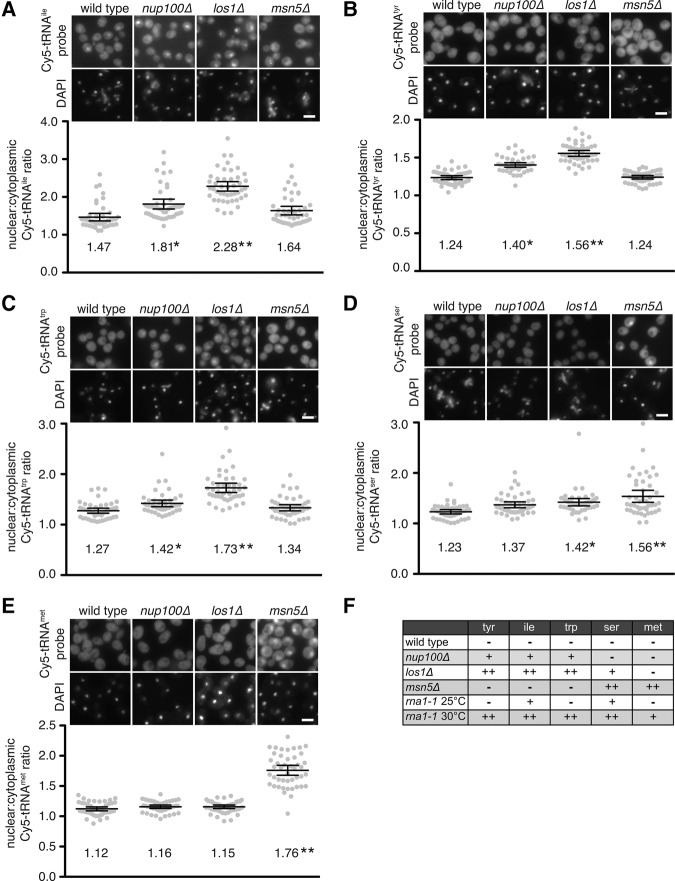
We hypothesized that longevity is increased in nup100Δ mutants because tRNA export is inhibited, which would result in the nuclear accumulation of at least some tRNAs. Fluorescence in situ hybridization (FISH) using Cy5-labeled probes that hybridize tRNAtyr, tRNAile, tRNAmet, tRNAtrp, or tRNAser was performed to assess whether these tRNAs are enriched in the nuclei of nup100Δ mutants relative to BY4741 wild-type cells. Mutant los1Δ and msn5Δ strains served as positive controls because of their thoroughly characterized defects in tRNA export (Sarkar and Hopper 1998; Grosshans et al. 2000; Murthi et al. 2010). Tyrosine, isoleucine, and methionine tRNAs were among those tested since their probes have been successfully utilized in past studies (Sarkar and Hopper 1998; Feng and Hopper 2002; Steiner-Mosonyi et al. 2003; Murthi et al. 2010). A significant enrichment of tRNAile, tRNAtyr, and tRNAtrp was observed in the nuclei of los1Δ and nup100Δ mutant cells (Fig. 1A–C). Unlike msn5Δ cells, which displayed methionine and serine tRNA nuclear export defects, neither tRNAser nor tRNAmet accumulated in nup100Δ nuclei (Fig. 1D,E). In addition to the los1Δ and msn5Δ strains, rna1-1 cells were used to demonstrate the probes were accurately hybridizing their target tRNAs. When shifted to nonpermissive temperatures, rna1-1 cells accumulate several types of RNA, including tRNA, in their nuclei (Sarkar and Hopper 1998). At 25°C, wild-type and rna1-1 cells exhibited similar phenotypes, but when shifted to 30°C for 3 h, all the Cy5 probes recognized transcripts that at least partially accumulated in the nuclei of rna1-1 cells (Supplemental Fig. 1A), suggesting the probes effectively hybridize their target tRNAs.
FIGURE 1.
Several tRNAs accumulate in the nuclei of nup100Δ cells. (A–E) (Top) Representative images of fixed cells from the indicated strains incubated with Cy5-labeled probes that hybridize tRNAile (A), tRNAtyr (B), tRNAtrp (C), tRNAser (D), or tRNAmet (E). DAPI was used to stain DNA and is shown below each Cy5 image. Scale bar, 5 µm. (Bottom) The nuclear:cytoplasmic Cy5 ratios of the cells shown above were determined as described in Materials and Methods. Each gray dot represents the ratio from one cell, and 45 cells were quantified for each strain (15 cells from three separate experiments). The large lines represent the averages for each strain, which are written below the dots, while the error bars represent the 95% CI. An asterisk indicates a P-value of less than 0.05 when the sample was compared to wild type using Tukey's post-hoc test following a one-way ANOVA, while two asterisks indicate a P-value of less than 0.05 when also compared to nup100Δ cells. (F) Table summarizing nuclear export defects for the listed tRNAs and strains. The lack of any clear defect is indicated using “−”, moderate defects are shown with a single “+”, while strong defects are shown with two “+” symbols.
Nuclear accumulation was quantified by determining the ratios of the nuclear versus cytoplasmic Cy5 probe intensities for at least 45 individual cells from each strain. The average nuclear:cytoplasmic (N:C) ratio for tRNAile was 1.47 in wild type, 1.81 in nup100Δ cells, 2.28 in los1Δ, and 1.64 in msn5Δ strains (Fig. 1A); the ratios of both los1Δ and nup100Δ mutants were statistically greater than the ratios of wild-type cells, although the tRNAile nuclear accumulation defect was more pronounced in los1Δ cells. Similar effects were observed when Cy5 signals from the tRNAtyr and tRNAtrp probes were quantified. For example, the average N:C ratio of the Cy5-tRNAtyr probe was 1.24 in wild type, 1.40 in nup100Δ, 1.56 in los1Δ, and 1.24 in msn5Δ strains (Fig. 1B). Both los1Δ and nup100Δ mutants also displayed statistically significant increases in the N:C ratios of tRNAtrp (Fig. 1C). Only msn5Δ cells displayed statistically increased N:C ratios when Cy5-tRNAmet accumulation was measured (Fig. 1E), while los1Δ and msn5Δ cells displayed increased amounts of nuclear tRNAser (Fig. 1D). Thus, tRNAtyr, tRNAile, and tRNAtrp accumulate in the nuclei of nup100Δ mutants, consistent with the hypothesis that Nup100 is required for proper tRNA export.
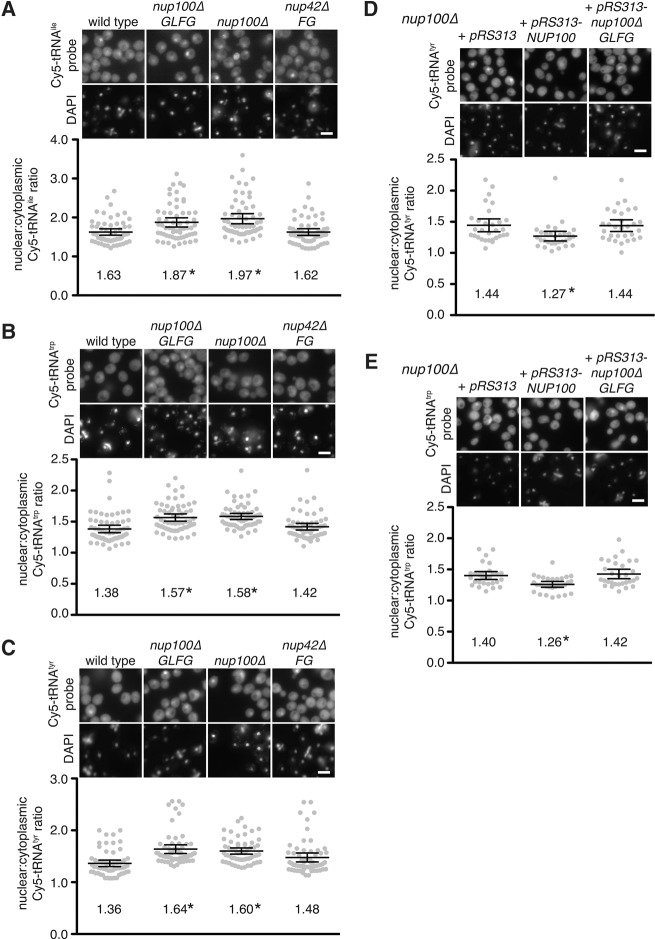
Since deleting the GLFG domain of Nup100 is sufficient to increase yeast life span (Lord et al. 2015), we tested whether tRNA export is inhibited in nup100ΔGLFG mutants, which specifically lack Nup100's GLFG domain and were previously generated in a W303 genetic background (Strawn et al. 2004). Compared to wild-type cells, nup100ΔGLFG and W303 nup100Δ cells exhibited nuclear accumulation of tRNAile, tRNAtrp, and tRNAtyr (Fig. 2A–C), similar to the phenotypes observed in nup100Δ BY4741 cells. We did not detect any significant nuclear enrichment of these tRNAs in nup42ΔFG cells (Fig. 2A–C), which lack the FG domain of the cytoplasmically oriented Nup42 (Strawn et al. 2004), suggesting the tRNA export defect specifically results from the loss of Nup100's GLFG domain. When the N:C Cy5 ratios were quantified for each of the tRNA probes, only nup100ΔGLFG and nup100Δ mutants exhibited statistically significant increases compared to wild-type cells (Fig. 2A–C). We also tested whether deletion of Nup100's GLFG domain is sufficient to cause nuclear tRNA enrichment in BY4741 cells by transforming nup100Δ mutants with pRS313, pRS313-NUP100, or pRS313-nup100ΔGLFG. Only nup100Δ cells transformed with pRS313-NUP100 exhibited a decrease in the amount of nuclear tRNAtrp and tRNAtyr (Fig. 2D,E). The fact that nup100Δ and nup100ΔGLFG cells both exhibited similar amounts of nuclear tRNA accumulation in different strain backgrounds suggests deletion of Nup100's GLFG domain is sufficient to inhibit tRNA export, and that other domains present in Nup100 do not significantly impact tRNA dynamics.
FIGURE 2.
tRNAs are enriched in the nuclei of cells specifically lacking the GLFG domain of Nup100. (A–C) (Top) Representative images of fixed cells from the listed W303 strains incubated with Cy5-labeled probes that hybridize tRNAile (A), tRNAtrp (B), or tRNAtyr (C). DAPI was used to stain DNA and is shown below each Cy5 image. Scale bar, 5 µm. (Bottom) Nuclear:cytoplasmic Cy5 ratios of the cells shown above were determined; each gray dot represents the ratio from one cell, and 60 cells were quantified for each strain (20 cells from three separate experiments). Large lines represent the averages for each strain, while error bars represent the 95% CI. Averages are listed below dots. An asterisk indicates a P-value of less than 0.05 when compared to wild type using Tukey's post-hoc test following a one-way ANOVA. (D,E) (Top) Representative images of S288C nup100Δ cells transformed with pRS313, pRS313-NUP100, or pRS313-nup100ΔGLFG and incubated with Cy5-labeled probes that hybridize tRNAtyr (D) or tRNAtrp (E). DAPI was used to stain DNA and is shown below each Cy5 image. Scale bar, 5 µm. (Bottom) Nuclear:cytoplasmic ratios for cells shown above were calculated; an asterisk indicates a P-value of less than 0.05 when compared to nup100Δ cells transformed with pRS313 using Tukey's post-hoc test following a one-way ANOVA with at least 30 cells.
Protein levels of Gcn4 are increased in nup100Δ and msn5Δ cells
Others have shown that protein levels of the transcription factor Gcn4 increase when tRNA export is inhibited (Qiu et al. 2000; Ghavidel et al. 2007; McCormick et al. 2015), and GCN4 is at least partially required for the increased life spans of los1Δ mutants (McCormick et al. 2015). Based on the observed tRNA export defects in nup100Δ cells, we measured cellular protein levels of Gcn4 in nup100Δ mutants using a well-established enzymatic reporter assay (Dever 1997). Wild type, los1Δ, msn5Δ, and nup100Δ cells were transformed with the vector p180, which expresses a truncated form of GCN4 fused to lacZ downstream from the four uORFs that regulate translation of GCN4 mRNA (Hinnebusch 1985). Lysates from nup100Δ and msn5Δ mutants had significantly increased levels of β-gal activity relative to wild type, though to a lesser extent than that observed in los1Δ mutants (Fig. 3A). β-Gal activity was also increased in nup100ΔGLFG mutants transformed with the GCN4-lacZ plasmid relative to wild-type cells (Supplemental Fig. 1B), indicating that Gcn4 protein levels are increased in both nup100Δ and nup100ΔGLFG cells.
FIGURE 3.
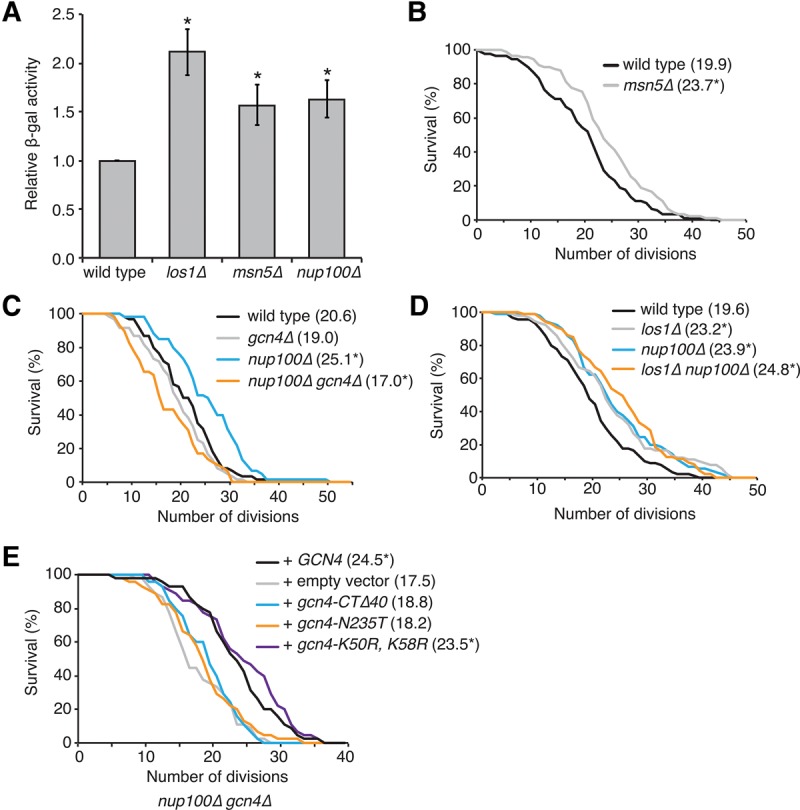
Gcn4 is activated in nup100Δ mutants and required for their increased longevity. (A) Relative levels of β-gal activity from the lysates of strains transformed with GCN4-lacZ (p180) (Hinnebusch 1985). An asterisk indicates a P-value of less than 0.05 when compared to wild type using a two-tailed Student's t-test with an n of at least six. (B–D) Survival curves for the indicated strains; average life spans for each are listed in parentheses next to the strain name. An asterisk indicates a P-value of less than 0.05 when the values were compared to wild type using a two-tailed Mann–Whitney U-test with an n of at least 60 cells per strain. (E) Survival curves for nup100Δ gcn4Δ cells transformed with the listed integrating vectors. An asterisk indicates a P-value of less than 0.05 when the values were compared to the strain transformed with the empty vector using a two-tailed Mann–Whitney U-test with an n of at least 45 cells per strain.
Based on their tRNA export and Gcn4 phenotypes, the life spans of msn5Δ cells were also measured. Consistent with the hypothesis that inhibiting tRNA export increases RLS, deletion of MSN5 significantly increased longevity (Fig. 3B). Elevated Gcn4 protein levels are observed when 60S ribosome subunit levels decrease (Steffen et al. 2008); however, polysome profiling experiments for nup100Δ and msn5Δ cells show 60S ribosome levels and translation are largely unaffected in either mutant strain (Chu and Hopper 2013; Lord et al. 2015). Together, this suggested that the increased Gcn4 protein levels in the nup100Δ and msn5Δ mutants are caused by nuclear tRNA accumulation.
It is also known that deleting GCN4 in los1Δ, rpl31aΔ, and rpl20bΔ mutants partially or fully suppresses the elevated life spans of these strains (Steffen et al. 2008; McCormick et al. 2015). Based on the increased protein levels of Gcn4-lacZ in nup100Δ cells, we hypothesized that nup100Δ gcn4Δ double mutants would have decreased life spans relative to nup100Δ single mutants. The RLSs of wild type, nup100Δ, gcn4Δ, and nup100Δ gcn4Δ cells were therefore measured. While deletion of GCN4 alone had no significant effect on life span relative to wild-type cells (Fig. 3C), the nup100Δ gcn4Δ double mutant exhibited a somewhat decreased RLS compared to wild-type and gcn4Δ cells (Fig. 3C). This indicated that increased longevity in nup100Δ cells is fully suppressed when GCN4 is deleted, further suggesting that NUP100 and LOS1 regulate life span through a similar GCN4-dependent signaling pathway. If NUP100 and LOS1 regulate longevity via related downstream signaling events, then a double nup100Δ los1Δ mutant should have a similar RLS compared to single nup100Δ and los1Δ mutants. When measured, there was no significant difference in the life spans of nup100Δ los1Δ cells compared to nup100Δ or los1Δ cells (Fig. 3D), consistent with the hypothesis that elevated Gcn4 protein levels mediate the increased life spans of nup100Δ and los1Δ mutants. Thus, Gcn4 protein levels are increased in nup100Δ mutant cells, and GCN4 activity is required for their increased life spans.
Several gcn4 mutants were tested for their ability to increase the life spans of nup100Δ gcn4Δ cells in order to define the aspects of GCN4 function necessary for its effects on longevity. A mutant lacking the 40 C-terminal amino acids of Gcn4 (gcn4-CTΔ40) and an N235T point mutant (gcn4-N235T) were tested to ascertain whether DNA binding is required (Hope and Struhl 1985; Suckow et al. 1993), and a gcn4-K50R, K58R mutant was also included to determine whether sumoylation (Rosonina et al. 2012) affected life span. Each mutant, as well as wild-type GCN4, was cloned into an integrating vector along with the ∼900 bp of the GCN4 promoter to ensure the constructs were regulated similar to endogenously expressed GCN4. These constructs were then transformed into nup100Δ gcn4Δ cells and life spans were measured. Cells transformed with an empty vector displayed decreased life spans similar to untransformed nup100Δ gcn4Δ mutants, while those transformed with GCN4 had an average life span of 24.5 divisions (Fig. 3E). Both the gcn4-CTΔ40 and gcn4-N235T mutants displayed life spans that were not significantly increased relative to the nup100Δ gcn4Δ cells, indicating DNA binding is necessary for Gcn4 to mediate its effects on life span. The life spans of the sumoylation-deficient strain were similar to cells transformed with GCN4 (Fig. 3E), suggesting sumoylation and promoter release does not significantly regulate Gcn4 function in aging.
Factors required for transcriptional memory do not affect life span
Another molecular process that requires NUP100 is transcriptional memory of the INO1 gene locus (Light et al. 2010). When S. cerevisiae cells are deprived of inositol, a defined sequence in the INO1 locus causes recruitment of the INO1 chromosomal region to the nuclear rim, where it physically associates with NPCs and is transcribed by RNA polymerase II (Ahmed et al. 2010). When inositol is reintroduced, transcription of INO1 is inhibited, yet the locus remains at the nuclear rim for up to 6 h and multiple cell divisions; this retention is considered a transcriptional memory event. NUP100, SET1, SET3, and HTZ1 are specifically required for retention of INO1 at the NPC, but not its initial recruitment following inositol deprivation (Light et al. 2010, 2013). While we hypothesized nuclear tRNA accumulation increases the life span of nup100Δ cells based on previous data linking tRNA export to yeast longevity (McCormick et al. 2015), we also tested whether transcriptional memory affects S. cerevisiae aging. If transcriptional memory regulates life span, then set3Δ and/or set1Δ nulls would have increased life spans and potentially Gcn4 protein levels. We found that neither set3Δ nor set1Δ mutants displayed increased life spans relative to wild-type cells (Fig. 4A), and the life spans of set1Δ cells actually decreased. Additionally, Gcn4-lacZ protein levels decreased in set1Δ and set3Δ mutants transformed with p180 (Fig. 4B). Since Set3 and Set1 are histone-regulating enzymes (Kim and Buratowski 2009) that regulate several other molecular events including telomere silencing (Krogan et al. 2002) and DNA replication (Rizzardi et al. 2012), these results did not definitively rule out that Nup100's role in longevity is unrelated to transcriptional memory, although this seems unlikely as neither set1Δ nor set3Δ mutants displayed any phenotypes similar to nup100Δ mutants.
FIGURE 4.
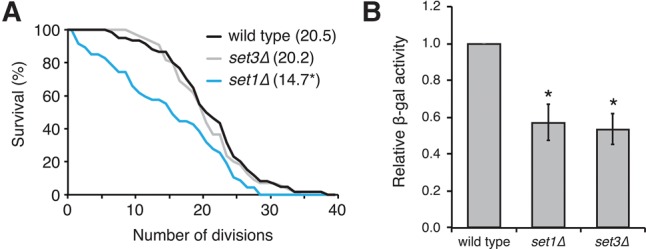
Transcriptional memory factors do not affect life span. (A) Survival curves of wild-type, set1Δ, and set3Δ strains; average life spans are listed in parentheses next to strain names. An asterisk indicates a P-value of less than 0.05 when compared to wild type using a Mann–Whitney U-test with an n of at least 50 cells per strain. (B) β-Gal activity of lysates from the listed strains transformed with p180; an asterisk indicates a P-value of less than 0.05 when compared to wild type using a two-tailed Student's t-test.
Spliced tRNAs largely accumulate in nup100Δ cells
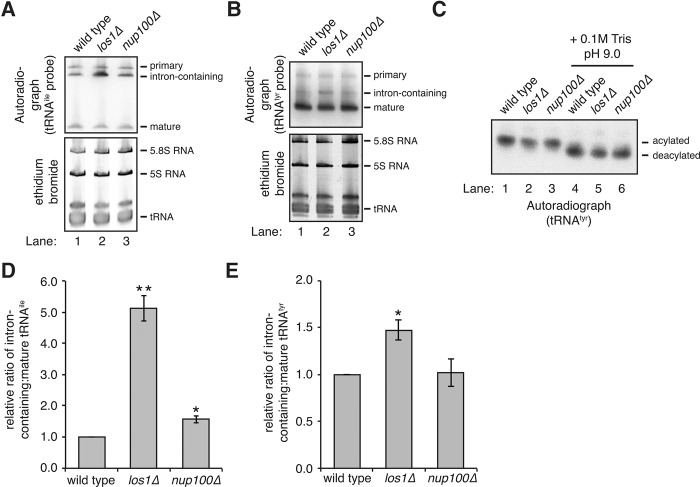
Northern blots were used to determine the splicing status of tRNAs in nup100Δ mutants. Since isoleucine, tryptophan, and tyrosine tRNAs all contain an intron prior to maturation, we initially tested whether intron-containing intermediates accumulate in nup100Δ cells. A radiolabeled probe complementary to an intron and exon region of tRNAile (Wu et al. 2015) was hybridized to total RNA from wild-type, nup100Δ, and los1Δ cells at 37°C; los1Δ mutants were used as a positive control because several studies demonstrate accumulation of intron-containing tRNAs (Sharma et al. 1996; Wu et al. 2015). Three different forms were detectable in all 3 strains (Fig. 5A): the primary transcript (top band), intron-containing tRNAile (middle band), and mature tRNAile (bottom band). There was a noticeable increase in the amount of intron-containing tRNAile relative to the mature form in los1Δ cells (Fig. 5A, cf. lane 1 to 2), indicating the probe clearly detects splicing defects. The ratios of the intron-containing:mature forms of tRNAile from three separate experiments were quantified using dosimetry (Fig. 5D). This revealed that nup100Δ cells had a small, but reproducible increase in the amount of intron-containing tRNAile (an ∼1.5-fold increase in nup100Δ cells), though this was significantly less than the amount observed in los1Δ cells (∼5.1-fold increase). Thus, while a small amount of the tRNAile that accumulates in the nuclei of nup100Δ cells may contain introns, the majority appears to be mature tRNAile. We also analyzed whether intron-containing pre-tRNAtyr accumulates in nup100Δ cells using a different probe. While the differences in the relative levels of intron-containing:mature tRNAtyr from wild type and los1Δ cells were not as robust as those observed with the tRNAile probe, we found that intron-containing pre-tRNAtyr did not significantly accumulate in nup100Δ mutants (Fig. 5B,E). This is consistent with the hypothesis that Nup100 largely regulates the nuclear export of mature tRNAs.
FIGURE 5.
Spliced tRNAs largely accumulate in nup100Δ mutants. (A,B) (Top) Northern blots of total RNAs from the listed strains. Membranes were incubated with a radiolabeled probe that hybridizes with tRNAile (A) or tRNAtyr (B); mature, intron-containing, and primary tRNA transcripts were detected and the locations of each are listed on the side of the image. (Bottom) RNAs were also stained with ethidium bromide to visualize total tRNA as well as 5S and 5.8S RNA. (C) RNAs isolated under acidic conditions from the listed strains were resuspended in 10 mM sodium acetate pH 4.5 (lanes 1–3) or 100 mM Tris pH 9.0 (lanes 4–6), and Northern blots were performed with a radiolabeled probe that hybridizes tRNAtyr. (D,E) The relative amounts of intron-containing:mature tRNAile (D) or tRNAtyr (E) were quantified from the listed strains from three different experiments. Error bars represent SEM, and an asterisk indicates a P-value of less than 0.05 when compared to wild-type RNA, while two asterisks indicate an additional P-value of less than 0.05 when compared to nup100Δ RNA.
Since inhibition of tRNA aminoacylation can significantly impair tRNA export (Gu et al. 2005), aminoacylation of tRNAtyr was then assessed using Northern blots with RNA isolated and electrophoresed under acidic conditions. Maintaining a pH of 4.5 during RNA isolation prevents hydrolysis of amino acids from tRNAs; aminoacylated tRNA can subsequently be separated from deacylated tRNA during electrophoresis (Chernyakov et al. 2008). Total RNA was isolated using a low pH buffer (Fig. 5C, lanes 1–3) or in a Tris pH 9.0 buffer (lanes 4–6) from wild-type, los1Δ, and nup100Δ cells. Treatment with Tris pH 9.0 prior to electrophoresis should fully deacylate all tRNA in a sample, providing a control to differentiate between aminoacylated and deacylated tRNA. Following transfer of the RNA, the membrane was incubated with a radiolabeled probe complementary to tRNAtyr. For all samples, there was a clear mobility shift when the RNAs were treated with Tris pH 9.0 (Fig. 5C, cf. lanes 1 and 4; 2 and 5; 3 and 6). Importantly, there was no deacylated tRNAtyr detected in nup100Δ mutants, indicating tRNAtyr aminoacylation is unaffected by Nup100.
Msn5 and Los1 localization are not regulated by Nup100
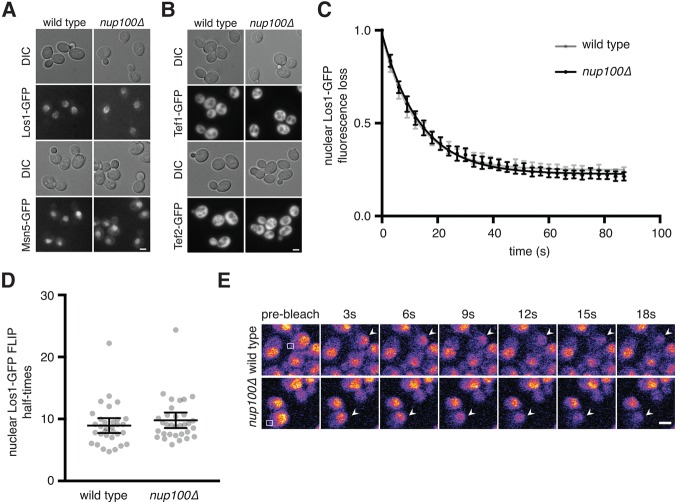
To better define the molecular basis for the nuclear accumulation of tRNAs in nup100Δ cells, we investigated whether inhibiting Nup100 function impairs NPC-mediated transport of either Los1 or Msn5. As Msn5 and Los1 are NTRs that bind tRNAs (Grosshans et al. 2000; Murthi et al. 2010; Huang and Hopper 2015), interaction with the GLFG-region of Nup100 might play a role in their export from nuclei. If the NPC-translocation of either NTR is mediated by Nup100, we predicted the NTR would be mislocalized at steady state in nup100Δ cells. The steady-state localizations of Los1- and Msn5-GFP were therefore examined using fluorescence microscopy when cells were grown in rich media. Los1-GFP primarily localized to the nuclear rim and nucleus in both wild-type and nup100Δ cells, while Msn5-GFP was enriched in the nucleus in both strains (Fig. 5A). The localizations of Tef1-GFP and Tef2-GFP were also analyzed; while these factors are not NTRs, they form a complex with Msn5 and Ran-GTP to allow export of spliced, processed tRNAs from the nucleus (Grosshans et al. 2000; Huang and Hopper 2015). Both Tef1-GFP and Tef2-GFP localized to the cytoplasm at steady-state in wild-type and nup100Δ mutants (Fig. 5B), consistent with their localization in wild-type cells in a previous study (Murthi et al. 2010). The subcellular localizations of the tRNA export factors Cex1-, Utp8-, and Utp22-GFP (Steiner-Mosonyi et al. 2003; McGuire and Mangroo 2007; Eswara et al. 2012) were also not significantly affected in nup100Δ cells (data not shown). These experiments therefore suggest that nuclear tRNA accumulation in nup100Δ mutants is not caused by disrupted localization of these tRNA export factors.
Subtle nucleocytoplasmic shuttling defects are not always apparent by simply examining the steady-state localizations of NTRs. Since los1Δ and nup100Δ mutants both display tRNAile, tRNAtyr, and tRNAtrp export defects (Fig. 1), we hypothesized that the export rate of Los1 could be decreased when NPCs lack Nup100. We therefore tested whether Los1-GFP shuttling is regulated by Nup100 using a fluorescence loss in photobleaching (FLIP) assay in los1-GFP::HIS3 and nup100Δ los1-GFP::HIS3 cells. When grown in synthetic media (to minimize autofluorescence), Los1-GFP mainly localizes to the nuclear rim and nucleus (Fig. 6E). In order to measure relative Los1 export rates in wild-type and nup100Δ cells, a small region of the cytoplasm from a cell expressing Los1-GFP was continuously bleached every ∼3 sec, with subsequent images taken to detect the loss of nuclear and rim GFP fluorescence (Fig. 6E). Since Los1-GFP is continuously exported from the nucleus, the nuclear GFP signal decreases as the cytoplasm is bleached. If Los1-GFP transport through NPCs is inhibited, then we expected fluorescence loss would take significantly longer in the nup100Δ mutant cells. We initially generated a best-fit one-phase decay curve for the averaged fluorescence loss over time for 30 individual cells per strain, which showed the curves were nearly identical (Fig. 6C). Curves were also generated to calculate half-times for fluorescence loss in individual cells (Fig. 6D). The average half-time for wild-type cells was 8.94 sec, while the average for nup100Δ mutants was 9.781 sec (Fig. 6D). These values were not statistically different using a two-tailed Student's t-test (P-value of 0.33), indicating the export rate of Los1-GFP was not affected by Nup100. Thus, Nup100 exerts its effects on tRNA export without mediating the export of Los1 through NPCs.
FIGURE 6.
Nup100 regulates tRNA export without affecting Los1 or Msn5 transport. (A) Los1- or Msn5-GFP were visualized in live wild-type or nup100Δ cells. DIC images of cells are shown above GFP images. Scale bar, 4 µm. (B) The localizations of Tef1- or Tef2-GFP were determined in wild-type or nup100Δ cells. Scale bar, 4 µm. (C) Graph of Los1-GFP nuclear and rim fluorescence loss from cells with cytoplasms that were continuously bleached every ∼3 sec, determined as described in Materials and Methods. At least 30 cells per strain were used for this analysis. The values at each time point were averaged, and a best-fit one-phase decay curve was produced and is shown on the graph. Error bars represent 95% CI. (D) Individual half-times for nuclear and rim Los1-GFP fluorescence loss with at least 30 cells (gray dots) per strain. Large lines represent the averages for each strain, while error bars represent the 95% CI. The P-value when comparing the two sets of data was 0.3268 using a two-tailed Student's t-test. (E) Representative Los1-GFP (top) and nup100Δ Los1-GFP (bottom) cells used to generate data in C and D, with cytoplasms that were bleached following the initial scan every ∼3 sec. Arrows point to the cells that were bleached, while the white box shows where cells were continuously bleached. Scale bar, 5 µm.
DISCUSSION
Here we report that several tRNAs specifically accumulate in the nuclei of nup100Δ and nup100ΔGLFG cells (Figs. 1, 2), revealing a novel role for Nup100 in the tRNA life cycle. The requirement for the GLFG domain correlates directly with the necessity of FG-NTR interactions during NPC translocation. Under our experimental conditions, Nup100 has a detectable impact only on a subset of tRNAs; deletion of NUP100 results in nuclear accumulation of tRNAtyr, tRNAile, and tRNAtrp without significantly affecting the localizations of tRNAser and tRNAmet (Fig. 1A–E). Given that los1Δ and msn5Δ mutants also display nuclear tRNA accumulation and elevated life spans (Fig. 3B; McCormick et al. 2015), we hypothesize RLS is increased in nup100Δ cells because tRNAs accumulate in their nuclei. Consistent with this hypothesis, los1Δ nup100Δ double mutants display similar life spans to both los1Δ and nup100Δ single mutants (Fig. 3D), suggesting LOS1 and NUP100 regulate longevity through similar downstream signaling pathways. Using a GCN4-lacZ reporter we show that protein levels of Gcn4 are elevated in nup100Δ and nup100ΔGLFG mutants (Fig. 3A; Supplemental Fig. 1B). Additionally, increased longevity in nup100Δ cells is GCN4-dependent, as nup100Δ gcn4Δ cells display significantly decreased life spans relative to nup100Δ mutants (Fig. 3C). Thus, Gcn4 plays an essential role in mediating the effects of nuclear tRNA accumulation on replicative life span.
Together, we propose that Nup100 is required for an important aspect of tRNA biology distinct from other well-characterized tRNA export factors. Our findings demonstrate that Nup100 does not impact Msn5-mediated nuclear tRNA export because different tRNA species accumulate in the nuclei of nup100Δ or msn5Δ cells (Fig. 1A–E). Although tRNAtyr, tRNAile, and tRNAtrp accumulate in the nuclei of both los1Δ and nup100Δ mutants, several pieces of data suggest Nup100 does not significantly affect Los1-mediated transport of tRNAs. First, Northern blots demonstrate that the majority of tRNAs that accumulate in nup100Δ cells are mature tRNAs, while a significant amount of intron-containing pre-tRNAile and pre-tRNAtyr accumulate in los1Δ cells (Fig. 5A–E). Additionally, the nuclear export rate of Los1-GFP is unaffected by deletion of NUP100 (Fig. 6C,D), indicating that Nup100 does not directly regulate the transport of this NTR through NPCs. The steady-state localizations of Cex1-, Utp8-, and Utp22-GFP are also unaffected in nup100Δ mutants (data not shown), suggesting the function of these factors is not regulated by Nup100 (Steiner-Mosonyi et al. 2003; McGuire and Mangroo 2007; Eswara et al. 2012).
Based on the lack of tRNA splicing and aminoacylation defects in nup100Δ cells, we hypothesize Nup100 is required for the re-export of a subset of mature tRNAs that are imported into the nucleus after being spliced in the cytoplasm (Fig. 7). tRNA re-export also requires Msn5 and Los1 (Murthi et al. 2010), which we propose function distinctly from Nup100. The accumulation of nuclear tRNA in los1Δ, nup100Δ, and msn5Δ cells results in elevated Gcn4 protein levels, which are necessary to regulate signaling pathways that impact RLS in these mutant strains. Based on our results and those from others (McCormick et al. 2015), we predict that inhibition of tRNA export is a general mechanism that can increase S. cerevisiae life span (Fig. 7). Although it is possible some other uncharacterized defect leads to GCN4-dependent increases in RLS, the lack of any translation or polysome defects in nup100Δ cells (Lord et al. 2015) is consistent with our hypothesis that nuclear tRNA accumulation in nup100Δ and nup100ΔGLFG mutants is largely responsible for their elevated life spans. Additionally, Nup100's role in transcriptional memory (Light et al. 2010) does not appear to influence RLS, as set1Δ and set3Δ mutants neither display increased life spans nor increased protein levels of Gcn4 (Fig. 4).
FIGURE 7.
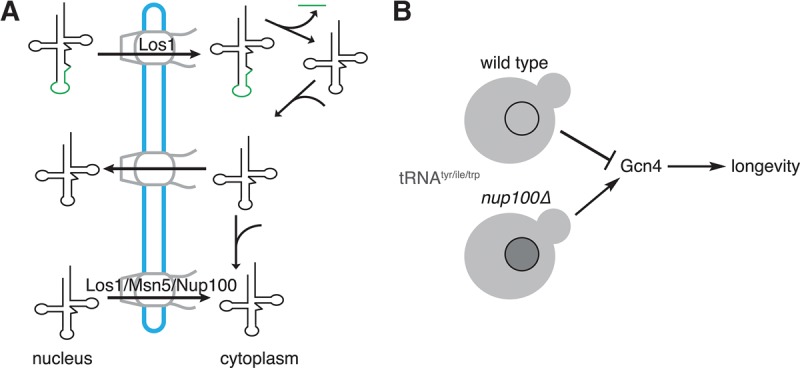
Model for how Nup100 potentially regulates S. cerevisiae replicative life span through its effect on tRNA export. (A) Nup100 is required for the re-export of specific tRNAs, including tRNAtyr, tRNAile, and tRNAtrp, following their splicing and aminoacylation. (B) Several tRNAs (gray) accumulate in the nuclei of nup100Δ cells. This tRNA export defect causes overproduction Gcn4, which may promote increased life span. Nuclear tRNA accumulation in other mutants, including los1Δ and msn5Δ cells, produces similar effects on Gcn4 protein levels and RLS.
Since different FG and GLFG domains facilitate the transport of specific NTRs and cargos through NPCs (Strawn et al. 2004; Terry and Wente 2007), we hypothesize Nup100 is necessary for the nuclear transport of a factor that influences tRNA export independent of Los1 or Msn5. Such a factor may be required for some final nuclear processing event for specific tRNAs, or it could directly bind these tRNAs in the nucleus to regulate their export. Another potential explanation for nuclear tRNA accumulation in nup100Δ cells is that this retention is caused by an indirect effect of a stress response that inhibits re-export of some tRNAs. Complete removal of amino acids or glucose from media causes significant accumulation of mature, aminoacylated tRNAs in wild-type yeast (Whitney et al. 2007; Huang and Hopper 2014; Pierce et al. 2014b). It is therefore possible that a nuclear transport or other type of defect in nup100Δ cells activates a signaling response that inhibits the export of specific tRNAs. We predict such a potential Nup100-dependent stress response is dissimilar to amino acid or glucose depletion responses since Los1 and Msn5 are mislocalized under either condition (Huang and Hopper 2014; Pierce et al. 2014b), a phenotype we did not observe in nup100Δ cells (Fig. 3A). Less likely possibilities are that Nup100 is directly necessary for an unidentified molecular event required for the export of specific spliced tRNAs, or that Nup100 may indirectly regulate the ability of Los1 or another NTR to physically interact with substrate tRNAs.
Other studies report nuclear tRNAtyr accumulation in both los1Δ and msn5Δ mutant strains (Murthi et al. 2010), as well as tRNAmet export defects in los1Δ mutants (Feng and Hopper 2002). The variance in our study could be due to differences in fixation or hybridization conditions during FISH, or potentially even yeast growth conditions. Interestingly, our results suggest that nuclear accumulation of several types of tRNA is sufficient to activate Gcn4 and regulate life span. For example, tyrosine, isoleucine, and tryptophan tRNAs are not efficiently exported in nup100Δ and los1Δ mutants, while tRNAmet and tRNAser accumulate in msn5Δ mutant nuclei. Furthermore, processed tRNAs as well as intron-containing pre-tRNAs accumulate in the nuclei of los1Δ cells, while spliced tRNAs specifically accumulate in the msn5Δ mutants (Murthi et al. 2010). The fact that unspliced pre-tRNAs also accumulate in los1Δ cells might result in further increases in Gcn4-lacZ protein levels observed in our assays compared to nup100Δ and msn5Δ mutants (Fig. 3A). Although elevated Gcn4 protein levels correlate with increased RLS in several yeast mutants (Steffen et al. 2008), it is currently unclear whether Gcn4 overexpression is sufficient to alter longevity in otherwise wild-type cells. Our gcn4 mutant data indicate that the DNA-binding activity of Gcn4 is required for its function in mediating life span extension, though it is unknown which Gcn4 target genes regulate longevity. More work will also be required to ascertain how Gcn4 protein levels are regulated by nuclear tRNA accumulation.
Overall our data are consistent with a model wherein Nup100 is required for an important aspect of tRNA re-export (Fig. 7) that affects longevity. Several tRNAs accumulate in nup100Δ nuclei, causing increased protein levels of the transcription factor Gcn4. Elevated Gcn4 levels might enhance RLS, and GCN4-mediated signaling is necessary for the increased life spans of nup100Δ mutants. Further experiments will be necessary to define how Nup100 regulates tRNA export and determine the precise role of GCN4-mediated signaling in life span regulation. Additionally, it will be important to test whether tRNA processing and/or export regulates protein levels of Gcn4 orthologs in higher eukaryotes, and if these factors influence metazoan aging.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains were derived from BY4741/4742 unless otherwise noted in the text or figure legend, and are listed along with their genotype in Supplemental Figure 2. Yeast were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) for all experiments except for ONPG substrate assays, where they were instead grown in SC-ura media, and FLIP assays, where they were grown in SC-his media. Yeast were grown at 30°C unless otherwise noted in figure legends.
Cloning and site-directed mutagenesis
Wild-type GCN4 and a mutant lacking its 40 C-terminal amino acids (gcn4-CTΔ40), along with 940 bp of its upstream promoter, were cloned into SnaBI- and XhoI-digested Addgene vector 41895 pAG306-GPD-empty chr I (Hughes and Gottschling 2012) using the Gibson cloning method. Digesting with SnaBI and XhoI removed the GPD promoter, allowing GCN4 and mutant forms to be regulated by the four uORFS similar to endogenously expressed GCN4. The two point mutants, gcn4-N235T and gcn4-K50R, K58R, were generated by site-directed mutagenesis of the wild-type construct.
Measurement of replicative life span
Replicative life span (RLS) was determined essentially as described elsewhere (Park et al. 2002; Steffen et al. 2009). Two to four yeast strains were grown in a small patch near the edge of YPD plate at 30°C overnight. The following day 30 cells from each strain were moved into a row on the plate using a dissection microscope. These cells were allowed to divide to produce virgin daughters, which were saved while the mothers were discarded. Daughters from these cells were counted and discarded to calculate each cell's RLS. RLS was considered complete when a cell had not divided for more than 12 h at 30°C. In Figures 3 and 4, mean RLS is shown in parentheses next to each strain name. Mann–Whitney U-tests were used to calculate statistical differences between survival curves.
Fluorescence in situ hybridization (FISH) detection of tRNAs
FISH to detect specific tRNAs was performed as previously described (Pierce et al. 2014a) with minor modifications. Fifty milliliters of cells were grown overnight in YPD to mid-log phase at 30°C. Thirty-six milliliters of each culture were mixed with 4 mL 37% formaldehyde in 50 mL conical tubes and incubated with shaking for 15 min at room temperature. Conicals were centrifuged for 5 min at 3000 rpm to pellet cells, which were resuspended in 10 mL fixation buffer (4% formaldehyde and 0.1 M KPi, pH 6.4) and nutated 2.75 h at room temperature. During this time, slides were prepared by adding 20 µL 0.1% poly-L-lysine to each well of a 15-well multitest slide (MP Biomedicals) for 5 min. Wells were aspirated and washed three times with 40 µL water for 5 min each, then the final wash was aspirated and the slide was left to dry. Conicals with fixed cells were centrifuged for 5 min at 3000 rpm and the pelleted cells were resuspended in 1 mL 0.1 M KPi, pH 6.4, and transferred to 1.5 mL tubes. Tubes were centrifuged at 13,000 rpm for 30 sec, and cells were resuspended in 1 mL 0.1 M KPi, pH 6.4. Cells were pelleted and resuspended in 1 mL wash buffer (0.1 M KPi, pH 6.4, and 1.2 M sorbitol), then pelleted and resuspended in 1 mL wash buffer. The cells were again pelleted and resuspended in 1 mL wash buffer along with 1 mg/mL zymolyase 20T and incubated 35 min at 30°C. Cells were centrifuged for 3 min at 3000 rpm and resuspended in 1 mL wash buffer. Finally, the cells were centrifuged for 3 min at 3000 rpm and resuspended in two times the cell pellet volume in wash buffer. Twenty microliters of this suspension was added to a prepared slide well and incubated for 5 min. After this point, the wells were not allowed to dry in between washes. Wells were aspirated and washed two times with 2× SSC for 5 min each. Wells were aspirated and 12 µL hybridization buffer (4× SSC, 50% formamide, 10% dextran sulfate, 125 µg/mL E. coli tRNA, 500 µg/mL herring sperm DNA, 0.5 units/µL RNasin, and 1× Denhardt's solution) was added to each. Slides were prehybridized by incubating them in a moist foil-wrapped plastic box for 1 h at 37°C. Of note, 0.5 µL of 10 pmol/µL Cy5-labeled tRNA probe was added to each well, and the slides were incubated overnight at 37°C in the humid foil-wrapped box. Wells were aspirated and washed sequentially with 25 µL of the following solutions for 5 min each: 2× SSC, 1× SSC + 0.1 µg/mL DAPI, 1× SSC, 1× SSC, and 0.5× SSC. About 2 µL of mounting media (1 mg/mL phenylenediamine and 90% glycerol) was added to each well, a cover slip was placed on top of the slides, and nail polish was applied to secure the cover slip. The following 5′ end-labeled Cy5 probes were used:
tRNAtyr: GCGAGTCGAACGCCCGATCTCAAGATTTACA GTCTTGCGCCTTAAACCAACTTGGCTACC
tRNAile: GTGGGGTTTGAACCCACGACGGTCGCGTTAT AAGCACGAAGCTCTAACCACTGAGCTAC
tRNAtrp: GGACAGGAATTGAACCTGCAACCCTTCGATT TGGAGTCGAAAGCTCTACCATTGAGCCACC
tRNAser: GACTCGAACCTGCGCGGGCAAAGCCCAAAAG ATTTCTAATCTTTCGCCTTAACCACTCGG
tRNAmet: CCAGGGGAGGTTCGAACTCTCGACCTTCAG ATTATGAGACTGACGCTCTTCCTACTGAGC
Quantification of nuclear:cytoplasmic tRNA ratios
FISH slides were visualized using an Olympus BX53 microscope equipped with a 100×/1.35 NA Olympus UPlan oil lens and Orca-R2 camera to detect Cy5 and DAPI signals. NIS Elements Advanced Research 3.2 was the software used to acquire images, while FIJI was used to quantify the average intensities of each cell nucleus and cytoplasm. The DAPI signal was used to make an ROI encompassing the nucleus for each cell that was quantified. Cytoplasmic ROIs were made by excluding the nuclear DAPI region and using the Cy5 signal to identify cell boundaries; vacuoles in the cytoplasm were excluded.
ONPG assays
ONPG substrate assays were performed as previously described (Dever 1997) with slight changes. Five milliliters of yeast strains transformed with p180 were grown overnight at 30°C in SC-ura to stationary phase. The stationary cultures (0.5–1 mL) were diluted into 30 mL SC-ura and grown at 30°C for 6 h. The cultures were transferred to 50 mL conical tubes and centrifuged for 5 min at 3000 rpm to pellet cells. Cells were resuspended in 250 µL breaking buffer (100 mM Tris, pH 8.0, 20% glycerol, 1 mM β-mercaptoethanol), transferred to 1.5 mL tubes, and 5 µL 100 mM PMSF was added. Acid-washed glass beads were added until they reached the top of cell suspensions and the tubes were vortexed on and off for 30 sec five times at 4°C. Breaking buffer (250 µL) was added to each tube, samples were quickly vortexed, and centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants (200 µL) were transferred to fresh 1.5 mL tubes and Bradford assays were performed to determine protein concentrations. Lysates (100 µL) were mixed with 900 µL Z buffer (100 mM NaPi, pH 6.4, 10 mM KCl, 1 mM magnesium sulfate, and 50 mM β-mercaptoethanol) in 2.0 mL tubes and incubated at 28°C for 5 min. Two hundred microliters of 4 mg/mL ONPG was added to reactions, which were then incubated at 28°C until they began to turn yellow. At this point 500 µL 1 M sodium carbonate was added, and the total reaction time was recorded. Once all reactions were complete their OD420 was determined. β-Gal activity was calculated using the following formula: (OD420 × 1.7)/(0.0045 × reaction time [min] × volume extract [mL] × protein concentration [mg/mL]).
Northern blots
Twenty-five OD599 units of yeast cultures grown overnight to mid-log phase in YPD were centrifuged for 5 min at 3000 rpm, washed in 1 mL ice-cold water, transferred to 1.5 mL tubes, then centrifuged at 13,000 rpm for 1 min. Supernatants were aspirated, and cell pellets were frozen using liquid nitrogen. Cells were resuspended in 500 µL TES buffer (10 mM Tris–HCl, pH 7.5, 10 mM EDTA, pH 8.0, and 0.5% SDS), 500 µL acid phenol was added, tubes were vortexed for 15 sec, then incubated at 60°C for 30 min with occasional vortexing. Tubes were incubated on ice for 5 min and centrifuged at 13,000 rpm for 5 min at 4°C. Supernatants were transferred to new tubes containing 500 µL acid phenol, which were then vortexed and centrifuged at 13,000 rpm for 5 min at 4°C. Supernatants were transferred to new tubes containing 500 µL chloroform, vortexed, and centrifuged at 13,000 rpm for 5 min at 4°C. Supernatants were transferred to new tubes, then one-tenth total volume 3 M sodium acetate, pH 5.3, and 2.5× volume ethanol were added. Tubes were incubated at −70°C for 45 min, then centrifuged at 13,000 rpm for 5 min at 4°C. Supernatants were aspirated, and RNA pellets were washed with 500 µL 70% ethanol and resuspended in 100 µL RNase-free water. Total RNA (10–15 µg) was mixed 1:1 with 2× formamide loading dye (95% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol, and 5 mM EDTA), then electrophoresed through a 7 M urea/10% acrylamide/1× TBE gel. RNAs were transferred onto Nytran SPC Supercharged membranes using a semi-dry apparatus, and then membranes were crosslinked twice at 1200 J. Membranes were incubated at least 4 h in 50 mL hyb buffer (4× SSC, 0.05% SDS, 2.5× Denhardt's solution, and 50 µg/mL SSDNA) at 37°C, then incubated with a radiolabeled probe overnight in 50 mL hyb buffer at 37°C. Membranes were washed three times with 2× SSC, then developed using a GE Typhoon FLA 7000IP Phosphoimager. ImageQuant TL software was used to quantify the intron-containing and mature tRNA for each strain, and the ratio of intron-containing:mature tRNA was determined and compared with wild type, which was considered to have a relative level of 1. The probes used for each Northern blot are listed below.
tRNAtyr: GCGAGTCGAACGCCCGATCTCAAGATTTACA GTCTTGCGCCTTAAACCAACTTGGCTACC
tRNAile: GGCACAGAAACTTCGGAAACCGAATGTTGCT ATAAGCACGAAGCTCTAACCACTGAGCTA
CACGAGC
Analysis of tRNA aminoacylation
Aminoacylated RNAs were isolated largely as previously described (Chernyakov et al. 2008). Twenty-five OD599 units of mid-log phase yeast were centrifuged for 5 min at 3000 rpm, washed in 1 mL ice-cold water, and transferred to 1.5 mL tubes. Cells were centrifuged at 13,000 rpm for 1 min, supernatants were aspirated, and the cell pellets were frozen in liquid nitrogen. Cells were resuspended in 200 µL AEB (0.3 M sodium acetate, pH 4.5, 10 mM EDTA), glass beads were added to saturation, and tubes were vortexed 15 sec four times every 3 min. One milliliter acetate-equilibrated phenol, pH 4.5, was added, and tubes were vortexed 15 sec three times every 3 min. Samples were centrifuged for 10 min at 5000 rpm at 4°C, and 150 µL of the aqueous phases were transferred to new 1.5 mL tubes with 350 µL ethanol. Samples were frozen using ethanol/dry ice, then centrifuged for 15 min at 13,000 rpm at 4°C. RNA pellets were resuspended in 200 µL cold AEB and mixed with 400 µL ethanol, then frozen using ethanol/dry ice. Tubes were then centrifuged for 15 min at 13,000 rpm at 4°C. The supernatants were aspirated, pellets were washed with 100 µL cold 70% ethanol, then resuspended in 80 µL 10 mM sodium acetate pH 4.5 and 1 mM EDTA. Half of each sample was incubated with 40 µL 100 mM Tris, pH 9.0, and 1 mM EDTA for 30 min at 37°C to deacylate tRNAs. Five micrograms of acylated and deacylated RNAs were mixed with an equal amount of 2× acidic loading dye (100 mM sodium acetate pH 4.5, 8 M urea, 0.05% bromophenol blue, and 0.05% xylene cyanol) and loaded onto an 8 M urea/7% acrylamide/100 mM sodium acetate pH 4.5 gel, then electrophoresed overnight at 4°C in 100 mM sodium acetate. Transfer and probing was performed using the same methods described above for Northerns.
Fluorescence loss in photobleaching (FLIP) experiments
los1-GFP::HIS3 and nup100Δ los1-GFP::HIS3 cells were grown overnight at 30°C in SC-his to mid-log phase, 1–2 mL were centrifuged for 3 min at 13,000 rpm at room temperature, the supernatants were aspirated, and the cells were resuspended in 10–20 µL of SC-his. Five microliters of of the suspension were dropped onto glass slides, a cover slip was added, and Los1-GFP was detected using an inverted confocal microscope (LSM510 META; Carl Zeiss) equipped with a 63×/1.40 NA Plan Apochromat oil lens (Carl Zeiss) at room temperature and photographed using a charge-coupled device camera (AxioCam; Carl Zeiss) with a 488 nm excitation laser. AxioVision version 3.0 SP software (Carl Zeiss) was used to acquire images. Following an initial image of the Los1-GFP, a small region of a cell's cytoplasm was bleached every 3 sec for 120 total seconds; an image was immediately recorded after each bleaching event. At least 30 cells for each strain were analyzed using this method. To quantify fluorescence loss, the following ROIs were measured for each time point: bleached cell's nucleus and rim signal (B); background ROI (BG); nonbleached cell's nucleus and rim signal (NB). The following formula was then used to calculate fluorescence loss for each time point (t), where 0 corresponds to the initial unbleached signal: [(Bt−BGt)/(NBt−BGt)]/[(B0−BG0)/(NB0−BG0)]. Half-times were then calculated for each cell based on using the Prism 7 best-fit one-phase decay function to generate data in Figure 6D. Average fluorescence loss over all the 30 cells was used to generate the data in Figure 6C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank members of the Wente laboratory for discussions about this manuscript. We also thank Parimal Samir and Andrew J. Link for technical assistance with RNA analysis, Katherine L. Friedman for use of her dissection microscope, Randi Ulbricht and Ron Emeson for their technical assistance with Northern blots as well as use of their equipment, Alan Hinnebusch for constructs used for β-gal assays, and Jason Brickner for helpful discussions regarding transcriptional memory. This work was supported by the National Institute on Aging award F32AG047737 (to C.L.L.) and National Institutes of Health award R37GM051219 (to S.R.W.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.057612.116.
REFERENCES
- Adams RL, Terry LJ, Wente SR. 2014. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics 197: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Baker MA, Grayhack EJ, Phizicky EM. 2008. Identification and analysis of tRNAs that are degraded in Saccharomyces cerevisiae due to lack of modifications. In Methods in enzymology (ed. Maquat LE, Kiledjian M), Vol. 449, pp. 221–237. Academic Press, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H-Y, Hopper AK. 2013. Genome-wide investigation of the role of the tRNA nuclear-cytoplasmic trafficking pathway in regulation of the yeast Saccharomyces cerevisiae transcriptome and proteome. Mol Cell Biol 33: 4241–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. 1997. Using GCN4 as a reporter of eIF2α phosphorylation and translational regulation in yeast. Methods 11: 403–417. [DOI] [PubMed] [Google Scholar]
- Eswara MBK, McGuire AT, Pierce JB, Mangroo D. 2009. Utp9p facilitates Msn5p-mediated nuclear reexport of retrograded tRNAs in Saccharomyces cerevisiae. Mol Biol Cell 20: 5007–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswara MBK, Clayton A, Mangroo D. 2012. Utp22p acts in concert with Utp8p to channel aminoacyl-tRNA from the nucleolus to the nuclear tRNA export receptor Los1p but not Msn5p. Biochem Cell Biol 90: 731–749. [DOI] [PubMed] [Google Scholar]
- Feng W, Hopper AK. 2002. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 99: 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. 2007. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131: 915–926. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt E, Simos G. 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 14: 830–840. [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. 2005. Depletion of Saccharomyces cerevisiae tRNAHis guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol Cell Biol 25: 8191–8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 5: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Struhl K. 1985. GCN4 protein, synthesize in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell 43: 177–188. [DOI] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-Y, Hopper AK. 2014. Separate responses of karyopherins to glucose and amino acid availability regulate nucleocytoplasmic transport. Mol Biol Cell 25: 2840–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-Y, Hopper AK. 2015. In vivo biochemical analyses reveal distinct roles of β-importins and eEF1A in tRNA subcellular traffic. Genes Dev 29: 772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Gottschling DE. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmann BB, Labokha AA, Görlich D. 2012. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150: 738–751. [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT. 2009. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. 2016. The nuclear pore complex as a flexible and dynamic gate. Cell 164: 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJH, Briggs JAG, Glavy JS, et al. 2016. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352: 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755. [DOI] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. 2012. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J 32: 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144: 539–550. [DOI] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D'Urso A, Horvath CM, Brickner JH. 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 11: e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. 2016. Architecture of the symmetric core of the nuclear pore. Science 352: aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. 2012. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16: 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CL, Timney BL, Rout MP, Wente SR. 2015. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J Cell Biol 208: 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC-Z, Ahmed U, Carr D, Murakami CJ, et al. 2015. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab 22: 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Mangroo D. 2007. Cex1p is a novel cytoplasmic component of the Saccharomyces cerevisiae nuclear tRNA export machinery. EMBO J 26: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang H-Y, Preston MA, Lai T-P, Phizicky EM, Hopper AK. 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 21: 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira T, Suzuki T. 2011. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc Natl Acad Sci 108: 10502–10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PU, McVey M, Guarente L. 2002. Separation of mother and daughter cells. Methods Enzymol 351: 468–477. [DOI] [PubMed] [Google Scholar]
- Pierce JB, Chafe SC, Eswara MBK, van der Merwe G, Mangroo D. 2014a. Strategies for investigating nuclear-cytoplasmic tRNA dynamics in yeast and mammalian cells. Methods Cell Biol 122: 415–436. [DOI] [PubMed] [Google Scholar]
- Pierce JB, van der Merwe G, Mangroo D. 2014b. Protein kinase A is part of a mechanism that regulates nuclear reimport of the nuclear tRNA export receptors Los1p and Msn5p. Eukaryot Cell 13: 209–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Anderson J, Björk GR, Sarkar S, Hopper AK, Hinnebusch AG. 2000. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 20: 2505–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi LF, Dorn ES, Strahl BD, Cook JG. 2012. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics 192: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL. 2012. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev 26: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK. 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 9: 3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Fabre E, Tekotte H, Hurt EC, Tollervey D. 1996. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol 16: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford DR, Whitney ML, Hurto RL, Eisaman DM, Shen W-C, Hopper AK. 2004. Division of labor among the yeast Sol proteins implicated in tRNA nuclear export and carbohydrate metabolism. Genetics 168: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. 2009. Measuring replicative life span in the budding yeast. J Vis Exp 28: e1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Mosonyi M, Leslie DM, Dehghani H, Aitchison JD, Mangroo D. 2003. Utp8p is an essential intranuclear component of the nuclear tRNA export machinery of Saccharomyces cerevisiae. J Biol Chem 278: 32236–32245. [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. 2008. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol 24: 29–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 6: 197–206. [DOI] [PubMed] [Google Scholar]
- Suckow M, von Wilcken-Bergmann B, Müller-Hill B. 1993. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J 12: 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T. 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol 178: 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 8: 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK. 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bao A, Chatterjee K, Wan Y, Hopper AK. 2015. Genome-wide screen uncovers novel pathways for tRNA processing and nuclear-cytoplasmic dynamics. Genes Dev 29: 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R, Osmanović D, Ehret S, Araya Callis C, Frey S, Stewart M, You C, Görlich D, Hoogenboom BW, Richter RP. 2016. A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. eLife 5: e14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.