Abstract
Although several studies have investigated the association between C4, C4A, and C4B gene copy number variations (CNVs) and susceptibility to autoimmune diseases, the results remain inconsistency for those diseases. Thus, in this study, a comprehensive meta-analysis was conducted to assess the role of C4, C4A, and C4B CNVs in autoimmune diseases in different ethnic groups. A total of 16 case-control studies described in 12 articles (8663 cases and 11099 controls) were included in this study. The pooled analyses showed that a low C4 gene copy number (GCN) (<4) was treated as a significant risk factor (odds ratio [OR] = 1.46, 95% confidence interval [CI] = 1.19–1.78) for autoimmune diseases compared with a higher GCN (>4). The pooled statistical results revealed that low C4 (<4) and low C4A (<2) GCNs could be risk factors for systemic lupus erythematosus (SLE) in Caucasian populations. Additionally, the correlation between C4B CNVs and all type of autoimmune diseases could not be confirmed by the current meta-analysis (OR = 1.07, 95% CI = 0.93–1.24). These data suggest that deficiency or absence of C4 and C4A CNVs may cause susceptibility to SLE.
The complement system, which is involved in both innate and adaptive immunity, is characterized by triggered-enzyme cascades activated by alternative, lectin or classical pathways1. As a necessary and important component of the complement system, complement component C4 plays a pivotal role in the activation of immune defenses and the clearance of immune complexes or apoptotic debris in vitro and in vivo2,3. If a structural variation such as fragmentary deficiency or deletion of a complement C4 gene occurs, it could cause an aberrant immune response and an autoimmune or inflammatory disorder4. Genetically, the complement component C4 gene is mapped in the class III region of the major histocompatibility complex (MHC) on chromosome 6p21.3 and there are two isotypes of C4 gene products, which are acidic C4A and basis C4B5. The C4A and C4B isotypes can be distinguished by four specific amino acids at positions 1101–1106 (or 1120–1125 if the protein numbering starts at the initiation codon for Methionine), which were the results of five nucleotide polymorphisms6,7. C4A prefers to bind to amino groups of immune complexes, while C4B binds to hydroxyl groups when activated8,9. Cumulative research has confirmed that gene expression as well as C4, C4A, and C4B CNVs could affect the strength of an individual’s immunity and susceptibility to autoimmune diseases8,10,11,12. The link between complement component C4 complete protein deficiency and autoimmune diseases was first observed among patients with systemic lupus erythematosus (SLE) in 197413.
CNVs in the human genome belong to a significant sequence variation, indicating that large stretches of DNA may exist in a number of different forms in different individuals14. CNVs in genes performing functions related to immunity can increase immunological diversity and genetic predisposition to autoimmune diseases15. It has been reported that partial C4 deficiency is one of the most frequent immunoprotein deficiencies in Homo sapiens10. Additionally, both C4A and C4B have various GCNs according to the Database of Genomic Variants (http://projects.tcag.ca/variation). The C4 gene encodes for either C4A or C4B, so the GCN of total C4 is equal to the sum of the GCNs of C4A and C4B16. CNV patterns in individuals belonging to different racial groups vary in terms of the number and size of C4 genes. C4 exists as two isoforms encoded by different genes, C4A and C4B. Together with three neighboring genes, C4 forms a genetic unit called the RCCX module (RP-C4A-CYP21-TNX or RP-C4B-CYP21-TNX). In the human diploid genome, the total C4 GCN varies from 2 to 6, with 4 being the most common. C4A GCN varies from 0 to 5, while that of C4B varies from 0 to 4, with 2 being the most common for both genes17. In Caucasian populations from the US and Europe, ~80% of individuals have three or four C4 GCNs, 20% have five or six, and <2% have only two8,10. In Chinese populations, about 60% of individuals have four C4 GCNs, and more than 63% have two C4A and C4B GCNs18,19. The most common GCNs of total C4, C4A and C4B in all racial groups are four, two, and two, respectively.
Deficiencies of C4 are strongly associated with increased risk of developing SLE20. However the association has failed to be confirmed in certain autoimmune diseases18. This inconsistency may be attributed to studies’ small sample size, genotyping method or different pathogeneses for different diseases and so on. Meta-analysis is an effective way to synthesize primary data from independent studies and generate more adequate and accurate statistical conclusions according to pooled analyses and results. We summarized current knowledge related to the role of C4, C4A, and C4B CNVs in SLE and other systemic autoimmune diseases and determined whether C4 genes are a genetic master key to autoimmunity. Subgroup analyses focusing on disease type and ethnicity were conducted to investigate potential sources of heterogeneity in the included studies.
Results
General characteristics of studies
The literature selection process is shown in Fig. 1. A total of 88 articles were initially identified using PubMed, Embase, Web of Science, and the China National Knowledge Infrastructure (CNKI). Sixty-six articles were excluded because they were duplicate studies, abstracts or not related to CNVs in autoimmune diseases. For detailed evaluation, another 10 articles without sufficient data and 1 article with overlapping data were ruled out. Additionally, a case-control study related to this subject was found by carefully screening the reference lists of each included article. Finally, 16 case-control studies from 12 articles were included in this meta-analysis. The studies’ characteristics and the extracted data are summarized in Table 1 17,18,19,21,22,23,24,25,26,27,28,29. As shown in Table 1, 8 studies focused on SLE, and 8 were mainly involved with GD, Crhon’s disease (CD), rheumatoid arthritis (RA), juvenile dermatomyositis (JDM), and uveitis. To identify differences between racial groups, among of 16 studies, 7 studies studying Asian populations and 9 studying Caucasian populations were enrolled in this analysis. Regarding genotyping method, a TaqMan-based quantitative real-time polymerase chain reaction (TaqMan) was applied in most studies (n = 10).
Figure 1. Flow diagram presenting the result of literature searching process in meta-analysis.
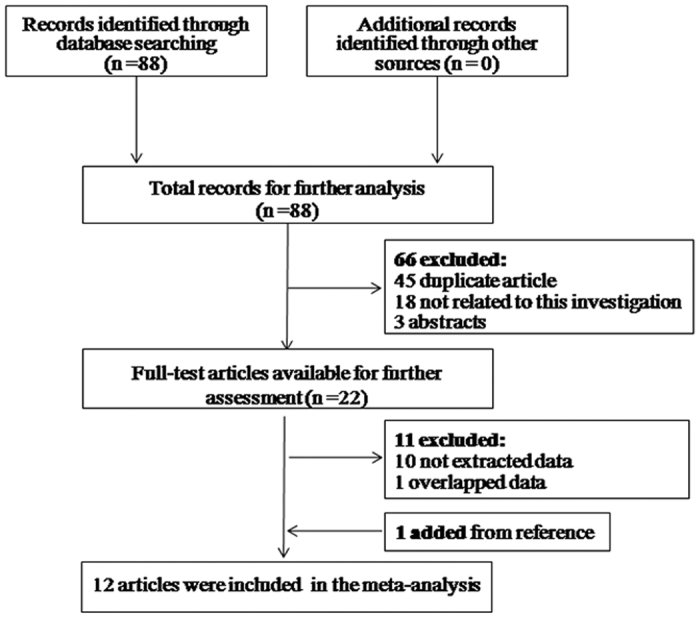
Table 1. The General Characteristics of All Studies Included in this Meta-Analysis.
| Refs | Year | Country | Ethnicity | Case/Control | Disease | Typing teaching | Study design |
|---|---|---|---|---|---|---|---|
| Liu et al.22 | 2011 | China | Asian | 624/160 | GD | TaqMan | Case-control,sex-, ethnic-, matched |
| Lv et al.23 | 2012 | China | Asian | 924/1007 | SLE | TaqMan | Case-control,age-, ethnic-, matched |
| Kim et al.21 | 2013 | Korea | Asian | 308/307 | SLE | TaqMan | Case-control,age-, ethnic-, matched |
| Yang et al.17 | 2007 | America | Caucasian | 216/517 | SLE | RFLP | Case-control,age-, ethnic-, matched |
| Boteva et al.24 | 2012 | UK | Caucasian | 501/719 | SLE | Four-digit genotyping | Case-control, ethnic-, matched |
| Boteva et al.24 | 2012 | Spanish | Caucasian | 464/449 | SLE | Four-digit genotyping | Case-control, ethnic-, matched |
| Cleynen et al.25 | 2015 | Belgium | Caucasian | 1887/1032 | CD | TaqMan | Case-control, ethnic-, matched |
| Hou et al.19 | 2014 | China | Asian | 1027/2083 | VKH | TaqMan | Case-control,age-, ethnic-, matched |
| Rigby et al.26 | 2012 | America | Caucasian | 160/51 | RA | Southern blot analyses | Case-control,age-, ethnic-, matched |
| Rigby et al.26 | 2012 | America | Caucasian | 88/51 | non-RA rheumatism | Southern blot analyses | Case-control,age-, ethnic-, matched |
| Pereira et al.27 | 2016 | Brazil | Caucasian | 90/200 | jSLE | TaqMan | Case-control, ethnic-, matched |
| Pereira et al.27 | 2016 | Brazil | Caucasian | 170/200 | SLE | TaqMan | Case-control, ethnic-, matched |
| Hou et al.18 | 2013 | China | Asian | 905/1238 | BD | TaqMan | Case-control,age-, ethnic-, matched |
| Hou et al.18 | 2013 | China | Asian | 205/1238 | AS + AAU+ | TaqMan | Case-control,age-, ethnic-, matched |
| Lintner et al.28 | 2016 | America | Caucasian | 95/500 | JDM | Southern blot analyses | Case-control,age, ethnic-, matched |
| Chen et al.29 | 2016 | China | Asian | 999/1347 | SLE | TaqMan | Case-control, ethnic-, matched |
Bias assessment of the included studies
The potential bias assessment of the included studies is presented in Table 2. Overall, the quality of the included studies was consistently good. Of the studies, there was no bias regarding selection, controls, genotyping controls, confounding, multiple tests, or selective outcome reports.
Table 2. Assessment of potential bias in enrolled studies.
| Year | First author | Bias in selection of cases | Bias in selection of controls | Bias in genotyping controls | Bias in population stratification | Confounding bias | Multiple test and Selective outcome reports |
|---|---|---|---|---|---|---|---|
| 2011 | Liu et al. | NO | NO | NO | NO | NO | NO |
| 2012 | Lv et al. | NO | NO | NO | NO | NO | NO |
| 2013 | Kim et al. | NO | NO | NO | NO | NO | NO |
| 2007 | Yang et al. | NO | NO | NO | NO | NO | NO |
| 2012 | Boteva et al. | NO | NO | NO | NO | NO | NO |
| 2015 | Cleynen et al. | NO | NO | NO | NO | NO | NO |
| 2014 | Hou et al. | NO | NO | NO | NO | NO | NO |
| 2012 | Rigby et al. | NO | NO | NO | NO | NO | NO |
| 2016 | Pereira et al. | NO | NO | NO | NO | NO | NO |
| 2013 | Hou et al. | NO | NO | NO | NO | NO | NO |
| 2016 | Lintner et al. | NO | NO | NO | NO | NO | NO |
| 2016 | Chen et al. | NO | NO | NO | NO | NO | NO |
Meta-analysis results
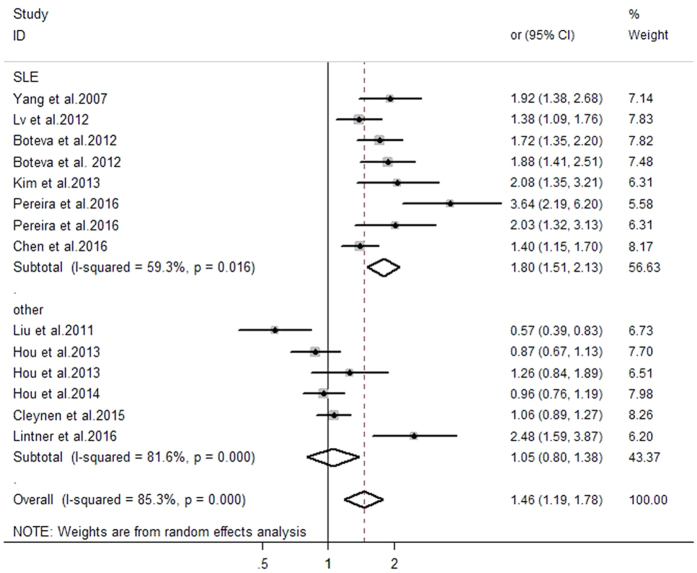
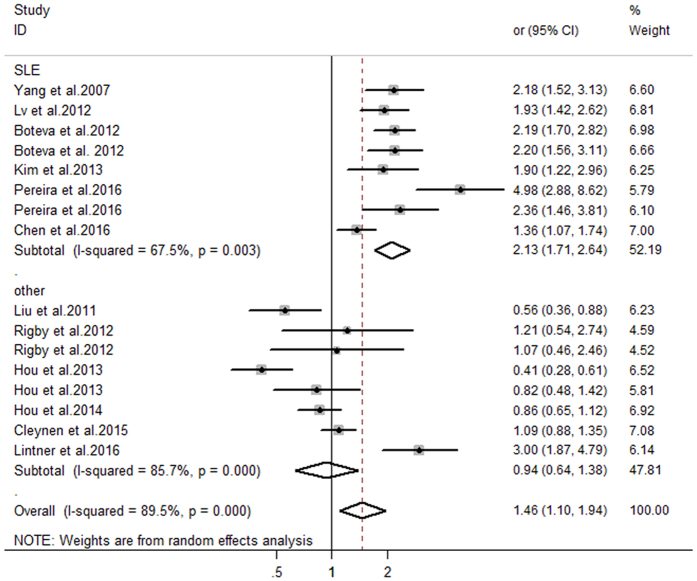
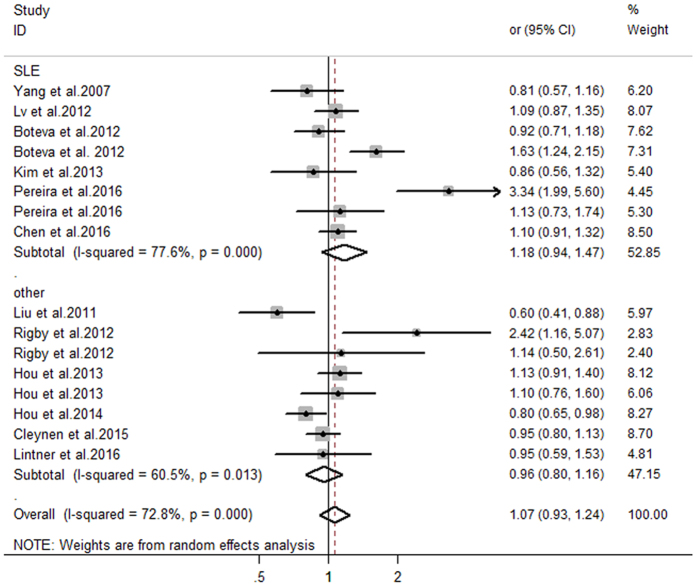
The relationship between C4, C4A, and C4B CNVs and autoimmune diseases was investigated, and the detailed results of pooled odds ratios (ORs) and a stratification analysis are presented in Table 3. The pooled analyses suggested significant between-study heterogeneity among the three genes (C4: I2 = 85.3%, PH < 0.001; C4A: I2 = 89.5%, PH < 0.001; C4B: I2 = 72.8%, PH < 0.001). Hence, the meta-analysis detected a random-effect model. As presented in Fig. 2, there was a significant association between C4 GCN and autoimmune diseases. Individuals with low C4 copy numbers (<4) were more likely to develop an autoimmune disease (pooled OR = 1.46, 95% CI: 1.19–1.78). In the subgroup analysis involving disease type, the risk of SLE was clearly increased in individuals with low C4 GCNs (pooled OR = 1.80, 95% CI: 1.51–2.13). Moreover, the subgroup analysis involving ethnicity revealed that low C4 GCNs were significantly associated with the risk of autoimmune diseases among Caucasian individuals (pooled OR = 1.91, 95% CI:1.42–2.56), while such an association was not confirmed in an Asian population (pooled OR = 1.13, 95% CI: 0.88–1.45). Furthermore, the results suggested that low C4A GCNs (<2) are more associated with apparent risk for autoimmune diseases compared with higher GCNs (≥2) (pooled OR = 1.46, 95% CI: 1.10–1.94) (Fig. 3). The subgroup analysis involving both disease type and ethnicity indicated that low C4A GCNs were significantly associated with susceptibility to SLE and a Caucasian population (pooled OR = 2.13, 95% CI: 1.71–2.64; pooled OR = 2.05, 95% CI: 1.49–2.83, respectively). No significant association between low C4B GCNs (<2) and autoimmune diseases was found in this meta-analysis (OR = 1.07, 95% CI: 0.93–1.24) (Fig. 4).
Table 3. Main Results of Pooled ORs and Analysis of C4,C4A and C4B CNV with Autoimmune Diseases in this Meta-Analysis.
| <4 VS ≥ 4 C4 | <2 VS ≥ 2 C4A | <2 VS ≥ 2 C4B | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Classification | N(Case/Control) | OR | 95%CI | PH | OR | 95%CI | PH | OR | 95%CI | PH |
| All disease | 1.46 | (1.19, 1.78) | <0.001 | 1.46 | (1.10, 1.94) | <0.001 | 1.07 | (0.93, 1.24) | <0.001 | |
| SLE | 8(3672/4746) | 1.80 | (1.51, 2.13) | 0.016 | 2.13 | (1.71, 2.64) | 0.003 | 1.18 | (0.94, 1.47) | <0.001 |
| Other | 8(4991/6353) | 1.05 | (0.80, 1.38) | <0.001 | 0.94 | (0.64, 1.38) | <0.001 | 0.96 | (0.80, 1.16) | 0.013 |
| Ethnicity | ||||||||||
| Asian | 7(4992/7380) | 1.13 | (0.88, 1.45) | <0.001 | 0.98 | (0.65, 1.49) | <0.001 | 0.96 | (0.82, 1.12) | 0.021 |
| Caucasian | 9(3671/3719) | 1.91 | (1.42, 2.56) | <0.001 | 2.05 | (1.49, 2.83) | <0.001 | 1.25 | (0.96, 1.63) | <0.001 |
PH: P value for heterogeneity.
Figure 2. Evaluation of the association between C4 gene CNVs with autoimmune diseases.
Figure 3. Assessment of the association between C4A gene CNVs with autoimmune diseases.
Figure 4. Estimation of the association between C4B gene CNVs with autoimmune diseases.
The fixed effect model was applied to these data in accordance with Chen et al.’s report30. In the subgroup analysis involving disease type, we found a significantly increased risk for SLE among carriers with low C4 or C4A GCNs (pooled OR = 1.68, 95% CI: 1.51–1.86; pooled OR = 1.99, 95% CI: 1.77–2.24, respectively). Finally, the studies were stratified by ethnicity, which found an apparently increased risk of autoimmune diseases associated with low C4 or C4A GCNs in Caucasians (pooled OR = 1.57, 95% CI: 1.41–1.75; pooled OR = 1.83, 95% CI: 1.62–2.06, respectively). These results suggest that, regardless of the random effect model or fixed effect model used for meta-analysis, the pooled data was consistent, believable, and stable.
Heterogeneity test and sensitivity analysis
As suggested in Table 3, there was significant heterogeneity between studies in terms of GCNs (p < 0.05). The results of our subgroup analysis confirmed that disease type and ethnicity were the main sources of heterogeneity. Additionally, a sensitivity analysis was conducted to evaluate the effect of individual studies on the pooled ORs by sequentially omitting each study. The pooled ORs were not affected by excluding any study (data not shown).
Publication bias
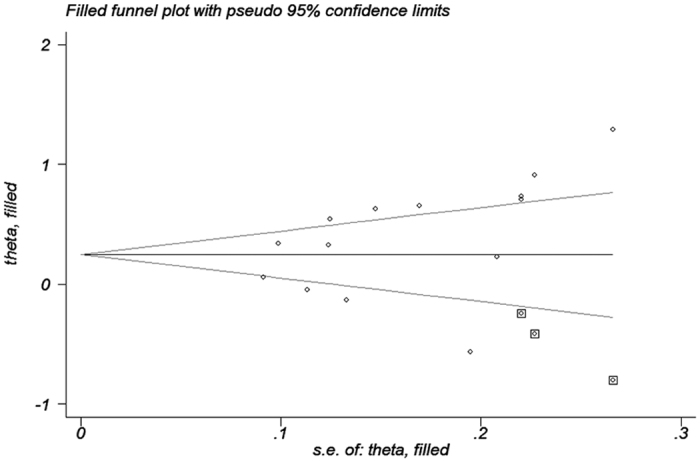
Begg’s funnel plots and Egger’s regression tests were applied to determine the potential publication bias for C4, C4A, and C4B CNVs. As presented in Table 4, there was no obvious publication bias among these detection. In order to validate whether there was potential publication bias regarding C4, we performed a funnel plot using the trim and fill method. LnOR and 95% CI were 0.376 (0.173, 0.579) and 0.246 (0.042, 0.450), respectively, before and after applying the trim and fill method, which indicated that publication bias was present in the meta-analysis (Fig. 5).
Table 4. Bias between C4,C4A and C4B CNV with Autoimmune Diseases in this Meta-Analysis.
| Gene | Number of publication | Publication bias |
|
|---|---|---|---|
| Begg’s test | Egger’s test | ||
| <4 VS ≥ 4 C4 | 14 | 0.063 | 0.096 |
| <2 VS ≥ 2 C4A | 16 | 0.893 | 0.720 |
| <2 VS ≥ 2 C4B | 14 | 0.444 | 0.300 |
Figure 5. Evaluation of the publication bias between C4 gene CNVs with autoimmune diseases.
Discussion
In our meta-analysis of 8663 cases and 11099 controls in 16 studies from 12 articles, we drew a general conclusion that C4 and C4A CNVs are tightly associated with autoimmune diseases, especially with SLE. Individuals with low GCNs of C4 (<4) or C4A (<2) are predisposed to autoimmune disorders in the presence of environmental triggers. Consistent with previous studies, both C4 and C4A CNVs were regarded as pivotal genetic factors in the pathogenesis of SLE. Furthermore, our meta-analysis demonstrated that low GCNs of C4 (<4) or C4A (<2) could lead to increased risk of autoimmune diseases among Caucasian populations. In other words, differences in the association between different ethnicities may result from other factors, such as geography, socioeconomic development, or race.
Complement C4 plays an essential role in innate and adaptive immune responses, which are involved in the classical and mannose-binding lectin complement activation pathways and help to direct against external attacks such as microbial infection, clearance of immune complexes, and removal of apoptotic cells31,32. Human C4 protein is encoded by two polymorphic genes, C4A and C4B, which are located in the MHC on chromosome 6. C4 deficiency and dysfunction are linked to the pathogenesis of many autoimmune and inflammatory diseases33. Several studies investigating different ethnic groups have shown that C4 deficiency may be one of the most penetrant genetic risk factors of SLE4,21,23. The results of the current meta-analysis were consistent with earlier observations.
C4A and C4B are two isotypes of C4. Although they share >99% of their amino acid sequences, their chemical reactions to substrates are remarkably different. C4A tends to combine amino group-containing antigens or immune complexes, while C4B combines hydroxyl group-containing antigens. Because C4A more efficiently handles immune complexes, deficiency of C4A while C4B affects the development of SLE34,35.
It is generally accepted that CNVs are a genetic determinant of phenotypic variation9. A CNV is a type of structural variation in which large segments of DNA are altered due to duplication, deletion, insertion, inversion, or complex combination or rearrangement36. Loci involved in immunity are prone to CNV, leading to differences in the intrinsic strength of the immune system and variations in susceptibility to immune disorders8,37. Haraksingh et al.38 found that CNVs may affect gene dosage due to various copies of a certain gene that is present in the genome. Yang et al.8 has reported a strong, positive correlation between C4 GCN and serum C4 protein concentration as well as C4A and C4B gene dosage and serum C4A and C4B concentration. In their study, a low GCN group (for C4, n < 4; for C4A, n < 2) had a significantly lower serum protein concentration than that of a medium GCN group (for C4, n = 4; for C4A, n = 2) and a high GCN group (for C4, n > 4; for C4A, n > 2)8. Since C4 plays a pivotal role in innate and adaptive immune responses, individuals with low GCNs of C4 (<4) or C4A (<2) could show reduced serum C4 or C4A concentrations, resulting in dysfunction in the ability to resist microbial infection, clear immune complexes, and remove apoptotic cells. Thus, low C4 CNV is a risk factor for SLE. As we know, SLE, RA, BD, and AS are immune-mediated autoimmune diseases. In this meta-analysis, we planned to summarize the current knowledge related to the relationship between C4 CNVs and SLE and other systemic autoimmune diseases and to determine whether C4 is a genetic master key to autoimmunity. Liu et al.22 revealed that individuals with less than 4 copies of C4 and those with less than 2 copies of C4A and C4B tended to be at less risk for GD. Liu et al.22 also found that less than 2 copies of C4A may be associated with high risk for vitiligo in patients with GD. Hou et al.19 found that patients with Vogt-Koyanagi-Harada (VKH) syndrome have decreased frequency of high C4 GCNs (>4) and decreased expression of serum C4 in patients. Hou et al.18 indicated that high GCNs (>2) of C4A lead to risk for BD but not acute anterior uveitis by modulating the expression of C4A and enhancing IL-6 production. In another study, Lintner et al.28 discovered that there were significant differences in the distribution of lower C4A and C4 GCNs (<2 and <4, respectively) among JDM patients and controls, suggesting that complement C4A deficiency appeared to be an important risk factor for JDM. Furthermore, Rigby et al.26 demonstrated that C4B deficiency resulted in increased risk for RA and had broad implications for the pathogenesis of RA. However, data from pooled analyses of C4 and non-SLE autoimmune diseases are relatively insufficient. Additionally, data from several published articles were only done by single method without validation using the other methods and therefore further studies with different methods and larger samples including individuals from different ethnic backgrounds would help reveal the role of C4 in GD, uveitis, JDM, and RA.
Some limitations of the current meta-analysis of the potential relationship between C4, C4A, and C4B CNVs and autoimmune diseases need to be addressed. First, heterogeneity among ethnic groups or disease types was discovered when the association between C4, C4A, and C4B CNVs and autoimmune diseases was investigated. However, based on the results of the sensitivity analysis, it is clear that the overall results were not affected by heterogeneity. Second, an obvious publication bias was detected in the comparison between C4 CNV and autoimmune diseases. Third, the size of the patient and control groups was relatively small in each study; therefore, future studies should employ a much larger sample size and include individuals from different ethnic populations. Fourth, the effects of common confounding factors, including sex, age, and medical condition, were not assessed in the present study because of insufficient data. Fifth, autoimmune diseases at various sites may vary widely in terms of the etiology of pathology, including genetic factors, immunologic factors, and environmental factors. Finally, the electronic databases from which we selected eligible studies were listed in English and Chinese; thus, the meta-analysis may have a language bias.
In conclusion, the present meta-analysis provides evidence-based pooled data revealing a significant association between low C4 or C4A CNVs and susceptibility to autoimmune diseases, especially for Caucasian individuals with SLE.
Materials and Methods
Literature search
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) criteria were using as a guide for this meta-analysis39. A systematic search of PubMed, Embase, Web of Science, the China Biomedical database, and CNKI was conducted with the following keywords: C4 OR complement C4 OR complement component 4 OR C4A OR complement C4A OR complement component 4A OR C4B OR complement C4B OR complement component 4B AND copy number variation OR copy number variations OR CNV OR CNVs OR gene copy number OR GCN AND autoimmune disease OR autoimmune disorder OR autoimmunity. These in silico literature searches were restricted to human studies, and we used studies published until September 20, 2016. No language restrictions were imposed. The reference lists of the retrieved articles were manually searched in order to identify more relevant studies.
Inclusion and exclusion criteria
Studies selected from electronic databases were included when they met the following criteria: (1) focus on the CNVs of total C4, C4A, or C4B in relation to autoimmune diseases; (2) a case-control study design; (3) presence of enough data to calculate an OR and corresponding 95% CI; and (4) inclusion of patients diagnosed according to international standards. The following items were regarded as the primary exclusion criteria: (1) focus on families or twins rather than random general populations; (2) inclusion of individuals with overlapping investigated diseases; (3) a case study design; (4) insufficient data applied after emailing the relevant corresponding author. When several articles with the same types of patients were identified, the most detailed article with the largest sample size and most comprehensive analysis was included in the meta-analysis.
Data extraction
Data from the retrieved studies were extracted independently by two authors (N.L. and J.Z.). The following information was collected from each eligible study: first author, year of publication, country of origin, ethnicity of subjects, type of disease, sample size, genotyping methods, and information about GCN distribution in cases and controls. Two authors carefully screened the collected data and reached agreement in all respects. When the authors disagreed, a third reviewer (D.L.) weighed the arguments and then helped reach a consensus.
Quality assessment
Evaluation of the quality of extracted studies was also performed by two authors (N.L. and J.Z.) based on the HuGENet Handbook40. Six bias assessment items referring to the association between genes and disease were derived from this handbook, including bias in selection of cases, bias in selection of controls, bias in genotyping cases, bias in genotyping controls, bias in population stratification, confounding bias, multiple tests, and selective outcome reports. The quality of every item was labeled as “Yes” or “No,” while “Unclear” was used if there was not enough information to make a determination. A correction and review were performed independently by the investigator (D.L.) if the two coauthors dissented. Consensus regarding all labels was achieved after discussion.
Statistical analysis
ORs and the corresponding 95% CIs were estimated to evaluate the amount of correlation between C4, C4A, and C4B CNVs and susceptibility to autoimmune diseases. The diversity of genotypes (for total C4, the number of subjects with <4 GCNs compared to those with ≥4 or >4, and for C4A and C4B, the number of subjects with <2 GCNs compared to those with ≥2 or >2) among cases and healthy controls was compared. The heterogeneity between studies was estimated and measured by Cochran’s Q statistic as well as the I2 statistic. When the p value of chi-square statistic was less than 0.05 or the I2 value was more than 50%, the random-effect model was adopted for meta-analysis, indicating that heterogeneity existed across studies41. In addition, a goodness of fit test proposed by Chen et al.30 was used to explore the adequacy of the model for our systematic meta-analysis. Sensitivity analyses were performed to assess the effect of individual studies on pooled ORs by omitting each study in turn. Publication bias was estimated by inspecting Begg’s funnel plots42 and Egger’s regression test43. Statistical data was analyzed using STATA 12.0 software (StataCorp LP, College Station, Texas, USA). A significant difference was estimated at p < 0.05 (a two-tailed p value). The final conclusions of the study were independently validated by two authors (J. Z. and S. H.).
Additional Information
How to cite this article: Li, N. et al. Association between C4, C4A, and C4B copy number variations and susceptibility to autoimmune diseases: a meta-analysis. Sci. Rep. 7, 42628; doi: 10.1038/srep42628 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by National Natural Science Foundation Project (81522013), Chongqing Outstanding Youth Grant (cstc2014jcyjjq10005), Chongqing Key Laboratory of Ophthalmology (CSTC, 2008CA5003), Chongqing Science & Technology Platform and Base Construction Program (cstc2014pt-sy10002), National Key Clinical Specialties Construction Program of China and Research and Cultivation Foundation Project of Chongqing Medical University (201407).
Footnotes
The authors declare no competing financial interests.
Author Contributions S.H. and Y.W. designed the study. N.L. and J.Z. collected and checked the information of eligible articles included in this meta-analysis. J.Z. and D.L. analyzed the data. L.Y. prepared the Figures 1–5 and Tables 1–4. N.L. wrote the main manuscript. S.H. and Y.W. revised the manuscript. All authors reviewed and approved the manuscript.
References
- Walport M. J. Complement. First of two parts. N Engl J Med 344, 1058–1066 (2001). [DOI] [PubMed] [Google Scholar]
- Carroll M. C. The role of complement and complement receptors in induction and regulation of immunity. Annu Rev Immunol 16, 545–568, doi: 10.1146/annurev.immunol.16.1.545 (1998). [DOI] [PubMed] [Google Scholar]
- Ross S. C. & Densen P. Complement deficiency states and infection: epidemiology, pathogenesis and consequences of neisserial and other infections in an immune deficiency. Medicine (Baltimore) 63, 243–273 (1984). [PubMed] [Google Scholar]
- Wu Y. L., Hauptmann G., Viguier M. & Yu C. Y. Molecular basis of complete complement C4 deficiency in two North-African families with systemic lupus erythematosus. Genes Immun 10, 433–445, doi: 10.1038/gene.2009.10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena K. et al. Great genotypic and phenotypic diversities associated with copy-number variations of complement C4 and RP-C4-CYP21-TNX (RCCX) modules: a comparison of Asian-Indian and European American populations. Mol Immunol 46, 1289–1303, doi: 10.1016/j.molimm.2008.11.018 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt K. T., Yu C. Y., Carroll M. C. & Porter R. R. Polymorphism of human complement component C4. Immunogenetics 21, 173–180 (1985). [DOI] [PubMed] [Google Scholar]
- Yu C. Y., Belt K. T., Giles C. M., Campbell R. D. & Porter R. R. Structural basis of the polymorphism of human complement components C4A and C4B: gene size, reactivity and antigenicity. Embo J 5, 2873–2881 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Diversity in intrinsic strengths of the human complement system: serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J Immunol 171, 2734–2745 (2003). [DOI] [PubMed] [Google Scholar]
- Flachsbart F. et al. Investigation of complement component C4 copy number variation in human longevity. PLoS One 9, e86188, doi: 10.1371/journal.pone.0086188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchong C. A. et al. Deficiencies of human complement component C4A and C4B and heterozygosity in length variants of RP-C4-CYP21-TNX (RCCX) modules in caucasians. The load of RCCX genetic diversity on major histocompatibility complex-associated disease. J Exp Med 191, 2183–2196 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E. K. et al. Determining the one, two, three, or four long and short loci of human complement C4 in a major histocompatibility complex haplotype encoding C4A or C4B proteins. Am J Hum Genet 71, 810–822 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Mendoza A. R., Welch T. R., Zipf W. B. & Yu C. Y. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem 274, 12147–12156 (1999). [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Grosshans E. & Heid E. Lupus erythematosus syndrome and complete deficiency of the fourth component of complement. Boll Ist Sieroter Milan 53, suppl: 228 (1974). [PubMed] [Google Scholar]
- Cantsilieris S. & White S. J. Correlating multiallelic copy number polymorphisms with disease susceptibility. Hum Mutat 34, 1–13, doi: 10.1002/humu.22172 (2013). [DOI] [PubMed] [Google Scholar]
- Olsson L. M. & Holmdahl R. Copy number variation in autoimmunity–importance hidden in complexity? Eur J Immunol 42, 1969–1976, doi: 10.1002/eji.201242601 (2012). [DOI] [PubMed] [Google Scholar]
- Wu Y. L. et al. Phenotypes, genotypes and disease susceptibility associated with gene copy number variations: complement C4 CNVs in European American healthy subjects and those with systemic lupus erythematosus. Cytogenet Genome Res 123, 131–141, doi: 10.1159/000184700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 80, 1037–1054 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S. et al. Copy number variations of complement component C4 are associated with Behcet’s disease but not with ankylosing spondylitis associated with acute anterior uveitis. Arthritis Rheum 65, 2963–2970, doi: 10.1002/art.38116 (2013). [DOI] [PubMed] [Google Scholar]
- Hou S. et al. High c4 gene copy numbers protects against vogt-koyanagi-harada syndrome in chinese han. Br J Ophthalmol 98, 1733–1737 (2014). [DOI] [PubMed] [Google Scholar]
- Lintner K. E. et al. Early Components of the Complement Classical Activation Pathway in Human Systemic Autoimmune Diseases. Front Immunol 7, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H. et al. Deletion variants of RABGAP1L, 10q21.3, and C4 are associated with the risk of systemic lupus erythematosus in Korean women. Arthritis Rheum 65, 1055–1063, doi: 10.1002/art.37854 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Y. H. et al. Association between copy number variation of complement component C4 and Graves’ disease. J Biomed Sci 18, 71, doi: 10.1186/1423-0127-18-71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y. et al. Confirmation of C4 gene copy number variation and the association with systemic lupus erythematosus in Chinese Han population. Rheumatol Int 32, 3047–3053, doi: 10.1007/s00296-011-2023-7 (2012). [DOI] [PubMed] [Google Scholar]
- Boteva L. et al. Genetically determined partial complement C4 deficiency states are not independent risk factors for SLE in UK and Spanish populations. Am J Hum Genet 90, 445–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleynen I. et al. Genome-Wide Copy Number Variation Scan Identifies Complement Component C4 as Novel Susceptibility Gene for Crohn’s Disease. Inflamm Bowel Dis 22, 505–515 (2015). [DOI] [PubMed] [Google Scholar]
- Rigby W. F. et al. Increased frequency of complement C4B deficiency in rheumatoid arthritis. Arthritis Rheum 64, 1338–1344, doi: 10.1002/art.33472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira K. M. et al. Low C4, C4A and C4B gene copy numbers are stronger risk factors for juvenile-onset than for adult-onset systemic lupus erythematosus. Rheumatology 55, 869–873, doi: 10.1093/rheumatology/kev436 (2016). [DOI] [PubMed] [Google Scholar]
- Lintner K. E. et al. Gene copy-number variations (CNVs) of complement C4 and C4A deficiency in genetic risk and pathogenesis of juvenile dermatomyositis. Ann Rheum Dis 75, 1599–1606, doi: 10.1136/annrheumdis-2015-207762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yih Chen J. et al. Effects of Complement C4 Gene Copy Number Variations, Size Dichotomy, and C4A Deficiency on Genetic Risk and Clinical Presentation of Systemic Lupus Erythematosus in East Asian Populations. Arthritis Rheumatol 68, 1442–1453, doi: 10.1002/art.39589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang G. & Li J. Goodness-of-fit test for meta-analysis. Sci Rep 5, 16983, doi: 10.1038/srep16983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacoff J. B. et al. Primate erythrocyte-immune complex-clearing mechanism. J Clin Invest 71, 236–247 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Koralov S. B. & Kelsoe G. Complement C4 inhibits systemic autoimmunity through a mechanism independent of complement receptors CR1 and CR2. J Exp Med 192, 1339–1352 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth D. J. Complement deficiency and disease. J Clin Pathol 61, 1013–1017, doi: 10.1136/jcp.2008.056317 (2008). [DOI] [PubMed] [Google Scholar]
- Law S. K., Dodds A. W. & Porter R. R. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J 3, 1819–1823 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M. C., Fathallah D. M., Bergamaschini L., Alicot E. M. & Isenman D. E. Substitution of a single amino acid (aspartic acid for histidine) converts the functional activity of human complement C4B to C4A. Proc Natl Acad Sci USA 87, 6868–6872 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., Carson A. R. & Scherer S. W. Structural variation in the human genome. Nat Rev Genet 7, 85–97, doi: 10.1038/nrg1767 (2006). [DOI] [PubMed] [Google Scholar]
- Huber A. K. et al. Analysis of immune regulatory genes’ copy number variants in Graves’ disease. Thyroid 21, 69–74, doi: 10.1089/thy.2010.0262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraksingh R. R. & Snyder M. P. Impacts of variation in the human genome on gene regulation. J Mol Biol 425, 3970–3977, doi: 10.1016/j.jmb.2013.07.015 (2013). [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. & Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6, e1000097, doi: 10.1371/journal.pmed.1000097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. & Higgins J. P. T. e. The HuGENetTM HuGE Review Handbook, version 1.0. http://www/hugenat.ca. (2006). [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560, doi: 10.1136/bmj.327.7414.557 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]