Abstract
Background
Salmonella pathogenicity island (SPI)-13 is conserved in many serovars of S. enterica, including S. Enteritidis, S. Typhimurium and S. Gallinarum. However, it is absent in typhoid serovars such as S. Typhi and Paratyphi A, which carry SPI-8 at the same genomic location. Because the interaction with macrophages is a critical step in Salmonella pathogenicity, in this study we investigated the role played by SPI-13 and SPI-8 in the interaction of S. Enteritidis and S. Typhi with cultured murine (RAW264.7) and human (THP-1) macrophages.
Results
Our results showed that SPI-13 was required for internalization of S. Enteritidis in murine but not human macrophages. On the other hand, SPI-8 was not required for the interaction of S. Typhi with human or murine macrophages. Of note, the presence of an intact copy of SPI-13 in a S. Typhi mutant carrying a deletion of SPI-8 did not improve its ability to be internalized by, or survive in human or murine macrophages.
Conclusions
Altogether, our results point out to different roles for SPI-13 and SPI-8 during Salmonella infection. While SPI-13 contributes to the interaction of S. Enteritidis with murine macrophages, SPI-8 is not required in the interaction of S. Typhi with murine or human macrophages. We hypothesized that typhoid serovars have lost SPI-13 and maintained SPI-8 to improve their fitness during another phase of human infection.
Electronic supplementary material
The online version of this article (doi:10.1186/s40659-017-0109-8) contains supplementary material, which is available to authorized users.
Keywords: Salmonella, Enteritidis, Typhi, SPI-13, SPI-8, Macrophages, RAW264.7, THP-1
Background
The genus Salmonella comprises more than 2500 serovars distributed in two species, S. bongori and S. enterica [1, 2]. Some serovars are able to infect a broad range of hosts (generalists), while others only cause disease in a particular host (specialists). S. Enteritidis is a generalist serovar that infects mammals and birds causing a typhoid-like infection in susceptible mice and asymptomatic carriage in hens. In humans, it represents a major cause of foodborne gastroenteritis [3, 4]. Conversely, S. Typhi is a specialist serovar that infects only humans, causing the systemic disease typhoid fever [5, 6].
Most genes required for Salmonella pathogenicity are clustered in genomic islands known as Salmonella pathogenicity islands (SPIs). So far, 23 SPIs have been described and characterized [7–11]. Five SPIs (SPI-1 to SPI-5) are common to all serovars of S. enterica, while the rest is distributed among different serovars and/or strains [7–11]. One such example is SPI-13, which is present in many S. enterica serovars including S. Enteritidis, S. Typhimurium, S. Cholerasuis and S. Gallinarum, but it is absent in S. Typhi and S. Paratyphi A, which carry SPI-8 in the same genomic location, adjacent to the tRNA-pheV gene [8, 9, 12] (Fig. 1). The number of genes within SPI-13 is not clearly defined; different authors have reported that it contains 4 [8], 18 [13] or 22 ORFs [9]. In this study we defined SPI-13 as the region including genes SEN2960 to SEN2966 of S. Enteritidis because they are flanked by nearly perfect direct repeats. In addition, these are the genes replaced in S. Typhi by SPI-8, which includes genes STY3273 to STY3292 [14].
Fig. 1.
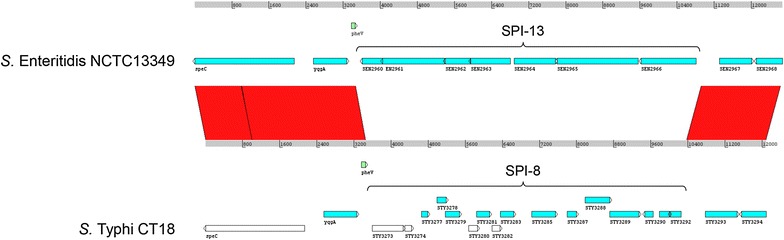
Comparison of the genomic region that includes SPI-13 in S. Enteritidis NCTC13349 and SPI-8 in S. Typhi CT18. A DNA-based comparison was performed by BLASTN analysis with WebACT (http://www.webact.org/WebACT/home) and visualized with ACT software. Homology between sequences (>98% identity) is highlighted in red. Green boxes correspond to tRNA-pheV genes and white boxes correspond to pseudogenes
It has been reported that genes in SPI-13 from S. Typhimurium [15–17] and S. Enteritidis [18] are expressed within macrophages in vitro, and are required for systemic infection in mice [9, 19–22]. These genes are homologues to the ripABC operon of Yersinia pestis, which encodes proteins required for the degradation of itaconate produced by macrophages during infection [23].
In comparison to SPI-13, much less information is available on SPI-8. In S. Typhi, this pathogenicity island contains a degenerate integrase gene, two bacteriocin pseudogenes and intact genes encoding proteins conferring immunity to these bacteriocins [14]. In addition, STY3280 has been shown to be expressed within human macrophages [24].
In this study, we investigated the roles of SPI-13 and SPI-8 in the interaction of S. Enteritidis and S. Typhi with cultured murine and human macrophages. Previously, other authors have demonstrated that both serovars are internalized by murine macrophages; but only S. Enteritidis is able to survive intracellularly [25]. In contrast, S. Enteritidis has a low ability to survive intracellularly in human macrophages, whereas S. Typhi survives and proliferates for long periods, showing less cytotoxicity than other serovars [25]. We hypothesized that the differential distribution of SPI-13 and SPI-8 in these two serovars might drive, in part, the different phenotypes observed in vitro in macrophages of different origins.
Methods
Bacterial strains, media, and growth conditions
Bacterial strains used in this study are listed in Table 1. All S. Enteritidis and S. Typhi strains are derivatives of wild-type strains NCTC13349 [13] and STH2370 [26], respectively. Bacteria were routinely grown in Luria–Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) at 37 °C with agitation. When required, LB medium was supplemented with ampicillin (Amp; 100 mg/L) or kanamycin (Kan; 50 mg/L). Media were solidified by the addition of agar (15 g/L).
Table 1.
Bacterial strains used in the present study
| Strain | Phenotype/genotype | Source or reference |
|---|---|---|
| Salmonella | ||
| S. Enteritidis NCTC13349 | Wild-type strain | [13] |
| S. Enteritidis ∆SPI-13 | NCTC13349 ∆(SEN2960-SEN2966)::FRT | This study |
| S. Enteritidis ∆SPI-13/pSPI-13 | NCTC13349 ∆(SEN2960-SEN2966)::FRT/pBAD-TOPO::(SEN2960-SEN2966) | This study |
| S. Typhi STH2370 | Wild-type strain | [26] |
| S. Typhi ∆SPI-8 | STH2370 ∆(STY3273-STY3292)::FRT | This study |
| S. Typhi ∆SPI-8/pSPI-8 | STH2370 ∆(STY3273-STY3292)::FRT/pBAD-TOPO::(STY3273-STY3292) | This study |
| S. Typhi ∆SPI-8/pSPI-13 | STH2370 ∆(STY3273-STY3292)::FRT/pBAD-TOPO::(SEN2960-SEN2966) | This study |
| Escherichia coli | ||
| TOP10 | K12 F− Ф80lacZ∆M15 ∆(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(r −k, m +k) phoA supE44 thi–1 gyrA96 relA1 λ− | Invitrogen |
Construction of mutant strains
All mutant strains were constructed by the Red-swap method [27] with modifications [21], using plasmid pCLF4 (KanR, GenBank accession number EU629214) as template and specific primers listed in Additional file 1: Table S1. The Kan-resistance cassette introduced by this procedure was eliminated by recombination using the FLP helper plasmid pCP20 [27, 28]. Each mutation was confirmed by PCR amplification using primers flanking the sites of substitution (Additional file 1: Table S1).
Construction of plasmids
SPI-13 and SPI-8 were amplified from strains NCTC13349 and STH2370, respectively, using PfuUltra II high-fidelity DNA polymerase (Stratagene) and specific primers listed in Additional file 1: Table S1. Each PCR product was purified from 1% agarose gels using the “QIAquick Gel Extraction Kit” (QIAGEN). Purified PCR products were adenylated at the 3′ end and cloned into pBAD-TOPO using the “pBAD-TOPO TA Expression Kit” (Invitrogen). Recombinant plasmids pSPI-13 and pSPI-8 were transformed into competent E. coli TOP10 (Invitrogen). The presence and orientation of the insert in each plasmid was confirmed by PCR amplification using primers listed in Additional file 1: Table S1. Finally, different strains of S. Enteritidis and S. Typhi were transformed by electroporation with these plasmids for complementation assays.
Cell lines and culture conditions
RAW264.7 murine macrophages were grown as monolayers in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). THP-1 human monocytes were grown in suspension using RPMI-1640 medium supplemented with 2 mM l-glutamine, 10% FBS and 50 μM β-mercaptoethanol. Both cell lines were maintained in an air-jacketed incubator (NuAire) at 37 °C in the presence of 5% CO2.
Bacterial infection assays
For infection assays using RAW264.7 macrophages, monolayers were prepared by seeding ~2.5 × 105 cells/well in 48-well plates (Nunc) and incubating for 20 h at 37 °C in a 5% CO2 atmosphere. From each bacterial culture grown overnight in LB at 37 °C with agitation, a 1/100 dilution in 5 mL of LB was prepared, and grown under the same conditions until reaching an OD600 = 0.4–0.6. Then, bacteria were centrifuged and washed with phosphate buffered saline (PBS), suspended in 100 μL of DMEM and added to the macrophages at a multiplicity of infection (MOI) of ~50 bacteria/cell. After 45 min of incubation at 37 °C, macrophages were washed three times with PBS containing 200 mg/L gentamicin and further incubated for 1 h in DMEM containing 200 mg/L gentamicin to kill extracellular bacteria. Then, cells from three wells were washed with PBS, and lysed with 0.5% sodium deoxycholate. Internalized bacteria were enumerated by serial dilution and plating on LB agar. The percentage of internalization was calculated as follows: 100 × (CFUinternalized/CFUinoculum). In the remaining six wells, medium was replaced by DMEM containing 20 mg/L gentamicin and cells were incubated for 2 h (three wells) or 20 h (three wells). Then, cells were washed, lysed and CFU were determined as described. The percentage of intracellular survival was calculated as follows: 100 × (CFU2 or 20h/CFUinternalized).
For infection assays using THP-1 monocytes, ~5 × 105 cells were seeded in 1.7 mL tubes and incubated for 20 h at 37 °C in a 5% CO2 atmosphere. Bacteria were grown as indicated above, washed with PBS and suspended in 100 μL of RPMI-FBS without β-mercaptoethanol. THP-1 cells were spun down and the bacterial suspension was added, resulting in a MOI of ~50 bacteria/cell. Internalization and intracellular survival were determined as described above.
Cytotoxicity assays
To determine the cytotoxicity produced by Salmonella strains during macrophage infection, lactate dehydrogenase (LDH) release was measured using “CytoTox96 Non-Radioactive Cytotoxicity Assay” (Promega), as recommended by the manufacturer.
Statistical analysis
Statistical significance of differences in the data was determined via one-way ANOVA with Dunnett’s test using the GraphPad Prism v6.0 software.
Results
SPI-13 is required for S. Enteritidis internalization in murine macrophages
It has been shown that several SPI-13 genes contribute to Salmonella pathogenesis in mice and chickens [9, 12, 19–22]. Because survival within macrophages is a crucial step in Salmonella systemic infection, we evaluated the contribution of SPI-13 in the interaction of S. Enteritidis with RAW264.7 macrophages. To this end, we tested the ability of the wild-type strain NCTC13349, an isogenic ∆SPI-13 mutant and the mutant complemented in trans with an intact copy of SPI-13, to be internalized by and survive within RAW264.7 macrophages. Our results showed that deletion of SPI-13 significantly impaired S. Enteritidis internalization in comparison to the wild-type strain. This phenotype was reverted by the introduction of a plasmid carrying an intact copy of SPI-13 (Fig. 2). On the other hand, deletion of SPI-13 did not affect S. Enteritidis survival after 2 or 20 h of infection (Fig. 2). The presence of the empty vector did not alter the phenotypes observed for the wild-type strain or the ∆SPI-13 mutant in our infection assays (data not shown). In addition, all strains showed similar cytotoxicity to RAW264.7 macrophages, as evaluated by LDH release during infections (data not shown).
Fig. 2.
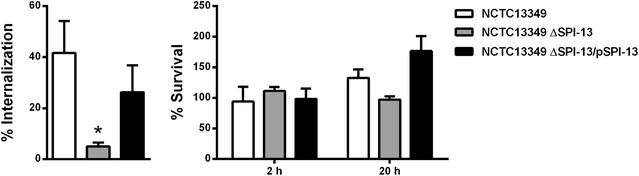
Contribution of SPI-13 to the internalization and intracellular survival of S. Enteritidis in murine macrophages. Infection assays were conducted as described in “Methods” section to determine the internalization and intracellular survival of S. Enteritidis strains in RAW264.7 macrophages. Graphics show mean values of at least 3 independent assays ± SEM. Statistical significance was determined using a one-way ANOVA with Dunnett’s test (*P < 0.05)
SPI-8 is not required for S. Typhi internalization or intracellular survival in murine macrophages
To determine the role played by SPI-8 in the interaction of S. Typhi with murine macrophages, we tested the ability of the wild-type strain STH2370, an isogenic ∆SPI-8 mutant and the mutant complemented in trans with an intact copy of SPI-8, to be internalized by and survive within RAW264.7 cells. No significant differences compared to the wild type were found when the ∆SPI-8 mutant and the complemented strain were analyzed (Fig. 3). In addition, the presence/absence status of SPI-8 did not affect the cytotoxicity of S. Typhi on RAW264.7 macrophages (data not shown).
Fig. 3.
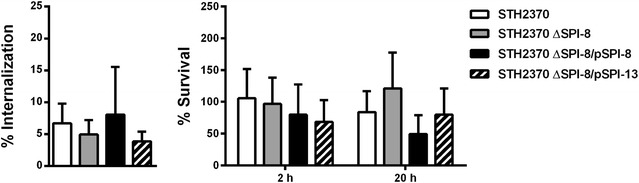
Contribution of SPI-8 and SPI-13 to the internalization and intracellular survival of S. Typhi in murine macrophages. Infection assays were conducted as described in “Methods” section to determine the internalization and intracellular survival of S. Typhi strains in RAW264.7 macrophages. Graphics show mean values of at least 3 independent assays ± SEM. Statistical significance was determined using a one-way ANOVA with Dunnett’s test
In view of our results with S. Enteritidis (Fig. 2), we tested if the presence of SPI-13 could improve the internalization or survival of S. Typhi ∆SPI-8 within murine macrophages. To do this, we performed infection assays using the S. Typhi ∆SPI-8 mutant carrying an intact copy of SPI-13. Our results showed that the presence of SPI-13 did not increase the internalization or intracellular survival of S. Typhi ∆SPI-8 in RAW264.7 macrophages (Fig. 3). Furthermore, the presence of this island did not affect the cytotoxicity of S. Typhi ∆SPI-8 on RAW264.7 macrophages (data not shown).
Neither SPI-8 nor SPI-13 contribute to the internalization or intracellular survival of S. Typhi and S. Enteritidis in human macrophages
Next, we aimed to determine whether SPI-13 and SPI-8 contribute to the internalization or intracellular survival of S. Enteritidis and S. Typhi, respectively, in human macrophages. To do this, we first performed infection assays in THP-1 human macrophages using wild-type S. Enteritidis, the ∆SPI-13 mutant, and the mutant complemented in trans with an intact copy of SPI-13. We did not observe differences in internalization, intracellular survival (Fig. 4a) and cytotoxicity (data not shown) between the wild-type strain and the mutants tested. Similar results were obtained when THP-1 macrophages were infected with wild-type S. Typhi, the ∆SPI-8 mutant, and the mutant complemented in trans with an intact copy of SPI-8 (Fig. 4b).
Fig. 4.
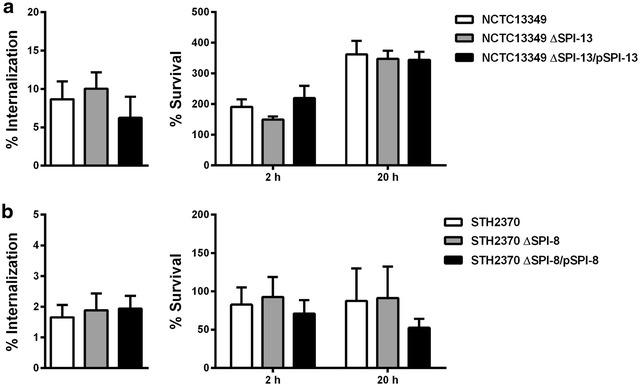
Contribution of SPI-13 and SPI-8 to the internalization and intracellular survival of S. Enteritidis and S. Typhi in human macrophages. Infection assays were conducted as described in “Methods” section to determine the internalization and intracellular survival of S. Enteritidis (a) and S. Typhi (b) strains in THP-1 macrophages. Graphics show mean values of at least 3 independent assays ± SEM. Statistical significance was determined using a one-way ANOVA with Dunnett’s test
Discussion
Although SPI-13 was identified in S. Gallinarum [12], it has been described in several members of S. enterica [8, 9, 13, 29]. The number of genes conforming SPI-13 is not clearly defined. In fact, different authors have reported that this island contains 4 [8], 18 [13] or 22 genes [9]. In this study, we defined SPI-13 as the region encompassing genes SEN2960 to SEN2966 in S. Enteritidis because these genes are adjacent to the tRNA-pheV gene and are flanked by nearly perfect direct repeats. Of note, these 7 genes are replaced in the genome of typhoid Salmonella serovars such as S. Typhi and S. Paratyphi A by genes composing SPI-8 [8, 9, 14]. We investigated whether this genetic difference contributes to the differential intracellular survival reported for S. Enteritidis and S. Typhi in macrophages of murine and human origin [25].
Our results indicate that SPI-13 is required for internalization of S. Enteritidis in RAW264.7 macrophages. In agreement with this observation, a null mutant of gene SEN2961 also presented an internalization defect in HD-11 chicken macrophages [18]. Reduced internalization by macrophages can affect S. Enteritidis pathogenicity because bacterial uptake by phagocytes is required for systemic spread of the pathogen and, as mentioned, SPI-13 is required for systemic colonization of mice and chicken by different Salmonella serovars [9, 12, 19–22].
Our data also indicate that SPI-13 is not involved in the intracellular survival of S. Enteritidis in RAW264.7 macrophages. These results do not agree with a recent study showing that deletion of SPI-13 (genes SEN2960 to SEN2977) resulted in increased uptake and impaired survival of S. Enteritidis in RAW264.7 macrophages [20]. The discrepancies between results from both studies may be due to differences in the S. Enteritidis strain employed and the number of SPI-13 genes deleted (7 vs. 18). Regarding this last point, the function of S. Enteritidis genes in the SEN2967-SEN2977 region (not included in our analysis) has been recently predicted and some of them may be required for intracellular survival in macrophages. For instance, SEN2973 encodes the antiadaptor protein IraD, which in S. Typhimurium has been reported to increase the stability of the sigma stress factor RpoS during oxidative stress [30]. In addition, transposon insertion mutants of S. Enteritidis in genes SEN2972, SEN2976 and SEN2977 present systemic colonization defects in mice [22]. Furthermore, the ∆SPI-13 mutant used by the authors of the other study kept the kanamycin-resistance cassette replacing the island, which may generate polar effects on genes surrounding the site of mutation.
Remarkably, genes SEN2961-SEN2963 of S. Enteritidis are homologous to the ripABC operon of Y. pestis. This operon was originally described as required for Y. pestis survival and replication within activated macrophages [31–33]. More recently, it was reported that Y. pestis ripABC operon encodes proteins with itaconate Co-A transferase, itaconil Co-A hydratase and citralil Co-A lyase activities, which together degrade itaconate and promote pathogenicity [23]. Itaconate is produced by macrophages in response to bacterial infection. This compound is a potent inhibitor of isocitrate lyase that blocks the glyoxylate cycle used by bacteria such as Salmonella and Mycobacterium to survive intracellularly [23, 34].
In contrast to our observations in RAW264.7 murine macrophages, SPI-13 was not required for internalization or survival of S. Enteritidis in THP-1 human macrophages. A possible explanation for these observations is that human macrophages produce itaconate at lower levels compared with mouse macrophages [34]. Hence, enzymes for itaconate degradation encoded in SPI-13 would not be needed in these macrophages. This hypothesis is supported by the fact that the genome of human pathogens S. Typhi and S. Paratyphi A do not contain SPI-13. Furthermore, our results showed that SPI-8 was not required for internalization or intracellular survival of S. Typhi in RAW264.7 murine macrophages or THP-1 human macrophages. We hypothesize that SPI-8 has been maintained in the genome of typhoid Salmonella serovars because it encodes factors required during other steps of human infection. For instance, proteins encoded in SPI-8 conferring immunity to bacteriocins [14] could improve bacterial fitness of typhoid serovars in the human gut. Further studies are required to confirm this hypothesis.
Conclusions
In this study we evaluated the contribution of pathogenicity islands SPI-13 and SPI-8 in the interaction of S. Enteritidis and S. Typhi with cultured murine (RAW264.7) and human (THP-1) macrophages. SPI-13 is present in the genome of many serovars of Salmonella enterica, including S. Enteritidis. However, this pathogenicity island is absent in the genome of typhoid serovars such as S. Typhi and S. Paratyphi C, which carry SPI-8 in the same genomic location. Our results indicate that SPI-13 is required for internalization of S. Enteritidis in murine but not in human macrophages. On the other hand, SPI-8 is not required for the internalization or survival of S. Typhi in human or murine macrophages. Thus, our results indicate different roles for SPI-13 and SPI-8 during Salmonella infection.
Authors’ contributions
RAE, IC and CAS conceived and designed the experiments. RAE, FAA and IMU carried out the experiments. IC, CAS, RAE, CAS-V, FAA and IMU analyzed and interpreted the results. IC, CAS, RAE and CAS-V wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by FONDECYT Grants 1110172 and 1140754 (to CAS). RAE, FAA and IMU were supported by CONICYT fellowships 21110114, 22151395 and 21150005, respectively.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon request.
Additional file
Additional file 1: Table S1. Primers used in the present study.
Contributor Information
Rodrigo A. Espinoza, Email: raetraet@gmail.com
Cecilia A. Silva-Valenzuela, Email: cecilia.silva@tufts.edu
Fernando A. Amaya, Email: fernando.amaya@ug.uchile.cl
Ítalo M. Urrutia, Email: iurrutia@ciq.uchile.cl
Inés Contreras, Email: inescontreraso@gmail.com.
Carlos A. Santiviago, Phone: (562) 2978-1681, Email: csantiviago@ciq.uchile.cl
References
- 1.Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38(7):2465–2467. doi: 10.1128/jcm.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves MW, Evins GM, Heiba AA, Plikaytis BD, Farmer JJ., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other Salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guard-Petter J. The chicken, the egg and Salmonella enteritidis. Environ Microbiol. 2001;3(7):421–430. doi: 10.1046/j.1462-2920.2001.00213.x. [DOI] [PubMed] [Google Scholar]
- 4.Patrick ME, Adcock PM, Gomez TM, Altekruse SF, Holland BH, Tauxe RV, Swerdlow DL. Salmonella Enteritidis infections, United States, 1985–1999. Emerg Infect Dis. 2004;10(1):1–7. doi: 10.3201/eid1001.020572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–353. [PMC free article] [PubMed] [Google Scholar]
- 6.House D, Bishop A, Parry C, Dougan G, Wain J. Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis. 2001;14(5):573–578. doi: 10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genom. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Bhonagiri-Palsikar V, Hallsworth-Pepin K, Clifton SW, Weinstock GM, McClelland M. Evolutionary genomics of Salmonella enterica subspecies. mBio. 2013;4(2):e00579-12. doi: 10.1128/mBio.00198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haneda T, Ishii Y, Danbara H, Okada N. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol Lett. 2009;297(2):241–249. doi: 10.1111/j.1574-6968.2009.01686.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayward MR, Jansen V, Woodward MJ. Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genom. 2013;14:365. doi: 10.1186/1471-2164-14-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensel M. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol. 2004;294(2–3):95–102. doi: 10.1016/j.ijmm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Shah DH, Lee MJ, Park JH, Lee JH, Eo SK, Kwon JT, Chae JS. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology. 2005;151(Pt 12):3957–3968. doi: 10.1099/mic.0.28126-0. [DOI] [PubMed] [Google Scholar]
- 13.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18(10):1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413(6858):848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47(1):103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 16.Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, Norbeck AD, Purvine SO, Manes NP, Smallwood HS, et al. Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J Biol Chem. 2006;281(39):29131–29140. doi: 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- 17.Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron AD, Hokamp K, Hinton JC. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog. 2015;11(11):e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Jansen R, Gaastra W, Arkesteijn G, van der Zeijst BA, van Putten JP. Identification of genes affecting Salmonella enterica serovar enteritidis infection of chicken macrophages. Infect Immun. 2002;70(9):5319–5321. doi: 10.1128/IAI.70.9.5319-5321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty S, Chaudhuri D, Balakrishnan A, Chakravortty D. Salmonella methylglyoxal detoxification by STM3117-encoded lactoylglutathione lyase affects virulence in coordination with Salmonella pathogenicity island 2 and phagosomal acidification. Microbiology. 2014;160(Pt 9):1999–2017. doi: 10.1099/mic.0.078998-0. [DOI] [PubMed] [Google Scholar]
- 20.Elder JR, Chiok KL, Paul NC, Haldorson G, Guard J, Shah DH. The Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog. 2016;8:16. doi: 10.1186/s13099-016-0098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009;5(7):e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva CA, Blondel CJ, Quezada CP, Porwollik S, Andrews-Polymenis HL, Toro CS, Zaldivar M, Contreras I, McClelland M, Santiviago CA. Infection of mice by Salmonella enterica serovar Enteritidis involves additional genes that are absent in the genome of serovar Typhimurium. Infect Immun. 2012;80(2):839–849. doi: 10.1128/IAI.05497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol. 2014;10(5):371–377. doi: 10.1038/nchembio.1482. [DOI] [PubMed] [Google Scholar]
- 24.Faucher SP, Curtiss R, 3rd, Daigle F. Selective capture of Salmonella enterica serovar Typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome. Infect Immun. 2005;73(8):5217–5221. doi: 10.1128/IAI.73.8.5217-5221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwan WR, Huang XZ, Hu L, Kopecko DJ. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun. 2000;68(3):1005–1013. doi: 10.1128/IAI.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela C, Ugalde JA, Mora GC, Alvarez S, Contreras I, Santiviago CA. Draft genome sequence of Salmonella enterica serovar Typhi strain STH2370. Genome Announc. 2014;2(1):e00104-14. doi: 10.1128/genomeA.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 29.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413(6858):852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 30.Battesti A, Tsegaye YM, Packer DG, Majdalani N, Gottesman S. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J Bacteriol. 2012;194(10):2470–2478. doi: 10.1128/JB.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pujol C, Grabenstein JP, Perry RD, Bliska JB. Replication of Yersinia pestis in interferon gamma-activated macrophages requires ripA, a gene encoded in the pigmentation locus. Proc Natl Acad Sci USA. 2005;102(36):12909–12914. doi: 10.1073/pnas.0502849102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres R, Chim N, Sankaran B, Pujol C, Bliska JB, Goulding CW. Structural insights into RipC, a putative citrate lyase beta subunit from a Yersinia pestis virulence operon. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68(Pt 1):2–7. doi: 10.1107/S1744309111048056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres R, Swift RV, Chim N, Wheatley N, Lan B, Atwood BR, Pujol C, Sankaran B, Bliska JB, Amaro RE, et al. Biochemical, structural and molecular dynamics analyses of the potential virulence factor RipA from Yersinia pestis. PLoS ONE. 2011;6(9):e25084. doi: 10.1371/journal.pone.0025084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci USA. 2013;110(19):7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon request.


