Abstract
Background
MicroRNAs (miRNAs) are a group of short non-coding RNAs involved in the inhibition of protein translation or in mRNA degradation. Although the regulatory roles of miRNAs in various biological processes have been investigated, there is as yet an absence of studies about the regulatory roles of miRNAs involved in the metabolism of plant allelochemicals in insects.
Results
We constructed five small RNA libraries from apterous Aphis gossypii adults that had fed on an artificial diet containing various allelochemicals. Using Illumina sequencing, a total of 73.27 million clean reads was obtained, and 292 miRNAs were identified from A. gossypii. Comparative analysis of read counts indicated that both conserved and novel miRNAs were differently expressed among the five libraries, and the differential expression was validated via qRT-PCR. We found that the transcript levels of several miRNAs were increased or decreased in all of the allelochemical treatment libraries compared to the control. The putative target genes of the miRNAs were predicted using in silico tools, and the target genes of several miRNAs were presumed to be involved in the metabolism of xenobiotic compounds. Furthermore, the target prediction results were confirmed using dual luciferase reporter assay, and Ago-miR-656a-3p was demonstrated to regulate the expression of CYP6J1 post-transcriptionally through binding to the 3′ UTR of CYP6J1.
Conclusion
Our research results indicate that miRNAs may be involved in the metabolism of plant allelochemicals in A. gossypii, and these results also represent an important new small RNA genomics resource for further studies on this topic.
Electronic supplementary material
The online version of this article (doi:10.1186/s12867-017-0080-5) contains supplementary material, which is available to authorized users.
Keywords: MicroRNAs, Plant allelochemicals, Aphis gossypii, Illumina sequencing, Regulatory roles
Background
MicroRNAs (miRNAs) are small non-coding RNAs (18-24 nucleotides in length) that regulate gene expression at the post-transcriptional level [1, 2]. Through binding complementarily to 3′ untranslated regions (UTRs), coding sequences, or 5′ UTRs, miRNA suppress the translation of target mRNA molecules, and thereby silence target gene expression [3–5]. miRNAs are generated in all eukaryotes and viruses [1, 6], and many miRNAs are conserved among related species [7]. Since the first miRNA was reported to regulate the timing of development in Caenorhabditis elegans [8], numerous miRNAs have been identified from animals, plants, and viruses. In the last two decades, thousands of miRNAs have been isolated from insect species, including Drosophila melanogaster [9], Bombyx mori [10, 11], Manduca sexta [12–14], Plutella xylostella [15, 16], and Helicoverpa armigera [17, 18]. Functional studies carried out in these insect species have demonstrated that insect miRNAs play very important regulatory roles in various biological processes, such as development, immune responses, metabolism, and host-pathogen interactions [1, 6, 19, 20].
The cotton aphid, Aphis gossypii Glover (Hemiptera: Aphididae), is an important insect pest in cotton and cucurbit crops that causes economic damage both through direct feeding and through the transmission of viruses [21, 22]. Given that A. gossypii has a very wide host range that encompasses least 300 species [23, 24], this pest encounters multiple plant toxic chemicals that are produced by host plants to defend against herbivores. These compounds have strong deleterious effects on herbivorous insects by affecting the growth and development or even by directly causing mortality [25, 26]. In humans and large mammals, increasing evidence suggests that miRNAs play very important roles in the metabolism of xenobiotic compounds [5, 27]. Such miRNAs can mediate the detoxification metabolism of xenobiotics by regulating the expression of xenobiotic-metabolizing enzymes and nuclear receptors [5]. For example, human P450 CYP1A1, which is involved in the metabolism of carcinogenic metabolites, was found to be post-transcriptionally regulated by miR-892a [28]. In addition, previous studies carried out in mosquitoes Culex pipiens suggested that miRNAs participate in the resistance to pyrethroid insecticides by mediating the expression levels of P450 genes [29, 30]. While there are a lot known about the miRNAs that participate in regulating the detoxification of xenobiotics in animals and miRNAs likely have essential roles in insecticide resistance, less is understood about the regulatory roles of miRNAs in the metabolism of plant allelochemicals in insects.
In the present study, five small RNA libraries were built from apterous A. gossypii adults fed on artificial diets that contained various plant allelochemicals (gossypol, 2-tridecanone, quercetin and tannic acid respectively, which are toxic chemicals found naturally in cotton plant or other host plants of the cotton aphid) and control. A total of 73.27 million clean reads was obtained by deep sequencing, and 292 miRNAs were identified from the five sample libraries. In order to identify putative allelochemical metabolism-related miRNAs, the expression levels of both conserved and novel miRNAs were compared among the five libraries, and the targets of the newly identified miRNAs were predicted. The results of this study deepen our understanding of the regulatory roles of miRNAs in A. gossypii and indicate that miRNAs are likely involved in the insect metabolism of plant allelochemicals.
Results
Deep sequencing of A. gossypii small RNA libraries
In order to examine the potential role of small RNAs in A. gossypii responses to plant allechemicals, we collected apterous A. gossypii adults that had been fed artificial diet containing various allechemicals for 24 h for small RNA sequencing. Five A. gossypii small RNA libraries were constructed and sequenced using the Illumina sequencing platform. A total of 76.96 million raw reads was obtained from the five libraries, and after filtering out sequences shorter than 18 nt and filtering the lower quality reads from the raw data, 73.27 million quality reads were obtained. The number of clean reads differed among the five libraries; more clean reads (16.09 million) were obtained from the 2-tridecanone-fed library than from any of the other four libraries (14.48, 13.96, 14.01, and 14.74 million, in the tannic acid, quercetin, gossypol, and control libraries respectively) (Table 1). The length of these small RNAs ranged from 18 to 30 nt. In all five libraries, the highest peak for nucleotide length distribution was that for 22 nt (Fig. 1; Additional file 1).
Table 1.
Total number of reads obtained from the small RNA libraries from aphids fed artificial diet containing various plant allechemicals
| Allechemicals | Raw reads | Clean reads |
|---|---|---|
| Control | 15,193,417 | 14,483,649 |
| 2-tridecanone | 16,819,205 | 16,090,539 |
| Tannic acid | 14,630,011 | 13,956,679 |
| Quercetin | 14,743,194 | 14,006,538 |
| Gossypol | 15,575,107 | 14,736,872 |
| Total | 76,960,934 | 73,274,277 |
Fig. 1.
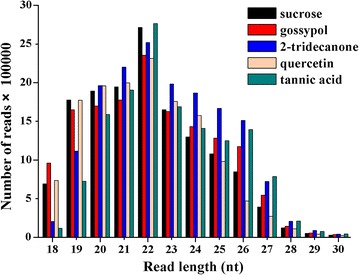
Length distribution of small RNAs from A. gossypii identified by deep sequencing. This length distribution was assessed using clean reads after filtering out the redundant small RNAs
Identification of conserved and novel miRNAs, and analysis of their features
To identify potential candidate miRNAs from the A. gossypii sample libraries, the raw sequencing data were analyzed with miRDeep2 software. The Acyrthosiphon pisum genome was used as a reference because the A. gossypii genome was not available at the time of the analysis. Total of 292 unique miRNA candidates were identified from among the five libraries, including 246 conserved miRNAs and 46 potentially novel miRNAs. The newly identified miRNAs of A. gossypii were named by prefixed with “Ago”, where “Ago” means A. gossypii. The length and copy number distribution analysis of these newly identified miRNAs indicated that the miRNAs of 22 nt in length were the most abundant (47.46%), and the miRNAs of 21–22 nt in length accounted for more than 81% of reads (Table 2). Further, we analyzed the common and unique distribution of the newly identified miRNAs from among the five libraries. The results demonstrated that 221 miRNAs were common among the five libraries; only a few of the miRNAs were uniquely expressed in a particular library (Additional file 2: Figure S1).
Table 2.
Length distribution and copy number of A. gossypii miRNAs
| miRNA length | Number of miRNAs | Copy number | Percentage (%) |
|---|---|---|---|
| 18 | 65 | 11,291 | 0.32 |
| 19 | 64 | 62,839 | 1.79 |
| 20 | 40 | 100,111 | 2.85 |
| 21 | 37 | 1,183,236 | 33.69 |
| 22 | 54 | 1,666,826 | 47.46 |
| 23 | 17 | 309,952 | 8.82 |
| 24 | 11 | 178,019 | 5.07 |
| 25 | 4 | 31 | 0.00 |
Since the dominance of uracil at the first position of the 5′ terminus terminal is considered to be one of the conserved features of mature miRNAs [31], and given that the first base toward the 5′ end of miRNAs is known to play very important roles in the interaction between miRNAs and argonaute complexes [32], the position-specific nucleotide occurrence of the candidate miRNA sequences was analyzed. A. gossypii miRNAs showed a nucleotide bias towards uracil (U) at the first nucleotide position (Fig. 2). In addition, the base composition of the miRNAs at each position was analyzed, and the nucleotide U was the most abundant nucleotide at most of the positions; this was especially pronounced at positions 1, 17, 22, 23, 25, and 30 (Fig. 2).
Fig. 2.
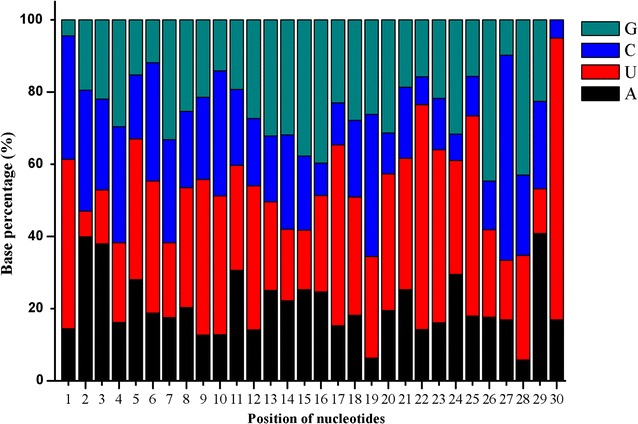
The position-specific nucleotide occurrence of A. gossypii mature miRNAs. Uracil dominated the first nucleotide position towards the 5′ end of miRNAs
A. gossypii miRNAs differentially expressed following allelochemical treatments
To identify the miRNAs that may play important roles in the responses of A. gossypii against plant alleochemicals, the differential expression of the identified miRNAs were analyzed using edgeR software. The expression values as assessed by miRDeep2 were used to analyze differential expression of A. gossypii miRNAs, and the miRNAs that had read counts of more than 10 in all five libraries were selected for differential expression analysis. Compared to the aphids fed on a 0.5 M sucrose solution (control), 134 miRNAs were found to be differentially expressed in the aphids fed on diet containing allelochemicals (Fig. 3). Of these, following for aphids feeding on gossypol, 2-tridecanone, quercetin, and tannic acid for 24 h, there were 33, 73, 59, and 42 differentially expressed miRNAs, respectively (Table 3). Interestingly, we found that most of the differentially expressed miRNAs were up-regulated in the aphids treated with gossypol, quercetin, and 2-tridecanone, but that the miRNAs from the tannic acid treated aphids possessed the tendency of down-regulation (Table 3).
Fig. 3.
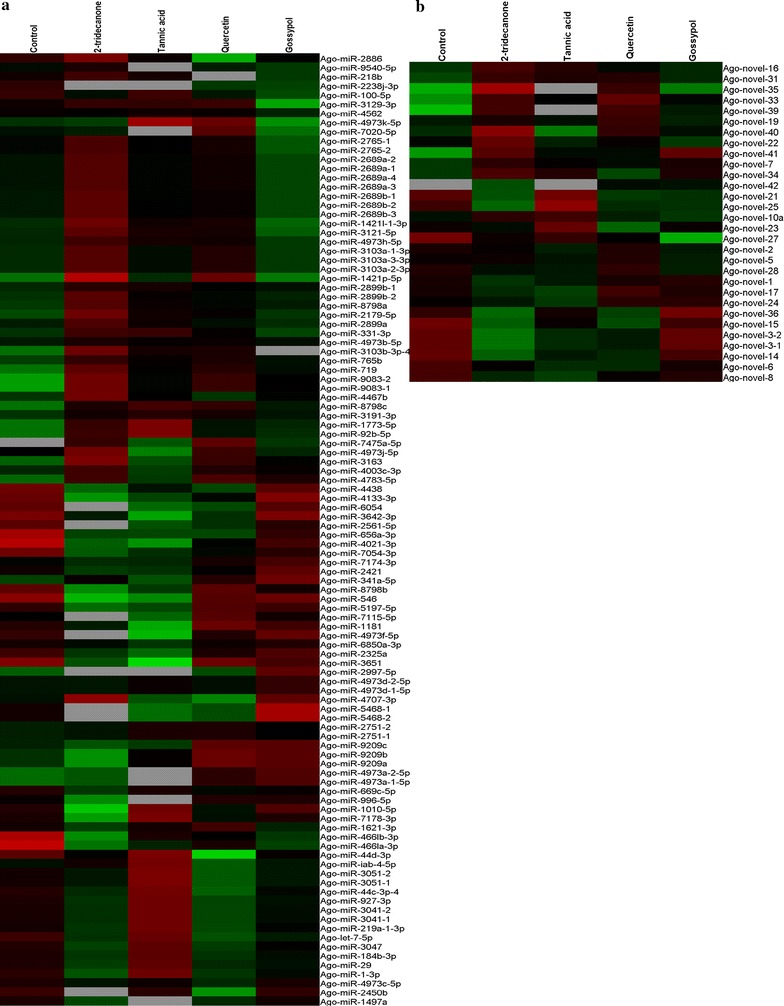
Differential expressions of A. gossypii miRNAs after feeding on various allelochemicals for 24 h. a Expression of conserved miRNAs. b Expression of novel miRNAs. Green color represents low expression levels and red color represents the high expression levels of the miRNAs
Table 3.
Number of miRNAs that were significantly differentially regulated after the different allelochemical treatments
| Allelochemicals | Number of up-regulated miRNAs | Number of down-regulated miRNAs |
|---|---|---|
| 2-tridecanone | 42 | 31 |
| Tannic acid | 4 | 38 |
| Quercetin | 49 | 10 |
| Gossypol | 29 | 4 |
The expression levels of these miRNAs were dysregulated among five libraries. For instance, Ago-miR-2997-5p expression was low in the control and quercetin treatment libraries and was not detected in the 2-tridecanone or tannic acid treatment, but was up-regulated in the gossypol library (Fig. 3). Although the expression patterns of most of the miRNAs differed among the various allelochemical treatments, several miRNAs were consistently up/down-regulated in all four of the allelochemical treatment libraries. For instance, compared to the control, the expressions of some miRNAs (Ago-miR-8798a, Ago-miR-331-3p, Ago-miR-3191-3p, Ago-miR-1773-5p, Ago-miR-2179-5p, Ago-miR-92b-5p, Ago-miR-9083-2, and Ago-miR-719) were up-regulated in all four of the allelochemical treatments (Table 4). While the expression levels of Ago-let-7-5p, Ago-miR-100-5p, Ago-miR-44b-3p, Ago-miR-7054-3p, Ago-miR-4021-3p, Ago-miR-656a-3p, Ago-miR-4661a-3p, and Ago-miR-2238j-3p were down-regulated in all four treatment libraries in compare with the control (Table 5). It is worth nothing that Ago-miR-7475a-5p expression was not detected in the control library, but was expressed in each of the allelochemical treatment libraries (Table 4).
Table 4.
The read counts and sequences of A. gossypii miRNAs that had increased expression following plant allelochemical treatment
| miRNA name | miRNA sequence | Control | 2-tridecanone | Tannic acid | Quercetin | Gossypol |
|---|---|---|---|---|---|---|
| Ago-miR-8798a | CGCGGTCGCCGCGCCGCC | 116 | 199 | 117 | 154 | 162 |
| Ago-miR-331-3p | GCCCCTGGTGGTCATGTTGGA | 39 | 58 | 56 | 57 | 42 |
| Ago-miR-3191-3p | GGGGGACGAGGTGGCCGAGCGGT | 37 | 39 | 45 | 60 | 54 |
| Ago-miR-1773-5p | GGGGGGAGGAGGAGGAGGA | 19 | 41 | 75 | 36 | 39 |
| Ago-miR-2179-5p | ATGCAAAATACATTTGTGTACT | 27 | 66 | 32 | 45 | 35 |
| Ago-miR-92b-5p | CGGGACGGCGAGGGTTGGGG | 16 | 36 | 64 | 30 | 31 |
| Ago-miR-719 | CTCTCGGCCGTCGGCGCGGC | 2 | 6 | 3 | 6 | 5 |
| Ago-miR-9083-2 | GGCCACGCCGCGCCGTCGG | 1 | 5 | 2 | 5 | 4 |
| Ago-miR-7475a-5p | CGCCACCGCCGCGCCGTCGT | 0 | 6 | 2 | 13 | 5 |
Table 5.
The read counts and sequences of A. gossypii miRNAs that had decreased expression following plant allelochemical treatment
| miRNA name | miRNA sequence | Control | 2-tridecanone | Tannic acid | Quercetin | Gossypol |
|---|---|---|---|---|---|---|
| Ago-let-7-5p | TGAGGTAGTTGGTTGTATAGT | 2943 | 673 | 2378 | 811 | 1586 |
| Ago-miR-100-5p | GACCCGTAGATCCGAACTTGTG | 2501 | 803 | 1521 | 1171 | 962 |
| Ago-miR-44b-3p | TGACTAGAGTTTATACTACCGA | 1475 | 497 | 1375 | 606 | 835 |
| Ago-miR-7054-3p | CCAACTTGGCAGCTTCTGA | 158 | 17 | 25 | 52 | 95 |
| Ago-miR-4021-3p | TAAGTATTTGGCTCTTGG | 52 | 3 | 2 | 10 | 22 |
| Ago-miR-656a-3p | CATATTATGGTCGTGAGTA | 24 | 2 | 2 | 3 | 10 |
| Ago-miR-466la-3p | TATAAATATTGTAGGTACC | 23 | 1 | 2 | 5 | 3 |
| Ago-miR-2238j-3p | TATGACGAGAGGGCAAAT | 16 | 0 | 0 | 6 | 7 |
To verify the expression analysis results from the sequencing experiments (read count), 6 differentially expressed miRNAs were selected, and their expression levels were measured via qRT-PCR analysis. Five of the selected miRNAs showed similar expression patterns as those assessed using sequencing read counts (Figs. 3, 4). In the sequencing results, the expression of Ago-miR-2179-5p was up-regulated, and Ago-let-7-5p was down-regulated in all of the allelochemicals treatment libraries, and the qRT-PCR analysis showed similar results (Fig. 4; Tables 4, 5). The qRT-PCR results for Ago-miR-3051-2 agreed with the sequencing results: it was highly expressed when apterous A. gossypii adults fed on an artificial diet containing 0.2% tannic acid (Figs. 3, 4). The expression level of Ago-miR-5468a as measured by qRT-PCR was different from that as assessed by the sequencing read count, the reason for this discrepancy is unknown.
Fig. 4.
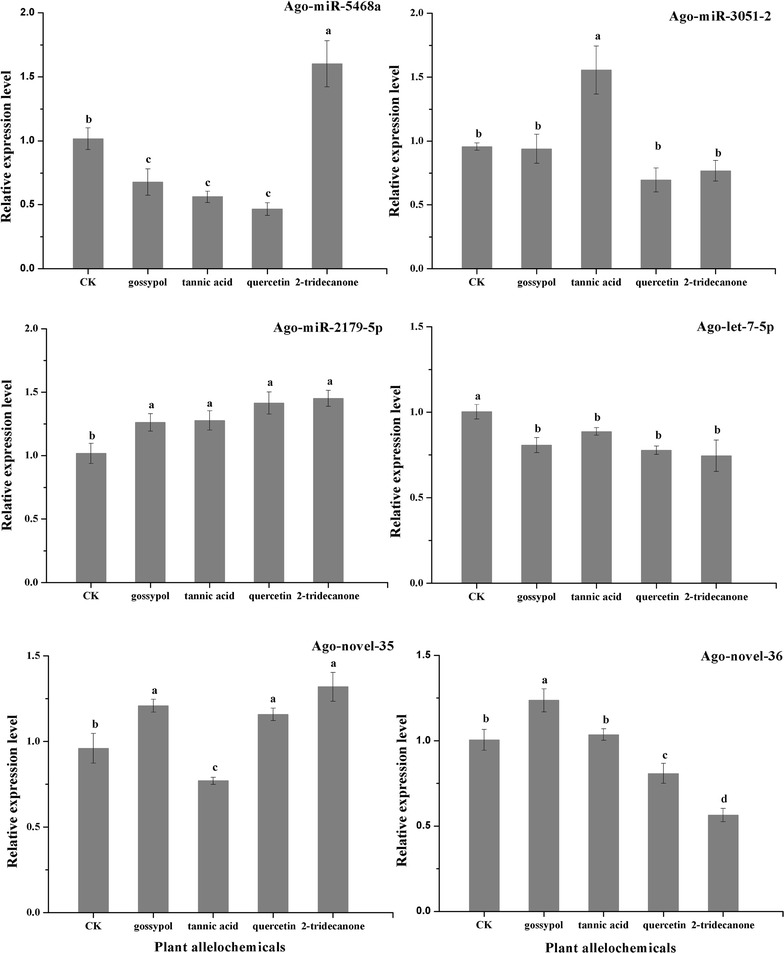
Differential expressions of miRNAs following plant allelochemical treatment. The results are presented as mean ± SD for three independent replicates. Different letters on the bars of the histogram indicate significant differences based on ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05)
Prediction of the putative target genes of the miRNAs
To further understand the role of the various miRNAs identified in A. gossypii, the putative targets of these identified miRNAs were computationally predicted. The miRanda and RNAhybrid programs were used to identify the targets of these miRNAs from an A. gossypii transcriptome database. Combining the results of these two target prediction programs, a total of 5929 genes were putatively targeted by 236 miRNAs from A. gossypii. GO annotation of these target genes included the following categories: cellular components, molecular function, and biological process. About 49.8% of the predicted target genes were classified into the biological process category, including cellular process (19.4%), metabolic process (17.6%), and biological regulation (10.8%) (Additional file 2: Figure S2).
To characterize the potential roles of A. gossypii miRNAs in the defense responses against allelochemicals, we focused our attention to the predicted target genes that were likely to be involved in xenobiotic metabolism. Interestingly, we found that several miRNAs were targeted to genes that are known to play very important roles in insect responses to xenobiotic stress, including cytochrome P450s, acetylcholinesterases, glutathione S-transferases, sodium channel proteins, etc. (Table 6). Several miRNAs were predicted to have many target genes, and numerous genes were putatively targeted by multiple miRNAs. For instance, Ago-miR-4467a-1 and Ago-miR-4973-5p-11 were found have many predicted target genes, and CYP6J1 was putatively targeted by Ago-miR-656a-3p, Ago-miR-669c-5p, and Ago-miR-4172-3p (Table 6). In addition, some of these miRNAs, which were putatively targeted to xenobiotic metabolism-related genes, were differently expressed following allelochemical treatment, including Ago-miR-3191-3p, Ago-miR-8798a, and Ago-miR-656a-3p (Table 6; Fig. 3).
Table 6.
Putative xenobiotic metabolism-related target genes of A. gossypii miRNAs
| miRNA | Xenobiotic metabolism related target genes |
|---|---|
| Ago-miR-1-3p | G-protein coupled receptor |
| Ago-miR-341b-5p | Acetylcholinesterase, CYP6A8, CYP6A14 |
| Ago-miR-656a-3p | CYP6J1, glutathione S-transferase sigma 1, carboxylesterase |
| Ago-miR-669c-5p | CYP6J1, glutathione S-transferase sigma 1 |
| Ago-miR-1181 | CYP6A13 |
| Ago-miR-1332-3p | UDP-glucuronosyltransferase, CYP6A8, CYP6K1, CYP4C1, CYP6A2, CYP6A13 |
| Ago-miR-1946a | Acetylcholinesterase, glutathione S-transferase omega 1 |
| Ago-miR-2886 | Acetylcholinesterase |
| Ago-miR-2899a | CYP18A1 |
| Ago-miR-3163 | Glutathione S-transferase sigma 1 |
| Ago-miR-3191-3p | Carboxylesterase |
| Ago-miR-3575 | CYP315A1, glutathione S-transferase sigma 2 |
| Ago-miR-4172-3p | CYP6J1, glutathione S-transferase sigma 1 |
| Ago-miR-4213-5p | Carboxylesterase, glutathione S-transferase sigma 2 |
| Ago-miR-466la-3p | Carboxylesterase |
| Ago-miR-4467a-1 | Sodium channel protein, calcium channel flower, CYP6A8, ryanodine receptor 44F, gamma-aminobutyric acid receptor, voltage-dependent calcium channel type A subunit alpha 1, nicotinic acetylcholine receptor alpha subunit |
| Ago-miR-4467a-2 | CYP6A2, CYP6A8, esterase FE4, ryanodine receptor 44F |
| Ago-miR-4783-5p | Glutamate-gated chloride channel, voltage-dependent T-type calcium channel subunit alpha 1 |
| Ago-miR-4973a-1-5p | Ryanodine receptor |
| Ago-miR-4973-5p-11 | Ryanodine receptor 44F, sodium channel protein para, voltage-dependent L-type calcium channel subunit beta 2, Nic acetylcholine receptor alpha 2, Glutamate-gated chloride channel, Voltage-dependent T-type calcium channel subunit alpha 1 |
| Ago-miR-4973 g-5p | Acetylcholinesterase, CYP6K1 |
| Ago-miR-6236 | Voltage-dependent calcium channel type A subunit alpha 1 |
| Ago-miR-6850-3p-1 | CYP6A8 |
| Ago-miR-7475a-5p | Venom carboxylesterase-6 |
| Ago-miR-8798a | CYP6A8 |
| Ago-miR-8798c | Voltage-dependent calcium channel type A subunit alpha 1 |
| Ago-novel-7 | Sodium channel protein 60E, CYP6A8, UDP-glucuronosyltransferase |
| Ago-novel-10a | Ryanodine receptor 44F |
| Ago-novel-16 | CYP18A1 |
| Ago-novel-19 | Acetylcholinesterase, CYP6A8, CYP6A2 |
| Ago-novel-24 | Acetylcholinesterase, CYP6A8 |
| Ago-novel-28 | Voltage-dependent calcium channel type A subunit alpha 1 |
Validation of the target prediction
The target prediction results were validated by selecting a cascading of Ago-miR-656a-3p and CYP6J1. To determine whether or not Ago-miR-656a-3p could bind to the 3′ UTR of CYP6J1 and suppress the expression of CYP6J1, the 3′ UTR of CYP6J1 containing the target site of Ago-miR-656a-3p was inserted into a pmirGLO vector to yield a recombined vector, pmirGLO-CYP6J1-UTR. The results of dual luciferase reporter assay showed that the firefly luciferase activity normalized to Renilla was significantly reduced after pmirGLO-CYP6J1-UTR was co-transfected with Ago-miR-656a-3p agomir in comparison with the negative mimic control; while the co-transfection of Ago-miR-656a-3p agomir with control vector (pmirGLO) did not decrease the relative activity of luciferase (Fig. 5).
Fig. 5.
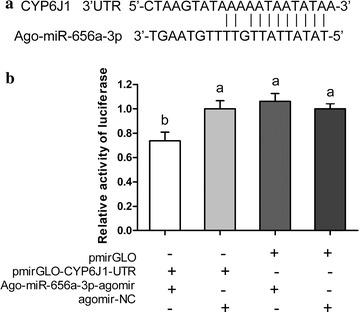
Interaction between Ago-miR-656a-3p and CYP6J1 using a dual fluorescent reporter system. a Predicted target sites of Ago-miR-656a-3p in the 3′ UTR of CYP6J1. b Luciferase reporter assays performed by co-transfection of Ago-miR-656a-3p agomir with a luciferase reporter gene linked to the 3′ UTR of CYP6J1. Firefly luciferase activity was normalized to Renilla luciferase activity and then normalized to the activity of the control group. The mathematical operators of “+”and “−” mean add and subtract. Different letters on the bars of the histogram indicate significant differences based on ANOVA followed by Tukey’s HSD multiple comparison test (P < 0.05)
Discussion
Allelochemicals are very important plant natural products that are known to play essential roles in plant defense responses to herbivores. Previous studies have shown that allelochemicals have great impacts on herbivorous insects. For example, detrimental effects were observed when H. armigera larvae were exposed to a high concentration of gossypol [33, 34]. Similarly, naringenin and quercetin have been reported to cause detrimental effects in the pea aphid, A. pisum, by effecting development, fecundity, and mortality [26]. As a very important polyphagous pest, cotton aphids encounter multiple allelochemicals in their life cycles, including gossypol, quercetin, and tannic acid. There is no doubt that allelochemicals have great effects on A. gossypii; for example, Gao et al. found that higher levels of gossypol adversely affected the development, longevity, and reproduction of A. gossypii [35]. miRNAs have been demonstrated to play very important regulatory roles in many biological processes over last few years. Therefore, it is reasonable to conjecture that small RNA molecules potentially function in regulatory roles in cotton aphid responses to plant allelochemicals. The identification and analysis of the expression profiles of miRNAs in allelochemical treated A. gossypii can potentially provide insight into the regulatory mechanisms underlying insect detoxification of plant allelochemicals. The present study was undertaken to identify the conserved and novel miRNAs of A. gossypii and to investigate the potential regulatory roles of these miRNAs in the metabolism of allelochemicals.
With the development of high throughput sequencing technology, small RNA sequencing has become a popular experimental approach to identifying miRNAs from a range of organisms. Sattar et al. [36] identified 102 miRNAs in A. gossypii fed on susceptible and resistant (Vat − and Vat +) melon plants. In the present study, we sequenced five small RNA libraries and identified 293 miRNAs from A. gossypii fed on various allelochemicals; our sequencing data greatly expands the scope of the resources available to study A. gossypii miRNAs. The size distribution pattern of the clean reads revealed that the five libraries were dominated by 22 nt sequences (Fig. 1), a result consistent with the known characteristics of miRNA [37]. The overall length distribution observed of the A. gossypii miRNA in this study was similar to distributions observed in several other insect species, including Blattella germanica [38] and Apis mellifera [39].
Our expression analysis based on read counts showed that 134 miRNAs were differentially expressed in the allelochemical-treated aphids, as compared to the control aphids, which clearly suggests that allelochemicals affect miRNA expression, thus implying a possible role for miRNAs in the regulation of the metabolism of allelochemicals in A. gossypii.
Our results showed that several miRNAs were up/down regulated in all four allelochemicals libraries, and these miRNAs may play important roles in the metabolism of plant allelochemicals in A. gossypii. For instance, miR-92b was up-regulated in all four allelochemical libraries, and suggests that miR-92b may involve in the response of A. gossypii to plant allelochemicals. In other insect species, miR-92b has been reported to be involved in multiple biological processes. In Drosophila, miR-92b plays an important role in muscle development [40], and is essential for neuroblast self-renewal [41]. Meanwhile, miR-92b was classified as a stress responsive marker in Eurosta solidaginis [42].
In the present study, the expression levels of Ago-let-7-5p and Ago-miR-100-5p were down-regulated following allelochemical treatment. Let-7 and miR-100 are basic components of the let-7-complex (let-7-C), which is required for the development of normal morphology in D. melanogaster [43]. Given that allelochemicals can have great effects on insect development [26, 34, 35], the differential expression of let-7 and miR-100 might be attributable to the influence of plant allelochemicals. The similar phenomenon was also observed in the host adaption of Myzus persicae to nicotianae by the let-7 and miR-100 participating in regulating of the expression of CYP6CY3 post-transcriptionally [44]. Further, Sattar et al. found that when apterous cotton aphids fed on Vat − melon with high susceptibility to aphids, the expression levels of let-7 and miR-100 were decreased in compare with the aphids that fed on resistant (Vat +) melon [36]. These results suggest that let-7 and miR-100 might be involved in the metabolism of xenobiotics of A. gossypii.
To further understand the possible roles of miRNAs in the metabolism of plant allelochemicals, the putative target genes of the newly identified miRNAs were predicted, and many of the predicted target genes were annotated to be involved in multiple biological processes. In addition, we found that several miRNAs were predicted to target genes from families known to be important in the metabolism of xenobiotics [45, 46]. Combining these results of the target prediction with our differential expression analysis, we found that some of the miRNAs predicted to target these xenobiotic metabolism-related genes were among the differentially express miRNAs. This suggested that miRNAs may be involved in insect metabolism of plant allelochemicals by regulating the expression of xenobitic metabolism genes.
Conclusions
A total of 292 miRNAs was identified from A. gossypii, and the expression analysis results demonstrated that the transcript levels of these miRNAs were changed depend on the plant allelochemicals feeding by A. gossypii. The results of target prediction suggest that miRNAs may be involved in the metabolism of plant allelochemicals of A. gossypii, and these results represent an important new small RNA genomics resource for further studies on this topic.
Methods
Cotton aphid strain and cell culture
The strain of A. gossypii used in this study was collected in 1999 from cotton fields in the Xinjiang Uygur Autonomous Region, China, and has been maintained in our laboratory for more than 15 years. The aphids were reared on cotton seedings in controlled conditions of 20–23 °C, 60% relative humidity, and a photoperiod of 16:8 h (L:D), as described previously [47]. The mammalian HEK293T cell line was maintained at 37 °C under 5% CO2 in DMEM high-glucose medium (Gibco) containing 10% fetal bovine serum (Gibco).
Chemicals
Gossypol, 2-tridecanone, quercetin, and tannic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sucrose was purchased from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China).
In vitro feeding assay
Sterilized glass tubes open at both ends (3 cm in length, 2 cm diameter) were used for the in vitro feeding assays. One end of each tube was covered by two layers of parafilm, with the following solution sandwiched between the two parafilm layers (artificial diet): 200 µl of a 0.5 M sterile sucrose solution that contained 0.2% gossypol, 2-tridecanone, quercetin, and tannic acid). One hundred healthy aperous adults were gently placed into the tube with a brush, and the tube was sealed with a piece of Chinese art paper with solid glue. Aphids were allowed to feed on this artificial diet for 24 h. The same system and solution, minus the alleochemicals, was used as a control. Following the 24 h of feeding, the live aphids were collected for RNA extraction.
RNA extraction and sequencing
Five samples of apterous A. gossypii adults collected from the feeding assays were used for the preparation of the small RNA libraries. Total RNA was isolated with TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The purity and the concentration of the RNA were assessed with a NAS-99 spectrophotometer (ACTGene, USA), and RNA integrity was evaluated with analysis on a 1% (w/v) agarose gel. About 10 µg of total RNA was isolated on a 15% denaturing polyacrylamide gel, and small RNA molecules ranging from 18 to 30 nt in length were purified and then ligated with 3′ and 5′ adapters. The ligated products were then reverse transcribed into cDNA with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol, and the resulting cDNA was amplified with PCR (15 cycles). Amplified cDNA products were purified with agarose gels and sent to the Beijing Genome Institute Inc. (China) for high-throughput sequencing with the Illumina Hiseq 2000 platform.
Identification of miRNAs from sequencing data
A. gossypii miRNA candidates were identified using miRDeep2 software [48]. Raw sequencing reads from the five libraries were submitted as input into miRDeep2, and the data from each library were analyzed separately. The miRDeep2 analysis was performed using the default options and settings. After trimming the adaptor sequences and discarding rRNA, tRNA, snRNA, and snoRNA, as well as the sequences containing polyA tails from the raw reads, the small RNAs between 18 and 30 nt in length were selected for further analysis. The Acyrthosiphon pisum genome sequence was used as the reference genome, since the A. gossypii genome sequence was not available at the time of analysis.
Expression profile of miRNAs in five libraries
In order to find the miRNAs that may play very important roles in the response of cotton aphids against plant alleochemicals, the differential expression of the identified miRNAs was analyzed. The read counts of the newly identified miRNA were analyzed using edgeR software; edgeR version 3.10.2 is available in Bioconductor version 3.1 (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) [49]. edgeR is a Bioconductor software package for analyzing the differential expression of digital gene expression data. Briefly, an overdispersed Poisson model is used to account for variability, and empirical Bayes methods are used to moderate the degree of overdispersion across the transcripts. The Benjamini-Hochberg method was used to adjust for multiple testing [50]. Only those miRNAs with a fold change ≥2 and a false discovery rate (FDR) <0.05 were considered to be significant.
Target gene prediction and analysis
Since the complete genome sequence of A. gossypii was not available at the time of analysis, the A. gossypii transcriptome database (unpublished) was used to predict the targets of the sequenced A. gossypii miRNAs. Two miRNA target prediction software programs were used, each with the default settings: miRanda (http://www.microrna.org/) [51] and RNAhybrid (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid/) [52]. The miRNA target genes commonly predicted by both programs were selected for further analysis. The predicted target genes were then aligned using the BLASTX program from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (e value cut-off used was 1.0E−5), and these genes were mapped and annotated by BLAST2GO [53]. The GO terms from this analysis were used to define the functional categories of the predicted miRNA target genes.
Quantitative real-time PCR
To validate the miRNA data obtained through our deep sequencing experiment, 4 conserved and 2 novel miRNA candidates were selected, and their expression was confirmed with quantitative real-time PCR (qRT-PCR) analysis. Aperous adults fed on an artificial diet (with 0.2% allechemicals) for 24 h were used for total RNA isolation; total RNA was extracted using miRNeasy Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. First strand cDNA was synthesized from 2 μg of total RNA using miScript II RT kit (Qiagen) following the manufacturer’s instructions. SYBR Green Master Mix (miScript SYBR Green PCR Kit, Qiagen) was used for miRNA expression assays, and qRT-PCR was performed as previously described [15]. Three biological replicates, with three technical replications for each, were evaluated for each sample. Analysis of the qRT-PCR data was carried out using the 2−∆∆Ct method of relative quantification [54]. As an endogenous control, U6 snRNA was used to quantify the expression level of miRNA. The primers used for the qRT-PCR analysis are listed in Additional file 2: Table S1.
Vector construction and dual luciferase reporter assay
The target prediction results were validated by selecting Ago-miR-656a-3p and CYP6J1 that has a target site of Ago-miR-656a-3p in the 3′ UTR. The CYP6J1 3′ UTR sequence was synthesized by GenePharm Co. Ltd (Shanghai, China), and was inserted into the pmirGLO vector, generating the pmirGLO-CYP6J1-UTR target construct. The agomir (mimics) of Ago-miR-656a-3p was synthesized by GenePharm Co. Ltd (Shanghai, China). The HEK293T cells were cultured in a 24-well plate and were transfected with target plasmids and agomir of Ago-miR-656a-3p or agomir-NC using a Calcium Phosphate Cell Transfection Kit (Beyotime, Nanjing, China) according to the manufacturer’s instruction. Each well contained 0.5 μg of the plasmid, with the final concentration of miRNA agomir reaching 100 nM/L. Luciferase assays were performed by using the Dual-Glo® Luciferase Assay System (Promega) at 24 h post-transfection. Normalized firefly luciferase activity (firefly luciferase activity/Renilla luciferase activity) was compared to that of the control pmirGLO Vector. The mean of the relative luciferase expression ratio (firefly luciferase/renilla luciferase) of the control was set to 1. For each transfection, luciferase activity was averaged from five replicates.
Authors’ contributions
KM and XG designed the research, analyzed the data and wrote the paper. KM, FL and PL performed the experiments. XC and YL participated in the data analysis. All authors read and approved the final the manuscript.
Acknowledgements
This work was supported by The National Natural Science Foundation of China (No. 31330064).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data and materials are presented in the main paper and additional supporting file.
Funding
The National Natural Science Foundation of China (No. 31330064).
Abbreviations
- miRNAs
microRNAs
- UTRs
untranslated regions
- P450
cytochrome p450
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real-time PCR
- U
uracil
- let-7-C
let-7-complex
Additional files
Additional file 1. Length distribution of unique small RNAs from A. gossypii identified by deep sequencing.
Additional file 2: Table S1. Primers used in the qRT-PCR analysis. Figure S1. The common and unique distribution of identified A. gossypii miRNAs among the five libraries. S01: CK; S02: 2-tridecanone; S03: Tannic acid; S04: Quercetin; S05: Gossypol. Figure S2. The GO annotation of A. gossypii miRNAs target genes.
Contributor Information
Kang-Sheng Ma, Email: txma1986@163.com.
Fen Li, Email: lifen2010happy@sina.com.
Ying Liu, Email: liuyingqh@sina.com.
Ping-Zhuo Liang, Email: liangpingzhu@126.com.
Xue-Wei Chen, Email: chen_xw2010@163.com.
Xi-Wu Gao, Email: gaoxiwu@263.net.cn.
References
- 1.Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2013;43:388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 5.Yokoi T, Nakajima M. microRNAs as mediators of drug toxicity. Annu Rev Pharmacol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 6.Asgari S. Role of microRNAs in insect host-microorganism interactions. Front Physiol. 2011;2:48. doi: 10.3389/fphys.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 9.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The Small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/S1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 10.Yu XM, Zhou Q, Li SC, Luo QB, Cai YM, Lin WC, Chen H, Yang Y, Hu SN, Yu J. The Silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS ONE. 2008;3:e2997. doi: 10.1371/journal.pone.0002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem Mol Biol. 2008;38:1066–1071. doi: 10.1016/j.ibmb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Zheng Y, Cao X, Ren R, Yu XQ, Jiang H. Identification and profiling of Manduca sexta microRNAs and their possible roles in regulating specific transcripts in fat body, hemocytes, and midgut. Insect Biochem Mol Biol. 2015;62:11–22. doi: 10.1016/j.ibmb.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification of conserved and novel microRNAs in Manduca sexta and their possible roles in the expression regulation of immunity-related genes. Insect Biochem Mol Biol. 2014;47:12–22. doi: 10.1016/j.ibmb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol. 2012;42:381–395. doi: 10.1016/j.ibmb.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang P, Feng B, Zhou XG, Gao XW. Identification and developmental profiling of microRNAs in diamondback moth, Plutella xylostella (L.) PLoS ONE. 2013;8:e78787. doi: 10.1371/journal.pone.0078787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etebari K, Hussain M, Asgari S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem Mol Biol. 2013;43:309–318. doi: 10.1016/j.ibmb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Agrawal N, Sachdev B, Rodrigues J, Sree KS, Bhatnagar RK. Development associated profiling of chitinase and microRNA of Helicoverpa armigera identified chitinase repressive microRNA. Sci Rep. 2013;3:2292. doi: 10.1038/srep02292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomate PR, Mahajan NS, Kale SM, Gupta VS, Giri AP. Identification and expression profiling of Helicoverpa armigera microRNAs and their possible role in the regulation of digestive protease genes. Insect Biochem Mol Biol. 2014;54:129–137. doi: 10.1016/j.ibmb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Lucas K, Raikhel AS. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 2013;43:24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain M, Asgari S. MicroRNAs as mediators of insect host-pathogen interactions and immunity. J Insect Physiol. 2014;70:151–158. doi: 10.1016/j.jinsphys.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Cao CW, Zhang J, Gao XW, Liang P, Guo HL. Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover) Pestic Biochem Physiol. 2008;90:175–180. doi: 10.1016/j.pestbp.2007.11.004. [DOI] [Google Scholar]
- 22.Blackman RL, Eastop VF. Aphids on the world’s crops: an identification guide. New York: John Wiley and Sons; 1984. [Google Scholar]
- 23.Li ZQ, Zhang S, Luo JY, Wang CY, Lv LM, Dong SL, Cui JJ. Ecological adaption analysis of the cotton aphid (Aphis gossypii) in different phenotypes by transcriptome comparison. PLoS ONE. 2013;8:e83180. doi: 10.1371/journal.pone.0083180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao DP, Liu Y, Walker WB, Li JH, Wang GR. Molecular characterization of the Aphis gossypii olfactory receptor gene families. PLoS ONE. 2014;9:e101187. doi: 10.1371/journal.pone.0101187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Despres L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol Evol. 2007;22:298–307. doi: 10.1016/j.tree.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Golawska S, Sprawka I, Lukasik I, Golawski A. Are naringenin and quercetin useful chemicals in pest-management strategies? J Pest Sci. 2014;87:173–180. doi: 10.1007/s10340-013-0535-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lema C, Cunningham MJ. MicroRNAs and their implications in toxicological research. Toxicol Lett. 2010;198:100–105. doi: 10.1016/j.toxlet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Choi YM, An S, Lee EM, Kim K, Choi SJ, Kim JS, Jang HH, An IS, Bae S. CYP1A1 is a target of miR-892a-mediated post-transcriptional repression. Int J Oncol. 2012;41:331–336. doi: 10.3892/ijo.2012.1418. [DOI] [PubMed] [Google Scholar]
- 29.Hong S, Guo Q, Wang W, Hu S, Fang F, Lv Y, Yu J, Zou F, Lei Z, Ma K, et al. Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem Mol Biol. 2014;55:39–50. doi: 10.1016/j.ibmb.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Z, Lv Y, Wang W, Guo Q, Zou F, Hu S, Fang F, Tian M, Liu B, Liu X, et al. MiR-278-3p regulates pyrethroid resistance in Culex pipiens pallens. Parasitol Res. 2015;114:699–706. doi: 10.1007/s00436-014-4236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 32.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 34.Celorio-Mancera MdlP, Ahn SJ, Vogel H, Heckel DG. Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genom. 2011;12:575. doi: 10.1186/1471-2164-12-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao F, Zhu S, Sun Y, Du L, Parajulee M, Kang L, Ge F. Interactive effects of elevated CO2 and cotton cultivar on tri-trophic interaction of Gossypium hirsutum, Aphis gossyppii, and Propylaea japonica. Environ Entomol. 2008;37:29–37. doi: 10.1093/ee/37.1.29. [DOI] [PubMed] [Google Scholar]
- 36.Sattar S, Addo-Quaye C, Song Y, Anstead JA, Sunkar R, Thompson GA. Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE. 2012;7:e48579. doi: 10.1371/journal.pone.0048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 38.Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X. Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae) PLoS ONE. 2011;6:e19350. doi: 10.1371/journal.pone.0019350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Peng W, Li Z, Li W, Li L, Pan J, Zhang S, Miao Y, Chen S, Su S. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol Biol. 2012;21:297–303. doi: 10.1111/j.1365-2583.2012.01135.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Liang S, Zhao Y, Han Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development. 2012;139:3543–3552. doi: 10.1242/dev.082719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuva-Aydemir Y, Xu XL, Aydemir O, Gascon E, Sayin S, Zhou W, Hong Y, Gao FB. Downregulation of the host gene jigr1 by miR-92 Is essential for neuroblast self-renewal in Drosophila. PLoS Genet. 2015;11:e1005264. doi: 10.1371/journal.pgen.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyons PJ, Storey KB, Morin PJ. Expression of miRNAs in response to freezing and anoxia stresses in the freeze tolerant fly Eurosta solidaginis. Cryobiology. 2015;71:97–102. doi: 10.1016/j.cryobiol.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139:1788–1797. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng TF, Pan YO, Gao XW, Xi JH, Zhang L, Ma KS, Wu YQ, Zhang JH, Shang QL. Reduced abundance of the CYP6CY3-targeting let-7 and miR-100 miRNAs accounts for host adaptation of Myzus persicae nicotianae. Insect Biochem Mol Biol. 2016;75:89–97. doi: 10.1016/j.ibmb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM. Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities. Toxicol Sci. 2011;120:49–75. doi: 10.1093/toxsci/kfq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 47.Ma KS, Li F, Liang PZ, Chen XW, Liu Y, Gao XW. Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae) J Insect Sci. 2016;16:1–9. doi: 10.1093/jisesa/iev152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 51.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials are presented in the main paper and additional supporting file.


